Abstract
Although influenza A viruses have been isolated from numerous shorebird species (Family: Scolopacidae) worldwide, our understanding of natural history of these viruses in this diverse group is incomplete. Gaining this information can be complicated by sampling difficulties related to live-capture, the need for large sample sizes related to a potentially low prevalence of infection, and the need to maintain flexibility in diagnostic approaches related to varied capabilities and resources. To provide information relevant to improving sampling and testing of shorebirds for influenza A viruses, we retrospectively evaluated a combined data set from Delaware Bay, USA collected from 2000–2009. Our results indicate that prevalence estimates and subtype diversity, can be effectively determined by either direct sampling of birds or indirect sampling of feces. Although predominant subtypes present in the populations also can be identified by either approach, the extent of detected subtype diversity is a function of the number of viruses recovered during that year. Even in cases where a large number of viruses are identified, this likely will result in an underestimation of actual subtype diversity. Influenza A virus isolation from Ruddy Turnstones can be enhanced by testing both cloacal and tracheal samples, and matrix real-time PCR can be used as an effective screening tool. Serologic testing to target species of interest also has application to shorebird surveillance. Overall, all of the sampling and diagnostic approaches have utility as applied to shorebird surveillance, but all are associated with inherent biases that need to be considered when comparing results from independent studies.
Keywords: Delaware Bay, influenza A virus, PCR, Ruddy Turnstone, serology, subtypes, shore birds, virus isolation, waders
INTRODUCTION
Shorebirds (Family: Scolopacidae) were first implicated in the natural history of influenza A viruses, when a high prevalence of infection was detected in migratory populations at Delaware Bay, USA (Kawaoka et al., 1988). Infections in shorebirds have been consistently detected at this site since this initial detection in 1987 (Krauss et al., 2010), but prevalence is variable related to species, time of sampling, and possibly location (Hanson et al., 2008; Krauss et al., 2010). Influenza A viruses have been reported from numerous shorebird species worldwide, but outside of Delaware Bay, reported prevalence estimates from these birds often have been very low (Olsen et al., 2006; Munster et al., 2007).
Because many shorebirds are long-distance migrants and could provide a vehicle for intercontinental movement of influenza A viruses, understanding the epidemiology of these viruses in these diverse species is potentially important. At present, however, there is limited information related to optimizing or improving sampling and testing approaches needed to efficiently advance this knowledge. In ducks, the ability to detect influenza A viruses is dependent upon the diagnostic approach (virus isolation versus molecular approaches, [Munster et al., 2009]) and sample selection (cloacal or orophayngeal [OP] swabs [Hoye et al., 2010]). Resulting inconsistencies in field sensitivity, related to either diagnostic performance or sampling approach, may bias results or create difficulties when comparing results from independent studies. In addition, it is often difficult to standardize virus detection and sampling protocols due to logistical constraints or limited resources related to both capture of birds and testing capabilities. In such cases, sampling and diagnostic approaches need to remain flexible but their limitations related to viral detection and isolation need to be clearly understood in order to adequately interpret results.
An additional problem associated with shorebird surveillance relates to a low infection prevalence, which appears to be the norm outside of Delaware Bay (Munster et al., 2007; Hanson et al., 2008) and it is not uncommon for influenza surveillance efforts directed at shorebirds, even with large sample sizes, to yield negative results (Hlinak et al., 2006; Winker et al., 2008). Serologic testing for influenza A virus antibodies has traditionally played a minor supportive role in surveillance especially with shorebirds. The potential application of this approach to surveillance has recently been demonstrated (Brown et al., 2010), and with shorebirds, this potential utility not only relates to providing an additional perspective for understanding epidemiology but also relates to the detection of species that are likely to be infected with these viruses. Such information can be utilized as an inexpensive means to target species for subsequent and efficient virus detection efforts and to maximize surveillance data return.
The objectives of this study are to: 1) retrospectively compare virus isolation results (prevalence and recovered subtype diversity) based on swab samples from live birds and fecal samples collected during the same weeks from 2000 to 2009 at Delaware Bay; 2) compare matrix (m) real-time (RT)-PCR and virus isolation approaches for influenza A virus detection in shorebirds; 3) compare isolation success as applied to paired cloacal and OP swabs; 4) further demonstrate the utility of using serologic testing to identify species that may be infected with influenza A viruses; and 5) based on these results and the existing literature, identify strengths and weaknesses associated with these surveillance approaches as applied to diverse shorebird species and populations.
