Abstract
Brucella is responsible for brucellosis, one of the most common zoonoses worldwide that causes important economic losses in several countries. Increasing evidence indicates that adhesion of Brucella spp. to host cells is an important step to establish infection. We have previously shown that the BmaC unipolar monomeric autotransporter mediates the binding of Brucella suis to host cells through cell-associated fibronectin. Our genome analysis shows that the B. suis genome encodes several additional potential adhesins. In this work, we characterized a predicted trimeric autotransporter that we named BtaE. By expressing btaE in a nonadherent Escherichia coli strain and by phenotypic characterization of a B. suis ΔbtaE mutant, we showed that BtaE is involved in the binding of B. suis to hyaluronic acid. The B. suis ΔbtaE mutant exhibited a reduction in the adhesion to HeLa and A549 epithelial cells compared with the wild-type strain, and it was outcompeted by the wild-type strain in the binding to HeLa cells. The knockout btaE mutant showed an attenuated phenotype in the mouse model, indicating that BtaE is required for full virulence. BtaE was immunodetected on the bacterial surface at one cell pole. Using old and new pole markers, we observed that both the BmaC and BtaE adhesins are consistently associated with the new cell pole, suggesting that, in Brucella, the new pole is functionally differentiated for adhesion. This is consistent with the inherent polarization of this bacterium, and its role in the invasion process.
INTRODUCTION
Brucella species are responsible for brucellosis, one of the most widespread zoonotic diseases in the world that affects a range of different mammals. Currently, there are 10 recognized species of Brucella, based on host preferences, pathogenicity, and other phenotypic differences. Brucellosis remains endemic in most areas of the world; several animal species with agricultural relevance are susceptible to the disease, and there are also wildlife reservoirs of Brucella, which makes eradication more complicated (1). In particular, Brucella abortus, Brucella melitensis, and Brucella suis are the most economically significant pathogens of the group. Human brucellosis is caused by accidental contamination with reproductive or mammary secretions from infected animals (2, 3) and is characterized by a strong undulating fever, which, if untreated, may lead to a chronic stage of the disease and the appearance of serious complications such as endocarditis, osteoarthritis, and neurological disorders (3).
The preferred niche of Brucella is intracellular; when brucellae are ingested or inhalated, they penetrate through mucosal surfaces, and subsequently are transported to the lymph nodes by macrophages. Brucella spp. are able to infect and survive within macrophages in a compartment derived from the endoplasmic reticulum (4–6). It was shown that Brucella also has the ability to infect a great variety of host cells, including epithelial cells, placental trophoblasts, neurons, and cells from male and female reproductive tissues, to name a few (6, 7). Brucella has a marked ability to elude some of the basic mechanisms of the host immune system, which partly explains the frequent relapse after treatment (8, 9).
It was shown that initial adhesion of several pathogens to the host cell surface is a critical step in the infection process that helps the pathogens to avoid rapid clearance from host tissues (10). Several bacterial filamentous or nonpolymeric adhesins have been shown to participate in the primary interaction with the host cells as well as with components of the extracellular matrix (ECM) (11). Cumulative evidence supports the idea that adhesion to the ECM or to host cells is also an important step for Brucella infection (12–16). It was proposed that binding of Brucella to the host is mediated by host molecules containing sialic acid and/or sulfated residues and by components of the ECM such as fibronectin, collagen, and vitronectin. By phage display, we have recently identified a unipolar fibronectin-binding protein belonging to the type I (monomeric) autotransporter family from B. suis, which we called BmaC (15). Autotransporters are a superfamily of proteins produced by Gram-negative bacteria that share a domain organization composed of an N-terminal functional domain named the “passenger” domain and a C-terminal translocator β-domain that exhibits a β-barrel structure with a hydrophilic pore. Autotransporters are transported by the Sec complex across the inner membrane, and the β-domain subsequently transports the covalently linked passenger domain across the outer membrane (17). Several sets of evidence indicated that BmaC is involved in the polar binding of single bacteria to epithelial cells (15). A comprehensive bioinformatics search allowed the identification of several additional putative adhesins or invasins. One such candidate corresponded to a protein named BtaE that is predicted to belong to the type II (trimeric) autotransporter family. In this work, we show that BtaE participates in the binding of B. suis to hyaluronic acid and epithelial cells and is required to achieve full infectivity in the mouse model. We also analyzed the localization of BtaE on the bacterial surface and found that it showed unipolar localization, which has not been previously reported for a trimeric autotransporter (TA). This observation, together with the previous evidence indicating that BmaC also shows unipolar localization (15), prompted us to determine whether BtaE or BmaC or both are localized at a particular pole, using markers of the new and old poles (18–20). Our observations show that BtaE and BmaC are exclusively associated with the new pole, suggesting that this pole in Brucella is specialized for adhesion. These data support the concept that bacterial polarity is an important feature of Brucella physiology.
MATERIALS AND METHODS
Bacterial strains, cell culture, and media.
Escherichia coli strains used in this study (DH5α, K-12, and derivatives) were grown at 37°C in Luria-Bertani (LB) medium. Antibiotics were added when needed: ampicillin at 200 μg/ml, chloramphenicol (Cm) at 50 μg/ml, and tetracycline at 5 μg/ml. Brucella suis M1330 (ATCC 23444) and derivative strains were grown at 37°C in Bacto tryptic soy broth (TSB; Bacto). When necessary, antibiotics were added: chloramphenicol at 6 μg/ml, kanamycin (Km) at 50 μg/ml, and nalidixic acid (Nal) at 10 μg/ml. HeLa cells and lung epithelial A549 (ATCC CCL-185) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) and murine J774 macrophages in RPMI (Gibco) media; both were supplemented with 5% fetal calf serum (PAA), at 37°C in a 5% CO2 atmosphere.
Molecular techniques.
All DNA manipulations were carried out by using standard procedures. Sequencing was done by cycle sequencing performed with a BigDye kit (Applied Biosystems) in a 3130 Genetic Analyzer (Applied Biosystems). Sequence data of B. suis were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org). PCR primers containing added restriction enzyme sites were designed to amplify the flanking regions of btaE. Primers F1 (5′-CGGGATCCATCGAGTGAGGCAAGAGTC-3′) and R1 (5′-GGCTGCAGCATGAGATATAATCTCTTAT-3′) amplified a region of 463 bp upstream of btaE, and primers F2 (5′-CGCTGCAGTGGAGCATAATCCGACCGCAGC-3′) and R2 (5′-GCGCATGCGTCACACACCAATGTCCAGT-3′) amplified a region of 332 bp downstream of the gene. New restriction sites are underlined. The generated PCR fragments were purified, cleaved with the corresponding enzymes, ligated together, and cloned into the pK18mobsacB mobilizable suicide vector (Km resistant) (21), obtaining pKmobsacΔbtaE plasmid. This plasmid was conjugated into a nalidixic-resistant derivative of the wild-type strain B. suis M1330, and double-recombinant clones were selected (Nalr, Kms, sucrose resistant). The deletion of the open reading frame (ORF) was confirmed by PCR and reverse transcription-PCR (RT-PCR).
