Abstract
Hypervirulent invasive group A streptococcus (GAS) isolates inhibit neutrophil infiltration more than pharyngitis isolates do, and the molecular basis of this difference is not well understood. This study was designed to first determine whether natural null mutation of the two-component regulatory system CovRS is responsible for the enhancement of the inhibition of neutrophil recruitment seen in hypervirulent GAS. Next, we examined the role of CovRS-regulated interleukin-8/CXC chemokine peptidase (SpyCEP), C5a peptidase (ScpA), and platelet-activating factor acetylhydrolase (SsE) in the enhanced innate immune evasion. Invasive isolate MGAS5005 induces less neutrophil infiltration and produced a greater lesion area than pharyngitis isolate MGAS2221 in subcutaneous infections of mice. It is known that MGAS5005, but not MGAS2221, has a natural 1-bp deletion in the covS gene. Replacement of covSΔ1bp in MGAS5005 with wild-type covS resulted in the MGAS2221 phenotype. Deletion of covS from MGAS2221 resulted in the MGAS5005 phenotype. Tests of single, double, and triple deletion mutants of the MGAS5005 sse, spyCEP, and scpA genes found that SsE plays a more important role than SpyCEP and ScpA in the inhibition of neutrophil recruitment and that SsE, SpyCEP, and ScpA do not have synergistic effects on innate immune evasion by MGAS5005. Deletion of sse, but not spyCEP or scpA, of MGAS2221 enhances neutrophil recruitment. Thus, covS null mutations can cause substantial inhibition of neutrophil recruitment by enhancing the expression of the chemoattractant-degrading virulence factors, and SsE, but not SpyCEP or ScpA, is required for CovRS-regulated GAS inhibition of neutrophil infiltration.
INTRODUCTION
Group A streptococcus (GAS) commonly causes relatively mild pharyngitis and superficial skin infections. This major human pathogen also causes approximately 10,000 cases of severe invasive infections, such as necrotizing fasciitis, sepsis, and toxic shock syndrome, annually in the United States (1). Necrotizing fasciitis is a rapidly progressive infection of the skin, subcutaneous and deep soft tissue, and muscle and leads to systemic dissemination (2). Innate immune invasion by hypervirulent GAS plays a critical role in severe invasive infections. Neutrophil infiltrate is sparse in streptococcal necrotizing fasciitis (3–5). This severe inhibition of neutrophil recruitment can be modeled in experimental animal infections with severe invasive GAS isolates (3, 6, 7) but not pharyngitis isolates (7). Peptidases ScpA and SpyCEP (also known as ScpC) produced by GAS degrade the chemotactic C5a peptide and interleukin-8 (IL-8)/CXC chemokines, respectively, and are believed to contribute to inhibition of neutrophil recruitment (3, 8–11). The secreted esterase SsE of GAS, a protective antigen (12), targets platelet-activating factor to critically contribute to GAS inhibition of neutrophil recruitment and skin invasion (7, 13). GAS also resists phagocytosis by neutrophils through the hyaluronic acid capsule and surface M protein (14, 15), kills neutrophils through streptolysins S and O (16, 17), and escapes neutrophil extracellular traps through DNases (18). Despite these advances, the molecular basis of innate immune evasion by hypervirulent GAS isolates is not fully understood. Furthermore, it is not known whether SpyCEP and ScpA also critically contribute to the inhibition of neutrophil infiltration by hypervirulent GAS isolates and if SpyCEP, ScpA, and SsE synergistically contribute to the inhibition of neutrophil recruitment in severe invasive infections.
Strains isolated from invasive infections have a high frequency of mutations in the two-component regulatory system CovRS (also known as CsrRS) (19, 20), and covRS mutations also readily arise during experimental animal infections (21, 22). Clinical isolates with a covRS mutation or deletion are usually hypervirulent. CovRS negatively regulates many virulence factors, including the capsule synthase HasA, streptolysin S, protease SpeB, DNase Sda1, IgG proteinase Mac, SpyCEP, ScpA, and SsE (13, 18, 22–26). Some covR mutations enhance virulence by relieving the CovR depression of virulence factor genes (27, 28). In contrast to the effects of CovR mutations on the expression of virulence genes, covS null mutations both up- and downregulate distinct subsets of CovR-repressed genes (26). Loss of SpeB production and enhancement of the production of the hyaluronic acid capsule and SsE, as results of covRS mutations/deletions, are critical factors in the progression of invasive GAS infections (13, 27, 28). Whether SpyCEP and ScpA are required for virulence and skin invasion of hypervirulent GAS isolates is not known.
We hypothesize that covS null mutation/deletion-enhanced expression of SpyCEP, ScpA, and SsE critically contributes to the enhanced innate immune evasion and virulence of GAS strains isolated from severe invasive infections. To test this hypothesis, we first performed a reciprocal analysis of the effect of covS deletion on neutrophil infiltration, virulence, and skin invasion by using two representative strains of a M1T1 subclone, MGAS5005, with a natural 1-bp deletion in covS, and MGAS2221, with the wild-type (WT) covS gene (22). We then examined the relative and synergistic contributions of SpyCEP, ScpA, and SsE to MGAS5005 inhibition of neutrophil infiltration, virulence, and skin invasion. We found that the 1-bp deletion in covS of MGAS5005 is the cause of the that strain's enhanced innate immune evasion, skin invasion, and virulence. SsE, but not SpyCEP or ScpA, is critical for the phenotype of MGAS5005. These results provide information about the basis of GAS innate immune evasion and the progression of invasive GAS infections.
MATERIALS AND METHODS
Bacterial strains and growth.
MGAS5005 and MGAS2221 are representative isolates of a prevalent M1T1 subclone from an invasive-infection case in Ontario and a scarlet fever patient in Australia, respectively (26). The sse gene deletion mutants of MGAS5005 (MGAS5005Δsse) and MGAS2221 (MGAS2221Δsse) have been described previously (7, 13). MGAS2221ΔcovS has also been described previously (26). These strains and their derivatives were grown in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) at 37°C in 5% CO2. Tryptose agar with 5% sheep blood and THY agar were used as solid media.
Generation of spyCEP, scpA, and sse deletion mutants.
Upstream and downstream flanking fragments of an internal 301-bp fragment of the spyCEP gene (bases 100 to 400) to be deleted were amplified by PCR by using MGAS5005 genomic DNA and primer pairs 5′-TTAAGCTTGTCGGTATGCCAATTGTTC-3′/5′-TTCTCGAGCTCTGTATTGGTGAGATGTTG-3′ and 5′-TTCTCGAGTGCGCATCAGTGATGTATC-3′/5′-TTGGATCCGGATCACGTTCAATTAAAGC-3′, respectively. The upstream and downstream PCR products were sequentially cloned into pGRV (29) at the HindIII/XhoI and XhoI/BamHI sites, respectively, producing pΔspyCEP. To delete a 1,240-bp fragment of the scpA gene (bases 357 to 1410), pΔscpA was similarly constructed by using primer pairs 5′-AGGATCCGTCAATCACAGCTTCCACTTG-3′/5′-ACTCGAGAACAGTCCCAGCTCCTTTG-3′ and 5′-ACTCGAGCAAAAGCAATATGAGACACAG-3′/5′-AAGATCTCAACATTTCCTTCTTTGTCC-3′ and the BamHI/XhoI and XhoI/BgIII sites of pGRV. pΔsse, which was used to delete the sse gene from the ΔspyCEP, ΔscpA, and ΔspyCEP ΔscpA mutants of MGAS5005, has been described previously (13).
These plasmids were used to generate single deletion mutants MGAS5005ΔspyCEP, MGAS2221ΔspyCEP, MGAS5005ΔscpA, and MGAS2221ΔscpA and spyCEP, scpA, and sse gene double and triple deletion mutants of MGAS5005 (ΔspyCEP ΔscpA, ΔspyCEP Δsse, ΔscpA Δsse, and ΔscpA ΔspyCEP Δsse mutants) by following the procedure that was used to generate the Δsse mutant of MGAS5005 (13). All of the mutants were confirmed by PCR and DNA sequencing. PCR confirmation of the spyCEP and scpA deletions was done with primer pairs 5′-GTTAAAACATTTAGGAGGG-3′/5′-GTATTTGGTGTCATCGTCTG-3′; and 5′-CCTGCTACCGAACAAGCTG-3′/5′-CTTTATCTGTCACATACATCG-3′, respectively. The deletion of spyCEP was further confirmed by Western blotting in which the smaller fragment of mature SpyCEP was extracted from GAS bacteria by using a saturated urea solution and detected by Western blotting using polyclonal rabbit antibodies raised against recombinant SP24F1, which was part of SpyCEP and was previously described (30).
