Abstract
Genome-wide association studies (GWAS) have identified hundreds of genomic regions associated with common human disease and quantitative traits. A major research avenue for mature genotype-phenotype associations is the identification of the true risk or functional variant for downstream molecular studies or personalized medicine applications. As part of the Population Architecture using Genomics and Epidemiology (PAGE) study, we as Epidemiologic Architecture for Genes Linked to Environment (EAGLE) are fine-mapping GWAS-identified genomic regions for common diseases and quantitative traits. We are currently genotyping the Metabochip, a custom content BeadChip designed for fine-mapping metabolic diseases and traits, in~15,000 DNA samples from patients of African, Hispanic, and Asian ancestry linked to deidentified electronic medical records from the Vanderbilt University biorepository (BioVU). As an initial study of quality control, we report here the genotyping data for 360 samples of European, African, Asian, and Mexican descent from the International HapMap Project. In addition to quality control metrics, we report the overall allele frequency distribution, overall population differentiation (as measured by FST), and linkage disequilibrium patterns for a select GWAS-identified region associated with low-density lipoprotein cholesterol levels to illustrate the utility of the Metabochip for fine-mapping studies in the diverse populations expected in EAGLE, the PAGE study, and other efforts underway designed to characterize the complex genetic architecture underlying common human disease and quantitative traits.
1. Introduction
In the last seven years, genome-wide association studies (GWAS) have been used extensively to identify common genetic variants associated with human diseases and quantitative traits. While there are many replicated and mature, known relationships between genomic regions and phenotypes, very few individual genetic variants have been identified as the risk variant for downstream molecular studies or personalized medicine applications. The lack of true functional variants revealed by GWAS stems from the fact that GWAS is based on linkage disequilibrium (LD), the non-random association of alleles at different variants along the chromosome. That is, GWAS fixed-content products mostly assay presumably neutral common genetic variants that are in LD or “tag” other genetic variants not directly assayed resulting in GWAS-identified regions that probably contain the true risk (unassayed) variant.
To identify the true risk variant, a major proposed activity in the “post-GWAS” era is fine mapping. In a fine-mapping experiment, the GWAS-identified region is densely interrogated via thousands of common and rare variants. Fine-mapping experiments can also take advantage of the known LD differences observed across populations. For example, populations of African-descent have lower levels of LD compared with populations of European-descent and therefore may be useful in identifying the risk variant masked by higher levels of LD in other populations. Fine mapping across populations is also useful for identifying population-specific variants associated with phenotypes.
In recognition for the need to fine-map mature GWAS-identified regions originally identified in European-descent populations, the National Human Genome Research Instituted established the Population Architecture using Genomics and Epidemiology (PAGE) study to genotype African American and Asian populations linked to phenotypes using the Illumina Metabochip, a custom iSelect BeadChip designed to fine-map GWAS-identified regions for metabolic diseases and traits. We as Epidemiologic Architecture for Genes Linked to Environment (EAGLE) are genotyping ~15,000 DNA samples linked to de-identified electronic medical records in the Vanderbilt University biorespository (BioVU) for fine mapping within the PAGE study. As the first step in quality control, EAGLE has genotyped 360 HapMap samples from European, African, Asians, and Mexican-descent populations. This short report describes the quality control, variant properties, and the potential for fine mapping of GWAS-identified regions in the anticipated populations within EAGLE and the PAGE study.
2. Methods
2.1. Study populations
DNA samples were obtained by the PAGE Coordinating Center from the Coriell Cell Repositories1. A total of 360 samples overlapping the International HapMap Project collection were obtained, including 30 trios of Northern and Western European ancestry from Utah from the Centre d’Etude du Polymorphisme Humain (CEPH) collection (CEU; catalog ID HAPMAPPT01), 90 unrelated individuals representing 45 individuals each from Tokyo, Japan and Beijing, China (ASN; catalog ID HAPMAPPT02), 30 trios from the Yoruba in Ibadan, Nigeria (YRI; catalog ID HAPMAPPT03), and 30 trios from communities of Mexican origin in Los Angeles, California (MEX; catalog ID HAPMAPV13). Samples were chosen to reflect the overall genetic ancestry of epidemiologic and clinical-based samples available in the PAGE study1.