METHODS
Sampling strategies
Independent virus isolation results from shorebirds sampled at Delaware Bay from 2000–2009 by St Jude Children’s Research Hospital (SJCRH) and the Southeastern Cooperative Wildlife Disease Study (SCWDS) were compared. Comparisons were limited to the same week of sampling during each year; sampling and testing protocols have been previously described (Krauss et al., 2004; Hanson et al., 2008). Three sampling approaches were evaluated in relation to influenza A virus prevalence and subtype diversity and included: 1) general shorebird sampling (all predominant species); 2) Ruddy Turnstone (Arenaria interpres) targeted sampling; and 3) fecal sampling directly from beach habitats utilized by these birds. Ruddy Turnstones were targeted because influenza A virus infections are routinely highest in this species at this location (Kawaoka et al; 1988; Hanson et al., 2008; Krauss et al., 2010).
Virus detection
To compare virus isolation results from cloacal and OP swabs of Ruddy Turnstones, 96 individual paired samples collected during 2009 were compared. These samples were tested by virus isolation only.
During 2007, 350 cloacal swab samples were collected as previously described from Ruddy Turnstones (Hanson et al., 2008) and tested by virus isolation and mRT-PCR. Briefly, samples that had been stored at −80 C following collection were thawed and inoculated into four 9 day old specific-pathogen-free embryonating chicken eggs for virus isolation as described (Hanson et al, 2008). Immediately following this procedure, RNA was extracted using a modified commercial protocol (Ambion MagMAX AI/ND Viral RNA Isolation Kit, Applied Biosystems, Foster City, California, USA) as previously described (Das et al., 2009). Extracted RNA was maintained at 4 C and tested within 24 hr by RRT-RCR on a SmartCycler PCR machine for amplification of the AIV M gene as previously described (Spackman et al., 2002; Das et al., 2006).
Serologic testing
In order to further determine the utility of serologic testing to support surveillance efforts, serum samples were collected from Ruddy Turnstones (n=160), Red Knots (Calidris canutus; n=56), and Sanderlings (Calidris alba, n=40) at Delaware Bay, and from 25 additional shorebirds from wintering areas in Georgia and Florida during 2010; these included Ruddy Turnstone (n=3), Sanderling (n=4), Red Knot (n=9), and Short-billed Dowitcher (Limnodromus griseus, n=9). All serologic testing was done using a commercially available type specific blocking enzyme-linked immunosorbent assay (bELISA) kit (FlockCheck AI MultiS-Screen Antibody Test Kit, IDEXX Laboratories, Westbrook, Maine, USA).
RESULTS
Sampling strategies
Prevalence data comparisons were limited to 2003–2009; the 2000–2002 samples from SJCRH were pooled (Table 1). As expected, prevalence estimates for the targeted Ruddy Turnstone sampling exceeded prevalence estimates based on sampling of multiple shorebird species during all years. With the exception of one year (2009), prevalence estimates based on cloacal swabs from Ruddy Turnstones also were higher than those derived from fecal sampling. With two exceptions, prevalence trends (an increase or decrease from the preceding year) were consistent between all sampling strategies. Exceptions included a decreased prevalence observed in the all bird sample during May 15–21, 2008 and increasing prevalence observed in the fecal samples during 2009 (Table 1).
Table 1.
Avian influenza prevalence estimates for total shorebirds, Ruddy Turnstones (RUTU), and fecal samples at Delaware Bay, USA based on independent virus isolation results at St Jude Children’s Research Hospital (SJCRH, fecal samples) and the Southeastern Cooperative Wildlife Disease Study, UGA (All bird and RUTU samples).
| Year | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | Totalc |
|---|---|---|---|---|---|---|---|---|
| All birdsa | 48/669 (7.2%) | 20/534 (3.7%) ↓ | 36/858 (4.2%) ↑ | 63/768 (8.2%) ↑ | 21/994 (2.1%) ↓ | 103/1645 (6.3%) ↑ | 132/1145 (11.5%) ↑ | 423/6613 (6.4%) |
| RUTUa | 46/441 (10.4%) | 19/256 (7.4%) ↓ | 33/245 (13.5%) ↑ | 53/293 (18.1%) ↑ | 19/441 (4.3%) ↓ | 95/584 (16.3%) ↑ | 111/847 (13.1%) ↓ | 376/3107 (12.1%) |
| Overlapping sampling dateb | May 15-21 | May 15-21 | May 15-21 | May 22-28 | May 22-28 | May 15-21 | May 15-21 | |
| All birdsb | 30/461 (6.5%) | 20/534 (3.7%) ↓ | 13/337 (3.9%) ↑ | 30/363 (8.3%) ↑ | 10/147 (6.8%) ↓ | 40/747 (5.4%) ↓ | 12/264 (4.5%) ↓ | 155/2853 (5.5%) |
| RUTUb | 29/294 (9.9%) | 19/256 (7.4%) ↓ | 13/116 (11.2%) ↑ | 30/224 (13.4%) ↑ | 10/96 (10.4%) ↓ | 35/204 (17.2%) ↑ | 9/158 (5.7%) ↓ | 145/1348 (10.8%) |
| Fecesb | 19/300 (6.3%) | 14/500 (2.8%) ↓ | 36/595 (6.1%) ↑ | 43/575 (7.5%) ↑ | 39/574 (6.8%) ↓ | 47/610 (7.7%) ↑ | 63/624 (10.1%) ↑ | 261/3778 (6.9%) |
These samples include all birds sampled during May of each year by UGA.