To provide a phenotypic confirmation of the B. suis ΔbtaE mutant, the entire btaE gene (including its own promoter) was cloned into a broad-host-range plasmid. A region of 2,839 bp containing 565 bp of the upstream region, the entire btaE ORF, and 54 bp corresponding to the downstream region was amplified using the following primers: FComp (5′-AGCTCGAGATGCCTGTTACCATGCGCCGCAGCG-3′) and RComp (5′-CTACTAGTGTCGGATTATGCTCCAAGACTGATCT-3′). New restriction sites are underlined. The amplified product was cloned into the pBBR1MCS-1 mid-copy-number plasmid (22), obtaining the pBBRbtaE plasmid, which was conjugated into the B. suis ΔbtaE strain. Complemented clones were selected (Cmr) and confirmed by PCR and RT-PCR.
E. coli K-12 was transformed with pBBRbtaE or empty pBBR1MCS vector as a control. Transformed clones were selected (Cmr) and confirmed by PCR.
Bioinformatics.
To identify putative adhesins, adhesion domains of characterized adhesins from other bacteria were compared to the entire Brucella suis 1330 genome using the Protein Motif Search Tool at the J. Craig Venter Institute Comprehensive Microbial Resource (JCVI CMR). Domains displayed by each candidate were analyzed with Pfam (23), BLAST (24), and daTAA (25), which is specific for trimeric autotransporters. Secretion signal was identified with the SignalP Server (26). Alignments of the proteins were made with ClustalX (1.81) (27).
Binding to ECM components.
Analysis of the affinity of BtaE-expressing bacteria to extracellular matrix (ECM) components was performed as described elsewhere (28). Briefly, 96-well plates (Nunc Maxisorp) were coated overnight at 4°C with 50 μl of 100 μg/ml solutions of the ligands, dissolved in phosphate-buffered saline (PBS). Bacteria were grown overnight, washed, and resuspended in PBS to a final concentration of 1 × 109 CFU/ml (optical density [OD] = 1 for E. coli and OD = 0.2 for Brucella). Wells were washed three times with 100 μl PBS to eliminate unbound ligand. Then, 50 μl of bacterial suspensions was added to each well and incubated at 37°C for 3 h. After incubation, wells were washed three times with PBS to remove nonadherent bacteria, and then adherent bacteria were harvested with trypsin-EDTA (0.05% trypsin [Gibco]–0.5% EDTA [USB]). After incubation for 10 min at 37°C, serial dilutions were done, followed by plating on LB or TSB agar with the appropriate antibiotic, and CFU were determined.
Antibodies.
Polyclonal antiserum was obtained by immunization of mice with a peptide corresponding to a portion of the BtaE passenger domain (Fig. 1, underlined region). The coding sequence was amplified using the primers FAb (5′-ATCATATGCTGCATGATATTGAAAGTGGC-3′) and RAb (5′-CTGTCGACTCACGCTTCACCAAGCG-3′) and cloned into pET28a (Novagen). New restriction sites are underlined. The recombinant peptide was purified by high-pressure liquid chromatography (HPLC) and then used for immunization of 10 C57 male mice. Animal procedures were performed in compliance with institutional as well as governmental rules and regulations. Anti-BmaC antibodies were already used in a previous publication (15).
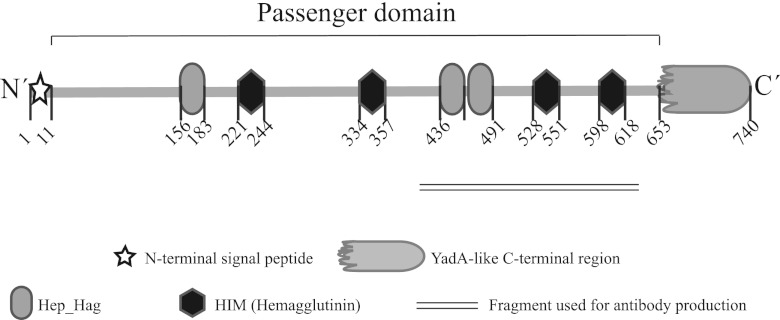
Fig 1.
BtaE domain organization. A schematic representation of BtaE showing the N-terminal signal, the conserved domains, and motifs predicted by bioinformatics (SignalP, Pfam, and BLAST) is presented. Numbers indicate amino acid positions within BtaE.
Western blot analysis.
The method used to obtain total membranes from Brucella strains was based on a protocol previously described (29). Briefly, 50-ml TSB cultures were harvested, resuspended in 1 ml of cold 20 mM Tris-HCl buffer (pH 8.0)–1 mM phenylmethylsulfonyl fluoride (PMSF), and disrupted using Precellys (Bertin Technologies). The homogenate was centrifuged twice at 4,700 relative centrifugal force (RCF) at 4°C for 10 min to remove unbroken cells and Precellys beads, and the supernatant was ultracentrifuged at 134,000 RCF for 90 min at 4°C. The pellet was suspended in 1% Sarkosyl (sodium lauroyl sarcosinate)–20 mM HEPES buffer and incubated for 30 min at room temperature, and then the suspension was centrifuged again at 134,000 RCF for 90 min. The pellet was resuspended in 300 μl 20 mM HEPES buffer, 700 μl of formic acid was added, and the suspension was incubated overnight at room temperature in darkness. One milliliter of distilled H2O was added, and the sample was lyophilized. Cracking buffer was used to resuspend the resulting powder, and the suspension was heated for 10 min at 95°C and subjected to sodium dodecyl sulfate (SDS)-PAGE. Proteins resolved by SDS-PAGE were transferred to polyvinylidene difluoride (PDVF) membranes (GE Healthcare) and blocked overnight with Tris-buffered saline (TBS)–5% (wt/vol) milk powder at 4°C with gentle agitation. Blots were probed with polyclonal mouse anti-BtaE serum (1:8,000) and a goat horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (1:30,000; Santa Cruz) and then revealed using ECL Plus (Amersham). BtaE antiserum was preadsorbed with E. coli carrying empty pET28a.
Immunofluorescence analysis.
Brucella was transformed with a derivative of pBBR1MCS-2 (30), which carries gfp constitutively expressed (excitation at 488 nm, emission at 509 nm; filters used a bandpass [BP] range of 505 to 530 nm) (15). The strains carrying polar markers fused to fluorescent proteins were obtained as previously described by biparental conjugation with E. coli S17.1 strains carrying either pdhS-egfp (19) or aidB-yfp (18).
To detect BtaE on the bacterial surface, 1 ml of a culture of fluorescent bacteria (green fluorescent protein [GFP]-tagged brucellae) or strains carrying either PdhS-enhanced GFP (PdhS-eGFP) or AidB-yellow fluorescent protein (AidB-YFP) was refreshed and grown for another hour in TSB media. The cells were then collected, washed once with PBS, and incubated with 3.7% paraformaldehyde for 15 min at 37°C. The cells were washed twice with PBS and once with PBS–1% bovine serum albumin (BSA) and then incubated 30 min with 40 μl of anti-BtaE or anti-BmaC antibodies (diluted 1:50 in PBS containing 0.5% BSA). The cells were washed again three times with PBS–1% BSA and incubated with 40 μl of IgG-CY3-conjugated donkey anti-mouse antibody preparation (Jackson ImmunoResearch) and diluted 1:250 in PBS containing 0.5% BSA for 40 min. In the case of the strains carrying the pole markers, DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA at a 1 μg/ml final concentration simultaneously with the secondary antibody used for the immunodetection of BtaE or BmaC. After two washes with 1 ml PBS–1% BSA and one wash with PBS, an aliquot of the cell suspension was mounted with Mowiol 4-88 (Calbiochem) on a microscope slide, or on agarose pads. Samples were covered with a coverslip and observed either with a Carl Zeiss LSM 5 Pascal laser scanning microscope, using a Plan-Aprochromat 100×/1.4 oil differential interference contrast (DIC) objective, or, in the case of fluorescent markers, with a Carl Zeiss LSM 510 Meta laser scanning microscope, using a Plan-Apochromat 60×/1.4 oil DIC objective. All incubations were carried out at room temperature.