2.2. Genotyping
Aliquots of HapMap DNA samples were distributed by the PAGE Coordinating Center to individual PAGE study sites. The Vanderbilt DNA Resources Core genotyped the Illumina Metabochip on the HapMap samples distributed by the PAGE Coordinating Center on the Illumina iScan (San Diego, California). The Metabochip is a custom BeadChip targeting 196,725 genetic variants. Common and less common genetic variants were chosen from among the first iteration of the 1000 Genomes Project and represent index GWAS-identified variants regardless of disease or phenotype as of 2009; regions targeted for fine-mapping for specific GWAS-identified regions associated with coronary artery disease, type 2 diabetes, QT-interval, body mass index/obesity, lipid traits, glycemic traits, and blood pressure; mitochondrial markers; HLA markers; sex chromosome markers; and ancestry informative markers2, 3. Illumina software GenomeStudio (v1.7.4) was used to determine the genotype calls for each variant for each sample, and manual re-clustering was performed on all mitochondrial and Y chromosome variants. Data were stored and accessed by the Vanderbilt Computational Genomics Core for quality control and downstream analyses using BC Platforms (Espoo, Finland).
2.3. Statistical methods
Standard quality control metrics were generated using PLINKv1.074 and PLATOv0.845. FST calculations were based on the Weir and Cockerham algorithm6 implemented in PLATO. Allele frequencies and FST were calculated for CEU, YRI, JPN and CHB combined (ASN), and MEX unrelated samples separately. Linkage disequilibrium (r2) was calculated using independent samples stratified by race/ethnicity using Haploviewv4.27.
3. Results
We genotyped 360 DNA samples from the International HapMap collection including 90 CEU, 90 YRI, 90 ASN, and 90 MEX on the Illumina Metabochip. From the 360 samples, 358 (99%) samples were successfully genotyped. And, out of the targeted 196,725 genetic variants on the Metabochip, we obtained data for 185,788 genetic variants for an overall pre-quality control call rate of 94.44%. From this initial dataset, we then performed quality control as outlined by Buyske et al2 (Table1).
Table 1.
Number of genetic variants removed from Metabochip dataset after quality control, by criteria and HapMap population. We performed quality control steps appropriate for a single dataset as outlined by Buyske et al.2. Lower genotyping call rates were observed for YRI compared with other HapMap populations consistent with our observations for targeted genotyping in EAGLE (data not shown).
| Criteria | SNP Failure Determination |
# SNPs removed | ||||
|---|---|---|---|---|---|---|
| CEU | YRI | CHB | JPN | MEX | ||
| Call Rate | < 0.95 | 14515 | 73445 | 11851 | 13585 | 14871 |
| Mendelian Errors | > 1 (out of 30 trios) |
97 | 10 | 0 | 0 | 144 |
| Replication Errors | < 2 | 0 | 0 | 0 | 0 | 0 |
| Hardy-Weinberg Equilibrium p- value |
< 1 × 10−6 | 11 | 1 | 11 | 10 | 19 |
| Discordant calls on EAGLE HapMap samples versus HapMap database |
> 3 (out of 90 samples) |
329 | 178 | 285 | 292 | 301 |
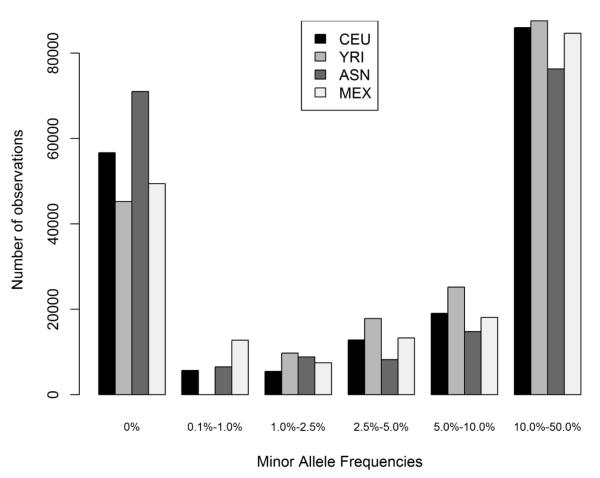
To examine potential population differences for genetic variants targeted by the Metabochip, we first determined minor allele frequencies for every variant by HapMap population. As shown in Figure 1, the majority of variants for this custom BeadChip are polymorphic. More than one half (58% for ASN) to up to three-quarters (75% for YRI) of the alleles assayed by the Metabochip occurred at greater than 1% frequency. Conversely, one quarter (24% for YRI) to more than one-third (38% for ASN) of the variants were monomorphic in this small sample set.
