Abstract
Coaggregation of Porphyromonas gingivalis and oral streptococci is thought to play an important role in P. gingivalis colonization. Previously, we reported that P. gingivalis major fimbriae interacted with Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and that amino acid residues 166 to 183 of GAPDH exhibited strong binding activity toward P. gingivalis fimbriae (H. Nagata, M. Iwasaki, K. Maeda, M. Kuboniwa, E. Hashino, M. Toe, N. Minamino, H. Kuwahara, and S. Shizukuishi, Infect. Immun. 77:5130–5138, 2009). The present study aimed to identify and characterize P. gingivalis components other than fimbriae that interact with S. oralis GAPDH. A pulldown assay was performed to detect potential interactions between P. gingivalis client proteins and S. oralis recombinant GAPDH with amino acid residues 166 to 183 deleted by site-directed mutagenesis. Seven proteins, namely, tonB-dependent receptor protein (RagA4), arginine-specific proteinase B, 4-hydroxybutyryl-coenzyme A dehydratase (AbfD), lysine-specific proteinase, GAPDH, NAD-dependent glutamate dehydrogenase (GDH), and malate dehydrogenase (MDH), were identified by two-dimensional gel electrophoresis followed by proteomic analysis using tandem mass spectrometry. Interactions between these client proteins and S. oralis GAPDH were analyzed with a biomolecular interaction analysis system. S. oralis GAPDH showed high affinity for five of the seven client proteins (RagA4, AbfD, GAPDH, GDH, and MDH). Interactions between P. gingivalis and S. oralis were measured by a turbidimetric method and fluorescence microscopy. RagA4, AbfD, and GDH enhanced coaggregation, whereas GAPDH and MDH inhibited coaggregation. Furthermore, the expression of luxS in P. gingivalis was upregulated by RagA4, AbfD, and GDH but was downregulated by MDH. These results indicate that the five P. gingivalis client proteins function as regulators in P. gingivalis biofilm formation with oral streptococci.
INTRODUCTION
Periodontal diseases are among the most common chronic human infections (1). A group of Gram-negative anaerobes is associated with the initiation and progression of severe manifestations of these diseases, and foremost among these is Porphyromonas gingivalis (2, 3, 4). Interaction of P. gingivalis with early plaque-forming bacteria plays an important role in the colonization of periodontal pockets (5, 6, 7). P. gingivalis interacts with a variety of other oral Gram-positive bacteria, including Actinomyces naeslundii (8), Actinomyces viscosus (9, 10, 11), Streptococcus gordonii (12, 13), Streptococcus oralis (13, 14), Streptococcus mutans (15), and Streptococcus sanguinis (13, 16). These intergeneric coaggregations may lead to the initial colonization of P. gingivalis in the oral cavity.
Previously, we reported that P. gingivalis FimA fimbriae mediated coaggregation with S. oralis, an early colonizer in dental plaque (17). Moreover, we demonstrated that oral streptococcal cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) bound to P. gingivalis FimA fimbriae, and that this interaction exhibited high affinity and specificity (18). The association constant (KA) was found to be 4.34 × 107 M−1 using a biomolecular interaction analysis system, suggesting that S. oralis GAPDH functions as a dominant receptor for P. gingivalis and contributes to P. gingivalis colonization.
GAPDH is a tetramic enzyme in the glycolytic pathway and is responsible for the phosphorylation of glyceraldehyde-3-phosphate, leading to the generation of 1,3-bisphosphoglycerate (19). However, multiple functions have been reported recently for GAPDH, including roles in membrane fusion, microtubule binding, phosphotransferase activity, nuclear RNA export, DNA replication and repair, apoptosis, and viral pathogenesis (20, 21). Our previous study demonstrated that the P. gingivalis FimA fimbria binding domain in S. oralis ATCC 9811 GAPDH was within amino acid residues 166 to 183, and that the peptide corresponding to this domain (DNFGVVEGLMTTIHAYTG) exhibited strong binding activity toward recombinant FimA fimbriae (rFimA) by BIAcore analysis (KA = 4.51 × 107 M−1) (22). The synthetic peptide inhibited biofilm formation between various streptococci and P. gingivalis strains with different types of FimA fimbriae in a dose-dependent manner. The high-affinity binding of the peptide to rFimA was confirmed by BIAcore analysis (KA = 3.84 × 108 M−1).
In this study, we used a proteomic analysis-based approach to identify new P. gingivalis client proteins that bind to S. oralis GAPDH. Interaction profiling of P. gingivalis client proteins revealed their significant affinity for S. oralis GAPDH and their participation in the regulation of biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis ATCC 33277 and S. oralis ATCC 9811 were maintained as frozen stocks in our laboratory. P. gingivalis was cultured in prereduced Trypticase soy broth (Becton, Dickinson and Company [BD], Sparks, MD) containing 1 mg/ml yeast extract (BD), 5 μg/ml hemin (Sigma-Aldrich Japan K. K., Tokyo, Japan), and 1 μg/ml menadione (Sigma-Aldrich) for 24 h in an anaerobic system 1025 (Forma, Marietta, OH) with an 80% N2-10% CO2-10% H2 atmosphere at 35°C. S. oralis was cultured at 37°C for 16 h in brain heart infusion broth (BD). Escherichia coli M15 (pREP4) (Qiagen GmbH, Hilden, Germany) was cultured in Luria-Bertani broth (BD), and when necessary, 100 μg/ml ampicillin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) was included.
Purification of S. oralis rGAPDH(Δ166-183).
The plasmid pQE30-Sogap, expressing S. oralis recombinant His-tagged GAPDH (HT-rGAPDH), was prepared as described previously (22). A mutation in the P. gingivalis FimA fimbria binding domain of rGAPDH [residues 166 to 183; termed rGAPDH(Δ166-183)] was introduced into pQE30-Sogap using the primers SogapdhΔ166-183F and SogapdhΔ166-183R (Table 1). The resulting plasmid, pQE30-Sogap(Δ166-183), was constructed according to the instructions for the QuikChange II-E site-directed mutagenesis kit (Stratagene, Agilent Technologies, Inc., Santa Clara, CA). The deletion was confirmed by DNA sequence analysis. E. coli M15(pREP4) was transformed with pQE-Sogap(Δ166-183). HT-rGAPDH(Δ166-183) was purified with a HisTrap HP column (GE Healthcare, Ltd., Buckinghamshire, United Kingdom) and subjected to SDS-PAGE using a 5 to 15% Ready Gel J (Bio-Rad Laboratories, Hercules, CA). The gel was stained with Bio-Safe Coomassie brilliant blue (CBB; Bio-Rad). A low-molecular-mass calibration kit (GE Healthcare) was used to estimate molecular masses.
Table 1.
Oligonucleotide primers used in this study
| Primer | Primer sequencea (5′–3′) | Reference or source |
|---|---|---|
| SogapdhΔ166-183F | AACTGCTTGGCTCCAATGGCTAAAGCTCTTCAAGACCAAATGATCCTTGACGGACCACAC | This study |
| SogapdhΔ166-183R | GTGTGGTCCGTCAAGGATCATTTGGTCTTGAAGAGCTTTAGCCATTGGAGCCAAGCAGTT | This study |
| rRagA4 SphI-F | GCGGCATGCAAAAGAATGACGCTATTCTTCCTTTGC | This study |
| rRagA4 SacI-R | CGCGAGCTCTTAGAAAGAAATCTGAATACCACC | This study |
| rRgpB BamHI-F | GCGGGATCCAAAAAGAATTTTAGCAGGATCGTT | This study |
| rRgpB SalI-R | CGCGTCGACTTACTTCACTATAACCTTTTCTGT | This study |
| rAbfD BamHI-F | GCGGGATCCACTAGCGAACAGTACGTAGAAAGT | This study |
| rAbfD SalI-R | CGCGTCGACTTACTTATCGAGTGATTCGTCGAT | This study |
| rKgp-cd SphI-F | GCGGCATGCGATGTTTATACAGATCATGGCGAC | This study |
| rKgp-cd SalI-R | CGCGTCGACTCAACGGGAAGCTTCTGCCTTCTT | This study |
| rGDH BamHI-F | GCGGGATCCAAGACCCAAGAAATTATGACAATG | This study |
| rGDH SalI-R | CGCGTCGACTTAGCAAACGCCCTGAGCTACCAT | This study |
| rGAPDH BamHI-F | GCGGGATCCACGAAAGTAGGTATTAACGGCTTT | This study |
| rGAPDH SphI-R | CGCGCATGCTTATGCGTTTACCTTAGCCATGTA | This study |
| rMDH BamHI-F | GCGGGATCCAGCTACTTAACAGAAGAGAAATTG | This study |
| rMDH SphI-R | CGCGCATGCTTAGAGGTTCGGATTCACCGTCTT | This study |
| Real-time RT-PCR | ||
| ragA4f | CCAGAATAGAACCGTGAAGG | This study |
| ragA4r | AAGCGTGAAGTTGCCATC | This study |
| rgpBf | TCGTAGCATTCTCCTCTC | This study |
| rgpBr | TGTGAACTGAAGATTGTCC | This study |
| abfDf | CAACAAGACTTCCGCCATCC | This study |
| abfDr | AACGCAGAACACGCATACG | This study |
| kgp-cdf | CGGTTCGTATGCTTGTTGTTG | This study |
| kgp-cdr | GATAGAGGCGTTTGTCGTTCC | This study |
| gdhf | AAGCCAGCGAATACTATGTAGC | This study |
| gdhr | GCAGCCACTTGTCCACTTC | This study |
| gapdhf | GCAGCAGGCGGCAATATC | This study |
| gapdhr | GGCACACGGAATGACATACC | This study |
| mdhf | GCCTTGTTACGCTTATCTAC | This study |
| mdhr | TGATGCCGAAGTGCTTAG | This study |
| fimAf | TTGTTGGGACTTGCTGCTCTTG | 29 |
| fimAr | TTCGGCTGATTTGATGGCTTCC | 29 |
| luxSf | GAATGAAAGAGCCCAATCG | 29 |
| luxSr | GTAATCGCCTCGCATCAG | 29 |
| 16Sf | AGGAACTCCGATTGCGAAGG | 29 |
| 16Sr | TCGTTTACTGCGTGGACTACC | 29 |
Underlined sequences represent restriction enzyme sites.