Cell infection assays.
HeLa or A549 (ATCC CCL 185) cells were seeded in 24-well plates (5 × 104 cells per well) and inoculated with the different strains (multiplicity of infection [MOI], 100:1). Plates were centrifuged for 10 min at 170 RCF and taken to a 5% CO2 atmosphere at 37°C for 1 h. To determine the total number of bacteria associated to the cells, wells were washed three times with PBS and treated for 10 min at 37°C with 0.1% Triton X-100 (in deionized sterile water). Serial dilutions of the obtained lysates were done in PBS and plated on TSA with the appropriate antibiotics, and CFU were determined. To determine the number of intracellular viable bacteria, a standard gentamicin protection assay was performed: 1 h postinfection (p.i.), the culture medium was changed to a fresh one containing 20 μg/ml gentamicin to eliminate remaining extracellular bacteria. After 1 h (5% CO2 atmosphere at 37°C), wells were washed three times with PBS and treated with 0.1% Triton X-100 as previously described. Lysates were serially diluted and plated on TSA with the appropriate antibiotics, and CFU were determined. The number of adherent bacteria was calculated as the difference between the total bacteria associated to the cells and the intracellular bacteria. All the results were expressed in reference to the wild-type strain, defined as 100%. For the inhibition experiment with hyaluronic acid, wild-type or mutant bacteria were preincubated with different concentrations of hyaluronic acid for 30 min at 37°C and bacterial adhesion to HeLa cells was determined as described above.
In the competition assay with the wild-type and mutant strains, the final MOI was maintained; for that, equal quantities of CFU of each strain were inoculated (MOI, 50:1 each). In order to determine the number of intracellular and total bacteria corresponding to each strain, the wild-type and mutant strains were labeled with different antibiotic resistances. The wild-type strain was transformed with a plasmid derivative of pBBR1MCS-2 that confers resistance to kanamycin (Km) (30). On the other hand, the gentamicin resistance (Gmr) gene from the miniTn7(Gm)PA1/04/03 HcRed-a plasmid (Gmr Cmr) (31) was removed by deletion using the PstI and ApaI restrictions sites, due to international restrictions with respect to antibiotic use for Brucella. The mini-Tn7 HcRed transposon (now Cmr, Gms) was inserted in the genome of the btaE mutant, generating a Cmr strain (31). Cellular lysates that included the bacteria were plated on TSA with either Cm or Km to determine the CFU of each strain. Both the wild-type and ΔbtaE B.suis strains, carrying the pBBR1MCS-2 (Kmr) and the miniTn7 (Cmr), respectively, were shown to grow normally in liquid or semisolid TSB media.
Virulence in BALB/c mice.
Six-to-8-week-old female BALB/c mice were purchased from the animal facility at Leloir Institute, Argentina. They were randomly distributed in experimental groups at least 1 week before being inoculated. The animals were housed in filter-ventilated containment in the Laboratory Animal Facility of the Facultad de Ciencias Veterinarias (Tandil, Argentina), receiving water and food ad libitum. All experimental protocols were performed according to the premise of minimizing the suffering to which animals are exposed and using the minimum number of experimental animals to ensure statistically significant results. Animal procedures and management protocols were approved by the local Institutional Animal Welfare Committee according to the Animal Welfare Policy (Act 087/02) of the Faculty of Veterinary Medicine (Universidad Nacional del Centro de la Provincia de Buenos Aires [U.N.C.P.B.A.], Tandil, Argentina; http://www.vet.unicen.edu.ar). BALB/c mice (10 per group) were inoculated by intragastric delivery of wild-type B. suis 1330 (1.3 × 108 CFU/mouse), its isogenic ΔbtaE mutant (1.3 × 108 CFU/mouse), or the complemented mutant (1.48 × 108 CFU/mouse) suspended in 300 μl of 10% sodium bicarbonate by use of a plastic feeding tube introduced through the mouth. Five mice from each group were sacrificed at 7 and 30 days postinfection (p.i.), and spleens were removed. Dilutions of spleen homogenates were plated in duplicate on TSA. After 4 days of incubation, CFU were counted and expressed as the log10 value per spleen. The CFU data were normalized by log transformation and evaluated by analysis of variance (ANOVA) followed by Dunnett's post hoc test (Prism 5.0; GraphPad Software, Inc.). The experiment was repeated twice with similar results.
RESULTS
BtaE: a putative adhesin from the trimeric autotransporter family.
Database searches within the NCBI genomic BLAST search engine showed the presence in the B. suis 1330 genome of 11 predicted ORFs that exhibit one or more conserved domains associated with adhesion, including BmaC (15). One of these candidates (BR0072) is annotated in the B. suis 1330 genome (32) as an “outer membrane protein” and has a predicted molecular mass of 75.45 kDa (740 amino acids). In silico analysis indicated that BR0072 displays an N-terminal signal peptide (26), a YadA-like C-terminal region (23), and a sequence of 642 amino acids between the two regions. The YadA-like and N-terminal regions (together with a central region or “passenger domain”) are the characteristic precursor domains of type II trimeric autotransporter (TA) proteins (17, 33, 34); therefore, we named BR0072 BtaE (for Brucella trimeric autotransporter). As mentioned earlier, TAs represent a subfamily of autotransporter proteins, which are defined by the presence of a distinctive C-terminal translocator domain that forms a highly stable trimer in the outer membrane (33). The characterized members of the TA subfamily have been found to mediate bacterial adherence to eukaryotic cells and components of the extracellular matrix (ECM) (33–35), as YadA of Yersinia enterocolitica (36) and UspA1 and UspA2 of Moraxella catarrhalis (37).
TAs have been found to exhibit similar architectural structures, consisting of a globular head and a stalk (both corresponding to the passenger domain) and an anchor domain (corresponding to the C-terminal region), similar to a lollipop-shaped structure (38). To analyze the modular organization, the entire BtaE sequence was entered into the Domain Annotation of Trimeric Autotransporter Adhesins (daTAA) (25). Three regions corresponding to the head (amino acids 154 to 222, 254 to 335, and 434 to 529) and four corresponding to the neck (amino acids 223 to 244, 336 to 357, 530 to 551, and 600 to 618) were identified. Interestingly, in silico analysis using the Pfam protein database (23) showed that two of the three regions predicted to be part of the head contain three Hep_Hag repeats, and the neck portions overlap four noncontiguous HIM repeat motifs (Fig. 1), which represent potential adhesion motifs. In addition, an anchor domain and connectors were also identified. As expected, the anchor domain matches the YadA-like C-terminal region (Fig. 1).
In vitro binding activities: BtaE is involved in adhesion to hyaluronic acid.