Figure 1. Distribution of minor allele frequencies of genetic variants assayed by the Metabochip, by HapMap population.
Allele frequencies were determined in the founder (unrelated) samples of Northern and Western European ancestry (CEU; n=60), West African ancestry (YRI; n=60), Asian ancestry (ASN; n=90), and Mexican ancestry (MEX; n=60). On the x-axis, genetic variant frequencies were binned as monomorphic, rare (0.1%-1-%), less common (1-2.5%), and common (2.5-5%, 5-10%, and 10-50%) by population. Number of observations for each bin is given on the y-axis.
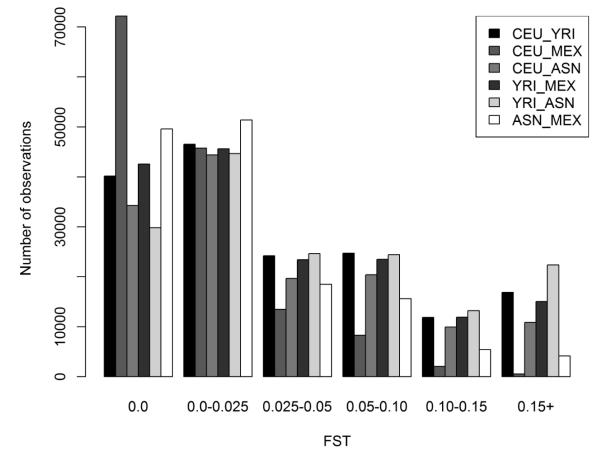
We also calculated a fixation index, FST, for all pair-wise population comparisons. FST is an estimate of population differentiation ranging from 0 (no measureable genetic differentiation) to 1.0 (very great genetic differentiation), and its distribution for Metabochip-targeted variants in HapMap samples is given in Figure 2. The majority (76%) of FST values are less than 15 for all genetic variant pair-wise population comparisons. The most population differentiation was observed between YRI and ASN. Conversely, the least population differentiation was observed between CEU and MEX.
Figure 2. Distribution of genetic differentiation (FST) by HapMap population pairwise comparison.
FST, a measure of population differentiation, was calculated per SNP in PLATO based on the Weir and Cockerham algorithm6 for each HapMap population pair. Calculations were performed on unrelated samples of Northern and Western European ancestry (CEU; n=60), West African ancestry (YRI; n=60), Asian ancestry (ASN; n=88), and Mexican ancestry (MEX; n=60). On the x-axis, FST values were binned no difference (zero), >0.0-0.25, >0.025-0.05, >0.05-0.10, >0.10-0.15, >0.15 by pair-wise population comparison. Number of observations for each bin is given on the y-axis.
Of the most highly differentiated SNPs (FST > 0.15), we examined the degree to which alleles altered the expression or function of genes using annotation resources from the Genome-Wide Annotation Repository (http://gwar.mc.vanderbilt.edu). We defined two categories of SNP annotation for this analysis: predicted changes to protein function via SIFT and PolyPhen2 algorithms 8, 9, and prior associations to expression levels of nearby genes 10, 11. The total number of SNP and gene annotations is shown in tables 2 and 3.
Table 2.
Number of differentiated SNPs showing functional effects
| Population Comparison |
SIFT (Deleterious) |
PolyPhen2 (Possibly or Probably Damaging) |
Significant eQTL |
Total functional SNPs* |
|---|---|---|---|---|
| ASN/MEX | 6 | 12 | 202 | 218 |
| YRI/ASN | 23 | 50 | 786 | 844 |
| YRI/MEX | 15 | 33 | 620 | 654 |
| CEU/ASN | 10 | 24 | 445 | 474 |
| CEU/YRI | 13 | 28 | 598 | 631 |
| CEU/MEX | 0 | 1 | 15 | 16 |
this total accounts for overlap between annotations
Table 3.