These samples were all collected during the same week where SJCRH and UGA sampling overlapped.
Differences in prevalence estimates for the total sample were detected by chi square with Yate’s correction between All birdsa and RUTUa (Χ2=92.295, df=1, P<0.0001), and within the overlapping sampling date between All birdsb and RUTUb (Χ2=37.71, df=1, P<0.0001), All birdsb and Fecesb (Χ2=5.48, df=1, P<0.0192), and RUTUb and Fecesb (Χ2=19.65, df=1, P<0.0001)
Overall, 628 influenza A viruses that were isolated during am overlapping one week period were identified to subtype; 199 by UGA and 429 by SJCRH. With the exception of one year (2003), the SJCRC samples (fecal) yielded the highest diversity of identified subtypes during an individual year (Table 2). The extent of detected subtype diversity, however, was a function of the number of viruses recovered during that year (Table 2, Figure 1). The ability to capture subtype diversity did not vary by sampling methods; based on the total results (2000–2009); cloacal swabs (UGA) and fecal sampling (SJCRH) yielded an average of .265 and .225 subtypes per isolated virus per year, respectively. Based on the combined sample, the number of detected subtypes ranged from 4–15/year (Table 3). In every year, the predominant HA/NA subtype combination (Table 3) and the predominant HA subtypes (Table 4) were detected by both UGA and SJCRH. In the case of HA diversity, viruses representing missed HA subtypes generally represented a very small proportion (≤8%) of the total isolates for that year.
Table 2.
Detection of type-A influenza virus subtypes by fecal and cloacal swab sampling of shorebirds during am overlapping one week period at Delaware Bay, USA, 2000–2009
| Year | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 |
|---|---|---|---|---|---|---|---|---|---|---|
| Week of sampling | May 15-21 | May 15-21 | May 15-21 | May 15-21 | May 15-21 | May 15-21 | May 22-28 | May 22-28 | May 15-21 | May 15-21 |
| Total no. subtypes (no. viruses) | 11 (n=60) | 9 (n=77) | 11 (n=88) | 12 (n=47) | 5 (n=34) | 4 (n=49) | 12 (n=73) | 15 (n=48) | 15 (n=84) | 13 (n=68) |
| UGAa total subtypes detected (no. viruses) % subtypes detected | 7 (n=23) 64% | 3 (n=8) 33% | 7 (n=17) 64% | 9 (n=28) 75% | 3 (n=20) 60% | 3 (n=13) 75% | 5 (n=30) 42% | 2 (n=9) 13% | 8 (n=40) 53% | 3 (n=11) 23% |
| SJCRHb % of total subtypes detected (no. viruses) % subtypes detected | 8 (n=37) 73% | 9 (n=69) 100% | 10 (n=71) 91% | 5 (n=19) 42%] | 5 (n=14) 100% | 4 (n=36) 100% | 10 (n=43) 83% | 15 (n=39) 100% | 11 (n=44) 73% | 12 (n=57) 92% |
| No. subtypes/isolate UGA | 0.30 | 0.38 | 0.41 | 0.32 | 0.15 | 0.23 | 0.17 | 0.22 | 0.20 | 0.27 |
| No. subtypes/isolate SJCRH | 0.21 | 0.13 | 0.14 | 0.26 | 0.36 | 0.11 | 0.23 | 0.38 | 0.25 | 0.21 |
Southeastern Cooperative Wildlife Disease Study, College of Veterinary Medicine, The University of Georgia
St Jude Children’s Research Hospital
Figure 1.
