Abstract
Influenza A virus (IAV) predisposes individuals to secondary infections with the bacterium Streptococcus pneumoniae (the pneumococcus). Infections may manifest as pneumonia, sepsis, meningitis, or otitis media (OM). It remains controversial as to whether secondary pneumococcal disease is due to the induction of an aberrant immune response or IAV-induced immunosuppression. Moreover, as the majority of studies have been performed in the context of pneumococcal pneumonia, it remains unclear how far these findings can be extrapolated to other pneumococcal disease phenotypes such as OM. Here, we used an infant mouse model, human middle ear epithelial cells, and a series of reverse-engineered influenza viruses to investigate how IAV promotes bacterial OM. Our data suggest that the influenza virus HA facilitates disease by inducing a proinflammatory response in the middle ear cavity in a replication-dependent manner. Importantly, our findings suggest that it is the inflammatory response to IAV infection that mediates pneumococcal replication. This study thus provides the first evidence that inflammation drives pneumococcal replication in the middle ear cavity, which may have important implications for the treatment of pneumococcal OM.
INTRODUCTION
Streptococcus pneumoniae is a Gram-positive bacterium that colonizes the nasopharynx of up to 80% of preschool children (1). Generally, S. pneumoniae colonization is transient and asymptomatic. However, in the presence of influenza A virus (IAV), S. pneumoniae is able to disseminate to other sites in the body and cause disease (2).
The genome of IAV consists of eight gene segments, two of which encode the surface glycoproteins the hemagglutinin (HA) and the neuraminidase (NA). The HA of influenza virus binds to terminal sialic acid residues on cell surface glycoproteins and glycolipids, facilitating internalization of the virus. In contrast, the NA of influenza virus cleaves terminal host sialic acid residues to facilitate the release of new virions. IAV strains are divided into subtypes based on the antigenically and structurally distinct HA and NA that they bear. H1N1 and H3N2 subtypes currently circulate in the community. Although there are limited data available, secondary bacterial disease appears to be more prevalent following outbreaks of H3N2 influenza viruses, compared to H1N1 virus outbreaks (3–7). Despite suggestions that this virus-subtype dependence of disease is associated with the increased activity of the viral NA in H3N2 strains (5), the mechanism by which IAV facilitates secondary pneumococcal disease largely remains unclear.
Recently, Shivshankar and colleagues (8) demonstrated that chronic inflammation in the lungs of the elderly facilitates pneumococcal pneumonia by upregulating host receptors which are then co-opted for pneumococcal adherence. These observations add further weight to previously noted associations of IAV-induced inflammation and secondary pneumococcal disease (9, 10). However, the role of inflammation remains controversial, as others have reported that inhibition of the immune response by IAV can also facilitate pneumococcal pneumonia (11). Moreover, it remains unclear as to to what extent these findings can be extrapolated to other pneumococcal disease phenotypes, such as otitis media (OM).
OM is the most common clinical consequence of pneumococcal-influenza virus coinfections in children and can lead to fatal meningitis and permanent hearing loss (12). Studies in the chinchilla suggested that the ability of IAV to perturb the function of neutrophils facilitated secondary pneumococcal OM, suggesting a role for virus-induced immunosuppression (rather than inflammation) in disease development (13). In contrast, secondary OM may be largely independent of IAV-induced immunosuppression and instead reflect the ability of the viral NA to cleave sialic acid and expose receptors for pneumococcal adherence (14). Herein, we have used our previously established infant mouse model of OM (designed to mimic the underdeveloped immune system of children) (15) and human middle ear epithelial cells (HMEECs) to provide the first evidence that it is the viral HA that dictates the likelihood of pneumococcal OM. This reflects the ability of the viral HA to affect the replication efficacy of IAV in the middle ear. Increased viral replication in the middle ear was associated with a proinflammatory response in this organ. We show that a proinflammatory response in the middle ear can then mediate secondary bacterial disease.
MATERIALS AND METHODS
Viral and bacterial strains.
The bioluminescent S. pneumoniae strain EF3030lux (type 19F) was used in all experiments (16). The preparation and storage of pneumococci prior to infection have been described elsewhere (17). The panel of different IAV strains used in this study is shown in Table 1. Select viruses were created using reverse genetics (RG), as described previously (18). Virus stocks were prepared in embryonated eggs, and titers of infectious virus were determined by three independent plaque assays on Madin-Darby canine kidney (MDCK) cells as described previously (17).
Table 1.