Number of distinct genes affected by differentiated SNPs
| Population Comparison |
SIFT (Deleterious) |
PolyPhen2 (Possibly or Probably Damaging) |
Significant eQTL |
Total Genes Affected* |
|---|---|---|---|---|
| ASN/MEX | 5 | 12 | 127 | 141 |
| YRI/ASN | 24 | 49 | 610 | 663 |
| YRI/MEX | 17 | 31 | 444 | 481 |
| CEU/ASN | 9 | 24 | 260 | 285 |
| CEU/YRI | 15 | 26 | 455 | 489 |
| CEU/MEX | 0 | 1 | 15 | 16 |
this total accounts for overlap between annotations
Using this collection of genes associated to differentiated SNPs through functional annotations, we performed gene enrichment analysis to identify specific biological mechanisms that likely have altered function between ethnic groups. This analysis revealed multiple pathways showing differences between CEU and MEX and CEU and ASN populations. KEGG pathways showing significant adjusted p-values (p < 0.05) are shown in Table 4.
Table 4.
Pathways with significant enrichment for highly differentiated functional alleles
| Population Comparison |
KEGG Pathway | Reference Genes |
Observed Genes |
Expected Genes |
P-value | P-value (adjusted for multiple testing) |
|---|---|---|---|---|---|---|
| CEU/MEX | Glutathione metabolism |
24 | 3 | 0.04 | 1.02E-05 | 9.47E-05 |
| CEU/MEX | Metabolism of xenobiotics by Cytochrome P450 |
30 | 3 | 0.06 | 2.03E-05 | 9.47E-05 |
| CEU/MEX | Drug metabolism - Cytochrome P450 |
29 | 3 | 0.05 | 1.83E-05 | 9.47E-05 |
| CEU/ASN | Allograft rejection | 26 | 6 | 0.83 | 0.0001 | 0.0007 |
| CEU/ASN | Graft-versus-host disease |
22 | 6 | 0.7 | 4.70E-05 | 0.0007 |
| CEU/ASN | Systemic lupus erythematosus |
54 | 9 | 1.71 | 4.35E-05 | 0.0007 |
| CEU/ASN | Arginine and proline metabolism |
17 | 5 | 0.54 | 0.0001 | 0.0007 |
| CEU/ASN | Autoimmune thyroid disease |
26 | 6 | 0.83 | 0.0001 | 0.0007 |
| CEU/ASN | Antigen processing and presentation |
29 | 6 | 0.92 | 0.0002 | 0.0013 |
| CEU/MEX | Asthma | 17 | 2 | 0.03 | 0.0004 | 0.0014 |
| CEU/ASN | Type I diabetes mellitus |
30 | 6 | 0.95 | 0.0003 | 0.0016 |
| CEU/MEX | Intestinal immune network for IgA production |
24 | 2 | 0.04 | 0.0009 | 0.0018 |
| CEU/MEX | Type I diabetes mellitus |
30 | 2 | 0.06 | 0.0013 | 0.0018 |
| CEU/MEX | Allograft rejection | 26 | 2 | 0.05 | 0.001 | 0.0018 |
| CEU/MEX | Graft-versus-host disease |
22 | 2 | 0.04 | 0.0007 | 0.0018 |
| CEU/MEX | Autoimmune thyroid disease |
26 | 2 | 0.05 | 0.001 | 0.0018 |
| CEU/MEX | Antigen processing and presentation |
29 | 2 | 0.05 | 0.0013 | 0.0018 |
| CEU/ASN | Intestinal immune network for IgA production |
24 | 5 | 0.76 | 0.0008 | 0.0039 |
| CEU/ASN | Riboflavin metabolism | 8 | 3 | 0.25 | 0.0016 | 0.007 |
Notably, the most significantly enriched pathways between CEU and MEX indicate a dramatic difference in the functional properties of glutathione and drug metabolism through cytochrome P450. Enrichment of these three pathways is the result of a single SNP – rs1010167 -- altering expression of three genes, GSTM1(p=3.88e-7), GSTM2(p=1.54e-7), and GSTM4(p=8.44e-7)11. This SNP falls within a region of chromatin that has been functionally categorized as an active promoter by the analysis of Ernst et al. in multiple cell types 12, and is confirmed to bind multiple proteins via ChIP-seq data as reported by the HaploREG database 13. rs1010167 was not previously genotyped by the HapMap phase III project.