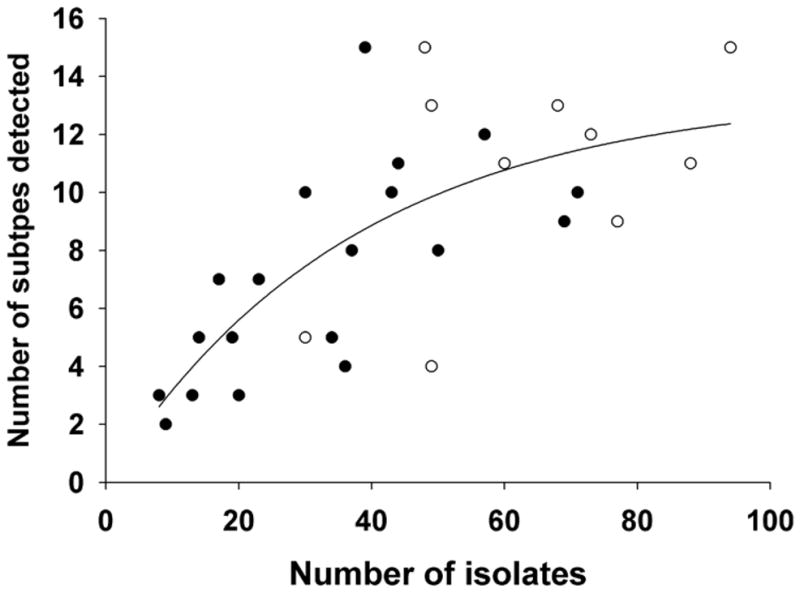
Detected subtype diversity as a function of sampling effort. Solid circles are results from individual sampling efforts by SCWDS and SJCRH. Open circles are the combined data for individual years. The nonlinear regressions line (R2= 0.514) was calculated based on an exponential, single, 2 parameter model and using SigmaPlot 10 software (Systat Software, Inc., Richmond, California, USA).
Table 3.
Subtypes diversity detected through fecal and cloacal swab sampling of shorebirds during am overlapping one week period at Delaware Bay, USA 2000–2009
| Year | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Week of sampling | May 15-21 | May 15-21 | May 15-21 | May15-21 | May 15-21 | May 15-21 | May 22-28 | May 22-28 | May 15-21 | May 15-21 |
|
| ||||||||||
| Subtypes detecteda | H12N5 (20)b | H10N7 (45) | H1N9 (45) | H9N2 (12) | H10N7 (18) | H3N6 (24) | H7N3 (48) | H12N5 (26) | H4N6 (31) | H10N7 (32) |
| H12N4 (13) | H12N7 (15) | H11N9 (12) | H9N5 (12) | H6N8 (10) | H3N8 (19) | H9N2 (5) | H12N4 (4) | H12N5 (25) | H10N1 (10) | |
| H10N7 (8) | H9N2 (4) | H1N4 (7) | H9N9 (6) | H5N7 (2) | H11N8 (4) | H1N4 (3) | H7N3 (4) | H10N7 (13) | H1N1 (6) | |
| H10N4 (5) | H6N2 (3) | H9N9 (7) | H9N1 (4) | H5N8 (2) | H11N6 (2) | H7N4 (3) | H5N4 (2) | H6N8 (3) | H6N1 (4) | |
| H5N4 (5) | H12N9 (3) | H11N4 (7) | H9N8 (4) | H11N9 (2) | H7N7 (3) | H12N1 (2) | H3N2 (2) | H11N1 (3) | ||
| H6N4 (3) | H9N7 (2) | H7N3 (3) | H9N4 (3) | H16N3 (3) | H4N9 (1) | H4N7 (1) | H1N8 (3) | |||
| H7N9 (2) | H11N2 (2) | H6N4 (2) | H5N9 (1) | H6N4 (2) | H5N1(1) | H4N8 (1) | H1N7 (2) | |||
| H5N3 (1) | H12N2 (2) | H11N2 (2) | H6N8 (1) | H13N9 (2) | H5N9 (1) | H6N2 (1) | H11N4 (2) | |||
| H9N7 (1) | H13N6 (1) | H9N4 (1) | H9N7 (1) | H6N2 (1) | H6N2 (1) | H10N6 (1) | H11N9 (2) | |||
| H11N6(1) | H1N5 (1) | H11N9 (1) | H6N7 (1) | H6N4 (1) | H10N8 (1) | H1N9 (1) | ||||
| H13N6 (1) | H6N9 (1) | H12N5 (1) | H6N8 (1) | H6N5 (1) | H10N9 (1) | H3N1 (1) | ||||
| H12N9 (1) | H7N5 (1) | H9N9 (1) | H11N9 (1) | H8N4 (1) | ||||||
| H11N9 (1) | H12N3 (1) | H11N8 (1) | ||||||||
| H13N9 (1) | H13N9 (1) | |||||||||
| H16N3 (1) | H16N3 (1) | |||||||||
Subtypes in bold and underlined were detected by the Southeastern Cooperative Wildlife Disease Study, College of Veterinary Medicine, The University of Georgia (UGA) and St Jude Children’s Research Hospital (SJCRH); Normal font= isolated by SJCRH only; Italics= isolated by UGA only;
Subtype (number of isolates)
Table 4.