It has been shown that Brucella spp. bind to components of the ECM, such as fibronectin and collagen. Indeed, as mentioned earlier, we have recently shown that the type I autotransporter BmaC protein is involved in the binding of B. suis to fibronectin (15). On the other hand, there are some examples of TAs mediating adherence to ECM components (34). In order to assess the affinity of BtaE for ECM components, we first performed a heterologous approach. The btaE gene was cloned with its own promoter into the pBBR1MCS vector, and the resulting plasmid (pBBRbtaE) was transferred into a nonadherent, noninvasive E. coli K-12-derivative strain. Binding of E. coli carrying pBBRbtaE plasmid or the empty vector to different immobilized ligands was evaluated. Expression of the BtaE protein in the heterologous host was confirmed by Western blotting (see below). E. coli pBBRbtaE was able to bind more efficiently to hyaluronic acid (190% ± 21%) and fibronectin (185% ± 22%) than the control strain, which was considered 100% (Fig. 2A). No differences were observed in the adhesion of the heterologous host expressing BtaE to collagen or fetuin (a sialic acid-rich protein) (Fig. 2A).
Fig 2.

Binding of BtaE-expressing bacteria to ECM components. Plastic wells were coated with collagen type I, hyaluronic acid, fetuin, and fibronectin and subsequently incubated with the different strains. After being washed, bound bacteria were harvested with trypsin-EDTA; then, serial dilutions were done and the bacteria were plated on appropriate media. (A) E. coli pBBRbtaE and E. coli pBBR1MCS (control strain) were assayed. (B) The wild-type (wt) strain of B. suis and the B. suis ΔbtaE and ΔbtaE pBBRbtaE isogenic strains were analyzed. Values correspond to the percentages of total bacteria recovered from the wells after incubation with reference to the control strain (E. coli pBBR1MCS in panel A and B. suis wt in panel B), to which a value of 100% was assigned. Data represent the means ± standard deviations (SD) of the results of a representative experiment done in triplicate. Three independent experiments were performed with similar results. Data were analyzed by Student's t test and by one-way ANOVA followed by a Tukey a posteriori test. *, significantly different from control (P < 0.05), with 95% confidence.
Autotransporters have been shown to mediate a variety of other functions. For example, YadA of Yersinia enterocolitica participates in the binding to hydrophobic abiotic surfaces, in cellular autoagglutination, and in serum resistance, in addition to adhesion to and invasion of host cells (36). E. coli expressing btaE did not show any differential phenotype regarding attachment to polystyrene or autoagglutination or even resistance to 8% serum (with complement activity) compared with the control strain (data not shown).
We next investigated the effect of a btaE deletion on the binding of B. suis to the immobilized ligands. No significant differences between the wild-type and mutant strains were observed in the binding to collagen, fetuin, or fibronectin. However, the B. suis ΔbtaE mutant showed a significant decrease in its ability to bind to hyaluronic acid (18% ± 8%) relative to the wild type (considered 100%). Wild-type binding levels were restored in the mutant strain complemented with the pBBRbtaE plasmid (Fig. 2B). Thus, although a contribution of BtaE to the interaction of B. suis with fibronectin cannot be ruled out, our observations support a role of BtaE in the binding of B. suis to hyaluronic acid.
The B. suis ΔbtaE mutant was not impaired in attachment to abiotic surfaces or in its ability to autoaggregate in liquid cultures, and was not more susceptible to 10% or 50% sera (both bovine and porcine, with complement activity) than the wild-type strain (data not shown). Hence, taken together, these observations suggest that, unlike other trimeric autotransporters, BtaE is not involved in any of these activities.
BtaE participates in the adhesion of B. suis to host cells and is required for full virulence in mice.
To analyze the contribution of BtaE to the interaction of B. suis with host cells, the ability of the B. suis ΔbtaE mutant to adhere to and invade HeLa cells (human epithelial carcinoma cell line) was compared with that of the wild-type strain. Cells were seeded in multiwell plates and inoculated with the different strains for 1 h. To determine the total number of cell-associated bacteria, cells were lysed with a nonionic detergent and CFU were determined by plating serial dilutions of cell lysates. To determine the number of intracellular viable bacteria, a standard gentamicin protection assay was performed. The number of adherent bacteria was calculated as the difference between total cell-associated and intracellular bacteria. B. suis ΔbtaE showed a significant reduction (by 63% ± 7%) in the ability to adhere to HeLa cells compared to the wild-type strain (Fig. 3A) as well as a decrease (by 70% ± 14%) in the ability to invade these cells (Fig. 3B). Both adhesion to and invasion of HeLa cells were restored by pBBRbtaE, confirming that the absence of btaE was responsible for the observed in vivo phenotypes. In order to determine if the diminished invasion was a consequence of the reduction in adherence or, alternatively, was due to a reduced invasion aptitude, the invasive cell/adherent cell ratio was calculated. As shown in Fig. 3C, the wild-type and mutant strains showed similar efficiencies of invasion, suggesting that BtaE is involved in the initial attachment of B. suis to HeLa cells rather than in host cell internalization. To further investigate the ability of the B. suis ΔbtaE mutant to attach to HeLa cells, a competitive infection assay was carried out by coinoculating equal amounts of B. suis wild-type and ΔbtaE bacteria into the HeLa cell monolayer. The BtaE-defective mutant was strongly outcompeted by the wild-type strain for adhesion to HeLa (Fig. 3D), thus reinforcing the idea that BtaE participates in the binding of B. suis to these cells.
Fig 3.
Adhesion to and invasion of HeLa cells. Approximately 5 × 106 bacteria were used to challenge 5 × 104 HeLa cells (MOI, 100:1). Total numbers of adherent bacteria (A) and intracellular or invasive bacteria (B) were determined, and the invasive cell/adherent cell ratio was calculated (C). Total numbers of adherent and intracellular bacteria are expressed relative to wild-type B. suis 1330, defined as 100%. (D) A competitive assay was carried out in which HeLa cells were coinoculated with wild-type and B. suis ΔbtaE bacteria. The number of adherent bacteria was expressed as CFU/ml. (E) Inhibition of bacterial binding to HeLa cells by preincubation with hyaluronic acid (HA); data are expressed as total CFU/ml. Values represent the means ± SD of the results of an experiment representative of three independent assays done in triplicate or quadruplicate. Data were analyzed by one-way ANOVA followed by a Tukey a posteriori test. *, significantly different from the wild type (P < 0.05).
We also investigated whether hyaluronic acid is able to compete the binding of B. suis to HeLa cells. We found that preincubation of wild-type bacteria with 25 μg/ml hyaluronic acid inhibited the binding to these cells by about 60%. In contrast, the binding of the btaE mutant was inhibited only slightly by hyaluronic acid (Fig. 3E). This observation provides further evidence that BtaE is involved in the binding of Brucella to hyaluronic acid.
To further investigate the in vivo adhesive phenotype of the btaE mutant, the A549 epithelial cell line from human lung was used as an alternative cellular model in the binding assay (39). B. suis ΔbtaE showed a significant reduction in the ability to adhere to (by 50% ± 8%) and invade (by 51% ± 13%) A549 cells compared with the wild type (considered 100%) (Fig. 4A and B). As predicted, the pBBRbtaE plasmid restored adhesion and invasion capacity to wild-type levels in the B. suis ΔbtaE mutant. Similar to the phenotypes in HeLa cells, our data indicate that the decrease in the invasion capacity of B. suis ΔbtaE was a consequence of a lower adhesion to A549 cells (Fig. 4C).
Fig 4.

Adhesion to and invasion of A549 lung epithelial cells. Approximately 5 × 106 bacteria were used to challenge 5 × 104 A549 cells (MOI, 100:1). Total numbers of adherent bacteria (A) and intracellular bacteria (B) were determined, and the invasive cell/adherent cell ratio was calculated (C). Total numbers of adherent and intracellular bacteria are expressed relative to the adhesion and invasion values of the wild type, defined as 100%. Values represent the means ± SD of the results of an experiment representative of three independent assays done in triplicate. Data were analyzed by one-way ANOVA and a Tukey a posteriori test. *, significantly different from the wild type (P < 0.05).