Remaining pathways showing high differentiation in the CEU/ASN and CEU/MEX comparisons are largely immune-related, and are driven mostly by functional changes to the Major Histocompatibility Complex (MHC) found on chromosome 6. Interestingly, there were no significant pathways found for differentiated functional SNPs involving YRI comparisons.
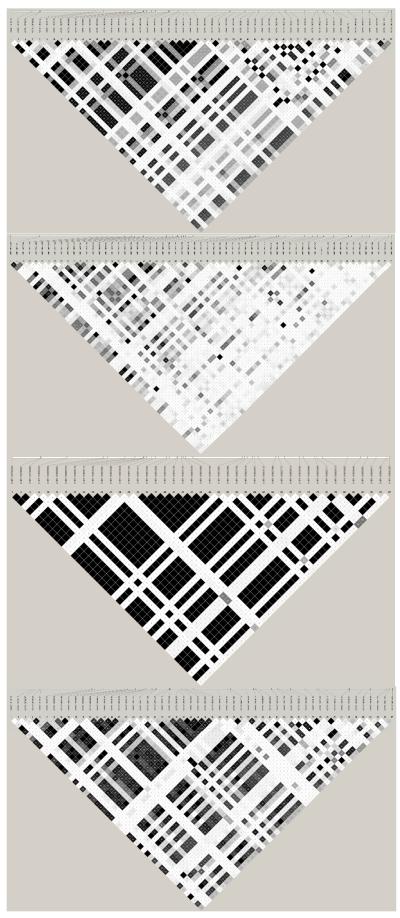
To illustrate the fine-mapping potential of densely targeted regions on the Metabochip, we calculated linkage disequilibrium (r2) by HapMap population for the CELSR2/PSRC1/SORT1 locus known to be associated with low-density lipoprotein cholesterol levels from GWA studies in European-descent populations14-16. Consistent with the observations of Buyske et al17 in samples from African American and Swedish participants, we observed less LD in YRI compared with CEU for this genomic region. To extend the observations made by Buyske et al, we examined LD for the same genomic region in HapMap samples of Asian and Mexican ancestry (Figure 3 c,d). As observed with minor allele frequency and FST, the CEU and MEX populations displayed similar levels of LD for this genomic region. In contrast, the ASN population had LD patterns that were distinct from CEU, YRI, and MEX LD patterns. For the ASN population, the CELSR2/PSRC1/SORT1 locus contained strong pair-wise LD statistics punctuated by weak LD.
Figure 3. Extent of linkage disequilibrium (r2) for 50kb region targeted by the Metabochip containing genome-wide association study (GWAS)-identified CELSR2/PSCR1/SORT1 by HapMap population.
Pair-wise linkage disequilibrium (LD) was calculated on unrelated samples using HaploView for European-descent [a)CEU; n=60], African [b)YRI; n=60], Asian [c)ASN; n=88], and Mexican [d)MEX; n=60] HapMap populations. For each LD plot, the genetic variants are labeled by chromosomal position at the top from 5′ to 3′. Each square represents a pair-wise LD statistic and they are coded on a gray scale where black is perfect LD (r2=1) and white to gray is weak LD. The numbers in select squares represent the LD metric for that pair-wise comparison (for example, 1 is r2=0.01).
4. Conclusions
We demonstrate here that the Metabochip custom BeadChip produces high-quality data for diverse populations from the International HapMap Project. We further show that the majority of variants observed in all populations considered were common and that a sizeable fraction of variants were monomorphic. Finally, we demonstrate population differences in both allelic diversity and LD patterns, both of which will impact the effectiveness of fine-mapping efforts that employ this BeadChip in the post-GWAS era.