Replacement of the covSΔ1bp pseudogene in MGAS5005 with the WT covS gene.
The covSΔ1bp pseudogene of MGAS5005 was replaced with the WT covS gene in two steps. First, a 1,290-bp internal fragment of covSΔ1bp in MGAS5005 was deleted. A plasmid (pΔcovSΔ1bp) used to generate MGAS5005ΔcovS was obtained by sequentially PCR cloning the upstream and downstream flanking fragments of the 1,290-bp fragment into pGRV at the BglII/XhoI and XhoI/BamHI restriction enzyme sites with paired primers 5′-TCGAGATCTGTTAGCTATTTCCGAAATCAG-3′/5′-GTCTCGAGGACTTCATAACCCTCATGTTG-3′ and 5′-GTCTCGAGCGCGGCAAAATTGACATTCCAG-3′/5′-GCGGATCCATTGCAACTTAGTATGTGTCTC-3′, respectively. In the second step, covS was knocked into MGAS5005ΔcovS. A DNA fragment containing covS and the flanking sequences was PCR amplified from MGAS2221. The PCR product was cloned into pGRV at the BglII and BamHI sites, yielding pCovS, which was introduced into MGAS5005ΔcovS by electroporation. The ΔcovS locus in MGAS5005ΔcovS was replaced with the WT covS gene in pCovS through two homologous crossovers, yielding an isogenic mutant of MGAS5005 that carried the WT covS gene (MGAS5005WTcovS), which was confirmed by DNA sequencing.
Mouse infections.
All animal experimental procedures were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (31). The protocol for the animal procedures was approved by the Institutional Animal Care and Use Committee at Montana State University (permit 2011-57).
GAS bacteria grown to mid-exponential phase in THY were harvested by centrifugation, washed three times with pyrogen-free Dulbecco's phosphate-buffered saline (DPBS), and resuspended in DPBS in the desired doses. Because outbred CD-1 Swiss mice were cheaper than BALB/c mice while MGAS5005 shows similar inhibition of neutrophil recruitment in both CD-1 and BALB/c mice, we used outbred, 5-week-old CD-1 female mice from Charles River Laboratories in this study. Groups of 5 or 10 mice were subcutaneously infected with 0.2 ml of GAS suspension with an optical density at 600 nm (OD600) of 0.9 or about 108 CFU of MGAS5005 and/or their isogenic mutant in 0.2 ml DPBS with an OD600 slightly higher or lower than 0.9, depending on the mutant. Actual inocula were determined by plating. For virulence comparisons, infected mice were monitored daily for 14 days to determine survival rates. For other analyses, mice infected were euthanized at 24 h after inoculation to collect skin, liver, and spleen samples. Liver and spleen samples were homogenized in DPBS by using a Kontes pestle. GAS bacterial numbers in the homogenized samples were determined by plating. To measure lesion sizes and neutrophil recruitment, the skin around the infection site was peeled off and the whole infection area was recognized by the boundary of the inflammation and excised; the area was traced on paper for measurement of the infection area by weighing the traced paper.
Quantification of neutrophil infiltration.
Numbers of recruited neutrophils in the excised skin were estimated by the myeloperoxidase assay as described previously (7, 32). Skin samples were ground in 0.5% hexadecyltrimethylammonium bromide in 50 mM potassium and sonicated on ice for 15 s to extract myeloperoxidase. The samples were frozen and thawed three times, sonicated, and centrifuged at 16,000 × g for 5 min, resulting in the supernatant for the following myeloperoxidase assay. The supernatant was added to 0.2 ml of 50 mM phosphate buffer, pH 6.0, containing the extracted myeloperoxidase, 0.167 mg/ml o-dianisidine dihydrochloride, and 0.001% hydrogen peroxide, and the change in absorption at 460 nm (ΔA460) with time was recorded with a SPECTRAmax 384 Plus spectrophotometer (Molecular Devices). The myeloperoxidase activity, ΔA460/min, was converted into the number of neutrophils by using a standard curve of myeloperoxidase activities versus known numbers of murine neutrophils isolated from bone marrow as previously described (33).
Histological analyses.
Skin infection sites were excised with a wide margin after the skin was peeled off and fixed in 10% neutral buffered formalin for 24 h. The samples were dehydrated with ethanol, cleared with xylene, and infiltrated with paraffin using a Tissue Embedding Console System (Sakura Finetek, Inc.). The paraffin blocks was processed to obtain 4-μm sections, which were stained with hematoxylin and eosin (H&E) or with a tissue Gram stain kit from Richard-Allan Scientific according to the manufacturer's protocol. Stained samples were examined by using a Nikon ECLIPSE 80i microscope.
GAS competitive growth assay.
A 0.2-ml volume of a 1:1 MGAS5005ΔspyCEP-MGAS5005, MGAS5005ΔscpA-MGAS5005, or ΔspyCEP ΔscpA Δsse-ΔspyCEP ΔscpA mutant mixture with 0.8 ml air was injected subcutaneously into mice. The mice were euthanized at 24 h after inoculation, and the air sac was lavaged with 1 ml PBS. The lavage samples were plated on THY agar plates. The ratio of the two strains in each lavage sample was determined by analyzing 48 colonies of each sample by colony PCR. The primers used to check ΔspyCEP/WT and ΔscpA/WT ratios were those that were used to confirm the spyCEP and scpA deletions. The primers used to check the ΔspyCEP ΔscpA Δsse/ΔspyCEP ΔscpA mutant ratio were 5′-ATAACATTTACATTAAGGAGATAC-3′ and 5′-CAGATTTGGTGTTTGAAAAAG-3′. In each PCR analysis, the PCR product of deletion mutant was smaller than that of the corresponding WT strain. The mutant/WT GAS ratio in the inoculum was determined by plating the individual GAS suspension prior to mixing. The competitive index was calculated by dividing the mutant/WT GAS ratio in the lavage samples by the ratio in the inoculum.
Other assays.
Quantitative reverse transcription (RT)-PCR analysis for spyCEP, hasA, and sse mRNAs was performed with a specific probe and gyrA as a control, as previously described (13). SpeB activity in the supernatant of overnight GAS cultures was detected by using the casein plate assay as previously described (34).
Statistical analyses.
The Prism software program (GraphPad Software, Inc.) was used for all statistical analyses. Survival data were analyzed by using the log-rank (Mantel-Cox) test. The data in Fig. 4C and D were analyzed by a one-way analysis of variance (ANOVA) Newman-Keuls multiple-comparison test. The data in Fig. 6 were analyzed using one-tailed Student t test. Other P values were obtained by using the two-tailed Mann-Whitney t test.
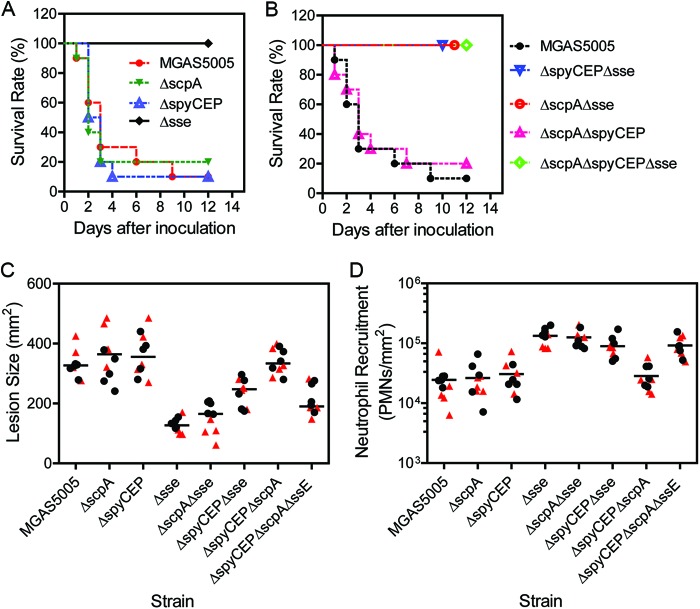
Fig 4.