To test the possibility that BtaE could participate in later steps of host cell infection, such as intracellular survival, a classical intracellular proliferation assay was carried out, using murine J774 macrophages and HeLa cells. In both cell models, the wild-type and mutant strains showed the same kinetics of intracellular replication (data not shown), indicating that the absence of BtaE does not affect later stages of cellular infection.
To evaluate the contribution of BtaE to the virulence of B. suis, we used the mouse infection model. Since it is possible that BtaE is involved in the initial stages of infection, we decided to inoculate mice through the intragastric route (40) and evaluate splenic infection at different times postinfection (p.i.). It was previously reported that the bacterial load in spleens and other organs of mice infected with B. abortus through the digestive track significantly increases at 7 days p.i. and stabilizes at later times (41). Therefore, we analyzed the spleen load after 7 and 30 days p.i. The B. suis ΔbtaE mutant and the complemented (B. suis ΔbtaE pBBRbtaE) and parental strains were administered intragastrically to mice, and CFU counts in spleens were determined. At 7 and 30 days p.i., the splenic infection by the B. suis ΔbtaE mutant was significantly reduced by 0.76 log and 1.24 log, respectively, compared with infection by the parental wild-type strain (Fig. 5A and B). As predicted, the complemented strain harboring the pBBRbtaE plasmid showed a splenic CFU count similar to that of the wild type. These results show that BtaE is required for full virulence of B. suis in mice infected through the oral route.
Fig 5.

Role of BtaE in the virulence of B. suis in BALB/c mice. BALB/c mice (10 mice per group) were inoculated by intragastric delivery of the B. suis wild type, B. suis ΔbtaE, or the complemented strains. Five mice from each group were sacrificed at 7 (A) and 30 (B) days postinfection, and then spleens were removed. Dilutions of spleen homogenates were plated in duplicate, and CFU were counted and expressed as the log10 value per spleen. The CFU data were normalized by log transformation and evaluated by one-way ANOVA followed by Dunnett's a posteriori test. The experiment was repeated twice with similar results. *, significantly different from the wild type (P < 0.05).
BtaE has a surface-exposed unipolar localization.
While the passenger domains of monomeric autotransporters may be cleaved after secretion, the passenger domains of characterized TAs remain intact after secretion (33). A recombinant peptide corresponding to a portion of the passenger domain (see Fig. 1, underlined region) was cloned, expressed in E. coli, and purified to generate polyclonal anti-BtaE antibodies in mice. To evaluate the membrane association of BtaE, Western blot analysis of whole membranes from the B. suis wild-type or ΔbtaE mutant strain obtained from exponential-phase cells was performed. Similar to other TAs (42, 43), we noted that BtaE is difficult to denature. It was suggested that the C-terminal domains of TAs form heat- and sodium dodecyl sulfate (SDS)-resistant trimers. Hence, the samples were submitted to a strong denaturing process in the presence of formic acid and resolved by SDS-PAGE. Western blot analysis using the anti-BtaE antibodies showed the presence of an ∼75-kDa protein band corresponding to monomeric BtaE, which was absent in the membrane fraction of the B. suis ΔbtaE strain (Fig. 6A).
Fig 6.
BtaE localization. (A and C) Western blot analysis of whole-membrane fractions from B. suis (A) and E. coli pBBRbtaE (C) was carried out. Membrane samples were submitted to a strong denaturing process. Samples were electrophoresed by SDS-PAGE and transferred to a PDVF membrane. Blots were incubated with anti-BtaE antisera and with anti-mouse HRP-conjugated secondary antibody and finally revealed using ECL Plus. On the right, a Coomassie blue pattern is shown as a loading control (ctrl). (B and D) Detection of BtaE (in red) on the B. suis surface (B) and on the E. coli pBBRbtaE surface (D) by immunofluorescence of GFP-tagged bacteria. Cultures of GFP-labeled strains were fixed, incubated with anti-BtaE antibodies, and then probed with a CY3-conjugated donkey anti-mouse antibody preparation. Samples were observed with a Plan-Aprochromat 100×/1.4 oil DIC objective on a Zeiss LSM 5 Pascal confocal microscope.
To assess the surface presentation of BtaE, late-exponential-phase cultures of the B. suis wild-type or ΔbtaE strain labeled with green fluorescent protein (GFP) were refreshed for 1 h in TSB medium and then fixed without permeabilization, incubated with anti-BtaE antibodies, and observed by confocal microscopy. Under the conditions assayed, we found that BtaE was detected in 1% to 3% of bacteria examined, but in all cases (n = 50), the red signal corresponding to BtaE exhibited unipolar localization (Fig. 6B). As expected, no red signal was observed on the surface of the B. suis ΔbtaE mutant but the unipolar signal was restored in the complemented strain harboring the pBBRbtaE plasmid (data not shown). To our knowledge, this is the first description of a TA with unipolar localization.
We also evaluated the BtaE localization in the E. coli heterologous host, carrying the pBBRbtaE plasmid. Western blot analysis indicated the presence of the 75-kDa protein corresponding to BtaE in the total membrane fraction of E. coli pBBRbtaE, which was absent in the control strain (Fig. 6C). Interestingly, immunofluorescence analysis of GFP-tagged E. coli carrying pBBRbtaE showed BtaE foci at one bacterial pole in 5% of bacteria examined (Fig. 6D), while no signal was observed in the control strain (data not shown). This observation suggests that the mechanism involved in polar targeting of the BtaE trimeric autotransporter is conserved in E. coli.
Brucella's new pole is functionally differentiated for adhesion.
Brucellae are proposed to be polarized bacteria, exhibiting asymmetric division (44) and unipolar growth, as seen with several other Rhizobiales (45). Moreover, proteins specifically recruited to the old pole or to the new pole have been reported. The PdhS histidine kinase is recruited to the old pole (19, 20), and the putative DNA repair protein AidB is recruited to the new pole (18). Since both the BmaC and BtaE adhesins showed unipolar localization, the question arises as whether these adhesins are localized at a specific pole and, in that case, if they are localized at the same pole. Therefore, to assess the relative localizations of BmaC and BtaE, we used the PdhS and AidB proteins as markers of the old and new poles, respectively.
Since the PdhS and AidB pole markers were identified in B. abortus, we first evaluated the localization of the BmaC homologue of B. abortus, which is 99.6% identical to BmaC from B. suis. By immunofluorescence, it was confirmed that in B. abortus the BmaC homologue consistently displays unipolar localization (data not shown). Next, we used the B. abortus strains carrying the fusion proteins encoded by either pdhS-egfp (19) or aidB-yfp (18) to evaluate by immunofluorescence the relative localization of BmaC. DAPI staining and phase contrast were used to distinguish the bacterial shapes. Analysis by confocal microscopy showed that every time BmaC was detected (red signal) on the surface of B. abortus carrying PdhS-eGFP protein (green), it was at the pole opposite PdhS-eGFP (29 occurrences) (Fig. 7A). It was never observed that BmaC and PdhS-eGFP colocalized. Conversely, every time BmaC was detected in the strain carrying the AidB-YFP protein (yellow), it was either at the same pole as the AidB-YFP marker (31 occurrences) or in cells with diffuse markers, but never at the opposite pole (Fig. 7B). As expected, a bmaC mutant of B. abortus did not show any red signal (data not shown).