Hemagglutinin subtype diversity detected in the combined sample during the overlapping one week period at Delaware Bay, USA 2000–2009. Values represent the percentage of total subtyped isolates
| HA Subtype | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 |
|---|---|---|---|---|---|---|---|---|---|---|
| H1 | 60% | 4% | 13%* | |||||||
| H2 | ||||||||||
| H3 | 88% | 2% | 1% | |||||||
| H4 | 2% | 39% | ||||||||
| H5 | 10%a | 2%* | 12%* | 8%* | ||||||
| H6 | 5% | 4% | 3% | 2%* | 29%* | 7% | 6%* | 5%* | 6%* | |
| H7 | 3% | 3%* | 75% | 8%* | ||||||
| H8 | 1% | |||||||||
| H9 | 2% | 8% | 9% | 89% | 7% | 1%* | ||||
| H10 | 22% | 58% | 53% | 19% | 62% | |||||
| H11 | 2%* | 3%* | 24% | 2% | 6% | 12%* | 2% | 1%* | 12% | |
| H12 | 55%b | 26% | 4%* | 67% | 31% | |||||
| H13 | 2% | 1% | 3% | 2% | 1%* | |||||
| H16 | 4% | 2% | 1% |
Shaded values represent HA subtypes detected by St Jude Children’s Research Hospital and the Southern Cooperative Wildlife Disease Study;
=HA detected by UGA in May of that year but outside of the overlapping sampling period.
Bold and underlined values represent the predominant HA subtype detected during that year.
Virus detection approaches
Virus isolation results from 96 paired samples collected from Ruddy Turnstones resulted in 13 isolates from cloacal swabs and 8 isolates from OP swabs. Only three birds tested positive on both cloacal and OP swabs. Prevalence estimates based on these results are 14% (95% CL= 6.8–20.38) for cloacal swabs only; 8% for OP swabs only (95% CL=2.8–13.9%); and 19% (95% CL= 10.9–26.6%) for the combined cloacal and OP results. The subtypes for the three viruses recovered from both cloacal and OP swabs from individual birds were identical for each positive bird.
Overall, the utilization of mRT-PCR resulted in increased detection of influenza A viruses in these samples; however, this increased sensitivity was dependent on the cutoff cycle threshold (ct) value (Table 5). This variation affected prevalence estimates as follows: 73/350 (20.9% [95% CL, 16.6–25.1%]) positive based on mRT-PCR (all ct-values ≤45); 49/350 (14.0%, [95% CL, 10.4–17.6%]) positive based on mRT-PCR (all ct-values <40); 20/350 (5.7% [95% CL, 3.3–8.1%] positive by virus isolation. There was only one virus isolation positive/mRT-PCR negative sample detected from these 350 samples.
Table 5.
Virus isolation and matrix real time polymerase chain reaction (mRT-PCR) results by critical threshold (Ct) value for cloacal swabs collected from ruddy turnstones at Delaware Bay, USA, 2007
| Ct-value rangea | Number positive by mRT-PCR | Number of mRT-PCR positive samples testing positive by virus isolation (% positive) |
|---|---|---|
| 25–29.9 | 11 | 9 (82%) |
| 30–34.9 | 18 | 4 (22%) |
| 35–39.9 | 20 | 6 (30%) |
| 40–44.9 | 24 | 0 (0%) |
| ALL | 73 | 19 (26%) |
Serologic testing
Antibody prevalence estimates for Ruddy Turnstone, Red Knot, and Sanderlings at Delaware Bay were 55% (95% CL=47.3–62.7%), 86% (95% CL= 76–95%), and 15% (95% CL=4–26%), respectively. Of the 25 birds sampled outside of Delaware Bay only Red Knots (five of nine, 56%, 95% CL= 23–88%) tested positive for antibodies to influenza A viruses.
DISCUSSION
At present, the epidemiology of influenza A viruses in shorebirds at Delaware Bay is unique as this is the only known “hotspot” where these viruses are reliably isolated from these species of birds (Krauss et al., 2010). While data obtained from this system provided an opportunity to evaluate surveillance and diagnostic approaches as applied to shorebirds, the application of these results to global surveillance also needs to consider the possibility of a very low prevalence of infection in these populations.
Our results from shorebirds at Delaware Bay indicate that virus isolation from cloacal swabs and fecal samples both are effective in providing reliable prevalence estimates in relation to detecting trends and changes in prevalence over time. Overall, there were few exceptions where prevalence trends did not agree related to sampling approach and this likely related to small sample size during the overlap period where results were compared. Such differences also may have resulted from spatial/temporal variation as there was no attempt to standardize collection to specific beach locations or sampling times within the week of data overlap. While differences in prevalence estimates derived from the three sampling approaches (all birds, Ruddy Turnstones, and fecal sampling) were relatively minor and never exceeded 10% in a given year (Table 1), such sampling related variation needs to be considered when utilizing published data in meta-analyses or when comparing prevalence estimates from independent studies. For example, although minor, prevalence estimates derived from the different sampling approaches were statistically different (Table 1), and if sampling bias were not considered in data analysis, this could lead to statistically supported but erroneous conclusions.