Preparation of P. gingivalis client proteins by pulldown assay.
P. gingivalis ATCC 33277 (1.5 × 109 CFU/ml) in broth (100 ml) was preincubated with rGAPDH(Δ166-183) (100 μg) at 35°C overnight. Cells were harvested and lysed by sonication in lysis buffer consisting of 10 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl, 0.1% SDS, 1% Triton X-100, 0.1% β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. After centrifugation, the supernatant was passed through a 0.45-μm-pore-size filter (Millipore, Bedford, MA). The lysate was subjected to a HisTrap HP column, and pulled-down proteins were eluted with elution buffer consisting of lysis buffer and 0.85 M NaCl.
Dot blot assay.
The eluate (15 μg) of the pulldown assay was immobilized on a nitrocellulose membrane (0.2-μm pore size; Bio-Rad) with a Bio-Dot apparatus (Bio-Rad). The membrane was blocked with 5% (vol/vol) Block Ace (a casein solution prepared from homogenized milk; Snow Brand Co., Ltd., Sapporo, Japan). The membrane was subsequently incubated with 1 mg/ml S. oralis rGAPDH at 4°C overnight. Following washing, the membrane was incubated with 1:1,000 mouse anti-HT-antibodies (Qiagen) at 4°C overnight. After washing, the membrane was incubated with 1:2,000 horseradish-peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (Zymed Laboratories Inc., San Francisco, CA) for 1 h at room temperature. The bound rGAPDH was visualized using a horseradish peroxidase (HRP) conjugate substrate kit (Bio-Rad).
2D gel electrophoresis.
The eluate from the pulldown assay was dialyzed against water, freeze-dried, and then redissolved in 7 M urea, 2 M thiourea, 2 mM tributylphosphine, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.2% Bio-Lyte 3/10NL Ampholyte (Bio-Rad), and 0.001% bromophenol blue. Isoelectric focusing was performed with immobilized pH 3 to 10 gradient strips (Bio-Rad). The strips were reduced by 2 mM tributylphosphine and alkylated by 2.5% (wt/vol) iodoacetamide. Two-dimensional (2D) SDS-PAGE was performed on 5 to 15% gradient acrylamide gels. The gels were stained with CBB.
Protein identification.
Following SDS-PAGE, the protein spots of interest were excised and digested with trypsin (modified sequencing grade; Promega Co., Madison, WI) overnight at 37°C. Tryptic peptides were extracted from each spot by previously described methods (23, 24) and analyzed with a 4800 matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometer (MALDI-TOF/TOF-MS; AB Sciex, Framingham, MA). Samples were analyzed in MS positive ion reflector mode in the mass range of 900 to 4,000 Da, and peak lists were generated as previously described (25). Peak lists were searched against the NCBInr Bacteria database (3,776,131 entries as of 16 November 2009) using Mascot (version 2.2), with trypsin specification. Carbamidomethylation of cysteine was set as a fixed modification. Peptides with a score above the identity threshold (corresponding to an expectation value below 0.05) were considered as identified.
Preparation of P. gingivalis recombinant client proteins.
The primers for the recombinant client proteins are listed in Table 1. The PCR fragments were confirmed by DNA sequencing and cloned into plasmid pQE-30 (Qiagen). E. coli M15 (pREP4) was transformed with the resulting plasmids. HT recombinant proteins were purified with HisTrap HP columns. Purified recombinant client proteins were subjected to SDS-PAGE. The gel was stained with CBB. Precision plus protein standards (Bio-Rad) were used to estimate molecular masses. Protein concentrations of the samples were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Recombinant proteins were analyzed with a 4800 MALDI-TOF/TOF-MS to confirm that the seven P. gingivalis proteins were correctly expressed as recombinant proteins.
Binding of P. gingivalis recombinant client proteins to S. oralis GAPDH.
S. oralis rGAPDH was prepared as reported previously (22). Interactions between P. gingivalis recombinant client proteins and S. oralis rGAPDH were analyzed with a BIAcore 2000 system (GE Healthcare). S. oralis rGAPDH in 10 mM sodium acetate buffer (pH 4.5) was immobilized at 250 resonance units (RU) on the matrix according to the manufacturer's instructions. Each recombinant client protein was injected across the active CM5 surface with immobilized S. oralis rGAPDH and an empty control CM5 surface at various concentrations (0.4 to 0.8 μM), a flow rate of 10 μl/min, and 37°C. Specific profiles of client proteins binding to immobilized S. oralis rGAPDH were obtained following subtraction of the control signal from the response signal. Analysis of these kinetic parameters was performed with BIAevaluation 3.1 (GE Healthcare).
Effects of recombinant client proteins on P. gingivalis biofilm formation. (i) Coaggregation turbidimetric assays.
Coaggregation assays were performed according to a previously reported method (26). Suspensions of P. gingivalis ATCC 33277 and S. oralis ATCC 9811 (0.5 ml; each containing 5 × 108 cells) were mixed in a total volume of 2 ml in a cuvette. The progress of coaggregation was monitored for 10 min and at 37°C with a UV-visible recording spectrophotometer (UV-1600; Shimadzu Co., Kyoto, Japan). The decrease in A550 was recorded, and dA/dt was continuously calculated. Coaggregation activity was calculated by subtraction of the −dA/dt of P. gingivalis alone from the maximum −dA/dt of both bacteria. The addition of each client protein to each bacterial suspension had no effect on the decrease in A550 during the assay. Inhibition was determined with the following equation: (coaggregation activity without protein − coaggregation activity with protein)/(coaggregation activity without protein) × 100%. Minus values indicated enhancement of coaggregation.
(ii) Heterotypic biofilm fluorescence microscopy analysis.
Biofilm formation of P. gingivalis with streptococci was conducted as described previously (22, 27). Briefly, samples of S. oralis ATCC 9811 (1.5 × 109 CFU/ml) were stained with 15 μg of hexidium iodide (HI; Life Technologies, Grand Island, NY). The stained cells (5 × 108 CFU/ml) were cultured anaerobically in a CultureWell chambered coverglass system (Life Technologies) at 37°C for 16 h. P. gingivalis ATCC 33277 (1.5 × 109 CFU/ml) was stained with 10 μg/ml of fluorescein isothiocyanate (FITC; Life Technologies). Samples of P. gingivalis (5 × 108 CFU/ml) and client proteins (5 μg) were subsequently added to the wells exhibiting streptococcal biofilm formation. The mixtures were incubated anaerobically at 37°C for 12 h in the dark on a rotator. Labeled bacterial communities were visualized using an Axiovert 200 M fluorescence microscope with an Apotome system and a 63× magnification, 1.4-numeric-aperture (NA) objective (Carl Zeiss Co., Ltd., Oberkochen, Germany), and the images were captured at autoexposure times using FITC and HI excitation filters (FITC, 475 nm; HI, 545 nm). Images were processed with AxioVision V4.8 (Carl Zeiss), ImageJ V1.33, and Imaris V5.7.1 (Bitplane, Zurich, Switzerland). The volumes of P. gingivalis and S. oralis were determined with Daime software (downloaded from the University of Vienna, Wien, Austria [http://www.microbial-ecology.net/daime/]).
(iii) Homotypic biofilm fluorescence microscopy analysis.
Homotypic biofilm formation was conducted as described previously (28). Briefly, P. gingivalis (1.5 × 109 CFU/ml) was stained with FITC as described above. Samples of P. gingivalis (5 × 108 CFU/ml) and recombinant client proteins (5 μg) were added to the chambered system and incubated anaerobically at 37°C for 12 h. Labeled bacterial communities were visualized using a fluorescence microscope as described above. P. gingivalis volume and maximum height were determined with Daime software.
Real-time quantitative RT-PCR.
Quantitative reverse transcription-PCR (RT-PCR) was used to determine the transcriptional activity of targeted genes after 3 h of contact between P. gingivalis (5 × 108 CFU/ml) and recombinant proteins (5 μg). Primers for TonB-dependent receptor protein (RagA4), arginine-specific proteinase B (RgpB), 4-hydroxybutyryl-coenzyme A (CoA) dehydratase (AbfD), lysine-specific proteinase (Kgp), GAPDH, NAD-dependent glutamate dehydrogenase (GDH), and malate dehydrogenase (MDH) were designed using Beacon Designer V7 software (Premier Biosoft International, Palo Alto, CA) (Table 1). The primer sequence information for luxS, fimA, and 16S rRNA was obtained from James et al. (29). RNA was extracted with a PureLink RNA Minikit (Life Technologies). An iScript cDNA synthesis kit (Bio-Rad) was used to generate cDNA from RNA templates. Real-time RT-PCR was performed on a Rotor Gene 6000 using a QuantiFast SYBR green kit (Qiagen).
