Abstract
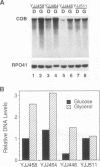
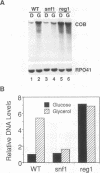
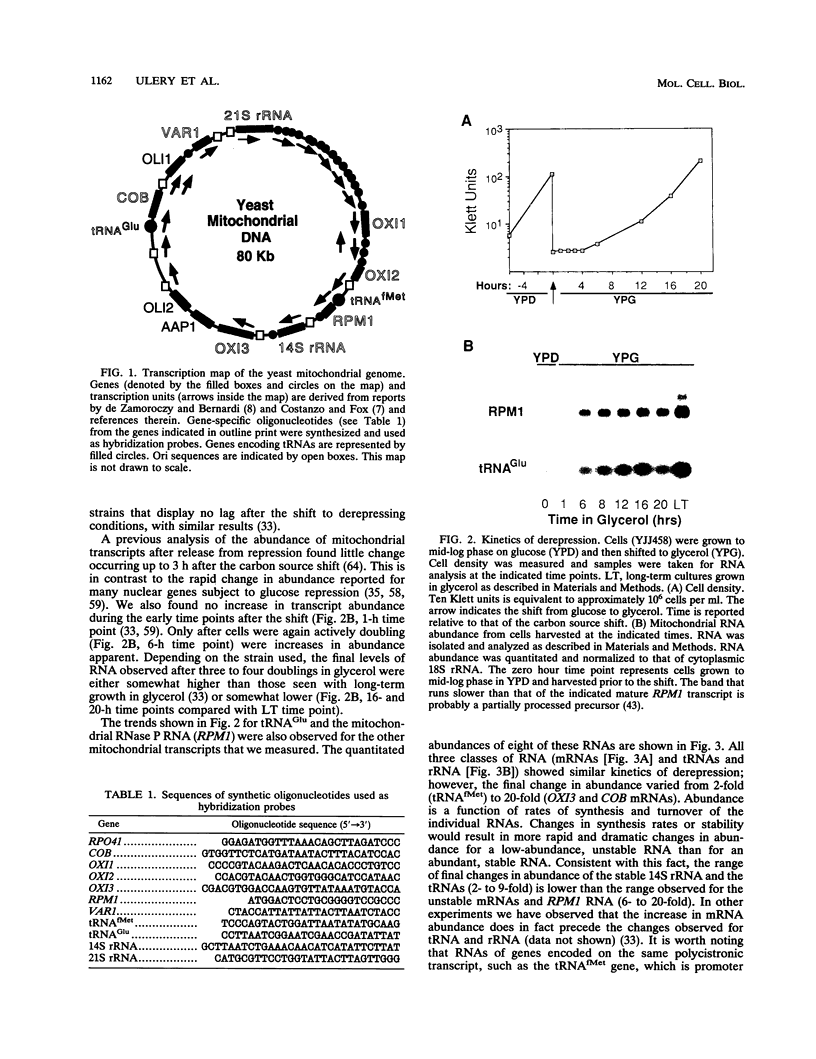
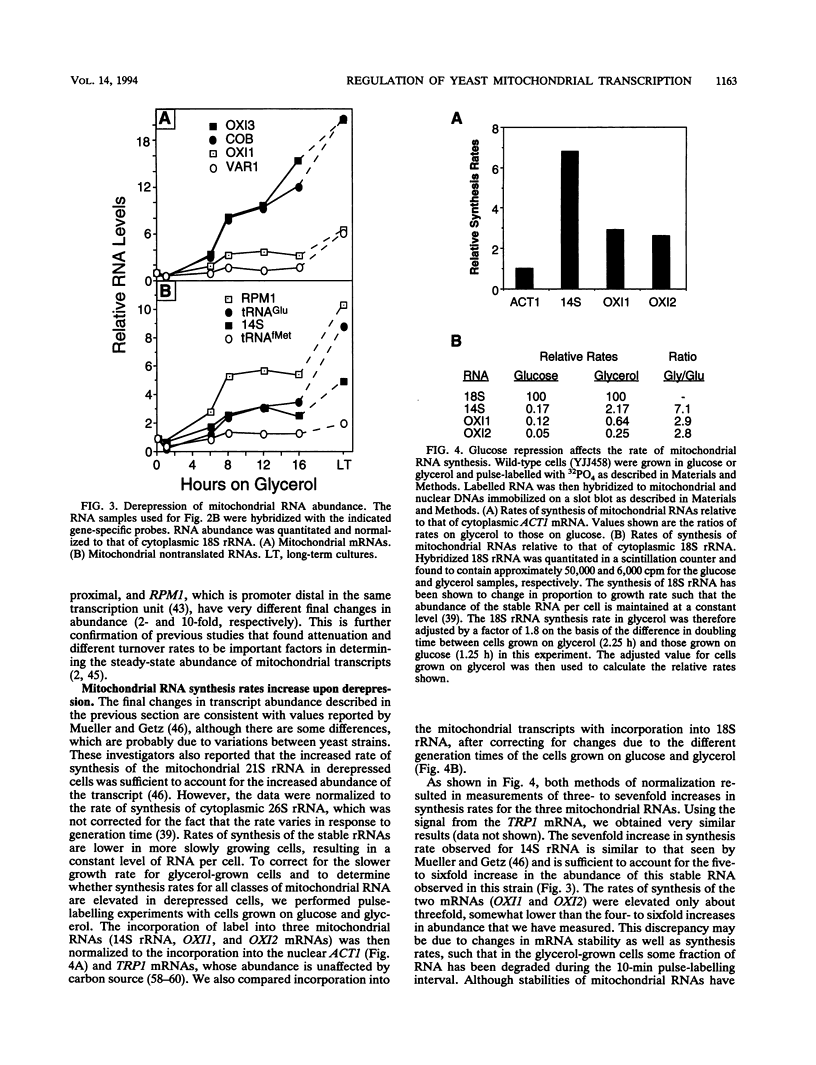
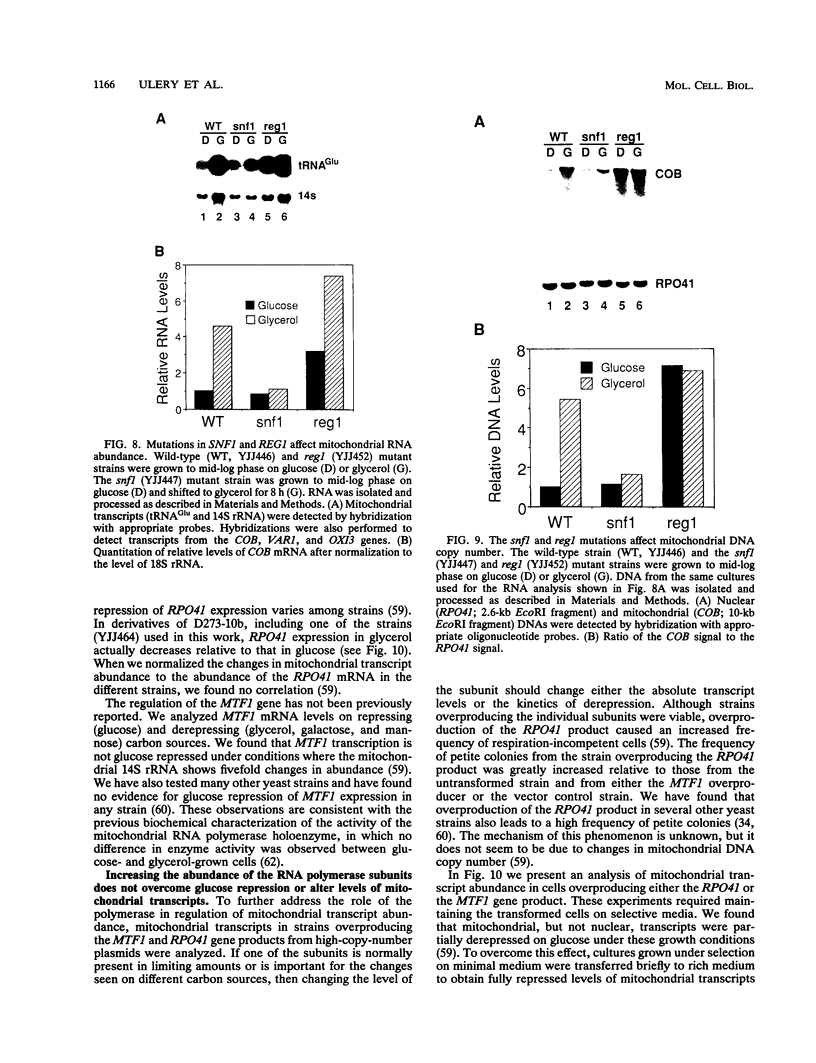
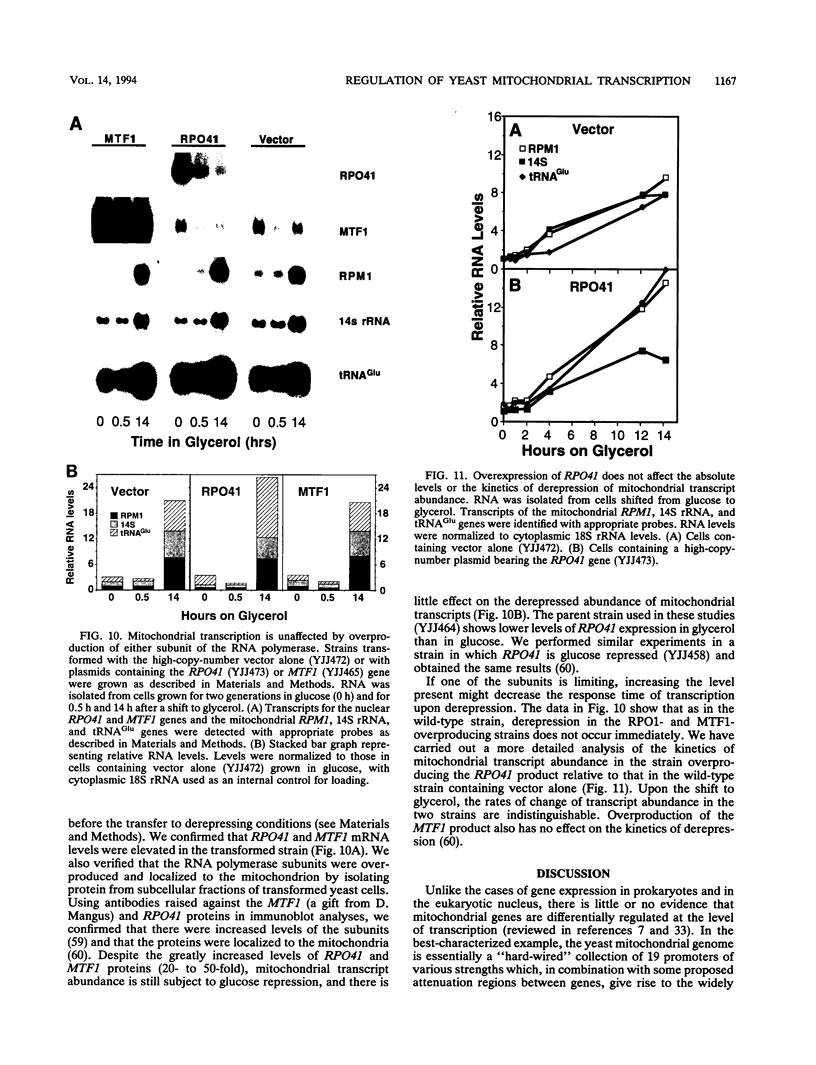
Yeast mitochondrial transcript and gene product abundance has been observed to increase upon release from glucose repression, but the mechanism of regulation of this process has not been determined. We report a kinetic analysis of this phenomenon, which demonstrates that the abundance of all classes of mitochondrial RNA changes slowly relative to changes observed for glucose-repressed nuclear genes. Several cell doublings are required to achieve the 2- to 20-fold-higher steady-state levels observed after a shift to a nonrepressing carbon source. Although we observed that in some yeast strains the mitochondrial DNA copy number also increases upon derepression, this does not seem to play the major role in increased RNA abundance. Instead we found that three- to sevenfold increases in RNA synthesis rates, measured by in vivo pulse-labelling experiments, do correlate with increased transcript abundance. We found that mutations in the SNF1 and REG1 genes, which are known to affect the expression of many nuclear genes subject to glucose repression, affect derepression of mitochondrial transcript abundance. These genes do not appear to regulate mitochondrial transcript levels via regulation of the nuclear genes RPO41 and MTF1, which encode the subunits of the mitochondrial RNA polymerase. We conclude that a nuclear gene-controlled factor(s) in addition to the two RNA polymerase subunits must be involved in glucose repression of mitochondrial transcript abundance.