Effects of single, double, and triple deletions of spyCEP, scpA, and sse on MGAS5005 virulence, skin invasion, and neutrophil recruitment in subcutaneously infection of mice. (A and B) Survival rates of 10 mice subcutaneously infected with 1.0 × 108 CFU MGAS5005, 1.0 × 108 CFU ΔscpA mutant bacteria, 1.5 × 108 CFU ΔspyCEP mutant bacteria, 1.4 × 108 CFU Δsse mutant bacteria, 1.6 × 108 CFU ΔspyCEP Δsse mutant bacteria, 1.6 × 108 CFU ΔscpA Δsse mutant bacteria, 1.4 × 108 CFU ΔscpA ΔspyCEP mutant bacteria, or 1.4 × 108 CFU ΔscpA ΔspyCEP Δsse mutant bacteria.(C and D) Lesion sizes (C) and neutrophil recruitment (D) at 24 h after subcutaneous infection of mice in two independent experiments. In experiment 1 (solid circles), mice were infected with 9.1 × 107 CFU MGAS5005, 1.1 × 108 CFU ΔspyCEP mutant bacteria, 9.2 × 107 CFU ΔscpA mutant bacteria, 1.7 × 108 Δsse mutant bacteria, 1.2 × 108 CFU ΔspyCEP Δsse mutant bacteria, 1.1 × 108 CFU ΔscpA Δsse mutant bacteria, 1.2 × 108 CFU ΔscpA ΔspyCEP mutant bacteria, or 1.2 × 108 CFU ΔscpA ΔspyCEP Δsse mutant bacteria. In experiment 2, mice were infected with 9.3 × 107 CFU MGAS5005, 1.2 × 108 CFU ΔspyCEP mutant bacteria, 1.0 × 108 CFU ΔscpA mutant bacteria, 1.5 × 108 CFU Δsse mutant bacteria, 1.1 × 108 CFU ΔspyCEP Δsse mutant bacteria, 1.0 × 108 CFU ΔscpA Δsse mutant bacteria, 1.2 × 108 CFU ΔscpA ΔspyCEP mutant bacteria, or 1.3 × 108 CFU ΔscpA ΔspyCEP Δsse mutant bacteria. One-way ANOVA of the PMN data: not significant, all pairs among MGAS5005 and the ΔspyCEP, ΔscpA, and ΔspyCEP ΔscpA mutants and pairs among the strains carrying Δsse; significant, all of the other pairs. One-way ANOVA of the lesion data: not significant, the Δsse mutant versus the ΔscpA Δsse mutant, the ΔspyCEP ΔscpA Δsse mutant versus the ΔspyCEP Δsse mutant, MGAS5005 versus the ΔspyCEP mutant, MGAS5005 versus the ΔscpA mutant, MGAS5005 versus the ΔspyCEP ΔscpA mutant, the ΔspyCEP ΔscpA mutant versus the ΔspyCEP mutant, the ΔspyCEP ΔscpA mutant versus the ΔscpA mutant, and the ΔscpA mutant versus the ΔspyCEP mutant; significant, all of the other pairs.
Fig 6.
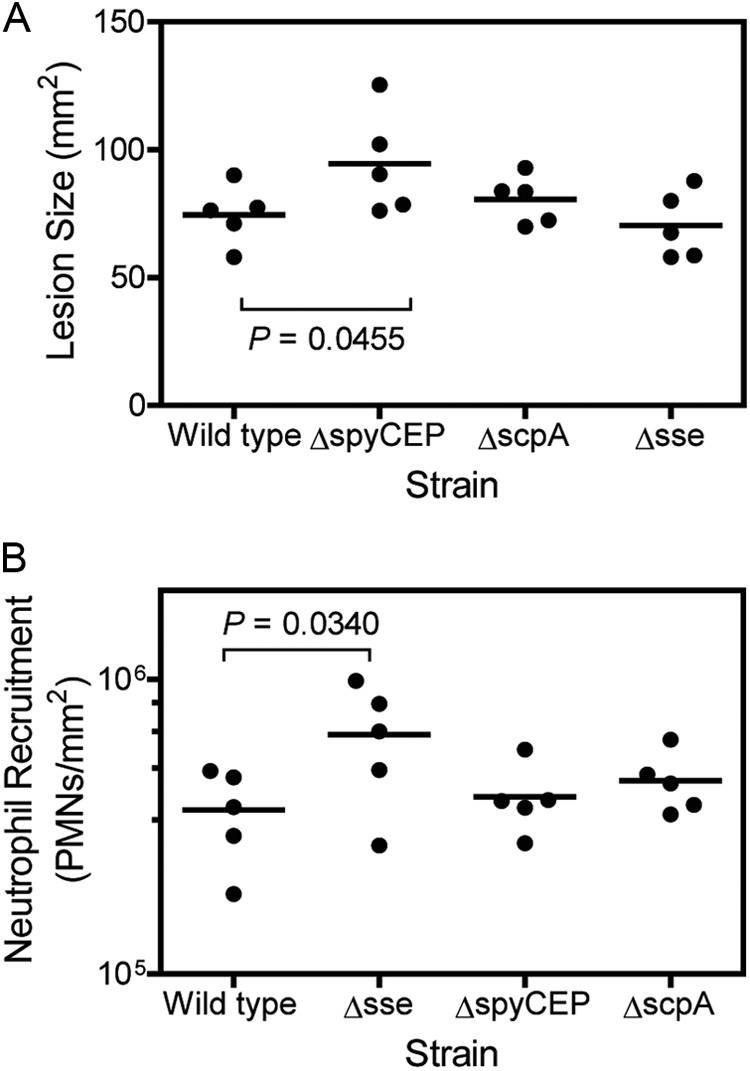
Effects of sse, spyCEP, and scpA deletions of MGAS2221 on skin lesion size (A) and neutrophil recruitment (B). The data were obtained at 24 h after the subcutaneous infection of mice with 1.5 × 108 CFU MGAS2221, 1.4 × 108 CFU MGAS2221ΔspyCEP, 1.4 × 108 CFU MGAS2221ΔscpA, or 1.6 × 108 MGAS2221Δsse.
RESULTS
Distinct neutrophil responses in subcutaneous MGAS2221 and MGAS5005 infections.
Although MGAS2221 and MGAS5005 have almost identical genetic contents (22), they display distinct phenotypes in subcutaneous infections of mice. Inside-out pictures of MGAS5005 and MGAS2221 infection sites display distinct overall pathologies with a greater infection area and less pus-like infiltrate for MGAS5005 than for MGAS2221 (Fig. 1A and B). Myeloperoxidase measurements found that MGAS2221 induced 25-fold greater neutrophil recruitment at the skin infection site than MGAS5005 did [mean neutrophil number ± standard deviation (SD): MGAS5005, (5.6 ± 1.4) × 103 neutrophils/mm2; MGAS2221, (1.4 ± 0.8) × 105 neutrophils/mm2 (P = 0.0046)] (Fig. 1C). The mean skin lesion size ± SD caused by MGAS5005 at 24 h after inoculation was 397 ± 27 mm2, which was 4.7-fold larger than the lesion size caused by MGAS2221 (85 ± 35 mm2) (P < 0.0001) (Fig. 1D). Consequently, MGAS5005 is more virulent than MGAS2221 in the skin infection model (Fig. 1E). The more severe infection phenotype of MGAS5005 is not due to a growth advantage because MGAS2221 grows faster than MGAS5005 in THY (Fig. 1F).
Fig 1.