Enzymatic activities. (i) P. gingivalis cell surface GAPDH activity.
GAPDH activity on P. gingivalis cell surfaces was measured spectrophotometrically at 340 nm according to the method of Pancholi and Fischetti (30). In brief, P. gingivalis cells (2.5 × 109 cells/ml) were incubated with glyceraldehyde-3-phosphate (49 mg/ml; Sigma-Aldrich) and 5 μg of rRagA4, rAbfD, or rGDH in the presence of NAD+ (10 mM; Wako). GAPDH activity was obtained by subtracting the response signal without glyceraldehyde-3-phosphate.
(ii) P. gingivalis cell surface GDH activity.
P. gingivalis cell surface GDH activity was measured spectrophotometrically at 340 nm with a GDH activity assay kit (BioVision Inc., Milpitas, CA) according to the manufacturer's instructions. In brief, P. gingivalis (2.5 × 109 cells/ml) was incubated with S. oralis rGAPDH (5 μg) and 2 M glutamate. GDH activity was obtained by subtracting the response signal without glutamate.
(iii) P. gingivalis cell surface MDH activity.
MDH activity was assayed by following the method of Molenaar et al. (31). In brief, P. gingivalis (2.5 × 109 cells/ml) was incubated with S. oralis rGAPDH (5 μg), 50 mM l-malate, and 2 mM NAD+. MDH activity was measured spectrophotometrically at 340 nm and 37°C by subtraction of the response signal without l-malate.
(iv) NADH oxidase activity of P. gingivalis cell surface AbfD.
Diaz et al. showed that AbfD had NADH oxidase activity in P. gingivalis cell extracts (32). Therefore, instead of measuring AbfD expression, NADH oxidase activity was assayed by following the methods of Higuchi et al. (33) by spectrophotometric monitoring at 340 nm and 25°C. The reaction mixture contained S. oralis rGAPDH (5 μg), 0.1 mM β-NADH, and P. gingivalis cells (2.5 × 109 cells/ml). NADH oxidase activity was obtained by subtraction of the response signal without β-NADH.
Statistical analysis.
Student's unpaired two-tailed t test was used to analyze the differences between groups. P values of <0.01 were considered statistically significant.
RESULTS
Identification of P. gingivalis components binding to S. oralis rGAPDH(Δ166-183).
HT-rGAPDH(Δ166-183) was constructed with the S. oralis ATCC 9811 GAPDH containing a deletion of amino acid residues 166 to 183. The rGAPDH(Δ166-183) plasmid was sequenced to confirm the deletion. The purified protein was subjected to SDS-PAGE and showed a single band with a molecular mass of approximately 38 kDa (Fig. 1A). S. oralis rGAPDH(Δ166-183) showed GAPDH enzyme activity (data not shown), suggesting that the mutant protein retains native conformation.
Fig 1.
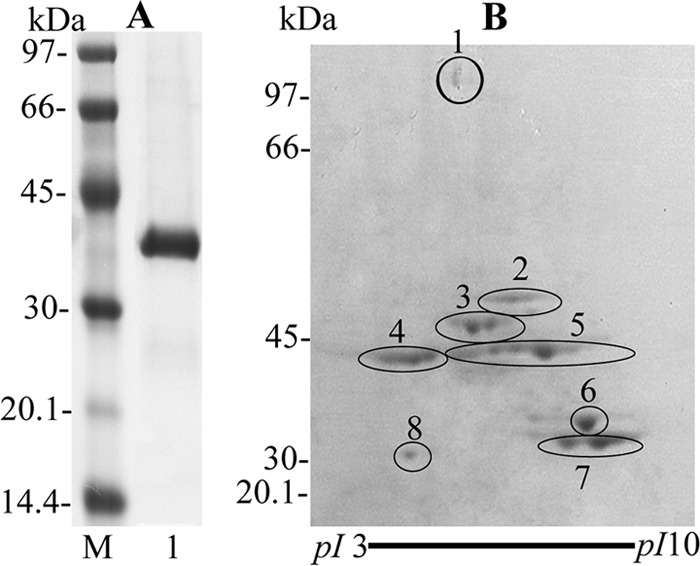
One-dimensional gel of S. oralis rGAPDH(Δ166-183) and 2D gel of P. gingivalis client proteins by a pulldown assay. (A) Purified S. oralis rGAPDH (Δ166-183) (3 μg) was subjected to SDS-PAGE (5 to 15% gradient gel) and stained with CBB. Lanes: M, molecular mass standard proteins; 1, S. oralis ATCC 9811 rGAPDH(Δ166-183). (B) P. gingivalis client proteins purified by a pulldown assay (100 μg) were separated by 2D gel electrophoresis and stained with CBB. The spots were identified as TonB-dependent receptor protein (RagA4; spot 1), 4-hydroxybutyryl-CoA dehydratase (AbfD; spot 2), lysine-specific proteinase (Kgp; spot 3), arginine-specific proteinase B (RgpB; spot 4), NAD-dependent glutamate dehydrogenase (GDH; spot 5), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; spots 6 and 8), and malate dehydrogenase (MDH; spot 7).
For identification of the P. gingivalis components interacting with S. oralis rGAPDH(Δ166-183), P. gingivalis cells were incubated with S. oralis rGAPDH(Δ166-183). The eluates of P. gingivalis components bound to S. oralis rGAPDH(Δ166-183) were purified by a pulldown assay and were shown to bind to S. oralis rGAPDH by dot blot assays (data not shown).
The eluate from the pulldown assay was subjected to 2D gel electrophoresis. Eight spots of molecular masses between approximately 30 and 110 kDa were detected (Fig. 1B). The eight spots were in-gel digested with trypsin and identified with a MALDI-TOF/TOF-MS. The spots were identified as TonB-dependent receptor protein (RagA4; spot 1), 4-hydroxybutyryl-CoA dehydratase (AbfD; spot 2), lysine-specific proteinase (Kgp; spot 3), arginine-specific proteinase B (RgpB; spot 4), NAD-dependent glutamate dehydrogenase (GDH; spot 5), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; spots 6 and 8), and malate dehydrogenase (MDH; spot 7) (Fig. 1B and Table 2). The pI of spot 8 was vastly less than that of spot 6 in the 2D gel. Therefore, calibration of pI and molecular mass, which were predicted from the potential arginine and lysine cleavage sites, was performed using the ExPASy compute pI/Mw tool (http://web.expasy.org/compute_pi/), and the theoretical protein pI and mass were similar to those in the apparent 2D gel. This result suggests that GAPDH on the P. gingivalis cell surface is digested with proteases such as Rgp and Kgp during the pulldown assay.
Table 2.
Identification of protein spots by MALDI-TOF/TOF-MS
| Spot no. | Name | TIGR IDa |
|---|---|---|
| 1 | TonB-dependent receptor protein (RagA4) | PG0185 |
| 2 | 4-Hydroxybutyryl-CoA dehydratase (AbfD) | PG0692 |
| 3 | Lysine-specific cysteine proteinase (Kgp) | PG1844 |
| 4 | Arginine-specific cysteine proteinase B (RgpB) | PG0506 |
| 5 | NAD-dependent glutamate dehydrogenase (GDH) | PG1232 |
| 6, 8 | Glyceraldehyde 3-phosphate dehydrogenase, type I (GAPDH) | PG2124 |
| 7 | Malate dehydrogenase (MDH) | PG1949 |
ID, identification; annotations are from the major P. gingivalis ATCC 33277 genome databases at TIGR (www.jcvi.org).
Purification of P. gingivalis recombinant proteins.
HT recombinant proteins of the identified components were purified. The seven purified P. gingivalis recombinant client proteins were subjected to SDS-PAGE and detected as single-molecular-mass bands of between approximately 35 and 110 kDa (Fig. 2). Each of the recombinant proteins was more than 97% pure as judged from the CBB-stained SDS-PAGE gel, coupled with densitometric analysis using Quantity One software (Bio-Rad). Each protein band was unambiguously identified with a MALDI-TOF/TOF-MS as the respective proteins listed in Table 2. P. gingivalis rAbfD, rGAPDH, rGDH, and rMDH exhibited their relevant enzyme activities, while rRgpB and rKgp had little enzyme activity (data not shown), suggesting that rAbfD, rGAPDH, rGDH, and rMDH retain native conformation and rRgpB and rKgp were not able to retain native conformation.
Fig 2.
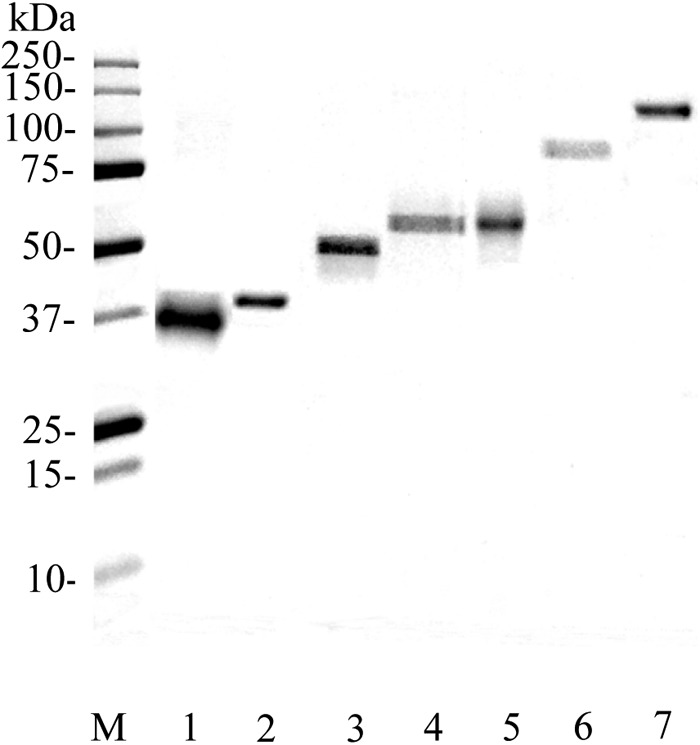
SDS-PAGE of P. gingivalis recombinant client proteins. Purified P. gingivalis recombinant client proteins (2 μg) were subjected to SDS-PAGE with a 5 to 15% gradient gel. The gel was stained with CBB. Lanes: M, molecular mass standard proteins; 1, MDH; 2, GAPDH; 3, GDH; 4, Kgp; 5, AbfD; 6, RgpB; 7, RagA4.