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Biswas T. K. In vitro transcription analysis of the region of Saccharomyces cerevisiae mitochondrial DNA containing the tRNA(fMet) gene. Nucleic Acids Res. 1991 Nov 11;19(21):5937–5942. doi: 10.1093/nar/19.21.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron V. L., Fox T. D., Poyton R. O. Isolation and characterization of a yeast strain carrying a mutation in the mitochondrial promoter for COX2. J Biol Chem. 1989 Aug 15;264(23):13391–13394. [PubMed] [Google Scholar]
- Cantwell R., McEntee C. M., Hudson A. P. Regulation of mitochondrial transcription during the stringent response in yeast. Curr Genet. 1992 Mar;21(3):241–247. doi: 10.1007/BF00336848. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992 Feb 15;267(5):3368–3374. [PubMed] [Google Scholar]
- Edwards J. C., Levens D., Rabinowitz M. Analysis of transcriptional initiation of yeast mitochondrial DNA in a homologous in vitro transcription system. Cell. 1982 Dec;31(2 Pt 1):337–346. doi: 10.1016/0092-8674(82)90127-1. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992 Jul;13(1):18–20. [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D. Glucose repression: a complex regulatory system in yeast. Microbiol Sci. 1986 Dec;3(12):366–371. [PubMed] [Google Scholar]
- Entian K. D., Zimmermann F. K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982 Sep;151(3):1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Henly J. W., Brewer B. J. RPO41-independent maintenance of [rho-] mitochondrial DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jan;10(1):10–15. doi: 10.1128/mcb.10.