The three sampling approaches all were effective at capturing annual subtype diversity present in the population. At Delaware Bay, annual subtype diversity appears to be dominated by a single HA type each year that accounts for 35–90% of the total HA diversity (Table 4). The dominant HA subtype usually is represented by multiple HA/NA subtypes suggesting that these viruses undergo extensive genetic reassortment. Most of the missed HA/NA subtypes (either by UGA or SJCRH) were viruses representing low frequency HA subtypes or low frequency HA/NA subtypes that probably represented recent reassortment viruses. The extensive collection of viruses and data made available for this study gave a unique opportunity to clearly demonstrate that subtype diversity will almost always be underestimated in field studies even those with a significant sample size and a high rate of virus recovery. As previously reported in Krauss et al. (2004) specific HA/NA combinations recovered from shorebirds and gulls often represent a very small proportion of the total viruses recovered. A similar result was recently demonstrated in duck populations in Minnesota, USA where intensive surveillance over a three month period that encompassed the entire staging and early migration periods during two years, resulted in isolation of most of the HA subtypes that are present in North America each year (Wilcox et al. in press). In that study, which included results from more than 600 viruses, subtype specific prevalence estimates ranged from 0.04–4.6%.
The predominant route of virus shedding of type-A influenza viruses in birds is variable depending on species (Hoye et al., 2010; Costa et al., 2011), and virus; there are consistent reports that highly pathogenic H5N1 influenza viruses are predominantly associated with OP rather than cloacal shedding (Sturm-Ramirez et al., 2004; Brown et al., 2006), while wild bird viruses in ducks are predominantly shed via the cloaca (Webster et al., 1978). Although recently evaluated in ducks (Parmley et al., 2011), there is little or no available information related to the detection of these viruses in cloaca and OP swabs of shorebirds. Our results with Ruddy Turnstones are similar to previous reports from Mallards (Anas platyrhynchos), that is, successful detection is more likely from cloacal swabs rather than OP swabs (Munster et al, 2009). With ducks, detection success can be improved by combined OP/cloacal sampling (Parmley et al., 2011), and based on the combined OP/cloacal results from our Ruddy Turnstone sample, prevalence estimates increased from 14% (cloacal swab results) to 19% (combined cloacal/OP results). Although this 5% increase is modest in relation to the prevalence estimates, it does represent a 36% error. This is another factor that needs to be considered when comparing results or utilizing data from independent studies. Due to potential species related differences in shedding patterns (Hoye et al., 2010), this type of error may be species dependent.
The virus isolation and mRT-PCR protocols that were compared in this study represent the predominant influenza virus detection approaches currently in use. It is well established that sensitivity can be enhanced with mRT-PCR, and this test is often used as a screening test for subsequent virus isolation attempts (Munster et al., 2009). The relationship between improved VI success and lower ct-values observed in our study is not surprising but underscores the need to clearly define positive threshold values and to use caution in the interpretation of positive results. For example, if we consider a ct-value of <40 as the positive threshold, prevalence estimates for our Ruddy Turnstone sample based on mRT-PCR (14%) would have been significantly higher than prevalence estimates based on virus isolation (5.7%; chi-square with Yate’s correction, Χ2=12.61, df=1, P<0.0004). Of the 24 samples testing positive by mRT-PCR at a ct-value ≥40, no viruses were isolated. Because the failure to isolate cannot in this case be attributed to sample degradation during processing, we can only assume that they most likely represented samples containing RNA and non-infective virus or an insufficient quantity of virus to infect eggs. If this is the case, prevalence estimates based on mRT-PCR may not provide an accurate estimate of influenza virus infection in the population. That shortcoming aside, the utility of using mRT-PCR as a screening tool is supported by our results, and if we had taken this approach, we would have missed only 1 of 20 (5%) of the viruses recovered in virus isolation attempts. Overall, we isolated virus from 19 of the 49 (39%) mRT-PCR positive (<40 ct) samples which is comparable to the 33.5 % isolation rate reported for mRT-PCR positive birds (primarily ducks and geese) sampled in Europe (Munster et al., 2007).
Serologic testing as currently applied to wild bird surveillance is underutilized especially with shorebirds. Our results further demonstrate the utility of this approach as applied to shorebirds in two ways. First, results from individual populations can be replicated and reflect consistent trends between years. Reported antibody prevalence estimates (combined 2007–2008 data) for Ruddy Turnstones, Red Knots, and Sanderlings sampled at Delaware Bay during May were 65%, 54%, and 3%, respectively (Brown et al., 2010). Although antibody prevalence was higher for all species tested during 2010 (Ruddy Turnstone [55%], Red Knot [86% ], Sanderling [15%]), results are consistent with high exposure rates in Ruddy Turnstones and Red Knots and limited exposure among Sanderlings. Secondly, although our winter testing was minimal, serologic testing did identify Red Knots as antibody positive prior to their arrival at Delaware Bay. This type of information has application to both identifying species that are commonly infected with influenza viruses (in the absence of direct virologic evidence) and in understanding population immunity and interpretation of virus detection results.