Binding of P. gingivalis recombinant client proteins to S. oralis GAPDH.
Interactions between P. gingivalis recombinant client proteins and S. oralis rGAPDH were analyzed with a BIAcore 2000 system. The KA values are shown in Table 3. The KA values of many antibody-protein antigen interactions are within the range of 106 to 1010 M−1 (34). The KA values of rRagA4, rAbfD, rGDH, rGAPDH, and rMDH toward S. oralis rGAPDH were 3.38 × 107, 8.44 × 107, 2.20 × 107, 1.49 × 107, and 1.78 × 107 M−1, respectively, indicating strong binding activity. In contrast, the KA values of rKgp and rRgpB toward S. oralis rGAPDH were 2.62 × 106 and 4.30 × 106 M−1, respectively, indicating weak binding activity.
Table 3.
Kinetic parameters for P. gingivalis client protein binding to immobilized S. oralis rGAPDH
| Analyte | KA (M−1) |
|---|---|
| rRagA4 | 3.38 × 107 |
| rAbfD | 8.44 × 107 |
| rKgp | 2.62 × 106 |
| rRgpB | 4.30 × 106 |
| rGDH | 2.20 × 107 |
| rGAPDH | 1.49 × 107 |
| rMDH | 1.78 × 107 |
Effects of P. gingivalis client proteins on the interaction between P. gingivalis and S. oralis. (i) Coaggregation turbidimetric assay.
We hypothesized that the P. gingivalis client proteins inhibit coaggregation between P. gingivalis and S. oralis.
Therefore, effects of the recombinant proteins on coaggregation were examined by a turbidimetric assay. P. gingivalis rGAPDH and rMDH inhibited coaggregation in a dose-dependent manner (Fig. 3). In contrast, P. gingivalis rRagA4, rAbfD, and rGDH greatly enhanced coaggregation in a dose-dependent manner. P. gingivalis rKgp and rRgpB had little effect on coaggregation.
Fig 3.
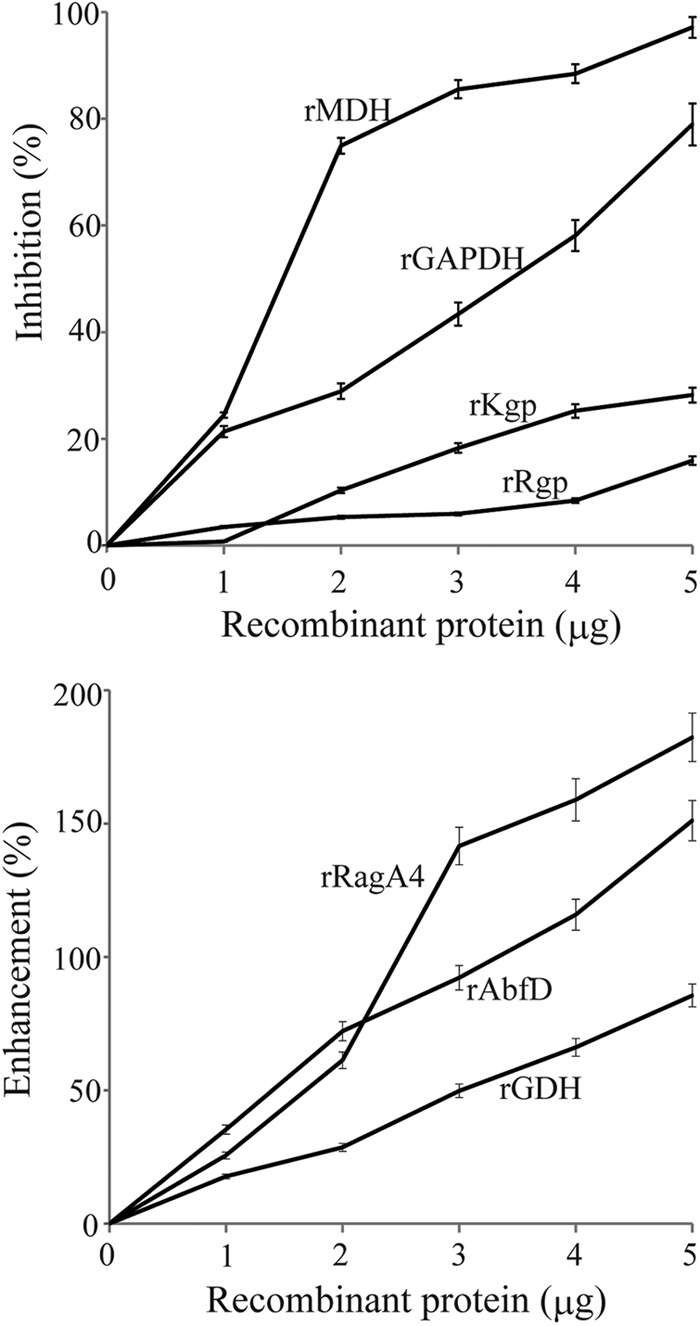
Effects of P. gingivalis recombinant client proteins on coaggregation between P. gingivalis and S. oralis by turbidimetric assay. Aliquots of suspensions of P. gingivalis ATCC 33277 (5 × 108 cells) and S. oralis ATCC 9811 (5 × 108 cells) with various concentrations of P. gingivalis recombinant client proteins were mixed simultaneously. The progress of coaggregation was monitored by measurement of the decrease in A550 at 37°C, and the −dA/dt was calculated continuously. Coaggregation activity was calculated by subtraction of the −dA/dt of P. gingivalis ATCC 33277 alone from the maximum −dA/dt of both bacteria. Inhibition was determined with the following formula: (coaggregation activity without protein − coaggregation activity with protein)/(coaggregation activity without protein) × 100%. Minus value indicated enhancement of coaggregation. Values represent means ± standard deviations (SD) from three replicates.
(ii) Heterotypic biofilm analysis by fluorescence microscopy.
Effects of the recombinant proteins on biofilm formation between P. gingivalis ATCC 33277 and S. oralis ATCC 9811 were examined by fluorescence microscopy. P. gingivalis rGAPDH and rMDH inhibited biofilm formation between P. gingivalis and S. oralis at a concentration of 5 μg/ml (P < 0.01) (Fig. 4A and B). In contrast, P. gingivalis rRagA4, rAbfD, and rGDH significantly enhanced the community phenotype compared to the control. P. gingivalis rKgp and rRgpB had little effect on heterotypic biofilm formation. The biovolume of S. oralis had little effect by the addition of recombinant proteins.
Fig 4.
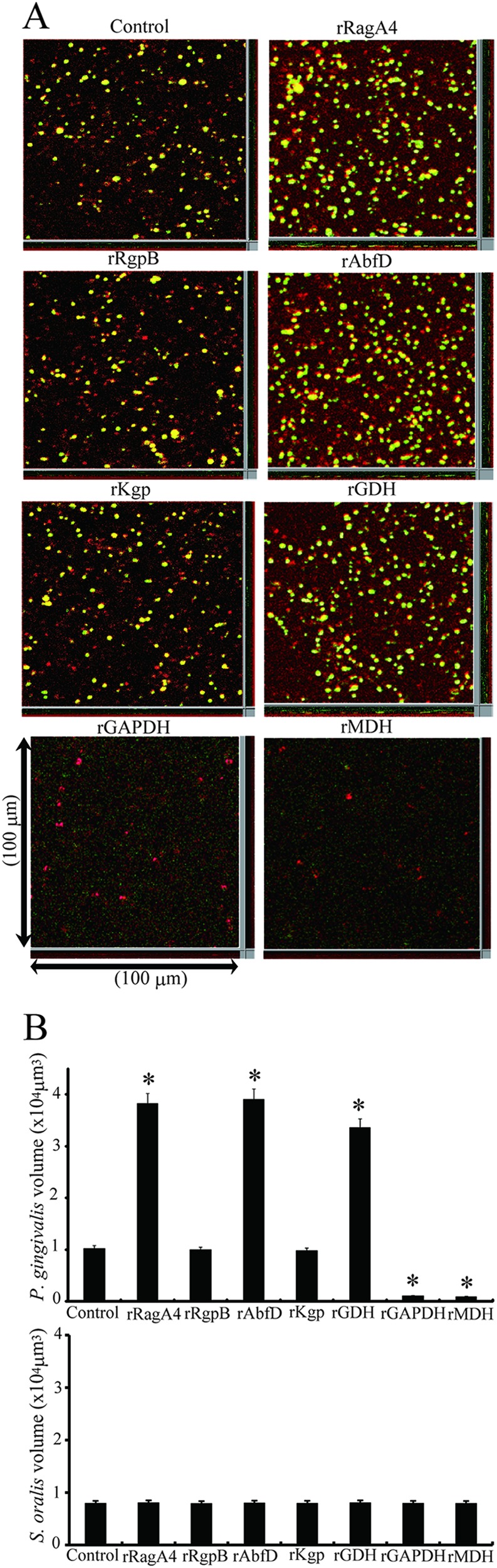
Effect of P. gingivalis recombinant proteins on biofilm formation between P. gingivalis and S. oralis by fluorescence microscopy. (A) Analysis of dual-species communities by fluorescence microscopy. Substrata of S. oralis cells (red) were reacted with P. gingivalis (green) and P. gingivalis recombinant proteins (5 μg) for 12 h. The resulting heterotypic community was observed on a fluorescence microscope with an Apotome system. A series of fluorescent optical x-y sections were collected to create digitally reconstructed 2D images (x-z section, y-z section, and x-y section) with ImageJ and Imalis software. Colocalized bacteria appear yellow. Biofilm formation without recombinant proteins served as a control. (B) P. gingivalis or S. oralis accumulation measured by green- or red-area analysis of fluorescence. Means and standard deviations of the total volume for x-y sections with a 5-μm-spaced z-series for the strains were processed with Daime software. An asterisk denotes a significant difference at P < 0.01 (t test) compared to the control.