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992 Feb 15;267(5):3358–3367. [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992 Feb;130(2):295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Fukuhara H. Relative proportions of mitochondrial and nuclear DNA in yeast under various conditions of growth. Eur J Biochem. 1969 Nov;11(1):135–139. doi: 10.1111/j.1432-1033.1969.tb00750.x. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldthwaite C. D., Cryer D. R., Marmur J. Effect of carbon source on the replication and transmission of yeast mitochondrial genomes. Mol Gen Genet. 1974;133(2):87–104. doi: 10.1007/BF00264830. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. L., Kelly J. L., Lehman I. R. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell L. A. Nucleo-mitochondrial interactions in yeast mitochondrial biogenesis. Eur J Biochem. 1989 Jul 1;182(3):477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. M., Nagley P., Linnane A. W. Biogenesis of mitochondria. XLII. Genetic analysis of the control of cellular mitochondrial DNA levels in Saccharomyces cerevisiae. Mol Gen Genet. 1976 May 7;145(2):169–175. doi: 10.1007/BF00269590. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Ibrahim N. G., Stuchell R. N., Beattie D. S. Formation of the yeast mitochondrial membrane. 2. Effects of glucose repression on mitochondrial protein synthesis. Eur J Biochem. 1973 Jul 16;36(2):519–527. doi: 10.1111/j.1432-1033.1973.tb02938.x. [DOI] [PubMed] [Google Scholar]
- Jaehning J. A. Mitochondrial transcription: is a pattern emerging? Mol Microbiol. 1993 Apr;8(1):1–4. doi: 10.1111/j.1365-2958.1993.tb01197.x. [DOI] [PubMed] [Google Scholar]
- Jang S. H., Jaehning J. A. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J Biol Chem. 1991 Nov 25;266(33):22671–22677. [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. A., Fangman W. L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992 Mar;6(3):380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowsky T., Michaelis G. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol Gen Genet. 1988 Oct;214(2):218–223. doi: 10.1007/BF00337714. [DOI] [PubMed] [Google Scholar]