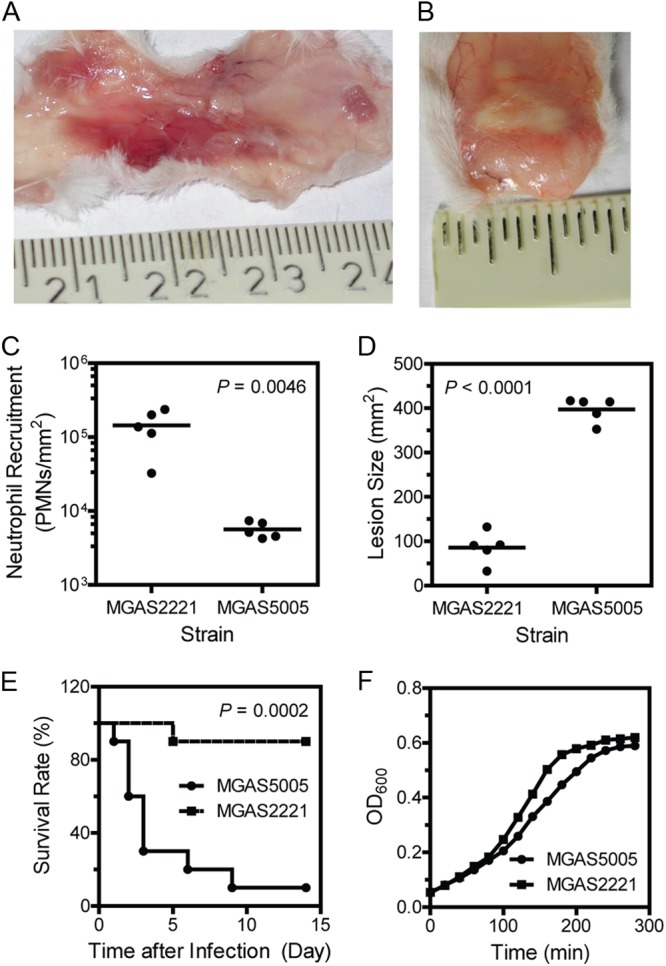
Distinct phenotypes of M1T1 GAS strains MGAS2221 and MGAS5005 in subcutaneous infections of mice. Inside-out pictures of MGAS5005 (A) and MGAS2221 (B) skin infection sites were taken at 24 h after inoculation. (C and D) Neutrophil recruitment at (C) and size of (D) the skin infection sites of mice infected with 1.0 × 108 CFU MGAS2221 or 9.1 × 107 CFU MGAS5005. (E) Survival rates of mice infected with 1.5 × 108 CFU MGAS2221 or 1.0 × 108 CFU MGAS5005. (F) Growth curves of MGAS5005 and MGAS2221 in THY. Each culture at the mid-exponential growth phase was diluted at time zero to start measurements of OD600 over time.
CovRS regulates inhibition of neutrophil infiltration.
Although MGAS5005 and MGAS2221 have almost identical genomes, MGAS5005 has a 1-bp deletion at base 83 of the covS gene, whereas MGAS2221 has the WT covS gene (22). To determine whether the 1-bp deletion in covS of MGAS5005 causes the distinct phenotypes of MGAS5005 and MGAS2221, we first knocked out the covSΔ1bp pseudogene of MGAS5005 and then knocked in the WT covS gene, resulting in MGAS5005WTcovS. covS null mutations cause loss of SpeB production (35, 36). MGAS5005 does not have detectable SpeB activity in vitro (13), and SpeB production should be restored if covSWT is successfully knocked in. Indeed, MGAS5005WTcovS, like MGAS2221, produced detectable SpeB activity (Fig. 2A). The transcription of spyCEP, sse, and hasA in MGAS5005WTcovS was 40-, 50-, and 133-fold lower than that in MGAS5005, respectively, and was similar to that in MGAS2221 (Fig. 2B), further confirming the replacement of covSΔ1bp with covSWT. Thus, we successfully replaced covSΔ1bp of MGAS5005 with the WT covS gene.
Fig 2.
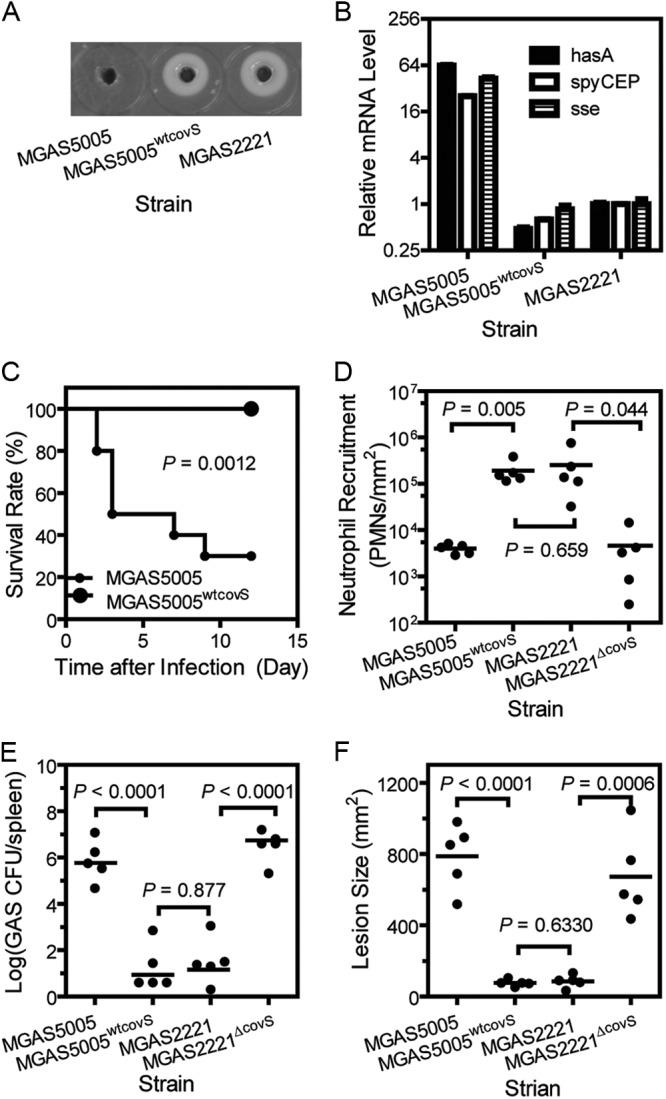
Replacement of covSΔ1bp with covSWT in MGAS5005 and covS deletion from MGAS2221 result in the MGAS2221 and MGAS5005 phenotypes, respectively, in subcutaneous GAS infections of mice. (A) SpeB activity in the culture supernatant of MGAS5005, MGAS5005WTcovS, and MGAS2221 as assessed by the casein hydrolysis plate assay. (B) Relative mRNA levels of the hasA, sse, and spyCEP genes in MGAS5005, MGAS5005WTcovS, and MGAS2221 determined by real-time RT-PCR. (C) Survival rates of mice infected with 9.1 × 107 CFU MGAS5005 or 9.5 × 107 CFU MGAS5005WTcovS. (D to F) Neutrophil recruitment (D), spleen GAS loads (E), and lesion sizes (F) in mice at 24 h after inoculation with 9.9 × 107 CFU MGAS5005, 1.0 × 108 CFU MGAS5005WTcovS, 1.1 × 108 CFU MGAS2221, or 9.5 × 107 CFU MGAS2221ΔcovS.
Next, we compared MGAS5005 and MGAS5005WTcovS for virulence, skin invasion, and neutrophil recruitment in subcutaneous infections of mice. Most of the mice infected with MGAS5005 died, whereas all of the mice infected with MGAS5005WTcovS survived (P = 0.0012) (Fig. 2C). At 1 day after inoculation, the area of MGAS5005WTcovS infection sites (76.5 ± 8 mm2) was 90% smaller than that of MGAS5005 infection sites (786 ± 82 mm2) (P < 0.0001) but was similar to that of MGAS2221 infection sites (85.5 ± 15.9 mm2) (P = 0.63) (Fig. 2F). In addition, the GAS load in the spleens of mice infected with MGAS5005WTcovS was 4.6 orders of magnitude lower than that in the spleens of MGAS5005-infected mice but was similar to that in the spleens of MGAS2221-infected mice (Fig. 2E). The level of neutrophils at MGAS5005WTcovS sites ([1.9 × 105 ± 0.04] neutrophils/mm2) was 34-fold higher than that at MGAS5005 sites ([4.0 × 103 ± 0.04] neutrophils/mm2) and similar to that at MGAS2221 sites ([2.5 × 105 ± 0.13] neutrophils/mm2) (P = 0.65) (Fig. 2D). Thus, the replacement of covSΔ1bp with covSWT enhances neutrophil ingress and reduces skin invasion and systemic dissemination, converting the MGAS5005 phenotype to the MGAS2221 phenotype.