(iii) Homotypic biofilm analysis by fluorescence microscopy.
Since rRagA4, rAbfD, and rGDH enhanced heterotypic biofilm formation, the effects of these recombinant proteins on P. gingivalis monospecies biofilms were examined. P. gingivalis rRagA4, rAbfD, and rGDH also enhanced the monospecies community phenotype compared to the control (Fig. 5A). The biovolume of P. gingivalis was significantly elevated by addition of rRagA4, rAbfD, and rGDH compared to that of the control (P < 0.01) (Fig. 5B). The maximum height of P. gingivalis accumulation was also elevated by the addition of rRagA4, rAbfD, and rGDH compared to that of the control (P < 0.01) (Fig. 5C).
Fig 5.
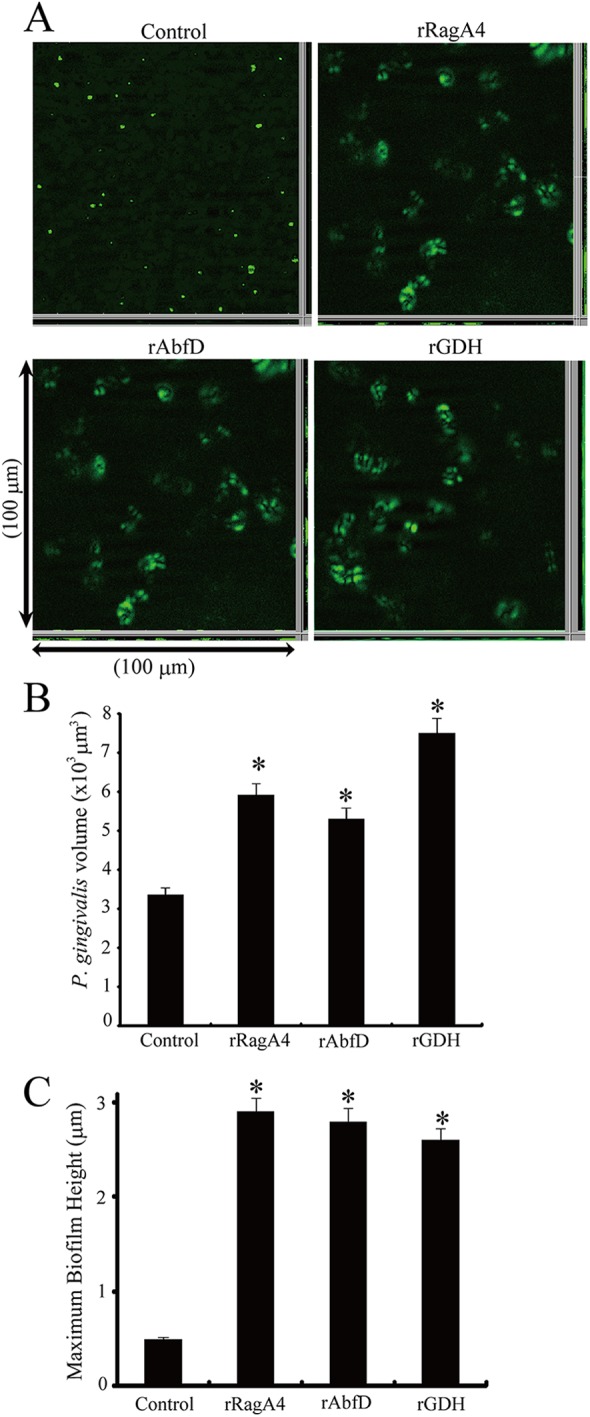
Effect of P. gingivalis recombinant proteins on homotypic P. gingivalis biofilm formation. (A) Analysis of monospecies community by fluorescence microscopy. P. gingivalis ATCC 33277 (green) cells were left unstimulated or were reacted with P. gingivalis recombinant proteins for 12 h. The resulting homotypic community was observed by a fluorescence microscope with an Apotome system. Images were digitally reconstructed (2D image; x-z section, y-z section, and x-y section) with ImageJ and Imaris software. Biofilm formation without recombinant proteins served as a control. (B) P. gingivalis accumulation measured by green-area analysis of fluorescence. Means and standard deviations of total volumes for x-y sections with a 5-μm-spaced z-series for the strains were processed with Daime software. An asterisk denotes a significant difference at P < 0.01 (t test) compared to the control. (C) Maximum extent and standard deviations of P. gingivalis accumulation in the z dimension measured across three random x-z sections. An asterisk denotes a significant difference at P < 0.01 (t test) compared to the control.
Modulation of P. gingivalis client protein expression by S. oralis GAPDH.
The expression level of representative genes encoding the client proteins in P. gingivalis cultured with S. oralis rGAPDH for 3 h was quantified by real-time quantitative RT-PCR (Fig. 6A). ragA4, abfD, and gdh showed significantly increased expression in P. gingivalis cultured with S. oralis rGAPDH compared to that in P. gingivalis alone (P < 0.01). In contrast, the expression of gapdh and mdh was significantly lower in P. gingivalis cultured with S. oralis rGAPDH than in the control (P < 0.01). NADH oxidase activity on the cell surface of P. gingivalis ATCC 33277 was increased 3-fold by the presence of S. oralis rGAPDH (P < 0.01) (Fig. 6B). GDH cell surface activity increased 2.4-fold when P. gingivalis was cultured with S. oralis rGAPDH (P < 0.01) (Fig. 6C). P. gingivalis MDH cell surface activity with S. oralis rGAPDH was 37.2% of the activity without S. oralis rGAPDH (P < 0.01) (Fig. 6D).
Fig 6.
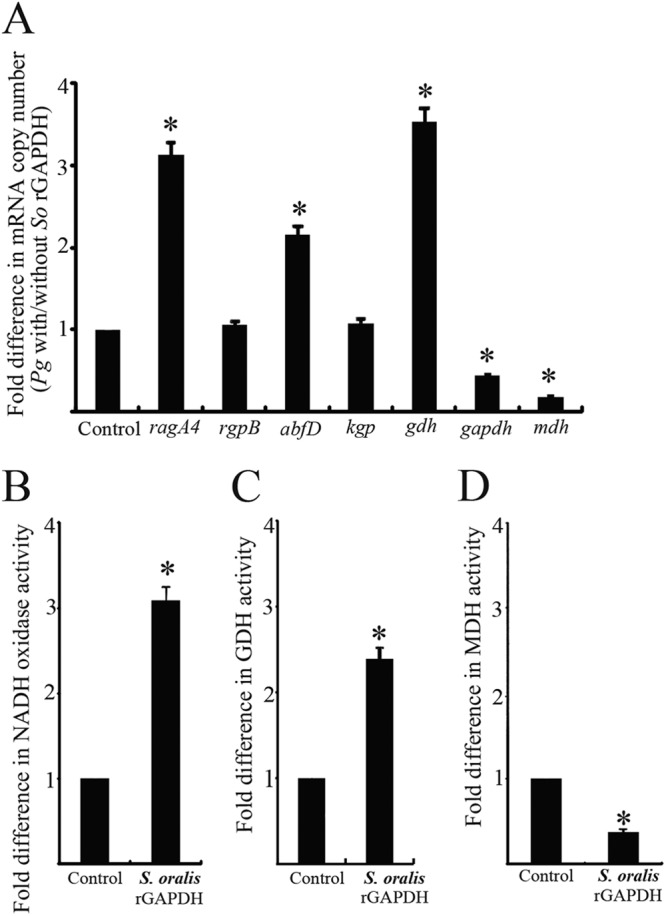
Modulation of P. gingivalis client protein expression by S. oralis rGAPDH. (A) Real-time quantitative RT-PCR of mRNA levels for genes involved in P. gingivalis client protein production and secretion. Transcript levels were normalized to 16S rRNA. P. gingivalis without S. oralis rGAPDH served as a control. The fold differences were expressed as targeted mRNA levels after exposure to S. oralis rGAPDH (5 μg) compared to the control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes a significant difference at P < 0.01 (t test) compared to the control. (B) Measurement of P. gingivalis cell surface NADH oxidase activity. NADH oxidase activity was measured at 25°C by monitoring the oxidation of β-NADH at 340 nm. (C) Measurement of P. gingivalis cell surface GDH activity. GDH activity was measured as the reduction of NAD by monitoring the oxidation of glutamate to α-ketoglutamate at 340 nm and 37°C. (D) Measurement of P. gingivalis cell surface MDH assays. MDH activity was measured by monitoring NADH at 340 nm and 37°C. P. gingivalis without S. oralis rGAPDH served as a control. The fold differences of NADH oxidase activity, GDH activity, and MDH activity were expressed as enzymatic activity after exposure to S. oralis rGAPDH (5 μg) compared to that of the control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone.