Fig 7.
BtaE and BmaC are localized to the new bacterial pole. (A and B) Immunofluorescence microscopy using anti-BmaC antibodies was carried out with fixed cultures of B. abortus strains expressing PdhS-eGFP (A) and AidB-YFP (B) as old and new pole markers, respectively. DAPI staining and phase contrast (PC) were used to visualize the bacterial DNA content and shape, respectively. Samples were observed with a confocal LSM 510 Meta microscope using a Plan-Apochromat 60×/1.4 oil DIC objective. Representative images are shown. BmaC, PdhS-eGFP, and AidB-YFP are indicated with red, green, and yellow arrows, respectively. A schematic representation is also displayed. An intensity profile of the channels (constructed through the cyan dotted arrow) is presented, expressed in arbitrary units. (C and D) The same analysis was carried out for BtaE in B. suis with both PdhS-eGFP (C) and AidB-YFP (D).
The pdhS-egfp and aidB-yfp fusions were introduced into the wild-type strain of B. suis and also into the isogenic B. suis ΔbmaC mutant as a negative control. The same analysis described above was carried out with these strains, leading to the same conclusion; i.e., BmaC is localized at the new pole of B. suis (data not shown). We next analyzed the relative localization of BtaE in B. suis. We found that every time BtaE was detected on bacteria expressing the old pole marker (PdhS-eGFP), it was at the opposite pole (30 occurrences) (Fig. 7C), but it was never observed at the same pole as PdhS-eGFP. Instead, in all the cases where BtaE was detected in bacteria carrying the aidB-yfp fusion, it was observed either at the same pole as the AidB-YFP fusion (17 occurrences) or in bacteria in which this marker was diffuse, but on no occasion was the BtaE fluorescent focus at the opposite pole (Fig. 7D). Taken together, our results show that under the conditions tested here both the BmaC and BtaE adhesins are localized at the new pole, suggesting that this pole is functionally differentiated for adhesion.
DISCUSSION
In microbial pathogens, adhesion to host cells is a critical step in the infection process. The importance of adhesion is reflected by the finding that several bacterial pathogens display multiple adhesins, which can act synergistically, thereby enhancing their adhesive capacity. Despite the relevance of adhesion in the interaction with the host, the impact of this step in Brucella pathogenesis was little explored. We presented evidence indicating that the monomeric BmaC autotransporter is required for an efficient adhesion of B. suis to host cells (15). In this work, we aimed to explore the role of BtaE, a protein that is predicted to belong to the TA family, in the interaction of B. suis with the host. Since it is expected that pathogens harbor several adhesins, probably exhibiting overlapping functions, we initially assessed the function of BtaE using a heterologous approach and found that this protein was able to increase the affinity of a “nonadherent” E. coli strain to immobilized hyaluronic acid and fibronectin. Analysis of the knockout btaE mutant confirmed the role of BtaE in the binding of B. suis to one of these ECM components, the hyaluronic acid glycosaminoglycan. The possibility exists that a fibronectin-binding-defective phenotype can be masked by other fibronectin-binding proteins, such as BmaC (15). The ability of BtaE to mediate the binding of Brucella to hyaluronic acid may be relevant at different stages of the infection. For example, it could facilitate initial bacterial adhesion to epithelial cells. It was previously described that hyaluronic acid is involved in adhesion of other pathogens such as Mycobacterium tuberculosis to the human lung epithelial cell line A549 (46), a cell type to which Brucella also adheres (47). Indeed, binding assays demonstrated that BtaE contributes to the adhesion of B. suis to both A549 and HeLa cells. Interestingly, addition of hyaluronic acid was able to compete the binding of the wild-type strain to HeLa cells, while adhesion of the btaE mutant was inhibited only slightly by hyaluronic acid. This observation provides further evidence that BtaE mediates the binding of Brucella to hyaluronic acid. Furthermore, taken together, our observations are consistent with the possibility that BtaE mediates adhesion to cells, at least in part, through hyaluronic acid. As hyaluronic acid is a component of the ECM of several tissues, BtaE could also be involved in Brucella dissemination to different target tissues such as cartilage, heart, and bone, which may result in serious complications of brucellosis.
The role of BtaE in the virulence of B. suis was studied by inoculating the wild type, the btaE deletion mutant, and the complementing strain in mice through the oral route. At 7 and 30 days p.i., splenic infection by B. suis ΔbtaE was significantly reduced, compared to infection by the wild type, while the complemented strain showed a restored phenotype. This result strongly suggests that the BtaE adhesin might be considered a virulence factor and highlights the importance of bacterial adhesion to host components during the Brucella infection process.
Immunofluorescence analysis showed that BtaE exhibits unipolar localization. To the best of our knowledge, this is the first report of unipolar localization for a TA. Yet several monomeric (type I) autotransporters from rod-shaped pathogens, including the BmaC adhesin of B. suis (15), the IcsA protein from Shigella flexneri (48), and the AIDA-I monomeric autotransporter that mediates adhesion to human cells by pathogenic diffusely adherent E. coli (DAEC) strains (49), were shown to localize at one bacterial pole. It was reported that IcsA localizes to the pole at which actin assembly occurs, which is the old pole (48). Remarkably, some of the paradigmatic type I autotransporters, including AIDA-I and IcsA, are polar in the cytoplasm prior to secretion, suggesting that secretion occurs at the pole (49, 50). Intriguingly, it was shown that NalP, a monomeric autotransporter of spherically shaped Neisseria meningitidis, contains the molecular information to localize to the pole of E. coli. In fact, the BtaE TA also showed unipolar localization in the E. coli heterologous host, supporting the notion that the polar targeting mechanism of proteins from the autotransporter families is conserved in several bacteria.
The observation that both BtaE and BmaC show unipolar localization prompted us to explore whether they are associated with a specific (new or old) pole and, if this is the case, whether they are at the same or a different one. Evidence presented by others and us indicates that the individual Brucella bacterium interacts with the host cell surface through one of its poles (15, 51). Furthermore, it seems that the pole where BmaC is located on single bacteria is the one that interacts with the cell surface (15). Therefore, it was also interesting to address the issue as to which could be the “adhesive” pole. To determine the pole where the adhesins localize, we used the PdhS-eGFP fusion as a marker of the old pole (19), and the AidB-YFP fusion as a marker of the new pole (18). We observed that every time BmaC or BtaE was detected, it localized to the pole opposite the pole where PdhS-eGFP was located. In contrast, on every occasion that the BmaC or BtaE fluorescent foci were observed in bacteria expressing the new pole fusion, they were found at the same pole as AidB-YFP. We conclude that under the conditions tested here, both BmaC and BtaE localize to the new pole generated after cell division. Accordingly, we propose that the new pole is functionally differentiated for adhesion.
BtaE was detected in a low proportion of cells. A similar observation was previously made for the unipolar BmaC adhesin (15). At present, we do not know the reason for these observations. One possibility is that the level of btaE or bmaC gene transcription in vitro is not high enough to allow detection of fluorescent foci in most cells. If this is the case, an increase in btaE or bmaC expression is predicted to occur within the host environment before bacterial internalization. Since the anti-BtaE and anti-BmaC antibodies used in the immunofluorescence assays recognize a peptide of the passenger domains, another nonexclusive explanation is that these peptides are not very accessible to the antibodies in the native β-roll structure exposed on the bacterial surface (17). Finally, another interesting hypothesis is that BtaE and BmaC are expressed only in a bacterial subpopulation. This possibility is supported by the finding that several cell types generated by asymmetric division and differentiation coexist in a culture of B. abortus (19, 20, 44). Furthermore, relatively low expression of btaE or bmaC in a small subpopulation of bacteria might be sufficient for bacterial internalization and a successful (but limited) infection. Further studies are required to explore these possibilities.