To further confirm this finding, we determined whether deletion of covS from MGAS2221 converts the MGAS2221 phenotype to the MGAS5005 phenotype. The level of neutrophils at MGAS2221ΔcovS sites ([4.6 ± 2.5] × 103 neutrophils/mm2) was 54-fold lower than that at MGAS2221 sites ([2.5 ± 1.3] × 105 neutrophils/mm2) (P = 0.0440) but was similar to that at MGAS5005 infection sites ([4.0 ± 0.4] × 103 neutrophils/mm2) (P = 0.4000) (Fig. 2D). The size of the MGAS2221ΔcovS infection sites (637 ± 107 mm2) was 7-fold larger than that of the MGAS2221 sites (85.5 ± 15.9 mm2) (P = 0.0006) and similar to that of MGAS5005 infection sites (786 ± 82 mm2) (P = 0.2100) (Fig. 2F). While the lesion sizes of MGAS2221 infections in Fig. 1D and 2F were similar, the lesion sizes of MGAS5005 infections in the two experiments were different. This difference was most likely due to the fluctuation of the actual inoculum size. An MGAS5005 suspension with an OD600 of 0.9 was used in both experiments, but the number of viable MGAS5005 bacteria in the inoculum for Fig. 2F was approximately 10% higher than that in the inoculum for Fig. 1D. Despite this difference, these results clearly indicate that the phenotype of MGAS5005 in skin infections is caused by the covS null deletion and that CovRS negatively regulates the inhibition of neutrophil recruitment by GAS.
Relative contributions of SpyCEP, ScpA, and SsE to MGAS5005 skin invasion, virulence, and inhibition of neutrophil recruitment.
Since the expression of spyCEP, scpA, and sse is enhanced by the covS deletion in MGAS5005, we hypothesize that SpyCEP, ScpA, and SsE contribute to the MGASA5005 phenotype. We first tested this hypothesis by determining the relative contributions of these hydrolases to MGAS5005 skin invasion, virulence, and inhibition of neutrophil recruitment. We deleted a 301-bp fragment of the spyCEP gene and a 1,240-bp fragment of the scpA gene. The mutants were identified by PCR and confirmed by DNA sequencing (data not shown). SpyCEP was detected in MGAS5005 by Western blotting but was not found in the ΔspyCEP mutant, confirming the spyCEP deletion (data not shown). Both the spyCEP and scpA deletion mutants produced the M protein at levels that were similar to that produced by MGAS5005, as judged by Western blotting (data not shown). MGAS5005ΔspyCEP and MGAS5005ΔscpA had competitive growth indexes of 0.78 and 0.94, respectively, against MGAS5005 in a mouse air sac infection model. Thus, the deletion of spyCEP or scpA did not have a growth issue or substantially alter emm expression. The competitive growth result of the ΔspyCEP mutant confirms the previous results (10, 37).
The MGAS5005ΔspyCEP and MGAS5005ΔscpA mutants were compared in subcutaneous infections of mice with the parent strain MGAS5005 and its Δsse mutant. MGAS5005ΔspyCEP, MGAS5005ΔscpA, and MGAS5005 infection sites had similar overall pathology, showing extensive GAS spreading and inflammation, whereas the MGAS5005Δsse site was small and appeared to have robust inflammatory cell infiltrate (Fig. 3). Quantitatively, the lesion sizes of MGAS5005ΔSpyCEP ([363 ± 72] mm2) and MGAS5005ΔscpA (358 ± 78) mm2) infections were not significantly different from that of MGAS5005 infections ([331 ± 43] mm2), whereas the MGAS5005Δsse lesion size was significantly smaller ([153 ± 48] mm2) (Fig. 4C). Mice infected with MGAS5005ΔspyCEP (P = 0.9037 versus the WT) or MGAS5005ΔscpA (P = 0.9524 versus the WT) had survival curves similar to that of MGAS5005 (Fig. 4A), whereas all of the mice infected with MGAS5005Δsse survived (P < 0.0001 versus the WT). These results indicate that deletion of spyCEP and scpA did not significantly attenuate MGAS5005 virulence and skin invasion but deletion of sse reduced virulence and skin infection.
Fig 3.
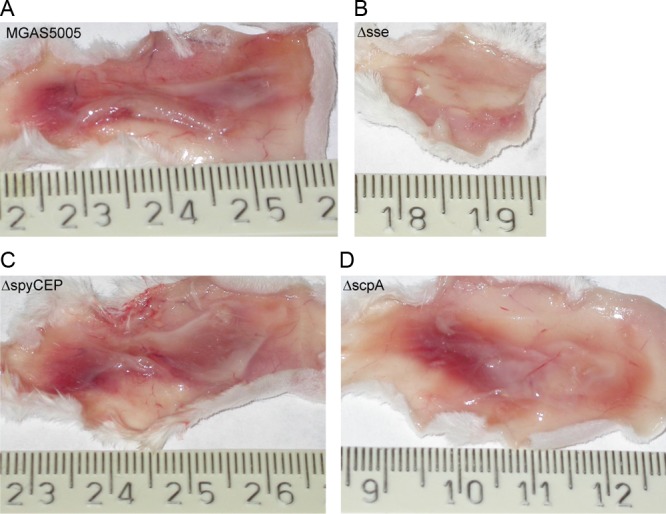
The sse gene, but not spyCEP or scpA, is required for skin invasion by MGAS5005. Shown are representative images of inside-out infection sites at 24 h after subcutaneous infection with 8.4 × 107 CFU MGAS5005 (A), 1.5 × 108 CFU MGAS5005Δsse (B), 8.4 × 107 CFU MGAS5005ΔspyCEP (C), or 1.1 × 108 CFU MGAS5005ΔscpA (D).
Consistent with the virulence and skin invasion results, SsE, but not SpyCEP or ScpA, is required for MGAS5005 inhibition of neutrophil recruitment. The levels of neutrophils at MGAS5005ΔspyCEP infection sites ([3.1 ± 1.8] × 104 neutrophils/mm2) and MGAS5005ΔscpA infection sites ([2.6 ± 1.7] × 104 neutrophils/mm2) were not different from those at MGAS5005 infection sites ([2.5 ± 1.8] × 104 neutrophils/mm2) (Fig. 4D). In contrast, deletion of sse significantly enhanced neutrophil ingress by 5.4-fold ([1.3 ± 0.4] × 105 neutrophils/mm2).