Effects of S. oralis rGAPDH and P. gingivalis client proteins on the production of P. gingivalis LuxS.
LuxS-dependent signaling is required for the development of P. gingivalis-S. gordonii biofilm communities (35). P. gingivalis is one of only a few known organisms in which LuxS production and AI-2 activity are regulated at the level of luxS transcription (29). However, little is known about luxS and the transcription of other P. gingivalis genes required for P. gingivalis-S. oralis heterotypic biofilm development. Therefore, we hypothesized that S. oralis GAPDH also controls expression of the LuxS enzyme that is responsible for AI-2 formation.
Quantitative RT-PCR was used to determine the transcriptional activity of luxS in P. gingivalis incubated with several proteins. As shown in Fig. 7A, the luxS mRNA level was significantly increased when P. gingivalis was cultured with S. oralis rGAPDH compared to that with P. gingivalis alone (P < 0.01). As shown in Fig. 7B, the luxS mRNA level was increased when P. gingivalis was incubated with rRagA4, rAbfD, or rGDH compared to that with P. gingivalis alone (P < 0.01), whereas the luxS mRNA level decreased when P. gingivalis was in contact with rMDH compared to that with P. gingivalis alone (P < 0.01).
Fig 7.
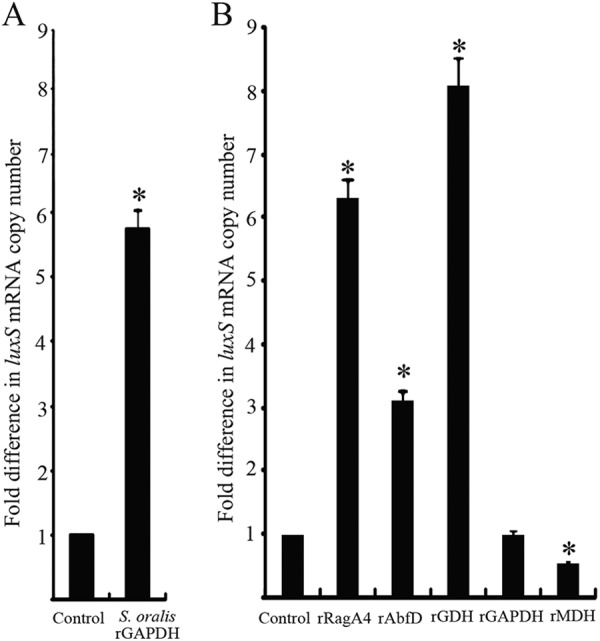
Effect of S. oralis rGAPDH and P. gingivalis client proteins on expression of P. gingivalis luxS. (A) luxS mRNA in P. gingivalis with S. oralis rGAPDH (5 μg) was measured by quantitative RT-PCR. (B) luxS mRNA in P. gingivalis with P. gingivalis client proteins (5 μg) was measured by quantitative RT-PCR. Transcript levels were normalized to 16S rRNA. P. gingivalis without recombinant protein served as a control. The fold differences were expressed as luxS mRNA levels after exposure to S. oralis rGAPDH and P. gingivalis client proteins compared to that of the control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone.
Effects of P. gingivalis client proteins on the expression of cell surface GAPDH.
Quantitative RT-PCR was used to determine the transcriptional activity of P. gingivalis gapdh in contact with the client proteins. As shown in Fig. 8A, the gapdh mRNA level was increased when P. gingivalis was in contact with rRagA4, rAbfD, and rGDH compared to that with P. gingivalis alone (P < 0.01). Compared to P. gingivalis alone, rRagA4, rAbfD, and rGDH increased GAPDH enzymatic activity on the cell surface of P. gingivalis by 1.5- to 4-fold (Fig. 8B) (P < 0.01).
Fig 8.
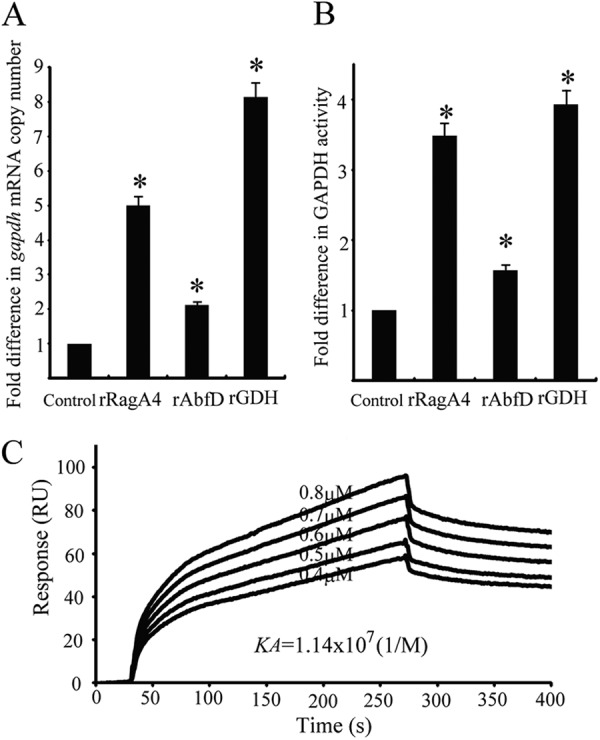
Effect of P. gingivalis client proteins on expression of P. gingivalis GAPDH. (A) gapdh mRNA in P. gingivalis was measured by quantitative RT-PCR. Transcript levels were normalized to 16S rRNA. P. gingivalis without recombinant proteins served as a control. The fold difference is expressed as gapdh mRNA levels after exposure to P. gingivalis client proteins (5 μg) compared to the control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes significant difference at P < 0.01 (t test) compared to P. gingivalis alone. (B) Measurement of P. gingivalis cell surface GAPDH activity. GAPDH activity was measured at 340 nm. The fold difference is presented as a ratio of GAPDH activity with P. gingivalis client proteins (5 μg) to that without the client proteins. P. gingivalis without recombinant protein served as a control. Data are means and standard deviations from three independent experiments performed in triplicate. An asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone. (C) Sensorgrams of P. gingivalis rFimA binding to immobilized P. gingivalis rGAPDH in kinetic studies. P. gingivalis rFimA fimbriae were injected over P. gingivalis rGAPDH on the sensor chip at various concentrations (0.4 to 0.8 μM).
Interactions between P. gingivalis rGAPDH and P. gingivalis rFimA were analyzed with a BIAcore 2000 system. P. gingivalis rFimA was injected across the active CM5 surface of P. gingivalis rGAPDH. The KA of P. gingivalis rFimA toward P. gingivalis rGAPDH was 1.14 × 107 M−1 (Fig. 8C), indicating strong binding activity. Quantitative RT-PCR was used to determine fimA transcriptional activity in P. gingivalis following 3 h of contact with recombinant client proteins (rRagA4, rAbfD, rGDH, rGAPDH, and rMDH). However, fimA mRNA levels were similar to those of the control, indicating that the recombinant client proteins had little effect on the expression of P. gingivalis FimA fimbriae (data not shown).
DISCUSSION
Previously, we demonstrated that S. oralis GAPDH bound to P. gingivalis fimbriae with high affinity. In this study, we investigated P. gingivalis components other than fimbriae that interact with S. oralis GAPDH. However, since S. oralis GAPDH binds strongly to P. gingivalis FimA fimbriae, it was difficult to identify new P. gingivalis client proteins other than fimbriae. Therefore, we constructed rGAPDH(Δ166-183), in which amino acid residues 166 to 183 that bind to FimA fimbriae were deleted, and used it to identify client proteins by a pulldown assay. Seven proteins, RagA4, RgpB, AbfD, Kgp, GAPDH, GDH, and MDH, were identified as client proteins that bind to rGAPDH(Δ166-183), indicating that these seven client proteins interact with domains other than that involving amino acid residues 166 to 183. Among these proteins, RagA4, RgpB, Kgp, GDH, and MDH have been reported to be localized at the surface of P. gingivalis (36, 37, 38). Gingipains, a unique class of cysteine proteinases composed of arginine-specific (RgpA and RgpB) and lysine-specific (Kgp) proteases, have been shown to be major proteins in the outer membrane (39). These proteinases are closely associated with the pathogenesis of periodontal diseases and presumably are important for obtaining nutrients from the environment and for the processing and maturation of various cell surface proteins (40). Using Kgp mutants, Kgp was shown to have a suppressive regulatory role during P. gingivalis-S. gordonii biofilm formation (28). Rgp controlled microcolony morphology and biovolume in the Rgp mutants. In this study, KA values for rRgpB and rKgp with S. oralis rGAPDH were lower than those of the other client proteins (KA = 4.30 × 106 and 2.62 × 106 M−1, respectively), and rRgpB and rKgp had little effect on P. gingivalis-S. oralis interaction (Fig. 3 and 4). Therefore, we focused on the other five client proteins (RagA4, AbfD, GAPDH, GDH, and MDH) and examined the effects of these proteins on P. gingivalis-S. oralis biofilm formation.
rRagA4, rGDH, and rAbfD enhanced P. gingivalis-S. oralis biofilm formation. In general, RagA has homology to TonB-linked outer membrane receptors, which are involved in the recognition and active transport of specific external ligands in a wide range of Gram-negative species (41, 42). There are four divergent alleles of the ragA locus (43). The original ragA allele from P. gingivalis W50 was renamed ragA1, whereas ragA4 was found in P. gingivalis ATCC 33277, which lacks ragA1. RagA4 has 70% amino acid homology with RagA1. In P. gingivalis ATCC 33277, an additional ragA4 open reading frame is found downstream of ragB. The protein encoded by ragA4 has no significant homology to known proteins, except for a C-terminal 80-amino-acid sequence that is 50% similar to the C-terminal domain shared by gingipain precursor proteins and hemagglutinins of P. gingivalis (44). Recently, chemical cross-linking and coimmunoprecipitation experiments revealed a physical association between RagA4 and RagB (45). In addition, cell surface labeling showed that both RagA4 and RagB were exposed on the cell surface. Moreover, RagA4 was identified as being involved in host cell invasion (46). Therefore, it is reasonable to deduce that RagA4 is associated with biofilm formation.