As previously stated, sampling and testing protocols associated with field research or surveillance need to be selected in relation to surveillance objectives and expected prevalence in the target population which may be very low in shorebird populations especially outside of Delaware Bay. Based on our work at Delaware Bay, sampling of avian communities (all birds), targeted surveillance (Ruddy Turnstones), or indirect sampling (feces) for virus detection and serologic testing all can be effectively applied (individually or in combination) to meet specific objectives under the varied logistical constraints expected in the field and support laboratory. The primary disadvantage of community-based surveillance relates to capture efforts but, as with Delaware Bay, this may be offset if coordinated with existing biological studies or banding efforts. A second disadvantage relates to scale. Avian assemblages often involve numerous species utilizing unique habitat components, and this can make capture efforts costly and challenging. Advantages include the ability to collect both cloacal and OP swabs, plasma or serum for supportive serologic testing, complete biological data on sampled birds, and data and isolates from interacting species at the community level. This can be especially important for understanding results in situations where shorebirds share habitats and potentially influenza A viruses with waterfowl. Although challenging, this type of study is necessary to fully understand the natural history of these viruses.
Surveillance targeted to specific species requires a priori species-related information. This approach has the same advantages and limitation as community based approaches, but as demonstrated in this study, can improve efficiency. It is ideally suited for long-term studies where precise prevalence estimates related to infection or immunity are needed or when efficient virus recovery is desired.
Although fecal sampling will provide less information related to the host population, it provides a very efficient approach for virus recovery, lower but reliable prevalence estimates, and does not require bird capture. This approach is ideally suited for testing species and populations of unknown status and the recent development of techniques to determine species from these fecal samples (Lee et al., 2010) has greatly increased its utility. With a priori information, as exists with Delaware Bay, this approach can be utilized at the community or individual species level. Although the most cost efficient approach, the major disadvantage relates to the inability to collect relevant biological data such as age, condition, or data retrieved from band recoveries.
As for testing, our results as applied to shorebirds support the use of mRT-PCR as an effective screening test; however, this should be confirmed by virus isolation when possible. This is important not only to provide subtype specific data and field isolates for genetic or phenotypic characterization, but also in cases of low detected prevalence, to confirm positive results. When sampling birds, the collection of combined cloacal and OP swabs to enhance virus recovery and serum for type-specific antibody testing should also be considered.
All of these surveillance and testing approaches have the potential to yield useful data related to understanding the natural history of influenza A viruses, but as recently reported by Hoye et al. (2010), success is highly dependent on appropriate sample size, especially when prevalence is low. Our results from Delaware Bay add another sample size consideration related to subtype detection; the prevalence of specific subtypes can be extremely low, and because of this, subtypes can be easily missed. As occurs in ducks (Stallknecht and Brown, 2008), prevalence estimates for avian influenza in shorebirds are highly dependent on species and temporal/spatial variables (Hanson et al., 2008; Krauss et al, 2010). Our results demonstrate that sampling and testing techniques as applied to shorebirds also affect prevalence estimates. These inherent biases do not negate the value of these data and cannot always be corrected due to sampling and testing constraints, but need to be considered when evaluating data from comprehensive data bases or published independent studies.
Acknowledgments
The authors gratefully acknowledge the field and lab efforts of K. Jones, B. Hanson, A. Maxted, J. Smith, and B. Wilcox. Funding for this work was provided by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under Contracts HHSN266200700005C, HHSN266200700007C, N01-AI95357, Cancer Center Support (CORE) grant CA-21765, the American Lebanese Syrian Associated Charities (ALSAC) and Specific Cooperative Agreement 58-6612-2-0220 (ARS, USDA). The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of any of the funding agencies.