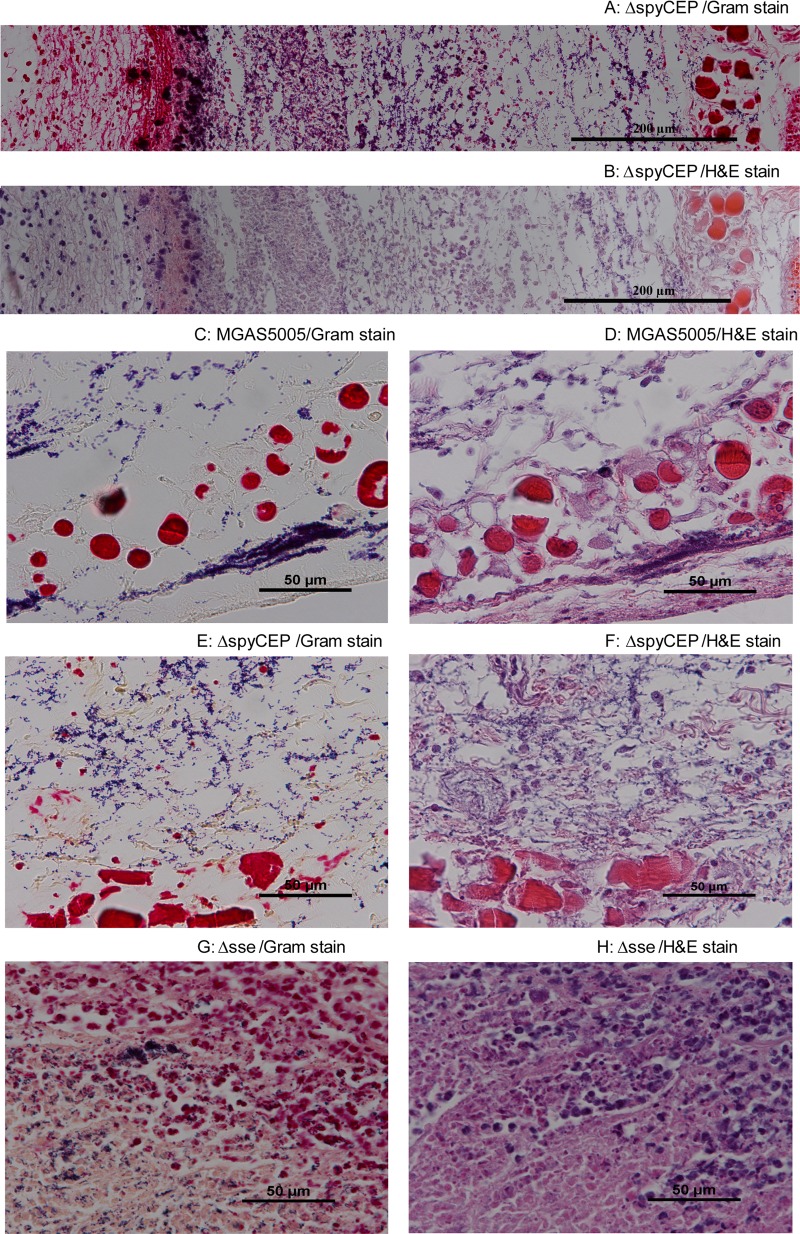
No alteration of the histological pattern by spyCEP deletion.
One feature of the innate immune evasion of MGAS5005 is that it can keep neutrophils at a distance. To determine whether spyCEP is required for this pattern of inhibition of neutrophil infiltration, we examined MGAS5005 and MGAS5005ΔspyCEP infection sites at 24 h after infection by H&E and Gram staining. The inoculation site of MGAS5005ΔspyCEP had a zone of scattered neutrophils at the inner side of the skin (the left side of Fig. 5A and B), which was followed by a band of amorphous material and then by a GAS-containing area (Fig. 5A and B). The bacterial territory had no neutrophils. This pattern is the same as one that was recently described at MGAS5005 infection sites (7). At the spread area, there were sparse neutrophils in both MGAS5005 (Fig. 5C and D) and MGAS5005ΔspyCEP (Fig. 5E and F) infections. This pattern of GAS and neutrophil distribution in MGAS5005 and MGAS5005ΔspyCEP is different from that in MGAS5005Δsse infection sites, where neutrophils march into the area containing bacteria (Fig. 5G and F). The histological findings on MGAS5005Δsse infection of CD-1 mice confirm our previous findings on MGAS5005Δsse infection of BALB/c mice (7). These data further indicate that SsE, but not SpyCEP, plays a critical role in innate immune evasion by MGAS5005.
Fig 5.
Histological analyses showing no difference in the pattern or level of neutrophil infiltration between MGAS5005 and MGAS5005ΔspyCEP. CD1 mice were subcutaneously inoculated in the back with 1.1 × 108 CFU MGAS5005, 1.0 × 108 CFU MGAS5005ΔspyCEP, or 1.3 × 108 CFU MGAS5005Δsse, and skin samples were collected at 24 h after inoculation. (A and B) Microscopic pictures of Gram (A)- and H&E (B)-stained MGAS5005ΔspyCEP samples at the inoculation site were each combined from three snapshots taken at a magnification of ×40. (C to F) Microscopic images of Gram (C and E)- and H&E (D and F)-stained skin samples at the spread areas of MGAS5005 (C and D) and MGAS5005ΔspyCEP (E and F) infection sites. (G and H) Microscopic images of Gram (G)- and H&E (H)-stained skin samples from an MGAS5005Δsse infection site.
No synergistic effect of SsE, SpyCEP, and ScpA on MGAS5005 inhibition of neutrophil infiltration.
Although SpyCEP and ScpA do not individually contribute to MGAS5005 inhibition of neutrophil recruitment, they may have additive effects on innate immune evasion and thus virulence and skin invasion. To test this idea, we also generated double and triple spyCEP, scpA, and sse mutants of MGAS5005. The sse gene was deleted after the spyCEP and/or scpA genes were deleted to obtain the double and triple mutants. The ΔspyCEP ΔscpA Δsse triple mutant had a competitive index of 0.98 against the ΔspyCEP ΔscpA double mutant (data not shown), indicating that the sse deletion in the background of no spyCEP or scpA had no effect on in vivo growth. The ΔspyCEP ΔscpA double deletion mutant induced (2.8 ± 1.4) × 104 neutrophils/mm2 and caused lesions of (341 ± 43) mm2, and these results were not significantly different from those of infections with MGAS5005, MGAS5005ΔspyCEP, and MGAS5005ΔscpA (Fig. 4D). The virulence of the ΔspyCEP ΔscpA mutant was similar to that of MGAS5005, MGAS5005ΔspyCEP, and MGAS5005ΔscpA as well (P = 0.6675 versus the WT, P = 0.5161 versus the ΔspyCEP mutant, and P = 0.6478 versus the ΔscpA mutant) (Fig. 4A and B).
In contrast, the ΔspyCEP Δsse, ΔscpA Δsse, and ΔspyCEP ΔscpA Δsse mutants all induced significantly higher levels of neutrophil recruitment (Fig. 4D), caused significantly larger lesions (Fig. 4C), and had significantly attenuated virulence (Fig. 4A and B) compared with MGAS5005 and its spyCEP and scpA single and double deletion mutants. In addition, the ΔspyCEP Δsse mutant caused significantly larger lesions than the Δsse and ΔscpA Δsse mutants did, suggesting that the spyCEP deletion in the absence of the sse gene enhanced the invasion of skin by MGAS5005. All of these results indicate that SpyCEP and ScpA did not additively contribute to a reduction of neutrophil infiltration by SsE.
Effects of sse, spyCEP, and scpA deletions on MGAS2221 skin invasion and inhibition of neutrophil recruitment.
The data on MGAS5005 inhibition of neutrophil recruitment suggest that CovRS regulates the inhibition of neutrophil recruitment and that SsE is an important factor in this regulation. To determine whether these findings are applicable to GAS with intact CovRS, we constructed MGAS2221ΔspyCEP and MGAS2221ΔscpA and compared them with MGAS2221 and MGAS2221Δsse for skin invasion and neutrophil infiltration in subcutaneous infections of CD-1 mice. All of these mutants had normal M protein production according to Western blotting (data not shown). MGAS2221Δsse induced (6.5 ± 2.8) × 105 neutrophils/mm2, which was 80% higher than the (3.6 ± 1.2) × 105 neutrophils/mm2 at MGAS2221 infection sites (P = 0.0340), whereas the levels of neutrophils at MGAS2221ΔspyCEP ([4.0 ± 1.1] × 105 neutrophils/mm2; P = 0.3068) and MGAS2221ΔscpA ([4.5 ± 1.1] × 105 neutrophils/mm2; P = 0.1225) infection sites were not significantly different from those at MGAS2221 infection sites (Fig. 6B). Deletion of the sse gene slightly decreased the lesion size in CD-1 mice, but the difference was not statistically significant. Deletion of scpA had no effect on skin invasion, whereas deletion of spyCEP significantly increased lesion size (Fig. 6A), confirming previous findings (11). These data indicate that the findings on invasive GAS isolates with nonfunctional CovRS regulation are qualitatively applicable to GAS with intact CovRS. Our results also suggest that CovRS can regulate neutrophil recruitment by regulating the sse gene.