In general, GDH is an enzyme that is present in most bacteria and in the mitochondria of eukaryotes, as are some of the other enzymes required for urea synthesis. GDH converts glutamate to α-ketoglutarate and vice versa, representing a key link between catabolic and anabolic pathways. Bacterial GDH enzymes are located in the cytoplasm or cytoplasmic membrane (47, 48); however, P. gingivalis GDH has been reported to be associated with the cell surface (37). Streptococcus suis GDH is also exposed on the surface (49). Our detection of GDH activity on the P. gingivalis cell surface is consistent with these results. P. gingivalis AbfD has NADH oxidase activity (32), and we detected NADH oxidase activity on the P. gingivalis surface (Fig. 6B). AbfD has a role in glutamate metabolism, and P. gingivalis cell extracts have most of the activities involved in this pathway (50). In particular, AbfD is involved in the dehydration of hydroxybutyryl-CoA to crotonyl-CoA (51). Moreover, the abfD gene of Clostridium aminobutyricum forms part of a genetic region containing other genes involved in this fermentation pathway (52). Comparison of the amino acid sequence of AbfD in P. gingivalis to that in C. aminobutyricum (GenBank accession number CAB60035) revealed high levels of homology (75%) (53). These observations support the identification of PG0625 (P. gingivalis W83 accession number according to http://www.oralgen.lanl.gov) as AbfD (32).
In this study, the KA values of P. gingivalis rRagA4, rGDH, and rAbfD toward S. oralis rGAPDH demonstrate high affinity (KA = 3.38 × 107, 2.20 × 107, and 8.44 × 107 M−1, respectively) (Table 3). The addition of rRagA4, rGDH, and rAbfD enhanced dual-species bacterial accumulation (Fig. 3 and 4) and enhanced monospecies biofilm formation (Fig. 5). Moreover, mRNA levels of ragA4, gdh, and abfD increased with the addition of S. oralis rGAPDH (Fig. 6A), and gapdh mRNA levels increased with the addition of rRagA4, rGDH, and rAbfD (Fig. 8A and B). These results indicate that expression of RagA4, GDH, and AbfD increases on P. gingivalis surfaces in contact with S. oralis GAPDH, and increased RagA4, GDH, and AbfD may enhance the formation of P. gingivalis-S. oralis biofilms and P. gingivalis monospecies biofilms.
Accumulation of P. gingivalis on streptococcal substrata requires adhesins, such as FimA fimbriae that bind to streptococcal GAPDH, and signaling molecules, such as the AI-2 family of compounds produced by the enzymatic action of LuxS (35). LuxS-dependent signaling stimulates the initial development of microcolonies. Therefore, we measured the fimA and luxS transcriptional activities following 3 h of contact between P. gingivalis and recombinant client proteins. P. gingivalis client proteins had little effect on P. gingivalis fimA mRNA levels; however, the luxS mRNA level increased 6-fold with the addition of S. oralis rGAPDH (Fig. 7A). S. oralis GAPDH may initially interact with P. gingivalis FimA and appropriately regulate the accumulation of P. gingivalis-S. oralis biofilms by LuxS to form stronger biofilms. Moreover, the luxS mRNA level was significantly increased by the addition of rRagA4, rAbfD, and rGDH (Fig. 7B), thereby facilitating P. gingivalis-S. oralis biofilm formation. In contrast, the luxS mRNA level was decreased by addition of rMDH and was unaffected by P. gingivalis rGAPDH. These results suggest that the client proteins other than P. gingivalis rGAPDH further regulate P. gingivalis-S. oralis biofilm formation and P. gingivalis monospecies biofilm formation through quorum-sensing-dependent signaling.
The gapdh mRNA level was increased by the addition of rRagA4, rAbfD, and rGDH (Fig. 8A), and P. gingivalis GAPDH activity was also increased by the addition of these proteins (Fig. 8B). We hypothesized that P. gingivalis GAPDH as well as S. oralis GAPDH bind to P. gingivalis FimA fimbriae. The representative sensorgrams shown in Fig. 8C reveal that the resonance response reflecting P. gingivalis rFimA-rGAPDH interaction occurred in an analyte concentration-dependent manner. The KA of P. gingivalis rGAPDH with rFimA demonstrated high affinity (1.14 × 107 M−1). Thus, GAPDH-FimA interaction might also function as an accumulator for homotypic P. gingivalis biofilm formation.
In this study, rMDH and rGAPDH inhibited P. gingivalis-S. oralis biofilm formation. MDH is an NAD-dependent malate dehydrogenase (EC 1.1.1.37) that participates in the malate oxidation reaction in the tricarboxylic acid cycle. P. gingivalis MDH has been shown to be surface associated (36), and P. gingivalis surface-associated MDH activity was also detected in this study (Fig. 6D). Acinetobacter baumannii has MDH in the outer membrane that may be involved in biofilm growth (54). In E. coli, MDH is known to be highly regulated for adaptation to changing conditions, such as aerobic and anaerobic growth, and to be involved in biofilm growth (55). Thus, it is logical that MDH also contributes to biofilm formation. GAPDH is generally a cytoplasmic glycolytic enzyme that has also been detected on the surfaces of several prokaryotic and eukaryotic organisms. P. gingivalis GAPDH has been reported to be involved in the regulation of host cell invasion (56). In this study, surface-associated GAPDH activity was detected in P. gingivalis (Fig. 6C). According to DDBJ BLAST searches, P. gingivalis GAPDH has 47% amino acid similarity to S. oralis GAPDH. The KA values of P. gingivalis rMDH and rGAPDH toward S. oralis rGAPDH demonstrated high affinity (KA = 1.78 × 107 and 1.49 × 107 M−1, respectively). The addition of P. gingivalis rMDH and rGAPDH inhibited dual-species bacterial accumulation (Fig. 3 and 4). Thus, the recombinant proteins may directly inhibit the interaction between P. gingivalis and S. oralis. As shown in Fig. 6A, mdh and gapdh mRNA levels were decreased by the addition of S. oralis rGAPDH. Moreover, MDH and GAPDH activities on the P. gingivalis cell surface were decreased by contact with S. oralis GAPDH. Therefore, decreased MDH and GAPDH may appropriately downregulate P. gingivalis-S. oralis biofilm formation.
In conclusion, S. oralis GAPDH may initially bind to P. gingivalis FimA fimbriae, and then RagA4, AbfD, GDH, GAPDH, and MDH may bind to S. oralis rGAPDH. The mRNA levels of ragA4, gdh, and abfD were upregulated with the existence of S. oralis GAPDH, while those of gapdh and mdh were downregulated. Therefore, when S. oralis GAPDH exists, expression of RagA4, AbfD, and GDH on the cell surface is increased, and biofilm formation between P. gingivalis and S. oralis is enhanced. On the other hand, when S. oralis GAPDH exists, expression of GAPDH and MDH on the cell surface is suppressed, and biofilm formation is inhibited. Subsequently, RagA4, AbfD, GDH, MDH, and GAPDH on the surface of P. gingivalis may appropriately regulate P. gingivalis-S. oralis biofilm formation. The role of the fimbriae is very important in P. gingivalis-S. oralis biofilm formation. However, the interaction between the client proteins and GAPDH is also significant in the biofilm formation, because the addition of the recombinant client proteins promoted or inhibited the biofilm formation between S. oralis and P. gingivalis possessing FimA fimbriae. Although a further study is necessary to understand the regulatory mechanisms of these client proteins in biofilm formation in detail, the present findings indicate that P. gingivalis client proteins play an important role in P. gingivalis biofilm formation.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Young Scientists (B) (20791638 and 22792114), a Grant-in-Aid for Scientific Research (B) (20390534), and a Grant-in-Aid for Scientific Research (C) (24593147) from the Japan Society for the Promotion of Science.