Many of the observations reported here were expected based on population genetics theory and recent empirical genome-wide data from the International HapMap Project18, 19 and 1000 Genomes Project20. That is, as expected, the greatest population differentiation (as measured by FST) was observed between African-descent and Asian-descent populations21. However, other observations such as the proportion of common and rare variants did not follow expectations given the bias in genetic variant selection for this custom BeadChip22. From our FST analysis, we also observe significant differentiation of functional alleles within drug metabolism and auto-immune associated pathways between CEU and ASN/MEX populations. These variants may explain some aspects of ethnic differences in HLA-based autoimmune disease susceptibility, and indicates that cytochrome P450 drug metabolism may be altered in individuals of Mexican ancestry.
A major limitation of this study is sample size. With only 60 to 90 independent samples per HapMap population, our ability to observe rare alleles targeted by the Metabochip was limited for any HapMap population. Indeed, although the shape of the allelic distribution was similar, proportionally more variants in our dataset were classified as common or monomorphic compared with Buyske et al reflecting our limited ability to observe rare variants. Larger sample sizes will be required to take advantage of the full range of the allelic spectrum targeted by the Metabochip for fine mapping.
A final observation made here that will impact fine-mapping efforts is the extent of LD for an LDL-C associated region across populations. As Buyske et al2 noted, the breakdown of LD in African Americans for this region (and West Africans here) will be useful in identifying the true risk variant in a region with high LD in European populations. However, we note in ASN that the same genomic region has very high LD and thus this custom BeadChip may not fine map equally well for all targeted GWAS-identified regions for all populations. Because this custom BeadChip was designed using early iterations of the 1000 Genomes Project data, additional iterations of chips designed for fine mapping will be required to capture the latest genetic diversity data now emerging in non-European descent populations from later releases of the 1000 Genomes Project.
Acknowledgements
This work was supported by NIH U01 HG004798 and its ARRA supplements. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
Contributor Information
DANA C. CRAWFORD, Department of Molecular Physiology and Biophysics, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA crawford@chgr.mc.vanderbilt.edu
ROBERT GOODLOE, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA robert.j.goodloe@vanderbilt.edu.
KRISTIN BROWN-GENTRY, Center for Human Genetics Research, Vanderbilt University, 1207 17th Avenue, Suite 300 Nashville, TN 37232, USA kristin.brown@chgr.mc.vanderbilt.edu.
SARAH WILSON, Center for Human Genetics Research, Vanderbilt University, 1207 17th Avenue, Suite 300 Nashville, TN 37232, USA sarah.wilson@chgr.mc.vanderbilt.edu.
JAMIE ROBERSON, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA jamie.l.roberson@vanderbilt.edu.
NILOUFAR B. GILLANI, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA nila.gillani@vanderbilt.edu
MARYLYN D. RITCHIE, Department of Biochemistry and Molecular Biology, Center for System Genomics, Pennsylvania State University, 512 Wartik Lab University Park, PA 16802, USA marylyn.ritchie@psu.edu
HOLLI H. DILKS, Department of Molecular Physiology and Biophysics, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA holli.dilks@chgr.mc.vanderbilt.edu
WILLIAM S. BUSH, Department of Biomedical Informatics, Center for Human Genetics Research, Vanderbilt University, 2215 Garland Avenue, 519 Light Hall Nashville, TN 37232, USA william.s.bush@vanderbilt.edu
References
- 1.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, Haiman CA, Heiss G, Kooperberg C, Marchand LL, Manolio TA, North KE, Peters U, Ritchie MD, Hindorff LA, Haines JL. The Next PAGE in Understanding Complex Traits: Design for the Analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. American Journal of Epidemiology. 2011;174(7):849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, Hindorff LA, et al. Evaluation of the Metabochip Genotyping Array in African Americans and Implications for Fine Mapping of GWAS-Identified Loci: The PAGE Study. PLoS ONE. 2012;7(4):e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Center for Statistical Genetics . MetaboChip SNP details. University of Michigan; Jul 26, 2012. 2012. [Google Scholar]
- 4.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in PLATO: a precursor to efficient interaction analysis in GWAS data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 6.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(1358):1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 7.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 8.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de GA, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):437–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, Hindorff LA, et al. Evaluation of the Metabochip Genotyping Array in African Americans and Implications for Fine Mapping of GWAS-Identified Loci: The PAGE Study. PLoS ONE. 2012;7(4):e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keinan A, Clark AG. Recent Explosive Human Population Growth Has Resulted in an Excess of Rare Genetic Variants. Science. 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]