Several surface virulence factors (including adhesive structures) were found to exhibit polar localization in other alphaproteobacteria closely related to Brucella. It was reported that all Agrobacterium tumefaciens VirB proteins are polarly localized (52). Furthermore, A. tumefaciens attaches efficiently to plant and abiotic surfaces through one of its poles in a process mediated by several structures, including a unipolar polysaccharide (UPP) (53). Very interestingly, evidence was presented indicating that cells of A. tumefaciens preferentially attach to surfaces via the old cell pole (45). Evidence presented in this work and other previously reported evidence (15) support the hypothesis that the unipolar BmaC and BtaE adhesins mediate Brucella binding to host cells at its new pole. It is possible that the characteristics of the polar asymmetry depend on the bacterial life style. In the case of A. tumefaciens, a sessile or attached mode of life would require that adhesion to abiotic or biotic surfaces is mediated by adhesins localized at the relatively stable old pole (53); in Brucella, initial binding of single bacteria to host cells prior to cellular internalization would be mediated by adhesins localized at the new pole of a subpopulation of infective bacteria.
In addition to BmaC and BtaE, other putative adhesins were shown to participate in the interaction of Brucella with the host. Recently, it was reported that a pathogenicity island of four genes contributes to the efficiency of attachment of B. abortus to both HeLa and J774 macrophage cells. One of the proteins encoded by the gene cluster was a hypothetical protein (Bab1_2009) that harbors an immunoglobulin-like BID_1 adhesion domain, found in adhesins such as the E. coli intimin (13). In addition, it was previously reported that a predicted adhesin from the monomeric autotransporter family from B. suis (OmaA) would be important during the chronic phase of infection (54). It will be interesting to determine whether Bab1_2009, OmaA, BtaE, and BmaC play redundant, synergistic, or complementary roles in the interaction of Brucella with different cell types and/or host tissues. Furthermore, different adhesins could exert their roles at different infection stages. Studies on gene regulation could give a hint regarding these possibilities. Through a transcriptomic analysis, it was previously shown that the expression of the btaE orthologue from B. melitensis (BMEI1873) is reduced in a mutant defective in the LuxR homologue VjbR regulator (55), which modulates the expression of transcripts required for intracellular survival (56). Besides, by microarray analysis it was found that transcription of the bmaC orthologue of B. abortus was downregulated by the BvrR/BvrS two-component system. This system plays a role in the adjustment of Brucella physiology to the shift expected to occur during the transition from the extracellular to the intracellular niche (57). Thus, one plausible hypothesis is that btaE expression and bmaC expression are induced under different stimulus conditions. Future studies will focus on the regulatory elements that control the expression of the autoransporter adhesins during the onset of cell infection.
Not only surface proteins were shown to participate in the adhesive properties of Brucella. Overexpression of an acyl homoserine-lactone (AHL) acylase (that degrades AHLs) in B. melitensis induced clumping and increased adhesion to both polystyrene and HeLa cells, probably due to the production of both some polymeric substances and outer membrane vesicles (16). It was argued that adhesion of the quorum-sensing-defective strain was somehow reminiscent of the adherence of localized bacterial colonies of B. abortus to epithelial cells (12). Thus, it is possible that initial and polar attachment of single bacteria to the host cell is mediated by proteinaceous adhesins, such as BtaE or BmaC and that, eventually, subsequent formation of bacterial microcolonies would be promoted by the production of other polymeric substances induced in contact with the cell surface.
Finally, host tropism is a very important feature of the Brucella genus, for which the concept of species is controversial, due to the high degree of similarity between the Brucella genomes (58). For example, comparison of the B. melitensis 16 M and B. suis 1330 genomes revealed extensive gene similarity and synteny, with the majority (>90%) of genes sharing 98% to 100% identity (32, 59). It is striking that most of the genes that were more variable were genes encoding surface-exposed proteins, leading to the hypothesis that they could contribute to differences in host preference and disease manifestation (32). In addition to B. suis and B. melitensis, we also found BtaE orthologues in B. abortus 2308 (BAB1_0069), B. canis ATCC 23365 (BCAN_A0073), B. ovis 25810 (BOV_0071), and B. microti CCM4915 (BMI_I75). Interestingly, in silico analysis indicated that there are significant differences either in protein lengths or in the number of repetitive motifs between the different ortologues. Therefore, variations found in BtaE and other adhesins such as BmaC (15) may well contribute to host preference or tissue tropism. It will be interesting, in the future, to assess this hypothesis.
ACKNOWLEDGMENTS
This work was supported by a grant from University of Buenos Aires (UBACyT, X-240) to A.Z. and by Concerted Research Action 08/13-015, FRFC (grants 2.4541.08 and 2.4510.12 from FNRS-FRS), Interuniversity Attraction Pole (P7/27, Belgian Science Policy Office), and the University of Namur to X.D.B. V.R.-R., D.M.P., G.M.A., and F.A.M. were supported by fellowships from CONICET, Argentina. A.Z., P.L.A., and R.S. are research fellows of CONICET. C.V.D.H. was FRIA-FNRS-FRS fellow.
We acknowledge Martín Rumbo and Dolores González Maciel for providing the A549 cell line, Susana Raffo and Marta Bravo for DNA sequencing and other technical support, and Máximiliano Neme for help with confocal microscopy. We also thank our colleagues at the laboratory for helpful discussions.