We thank Mitsuko Nakatani of the National Cerebral and Cardiovascular Center Research Institute for her expert technical assistance.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1.Albandar JM. 2005. Epidemiology and risk factors of periodontal diseases. Dent. Clin. North Am. 49: 517– 532 [DOI] [PubMed] [Google Scholar]
- 2.Haffajee AD, Socransky SS. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 5: 78– 111 [DOI] [PubMed] [Google Scholar]
- 3.Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38: 72– 122 [DOI] [PubMed] [Google Scholar]
- 4.Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. 1988. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 239: 55– 57 [DOI] [PubMed] [Google Scholar]
- 5.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54: 413– 437 [DOI] [PubMed] [Google Scholar]
- 6.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11: 94– 100 [DOI] [PubMed] [Google Scholar]
- 7.Slots J, Gibbons RJ. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19: 254– 264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi T, Kasamo K, Chuman M, Machigashira M, Inoue M, Sueda T. 1998. Preparation and characterization of an Actinomyces naeslundii aggregation factor that mediates coaggregation with Porphyromonas gingivalis. J. Periodont. Res. 33: 460– 468 [DOI] [PubMed] [Google Scholar]
- 9.Abe N, Baba A, Takii R, Nakayama K, Kamaguchi A, Shibata Y, Abiko Y, Okamoto K, Kadowaki T, Yamamoto K. 2004. Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol. Chem. 385: 1041– 1047 [DOI] [PubMed] [Google Scholar]
- 10.Goulbourne PA, Ellen RP. 1991. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 173: 5266– 5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiratsuka K, Abiko Y, Hayakawa M, Ito T, Sasahara H, Takiguchi H. 1992. Role of Porphyromonas gingivalis 40-kDa outer membrane protein in the aggregation of P. gingivalis vesicles and Actinomyces viscosus. Arch. Oral Biol. 37: 717– 724 [DOI] [PubMed] [Google Scholar]
- 12.Lamont RJ, Hersey SG, Rosan B. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7: 193– 197 [DOI] [PubMed] [Google Scholar]
- 13.Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, Shizukuishi S. 2004. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 6: 1163– 1170 [DOI] [PubMed] [Google Scholar]
- 14.Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, Shizukuishi S. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72: 1341– 1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamaguchi A, Baba H, Hoshi M, Inomata K. 1995. Effect of Porphyromonas gingivalis ATCC 33277 vesicle on adherence of Streptococcus mutans OMZ 70 to the experimental pellicle. Microbiol. Immunol. 39: 521– 524 [DOI] [PubMed] [Google Scholar]
- 16.Stinson MW, Safulko K, Levine MJ. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59: 102– 108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar HT, Genco RJ, Shizukuishi S. 1997. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76: 852– 857 [DOI] [PubMed] [Google Scholar]
- 18.Maeda K, Nagata H, Kuboniwa M, Kataoka K, Nishida N, Tanaka M, Shizukuishi S. 2004. Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect. Immun. 72: 5475– 5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winram SB, Lottenberg R. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142: 2311– 2320 [DOI] [PubMed] [Google Scholar]
- 20.Oliveira L, Madureira P, Andrade EB, Bouaboud A, Morello E, Ferreira P, Poyart C, Trieu-Cuot P, Dramsi S. 2012. Group B streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS One 7: e29963 doi:10.1371/journal.pone.0029963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirover MA. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432:159– 184 [DOI] [PubMed] [Google Scholar]
- 22.Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, Minamino N, Kuwahara H, Shizukuishi S. 2009. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect. Immun. 77: 5130– 5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenfeld J, Capdeville J, Guillemot JC, Ferrara P. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203: 173– 179 [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856– 2860 [DOI] [PubMed] [Google Scholar]
- 25.Osaki T, Sasaki K, Minamino N. 2011. Peptidomics-based discovery of an antimicrobial peptide derived from insulin-like growth factor-binding protein. J. Proteome Res. 10: 1870– 1880 [DOI] [PubMed] [Google Scholar]
- 26.Nagata H, Murakami Y, Inoshita E, Shizukuishi S, Tsunemitsu A. 1990. Inhibitory effect of human plasma and saliva on co-aggregation between Bacteroides gingivalis and Streptococcus mitis. J. Dent. Res. 69: 1476– 1479 [DOI] [PubMed] [Google Scholar]
- 27.Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, Demuth DR, Lamont RJ. 2008. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol. 69: 1153– 1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuboniwa M, Amano A, Hashino E, Yamamoto Y, Inaba H, Hamada N, Nakayama K, Tribble GD, Lamont RJ, Shizukuishi S. 2009. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 9: 105 doi:10.1186/1471-2180-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, Demuth DR, Lamont RJ. 2006. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect. Immun. 74:3834– 3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancholi V, Fischetti VA. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. U. S. A. 90: 8154– 8158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molenaar D, van der Rest ME, Petrović S. 1998. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) from Corynebacterium glutamicum. Eur. J. Biochem. 254: 395– 403 [DOI] [PubMed] [Google Scholar]
- 32.Diaz PI, Zilm PS, Wasinger V, Corthals GL, Rogers AH. 2004. Studies on NADH oxidase and alkylhydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol. Immunol. 19: 137– 143 [DOI] [PubMed] [Google Scholar]
- 33.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J. Gen. Microbiol. 139: 2343– 2351 [DOI] [PubMed] [Google Scholar]
- 34.Karlsson R, Falt A. 1997. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J. Immunol. Methods 200: 121– 133 [DOI] [PubMed] [Google Scholar]
- 35.McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185: 274– 284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deslauriers M, ní Eidhin D, Lamonde L, Mouton C. 1990. SDS-PAGE analysis of protein and lipopolysaccharide of extracellular vesicles and Sarkosyl-insoluble membranes from Bacteroides gingivalis. Oral Microbiol. Immunol. 5: 1– 7 [DOI] [PubMed] [Google Scholar]
- 37.Joe A, Murray CS, McBride BC. 1994. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect. Immun. 62: 1358– 1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura F, Murakami Y, Nishikawa K, Hasegawa Y, Kawaminami S. 2009. Surface components of Porphyromonas gingivalis. J. Periodont. Res. 44: 1– 12 [DOI] [PubMed] [Google Scholar]
- 39.Murakami Y, Imai M, Nakamura H, Yoshimura F. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110: 157– 162 [DOI] [PubMed] [Google Scholar]
- 40.Kadowaki T, Takii R, Yamatake K, Kawakubo T, Tsukuba T, Yamamoto K. 2007. A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis-infected cells. Front. Biosci. 12: 4800– 4809 [DOI] [PubMed] [Google Scholar]
- 41.Postle K, Kadner RJ. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49: 869– 882 [DOI] [PubMed] [Google Scholar]
- 42.Wiener MC. 2005. TonB-dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15: 394– 400 [DOI] [PubMed] [Google Scholar]
- 43.Hall LM, Fawell SC, Shi X, Faray-Kele MC, Aduse-Opoku J, Whiley RA, Curtis MA. 2005. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect. Immun. 73: 4253– 4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodont. Res. 34: 464– 472 [DOI] [PubMed] [Google Scholar]
- 45.Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. 2007. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56: 1536– 1548 [DOI] [PubMed] [Google Scholar]
- 46.Dolgilevich S, Rafferty B, Luchinskaya D, Kozarov E. 2011. Genomic comparison of invasive and rare non-invasive strains reveals Porphyromonas gingivalis genetic polymorphisms. J. Oral Microbiol. 3: 5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gore MG. 1981. L-glutamic acid dehydrogenase. Int. J. Biochem. 13:879– 886 [DOI] [PubMed] [Google Scholar]
- 48.Joannou CL, Brown PR. 1992. NAD-dependent glutamate dehydrogenase from Pseudomonas aeruginosa is a membrane-bound enzyme. FEMS Microbiol. Lett. 90: 205– 210 [DOI] [PubMed] [Google Scholar]
- 49.Okwumabua O, Persaud JS, Reddy PG. 2001. Cloning and characterization of the gene encoding the glutamate dehydrogenase of Streptococcus suis serotype 2. Clin. Diagn. Lab. Immunol. 8: 251– 257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi N, Sato T, Yamada T. 2000. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J. Bacteriol. 182: 4704– 4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckel W. 2001. Unusual enzymes involved in five pathways of glutamate fermentation. Appl. Microbiol. Biotechnol. 57: 263– 273 [DOI] [PubMed] [Google Scholar]
- 52.Scherf U, Buckel W. 1993. Purification and properties of an iron-sulfur and FAD-containing 4-hydroxybutyryl-CoA dehydratase/vinylacetyl-CoA delta 3-delta 2-isomerase from Clostridium aminobutyricum. Eur. J. Biochem. 215: 421– 429 [DOI] [PubMed] [Google Scholar]
- 53.Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. 2000. Fermentation of 4-aminobutyrate by Clostridium aminobutyricum: cloning of two genes involved in the formation and dehydration of 4-hydroxybutyryl-CoA. Arch. Microbiol. 174: 189– 199 [DOI] [PubMed] [Google Scholar]
- 54.Shin JH, Lee HW, Kim SM, Kim J. 2009. Proteomic analysis of Acinetobactor baumannii in biofilm and planktonic growth mode. J. Microbiol. 47: 728– 735 [DOI] [PubMed] [Google Scholar]
- 55.Trémoulet F, Duché O, Namane A, Martinie B, Labadie JC. 2002. A proteomic study of Escherichia coli O157:H7 NCTC 12900 cultivated in biofilm or in planktonic growth mode. FEMS Microbiol. Lett. 215: 7– 14 [DOI] [PubMed] [Google Scholar]
- 56.Tribble GD, Mao S, James CE, Lamont RJ. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. U. S. A. 103: 11027– 11032 [DOI] [PMC free article] [PubMed] [Google Scholar]