DISCUSSION
This report describes two findings on the innate immune evasion of GAS: (i) the natural covS null deletion in MGAS5005 confers its innate immune evasion phenotype, and (ii) SsE is a dominant factor in MGAS5005 evasion of innate immunity, and SpyCEP and ScpA alone and in combination do not significantly contribute to the inhibition of neutrophil infiltration of MGAS5005. In addition, the relative contributions of SsE, SpyCEP, and ScpA to the inhibition of neutrophil recruitment is correlated with their relative importance for GAS virulence and skin invasion, suggesting that reduced neutrophil ingress is a critical factor in hypervirulence and skin invasion. These findings provide insight into the molecular basis of the regulation of innate immune evasion by CovRS and innate immune evasion by hypervirulent M1T1 GAS isolates.
Sparse neutrophil infiltrate has been documented in necrotizing fasciitis patients (3–5). This phenotype of innate immune evasion can be mimicked in the mouse model of necrotizing fasciitis using invasive isolates (3, 7) but not pharyngitis isolates (7). A novel finding of this study is that covS deletion can result in the phenotype of the severe innate immune evasion, which is correlated with the severity of skin invasion and hypervirulence. This finding indicates that CovRS regulates the inhibition of neutrophil infiltration and covRS null mutations maximize the inhibition of neutrophil infiltration by releasing CovRS repression of virulence factors involved in innate immune evasion. Thus, covS null mutation-enhanced inhibition of neutrophil recruitment is an addition to the list of the determinants of CovRS mutation-mediated progression of invasive GAS infection, which include loss of SpeB production and enhanced production of the hyaluronic acid capsule and DNase Sda1 in covS null mutants (18, 27, 28).
The spyCEP and sse genes are negatively regulated by CovRS. Deletion of covS enhances the expression of spyCEP and sse by ≥40-fold (Fig. 2B), confirming the previous observations (13, 38). Even though scpA is regulated by the positive regulator Mga (39), its expression is also upregulated by covS deletion (22). Thus, the relief of the CovR repression of inhibitors of neutrophil infiltration as a result of covS null mutations is expected to be the reason for the sparse neutrophil infiltrate in hypervirulent GAS infections. We previously showed that deletion of sse enhances neutrophil recruitment and the function of SsE is partly mediated by its platelet-activating factor acetylhydrolase activity (7). Thus, it is not surprising that deletion of sse from ΔspyCEP, ΔscpA, and ΔspyCEP ΔscpA mutants enhanced neutrophil recruitment. However, it is unexpected that SpyCEP and ScpA, both alone and in combination, did not significantly contribute to the inhibition of neutrophil recruitment and SsE is a dominant factor in MGAS5005 inhibition of neutrophil infiltration. Nonetheless, it appears to be true that the relief of CovRS repression of the sse gene as a result of the covS deletion critically contributes to the inhibition of neutrophil recruitment. Deletion of sse, but not spyCEP or scpA, of MGAS2221 significantly enhanced neutrophil recruitment, supporting the idea that the findings associated with invasive GAS isolates with nonfunctional CovRS regulation are qualitatively applicable to GAS with intact CovRS.
ScpA degrades the C5a peptide, and immunization with ScpA prevents nasopharyngeal GAS colonization of mice (40). However, scpA deletion does not affect GAS virulence in subcutaneous infection of mouse tissue (41). Therefore, the insignificant contribution of ScpA to MGAS5005 inhibition of neutrophil recruitment, virulence, and skin invasion is not surprising. However, the insignificant involvement of SpyCEP in MGAS5005 innate immune evasion is a surprise. SpyCEP degrades IL-8/CXC chemokines (3, 8, 10, 11, 36, 42). This protein reduces IL-8/CXC-induced neutrophil transmigration in vitro (10, 19) and confers resistance of GAS to killing by isolated neutrophils (10). Three studies have investigated the contribution of SpyCEP to GAS pathogenesis and inhibition of neutrophil infiltration by using the mouse model of subcutaneous infection, proposing that SpyCEP contributes to the inhibition of neutrophil recruitment (3, 10, 11). However, these studies lack quantitative data on the effect of spyCEP deletion on neutrophil ingress into GAS infection sites. Furthermore, a single spyCEP deletion mutant was not available for one of these studies (3). Thus, whether SpyCEP is a critical factor in GAS inhibition of neutrophil recruitment has not been firmly established. Our quantitative analyses of neutrophil ingress indicate that SpyCEP is dispensable to the inhibition of neutrophil recruitment by the hypervirulent M1T1 isolate. Our results suggest that SpyCEP is not critical for covS null mutation/deletion-induced enhancement of the inhibition of neutrophil recruitment.
Sumby et al. found that skin lesion size was increased following infection with a ΔspyCEP mutant of MGAS2221, a M1T1 isolate with the covRS genes intact (11). Our test, which used MGAS2221 and our own MGAS2221ΔspyCEP mutant, confirmed the findings of Sumby et al. In a similar dermonecrosis model, two other groups found that the lesion size was reduced with a spyCEP mutant (3, 10). The discrepancy could be due to the use of different mice and GAS isolates in these studies. Although the deletion of spyCEP in MGAS5005 did not significantly affect lesion size, the deletion of spyCEP in sse-lacking MGAS5005 significantly increased skin invasion. The impact of the spyCEP deletion on skin invasion by MGAS5005 could be masked by the high capacity of MGAS5005 to invade skin. Sumby et al. proposed that the increased lesion size in a ΔspyCEP mutant infection is caused by enhanced neutrophil infiltration as a result of spyCEP deletion (11). Our neutrophil influx data are not consistent with this proposal, suggesting that SpyCEP has another functional mechanism in addition to IL-8/CXC degradation. SpyCEP has been shown to be sufficient for GAS dissemination in mouse models of muscular and intranasal infections by heterologous expression of SpyCEP in Lactococcus lactis (42). The enhanced skin invasion in the absence of SpyCEP and the persistence of SpyCEP-expressing L. lactis might be due to another function of SpyCEP in promoting GAS uptake by endothelial cells (43).
MGAS5005 grows more slowly than MGAS2221 in vitro. This growth difference is likely due to the higher consumption of energy because of the enhanced production of virulence factors by MGAS5005 as a result of the covS deletion. Although MGAS2221 grows faster, it is less virulent than MGAS5005. Thus, the ability to evade the innate immune system for in vivo survival appears to be more important for GAS virulence than the capacity to grow.
In summary, a natural covS null deletion is shown to greatly enhance the inhibition of neutrophil infiltration, skin invasion, and GAS dissemination, and Sse, but not SpyCEP or ScpA, plays a dominant role in the covS deletion-caused enhancement of GAS inhibition of neutrophil infiltration, skin invasion, and virulence. The findings indicate that CovRS regulates neutrophil infiltration and that covS deletion-enhanced expression of sse, but not the enhanced expression of spyCEP, is a critical factor in the severe inhibition of neutrophil recruitment and hypervirulence, thereby advancing our understanding of the molecular basis of innate immune evasion by GAS and the progression of invasive GAS infections.
ACKNOWLEDGMENTS
This work was supported in part by grants AI095704, AI097703, and GM103500-09 from the National Institutes of Health and the Montana State Agricultural Experimental Station. J.L. was supported by a Ph.D. student exchange scholarship from the Ministry of Education, China. The work done at Harbin Medical University was supported by grant LC2011C02 from the Natural Science Foundation of Heilongjiang Province and a grant from the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State of Education Ministry, China.