- Marczynski G. T., Schultz P. W., Jaehning J. A. Use of yeast nuclear DNA sequences to define the mitochondrial RNA polymerase promoter in vitro. Mol Cell Biol. 1989 Aug;9(8):3193–3202. doi: 10.1128/mcb.9.8.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983 Oct;34(3):911–917. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Min J., Zassenhaus H. P. A nucleoside triphosphate-regulated, 3' exonucleolytic mechanism is involved in turnover of yeast mitochondrial RNAs. J Bacteriol. 1993 Oct;175(19):6245–6253. doi: 10.1128/jb.175.19.6245-6253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisiae under catabolite repression and derepression. J Biol Chem. 1986 Sep 5;261(25):11816–11822. [PubMed] [Google Scholar]
- Mueller D. M., Getz G. S. Transcriptional regulation of the mitochondrial genome of yeast Saccharomyces cerevisiae. J Biol Chem. 1986 Sep 5;261(25):11756–11764. [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Characterization of Hex2 protein, a negative regulatory element necessary for glucose repression in yeast. Eur J Biochem. 1991 Sep 1;200(2):311–319. doi: 10.1111/j.1432-1033.1991.tb16187.x. [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Xu B., Clayton D. A. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993 Mar;13(3):1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel A. H., Koerkamp M. J., Touw E. P., Tabak H. F. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J Biol Chem. 1987 Sep 15;262(26):12785–12791. [PubMed] [Google Scholar]