Footnotes
Published ahead of print 14 January 2013
REFERENCES
- 1. Thorne ET. 2001. Brucellosis, p 372–395 In Williams ES, Barker IK. (ed), Infectious diseases of wild mammals, 3rd ed Manson Publishing, London, United Kingdom [Google Scholar]
- 2. Boschiroli ML, Foulongne V, O'Callaghan D. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4: 58–64 [DOI] [PubMed] [Google Scholar]
- 3. Seleem MN, Boyle SM, Sriranganathan N. 2010. Brucellosis: a re-emerging zoonosis. Vet. Microbiol. 140: 392–398 [DOI] [PubMed] [Google Scholar]
- 4. Ko J, Splitter GA. 2003. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin. Microbiol. Rev. 16: 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Celli J, Gorvel JP. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7: 93–97 [DOI] [PubMed] [Google Scholar]
- 6. Roop RM, II, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med. Microbiol. Immunol. 198: 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel JP. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66: 5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martirosyan A, Moreno E, Gorvel JP. 2011. An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol. Rev. 240: 211–234 [DOI] [PubMed] [Google Scholar]
- 9. Gorvel JP. 2008. Brucella: a Mr “Hide” converted into Dr Jekyll. Microbes Infect. 10: 1010–1013 [DOI] [PubMed] [Google Scholar]
- 10. Pizarro-Cerda J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124: 715–727 [DOI] [PubMed] [Google Scholar]
- 11. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5: 580–592 [DOI] [PubMed] [Google Scholar]
- 12. Castaneda-Roldan EI, Avelino-Flores F, Dall'Agnol M, Freer E, Cedillo L, Dornand J, Giron JA. 2004. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6: 435–445 [DOI] [PubMed] [Google Scholar]
- 13. Czibener C, Ugalde JE. 2012. Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect. 14: 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hernandez-Castro R, Verdugo-Rodriguez A, Puente JL, Suarez-Guemes F. 2008. The BMEI0216 gene of Brucella melitensis is required for internalization in HeLa cells. Microb. Pathog. 44: 28–33 [DOI] [PubMed] [Google Scholar]
- 15. Posadas DM, Ruiz-Ranwez V, Bonomi HR, Martin FA, Zorreguieta A. 2012. BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell. Microbiol. 14: 965–982 [DOI] [PubMed] [Google Scholar]
- 16. Godefroid M, Svensson MV, Cambier P, Uzureau S, Mirabella A, De Bolle X, Van Cutsem P, Widmalm G, Letesson JJ. 2010. Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol. Med. Microbiol. 59: 364–377 [DOI] [PubMed] [Google Scholar]
- 17. Dautin N, Bernstein HD. 2007. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 61: 89–112 [DOI] [PubMed] [Google Scholar]
- 18. Dotreppe D, Mullier C, Letesson JJ, De Bolle X. 2011. The alkylation response protein AidB is localized at the new poles and constriction sites in Brucella abortus. BMC Microbiol. 11: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, Letesson JJ, Jacobs-Wagner C, De Bolle X. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 26: 1444–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van der Henst C, Beaufay F, Mignolet J, Didembourg C, Colinet J, Hallet B, Letesson JJ, De Bolle X. 2012. The histidine kinase PdhS controls cell cycle progression of the pathogenic alphaproteobacterium Brucella abortus. J. Bacteriol. 194: 5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69–73 [DOI] [PubMed] [Google Scholar]
- 22. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176 [DOI] [PubMed] [Google Scholar]
- 23. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szczesny P, Lupas A. 2008. Domain annotation of trimeric autotransporter adhesins—daTAA. Bioinformatics 24: 1251–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- 27. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- 28. Serruto D, Spadafina T, Scarselli M, Bambini S, Comanducci M, Hohle S, Kilian M, Veiga E, Cossart P, Oggioni MR, Savino S, Ferlenghi I, Taddei AR, Rappuoli R, Pizza M, Masignani V, Arico B. 2009. HadA is an atypical new multifunctional trimeric coiled-coil adhesin of Haemophilus influenzae biogroup aegyptius, which promotes entry into host cells. Cell. Microbiol. 11: 1044–1063 [DOI] [PubMed] [Google Scholar]
- 29. Klingman KL, Murphy TF. 1994. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect. Immun. 62: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16: 800–802 [PubMed] [Google Scholar]
- 31. Lambertsen L, Sternberg C, Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6: 726–732 [DOI] [PubMed] [Google Scholar]
- 32. Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ, Daugherty SC, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Nelson WC, Ayodeji B, Kraul M, Shetty J, Malek J, Van Aken SE, Riedmuller S, Tettelin H, Gill SR, White O, Salzberg SL, Hoover DL, Lindler LE, Halling SM, Boyle SM, Fraser CM. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. U. S. A. 99: 13148–13153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cotter SE, Surana NK, St Geme JW., III 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13: 199–205 [DOI] [PubMed] [Google Scholar]
- 34. Lyskowski A, Leo JC, Goldman A. 2011. Structure and biology of trimeric autotransporter adhesins. Adv. Exp. Med. Biol. 715: 143–158 [DOI] [PubMed] [Google Scholar]
- 35. Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14: 264–270 [DOI] [PubMed] [Google Scholar]
- 36. El Tahir Y, Skurnik M. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291: 209–218 [DOI] [PubMed] [Google Scholar]
- 37. Cope LD, Lafontaine ER, Slaughter CA, Hasemann CA, Jr, Aebi C, Henderson FW, McCracken GH, Jr, Hansen EJ. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181: 4026–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19: 5989–5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferrero MC, Fossati CA, Baldi PC. 2009. Smooth Brucella strains invade and replicate in human lung epithelial cells without inducing cell death. Microbes Infect. 11: 476–483 [DOI] [PubMed] [Google Scholar]
- 40. Delpino MV, Marchesini MI, Estein SM, Comerci DJ, Cassataro J, Fossati CA, Baldi PC. 2007. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 75: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paixao TA, Roux CM, den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, Tsolis RM. 2009. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect. Immun. 77: 4197–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann J. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185: 3735–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Surana NK, Cutter D, Barenkamp SJ, St Geme JW., III 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 279: 14679–14685 [DOI] [PubMed] [Google Scholar]
- 44. Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12: 361–365 [DOI] [PubMed] [Google Scholar]
- 45. Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci. U. S. A. 109: 1697–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aoki K, Matsumoto S, Hirayama Y, Wada T, Ozeki Y, Niki M, Domenech P, Umemori K, Yamamoto S, Mineda A, Matsumoto M, Kobayashi K. 2004. Extracellular mycobacterial DNA-binding protein 1 participates in Mycobacterium-lung epithelial cell interaction through hyaluronic acid. J. Biol. Chem. 279: 39798–39806 [DOI] [PubMed] [Google Scholar]
- 47. Ferrero MC, Fossati CA, Baldi PC. 2010. Direct and monocyte-induced innate immune response of human lung epithelial cells to Brucella abortus infection. Microbes Infect. 12: 736–747 [DOI] [PubMed] [Google Scholar]
- 48. Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J. Bacteriol. 175: 2189–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jain S, van Ulsen P, Benz I, Schmidt MA, Fernandez R, Tommassen J, Goldberg MB. 2006. Polar localization of the autotransporter family of large bacterial virulence proteins. J. Bacteriol. 188: 4841–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steinhauer J, Agha R, Pham T, Varga AW, Goldberg MB. 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol. Microbiol. 32: 367–377 [DOI] [PubMed] [Google Scholar]
- 51. Pizarro-Cerdá J, Moreno E, Gorvel JP. 1999. Brucella abortus: invasion and survival within professional and non-professional phagocytes, vol 6 Jai Press Inc., Greenwich, CT [Google Scholar]
- 52. Judd PK, Kumar RB, Das A. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55: 115–124 [DOI] [PubMed] [Google Scholar]
- 53. Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr. Opin. Microbiol. 12: 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bandara AB, Sriranganathan N, Schurig GG, Boyle SM. 2005. Putative outer membrane autotransporter protein influences survival of Brucella suis in BALB/c mice. Vet. Microbiol. 109: 95–104 [DOI] [PubMed] [Google Scholar]
- 55. Weeks JN, Galindo CL, Drake KL, Adams GL, Garner HR, Ficht TA. 2010. Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol. 10: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, Bonnot S, Ferooz J, Tibor A, De Bolle X, Letesson JJ. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7: 1151–1161 [DOI] [PubMed] [Google Scholar]
- 57. Viadas C, Rodriguez MC, Sangari FJ, Gorvel JP, Garcia-Lobo JM, Lopez-Goni I. 2010. Transcriptome analysis of the Brucella abortus BvrR/BvrS two-component regulatory system. PLoS One 5: e10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Moreno E, Cloeckaert A, Moriyon I. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90: 209–227 [DOI] [PubMed] [Google Scholar]
- 59. DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, Reznik G, Jablonski L, Larsen N, D'Souza M, Bernal A, Mazur M, Goltsman E, Selkov E, Elzer PH, Hagius S, O'Callaghan D, Letesson JJ, Haselkorn R, Kyrpides N, Overbeek R. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. U. S. A. 99: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]