LITERATURE CITED
- Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerging Infectious Diseases. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Luttrell MP, Berghaus RD, Kistler W, Keeler S, Wilcox B, Hall J, Niles L, Niles M, Knutsen G, Fritz K, Stallknecht DE. Prevalence of antibodies to type A influenza virus in wild avian species using two serologic assays. Journal of Wildlife Diseases. 2010;46:896–911. doi: 10.7589/0090-3558-46.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TP, Brown JD, Howerth EW, Stallknecht DE. Variation in viral shedding patterns between different wild bird species infected experimentally with low-pathogenicity avian influenza viruses that originated from wild birds. Avian Pathology. 2011;40:119–124. doi: 10.1080/03079457.2010.540002. [DOI] [PubMed] [Google Scholar]
- Das A, Spackman E, Senne D, Pedersen J, Suarez DL. Development of an internal positive control for rapid diagnosis of avian influenza virus infections by real-time reverse transcription-PCR with lyophilized reagents. Journal of Clinical Microbioly. 2006;44:3065–73. doi: 10.1128/JCM.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Spackman E, Pantin-Jackwood MJ, Suarez DL. Removal of real-time reverse transcription polymerase chain reaction (RT-PCR) inhibitors associated with cloacal swab samples and tissues for improved diagnosis of Avian influenza virus by RT-PCR. Journal of Veterinary Diagnostic Investigation. 2009;21:771–8. doi: 10.1177/104063870902100603. [DOI] [PubMed] [Google Scholar]
- Hanson BA, Luttrell MP, Goekjian VH, Beck JD, Niles L, Swayne DE, Senne DA, Stallknecht DE. Is the occurrence of avian influenza virus in Charadriiformes species and location dependent? Journal of Wildlife Diseases. 2008;44:351–361. doi: 10.7589/0090-3558-44.2.351. [DOI] [PubMed] [Google Scholar]
- Hlinak A, Mühle RU, Werner O, Globig A, Starick E, Schirrmeier H, Hoffmann B, Engelhardt A, Hübner D, Conraths FJ, Wallschläger D, Kruckenberg H, Müller T. A virological survey in migrating waders and other waterfowl in one of the most important resting sites in Germany. Journal of Veterinary Medicine B. 2006;53:105–110. doi: 10.1111/j.1439-0450.2006.00935.x. [DOI] [PubMed] [Google Scholar]
- Hoye BJ, V, Munster J, Nishiure H, Klaasen M, Fouchier RAM. Surveillance of wild birds for avian influenza virus. Emerging Infectious Diseases. 2010;16:1827–1834. doi: 10.3201/eid1612.100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Chambers TM, Sladen WL, Webster RG. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247–250. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Krauss S, Walker D, Pryor P, Niles L, Chenghong L, Hinshaw VS, Webster RG. Influenza A viruses of migrating wild aquatic birds in North America. Vector-Borne and Zoonotic Diseases. 2004;4:177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG. Coincident ruddy turnstone migration and horseshoe crab spawning creates and ecological “hot spot” for influenza viruses. Proceedings of the Royal Society B, Biological Sciences. 2010;1699:3373–3379. doi: 10.1098/rspb.2010.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Lee HJ, Kang HM, Jeong OM, Kim MC, Kwon JS, Kim CB, Lee JB, Park SY, Choi IS, Song CS. DNA barcoding techniques for avian influenza virus surveillance in migratory bird habitats. Journal of Wildlife Diseases. 2010;46:649–654. doi: 10.7589/0090-3558-46.2.649. [DOI] [PubMed] [Google Scholar]
- Munster VJ, Baas C, Lexmond P, TM, Wallenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus ADME, Fouchier RM. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathogens. 2007;35(5):e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Bestebroer TM, Guldemeester J, Beyer WEP, De Wit E, Schutten M, Rimmelzwann GF, Osterhaus ADME, Fouchier RM. Practical Consideration for high-throughput influenza A virus surveillance studies of wild birds by use of molecular diagnostic tests. Journal of Clinical Microbiology. 2009;47:666–673. doi: 10.1128/JCM.01625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Munster V, Wallensten A. Global patterns of influenza A virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Parmley EJ, Soos C, Breault A, Fortin M, Jenkins E, Libenge F, King R, Mcaloney K, Pasick J, Pryor SP, Robinson J, Rodrique J, Leighton FA. Detection of low pathogenic avian influenza viruses in wild ducks in Canada: comparison of tow sampling methods. Journal of Wildlife Diseases. 2011;47:466–470. doi: 10.7589/0090-3558-47.2.466. [DOI] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. Journal of Clinical Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht DE, Brown JD. Ecology of Avian Influenza. In: Swayne DE, editor. Avian Influenza. Blackwell Publishing; Ames, Iowa: 2008. pp. 43–58. [Google Scholar]
- Sturm-Ramirez KM, Ellis T, Bousfield B, Bissett L, Dyrting K, Rehq JE, Poom L, Guan Y, Peiris M, Webster RG. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. Journal of Virology. 2004;78:4892–4901. doi: 10.1128/JVI.78.9.4892-4901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox BR, Knutsen G, Anderson M, Berdeen J, Goekjian V, Poulson R, Goyal S, Sreevatsan S, Cardona C, Berghaus R, Swayne D, Yablsley M, Stallknecht D. Influenza A viruses in ducks in Northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLos One. doi: 10.1371/journal.pone.0024010. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K, Spackman E, Swayne DE. Rarity of influenza A virus in spring shorebirds, southern Alaska. Emerging Infectious Diseases. 2008;14:1314–1316. doi: 10.3201/eid1408.080083. [DOI] [PMC free article] [PubMed] [Google Scholar]