Footnotes
Published ahead of print 14 January 2013
REFERENCES
- 1. O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000-2004. Clin. Infect. Dis. 45: 853–862 [DOI] [PubMed] [Google Scholar]
- 2. Olsen RJ, Musser JM. 2010. Molecular pathogenesis of necrotizing fasciitis. Annu. Rev. Pathol. 5: 1–31 [DOI] [PubMed] [Google Scholar]
- 3. Hidalgo-Grass C, Mishalian I, Dan-Goor M, Belotserkovsky I, Eran Y, Nizet V, Peled A, Hanski E. 2006. A streptococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 25: 4628–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bakleh M, Wold LE, Mandrekar JN, Harmsen WS, Dimashkieh HH, Baddour LM. 2005. Correlation of histopathologic findings with clinical outcome in necrotizing fasciitis. Clin. Infect. Dis. 40: 410–414 [DOI] [PubMed] [Google Scholar]
- 5. Cockerill FR, Thompson RL, Musser JM, Schlievert PM, Talbot J, Holley KE, Harmsen WS, Ilstrup DM, Kohner PC, Kim MH, Frankfort B, Manahan JM, Steckelberg JM, Roberson F, Wilson WR. 1998. Molecular, serological, and clinical features of 16 consecutive cases of invasive streptococcal disease. Clin. Infect. Dis. 26: 1448–1458 [DOI] [PubMed] [Google Scholar]
- 6. Taylor FB, Bryant AE, Blick KE, Hack E, Jansen PM, Kosanke SD, Stevens DL. 1999. Staging of the baboon response to group A streptococci administered intramuscularly: a descriptive study of the clinical symptoms and clinical chemical response patterns. Clin. Infect. Dis. 29: 167–177 [DOI] [PubMed] [Google Scholar]
- 7. Liu M, Zhu H, Li J, Garcia CC, Feng W, Kirpotina LN, Hilmer J, Tavares LP, Layton AW, Quinn MT, Bothner B, Teixeira MM, Lei B. 2012. Group A Streptococcus secreted esterase hydrolyzes platelet-activating factor to impede neutrophil recruitment and facilitate innate immune evasion. PLoS Pathog. 8: e1002624 doi:10.1371/journal.ppat.1002624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edwards RJ, Taylor GW, Ferguson M, Murray S, Rendell N, Wrigley A, Bai Z, Boyle J, Finney SJ, Jones A, Russell HH, Turner C, Cohen J, Faulkner L, Sriskandan S. 2005. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J. Infect. Dis. 192: 783–790 [DOI] [PubMed] [Google Scholar]
- 9. Wexler DE, Chenoweth DE, Cleary PP. 1985. Mechanism of action of the group A streptococcal C5a inactivator. Proc. Natl. Acad. Sci. U. S. A. 82: 8144–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, Mishalian I, Sriskandan S, Hanski E, Nizet V. 2008. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe 4: 170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sumby P, Zhang S, Whitney AR, Falugi F, Grandi G, Graviss EA, Deleo FR, Musser JM. 2008. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect. Immun. 76: 978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M, Zhu H, Zhang J, Lei B. 2007. Active and passive immunizations with the streptococcal esterase Sse protect mice against subcutaneous infection with group A streptococci. Infect. Immun. 75: 3651–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu H, Liu M, Sumby P, Lei B. 2009. The secreted esterase of group A Streptococcus is important for invasive skin infection and dissemination in mice. Infect. Immun. 77: 5225–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perez-Casal J, Caparon MG, Scott JR. 1992. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res. Microbiol. 143: 549–558 [DOI] [PubMed] [Google Scholar]
- 15. Ashbaugh CD, Moser TJ, Shearer MH, White GL, Kennedy RC, Wessels MR. 2000. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell. Microbiol. 2: 283–292 [DOI] [PubMed] [Google Scholar]
- 16. Timmer AM, Timmer JC, Pence MA, Hsu LC, Ghochani M, Frey TG, Karin M, Salvesen GS, Nizet V. 2009. Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J. Biol. Chem. 284: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyoshi-Akiyama T, Takamatsu D, Koyanagi M, Zhao J, Imanishi K, Uchiyama T. 2005. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J. Infect. Dis. 192: 107–116 [DOI] [PubMed] [Google Scholar]
- 18. Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13: 981–985 [DOI] [PubMed] [Google Scholar]
- 19. Ato M, Ikebe T, Kawabata H, Takemori T, Watanabe H. 2008. Incompetence of neutrophils to invasive group A Streptococcus is attributed to induction of plural virulence factors by dysfunction of a regulator. PLoS One 3: e3455 doi:10.1371/journal.pone.0003455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. 2010. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 6: e1000832 doi:10.1371/journal.ppat.1000832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Engleberg NC, Heath A, Miller A, Rivera C, DiRita VJ. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183: 1043–1054 [DOI] [PubMed] [Google Scholar]
- 22. Sumby P, Whitney AR, Gravis EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2: e5 doi:10.1371/journal.ppat.0020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heath A, DiRita VJ, Barg NL, Engleberg NC. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67: 5298–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Federle MJ, McIver KS, Scott JR. 1999. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J. Bacteriol. 181: 3649–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lei B, DeLeo FR, Hoe NP, Graham MR, Mackie SM, Cole RL, Liu M, Hill HR, Low DE, Federle MJ, Scott JR, Musser JM. 2001. Evasion of human innate and acquired immunity by a bacterial homolog of CD11b that inhibits opsonophagocytosis. Nat. Med. 7: 1298–1305 [DOI] [PubMed] [Google Scholar]
- 26. Treviño J, Perez N, Ramirez-Peña E, Liu Z, Shelburne SA, III, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect. Immun. 77: 3141–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miyoshi-Akiyama T, Ikebe T, Watanabe H, Uchiyama T, Kirikae T, Kawamura Y. 2006. Use of DNA arrays to identify a mutation in the negative regulator, csrR, responsible for the high virulence of a naturally occurring type M3 group A streptococcus clinical isolate. J. Infect. Dis. 193: 1677–1684 [DOI] [PubMed] [Google Scholar]
- 28. Horstmann N, Sahasrabhojane P, Suber B, Kumaraswami M, Olsen RJ, Flores A, Musser JM, Brennan RG, Shelburne SA., III 2011. Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog. 7: e1002311 doi:10.1371/journal.ppat.1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu M, Hanks TS, Zhang J, McClure MJ, Siemsen DW, Elser JL, Quinn MT, Lei B. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lei B, Mackie S, Lukomski S, Musser JM. 2000. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun. 68: 6807–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. National Research Council 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC [Google Scholar]
- 32. Bradley PP, Priebat DA, Christensen RD, Rothstein G. 1982. Measurement of cutaneous inflammation: estimation of neutrophil content with an enzyme marker. J. Investig. Dermatol. 78: 206–209 [DOI] [PubMed] [Google Scholar]
- 33. Siemsen DW, Schepetkin IA, Kirpotina LN, Lei B, Quinn MT. 2007. Neutrophil isolation from nonhuman species. Methods Mol. Biol. 412: 21–34 [DOI] [PubMed] [Google Scholar]
- 34. Ma Y, Bryant AE, Salmi DB, Hayes-Schroer SM, McIndoo E, Aldape MJ, Stevens DL. 2006. Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J. Bacteriol. 188: 7626–7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51: 123–134 [DOI] [PubMed] [Google Scholar]
- 36. Engleberg NC, Heath A, Vardaman K, DiRita VJ. 2004. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect. Immun. 72: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiappini N, Seubert A, Telford JL, Grandi G, Serruto D, Margarit I, Janulczyk R. 2012. Streptococcus pyogenes SpyCEP influences host-pathogen interactions during infection in a murine air pouch model. PLoS One 7: e40411 doi:10.1371/journal.pone.0040411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turner CE, Kurupati P, Jones MD, Edwards RJ, Sriskandan S. 2009. Emerging role of the interleukin-8 cleaving enzyme SpyCEP in clinical Streptococcus pyogenes infection. J. Infect. Dis. 200: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McIver KS, Heath AS, Green BD, Scott JR. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A Streptococcus. J. Bacteriol. 177: 6619–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji Y, Carlson B, Kondagunta A, Cleary PP. 1997. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65: 2080–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ji Y, McLandsborough L, Kondagunta A, Cleary PP. 1996. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect. Immun. 64: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kurupati P, Turner CE, Tziona I, Lawrenson RA, Alam FM, Nohadani M, Stamp GW, Zinkernagel AS, Nizet V, Edwards RJ, Sriskandan S. 2010. Chemokine-cleaving Streptococcus pyogenes protease SpyCEP is necessary and sufficient for bacterial dissemination within soft tissues and the respiratory tract. Mol. Microbiol. 76: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaur SJ, Nerlich A, Bergmann S, Rohde M, Fulde M, Zähner D, Hanski E, Zinkernagel A, Nizet V, Chhatwal GS, Talay SR. 2010. The CXC chemokine-degrading protease SpyCep of Streptococcus pyogenes promotes its uptake into endothelial cells. J. Biol. Chem. 285: 27798–82705 [DOI] [PMC free article] [PubMed] [Google Scholar]