- Sena E. P., Revet B., Moustacchi E. In vivo homologous recombination intermediates of yeast mitochondrial DNA analyzed by electron microscopy. Mol Gen Genet. 1986 Mar;202(3):421–428. doi: 10.1007/BF00333272. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R., Woontner M., Scott J. F. Mutations in ARS1 increase the rate of simple loss of plasmids in Saccharomyces cerevisiae. Yeast. 1986 Sep;2(3):169–178. doi: 10.1002/yea.320020305. [DOI] [PubMed] [Google Scholar]
- Szekely E., Montgomery D. L. Glucose represses transcription of Saccharomyces cerevisiae nuclear genes that encode mitochondrial components. Mol Cell Biol. 1984 May;4(5):939–946. doi: 10.1128/mcb.4.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C., Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975 Jun;122(3):855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxen S. E., Peterson C. R., Winkley C. S., Keller M. J., Jaehning J. A. Two forms of RPO41-dependent RNA polymerase. Regulation of the RNA polymerase by glucose repression may control yeast mitochondrial gene expression. J Biol Chem. 1988 Sep 5;263(25):12346–12351. [PubMed] [Google Scholar]
- Winkley C. S., Keller M. J., Jaehning J. A. A multicomponent mitochondrial RNA polymerase from Saccharomyces cerevisiae. J Biol Chem. 1985 Nov 15;260(26):14214–14223. [PubMed] [Google Scholar]
- Zennaro E., Grimaldi L., Baldacci G., Frontali L. Mitochondrial transcription and processing of transcripts during release from glucose repression in 'resting cells' of Saccharomyces cerevisiae. Eur J Biochem. 1985 Feb 15;147(1):191–196. doi: 10.1111/j.1432-1033.1985.tb08736.x. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae--a review. Gene. 1986;47(2-3):155–177. doi: 10.1016/0378-1119(86)90060-0. [DOI] [PubMed] [Google Scholar]