Viral strains used
| Virus name | Subtype | Description |
|---|---|---|
| Mt. Sinai lineage of A/PR/8/34 (PR8/34) | H1N1 | Mouse-adapted IAV strain grown in embryonated eggs |
| Cambridge lineage of A/PR/8/34 (Cambridge/34) | H1N1 | Alternative lineage of PR8/34 |
| A/Memphis/1/71 (Mem/71) | H3N2 | Human isolate grown in embryonated eggs |
| A/Udorn/307/72 (Udorn/72) | H3N2 | Human isolate grown in embryonated eggs |
| A/Port Chalmers/1/73 (Port Chalmers/73) | H3N2 | Human isolate grown in embryonated eggs |
| A/Brazil/11/78 (Brazil/78) | H1N1 | Human isolate grown in embryonated eggs |
| A/WSN/33 (WSN/33) | H1N1 | Mouse-adapted IAV strain grown in embryonated eggs |
| A/Netherlands/246/08 (NL/08) | H1N1 | Recent human isolate |
| A/Netherlands/364/06 (NL/06) | H1N1 | Recent human isolate |
| A/Netherlands/602/09 (NL/09) | H1N1 | Recent human isolate |
| A/Netherlands/348/07 (NL/07) | H3N2 | Recent human isolate |
| A/Netherlands/312/03 (NL/03) | H3N2 | Recent human isolate |
| HKx31 | H3N2 | Reassortant of PR8/34 and A/Aichi/2/68(H3N2) bearing the H3N2 surface antigens |
| PR8-HKx31 HA | H3N1 | RG virus with the HA gene of HKx31 and other genes from PR8/34 |
| PR8-HKx31 NA | H1N2 | RG virus with the NA gene of HKx31 and other genes from PR8/34 |
| Udorn-PR8 HA | H1N2 | RG virus with the HA gene of PR8/34 and other genes from Udorn/72 |
| Udorn-PR8 NA | H3N1 | RG virus with the NA gene of PR8/34 and other genes from Udorn/72 |
| RG PR8/34 | H1N1 | RG virus with all the genes of PR8/34 |
| RG Udorn/72 | H3N2 | RG virus with all the genes of Udorn/72 |
| PR8-Auck HA | H1N1 | RG virus with the HA gene of A/Auckland/1/09 (H1N1) and other genes from PR8/34 |
| PR8—Udorn HA | H3N1 | RG virus with the HA gene of Udorn/72 and other genes from PR8/34 |
| PR8—Udorn NA | H1N2 | RG virus with the NA gene of Udorn/72 and other genes from PR8/34 |
BPL inactivation of IAV.
Udorn/72 was grown in the allantoic fluid of embryonated chicken eggs as described previously (17). A final concentration of 0.1% β-propiolactone (BPL; Acros Organics, Morris Plains, NJ) in citrate buffer (125 mM sodium citrate, 150 mM sodium chloride, pH 8.2) was added to the virus. Virus was subsequently incubated overnight at 20°C under conditions of continuous slow shaking and then purified by dialysis (Slide-A-Lyzer Dialysis Cassettes; Thermo Scientific Inc., Bremen, Germany). The structural integrity of the HA postpurification was confirmed using a standard hemagglutination assay (19). Virus inactivation was confirmed by three passages in MDCK cells, where no virus could be detected by plaque assay following the third passage.
Infection of mice.
Animal experiments were approved by the Animal Ethics Committee of the University of Melbourne. C57BL/6 mice were bred and housed under specific pathogen-free (SPF) conditions at the Department of Microbiology and Immunology at The University of Melbourne, Australia. Five-day-old C57BL/6 mice were colonized intranasally (i.n.) with 2 × 103 CFU of S. pneumoniae EF3030Lux in 3 μl of phosphate-buffered saline (PBS). Alternatively, mice were mock treated with an equivalent volume of PBS. At 14 days of age, infant mice were infected i.n. with 20 PFU (strains PR8/34, Cambridge/34, and WSN/33) or 102.5 PFU (all other virus strains) of egg-grown IAV in 3 μl of PBS. Six days post-IAV infection, mice were euthanized and organs were collected for analysis. Where relevant, Salmonella lipopolysaccharide (LPS; Sigma) (0.05 mg) or PBS was administered to the middle ear of 14-day-old mice via a transtympanic injection, and middle ears were collected 2 days later for histology.
Experiments using the pressure cabin were performed with 14-day-old, SPF C57BL/6 mice (Harlan, The Netherlands) and were approved by the Animal Ethics Committee of the Radboud University. The pressure cabin was used essentially as described previously (20). Briefly, 10 μl BPL-inactivated Udorn/72 diluted in PBS was applied to both nostrils of anesthetized mice and the pressure was raised to 40 kPa. Four days after challenge with BPL-inactivated virus, mice were euthanized and middle ears were collected. The dose of BPL-inactivated Udorn/72 used in these experiments was the dilution at which the BPL-inactivated virus had an HA titer which was equivalent to the HA titer of 104 PFU of live Udorn/72.
Enumeration of bacterial and viral load and in vivo imaging.
Tissues used to quantify bacterial and viral load were collected and processed as described previously (15–17). All CFU and PFU data are expressed per organ. In vivo imaging was performed using an IVIS-Spectrum system (Caliper LifeSciences) as described previously (15, 17).
RNA extraction, cDNA synthesis, and qPCR.
RNA was extracted using TRIzol (Invitrogen), purified using an RNAeasy minielute kit (Qiagen), and DNA-se treated (Turbo DNase; Ambion). Each sample consisted of the two middle ear bullas taken from one mouse. cDNA for host expression analysis and viral mRNA quantification was synthesized using oligo(dT)18 (Roche) and a Transcriptor High Fidelity cDNA synthesis kit (Roche). Alternatively, cDNA for viral RNA (vRNA) quantification was synthesized using the Uni12 primer (21) and a Superscript II kit (Invitrogen). Quantitative PCR (qPCR) was performed using SYBR green (Quantance), a Stratagene Mx3005 qPCR Thermocycler (Agilent Technologies), and the relevant primers against either the relevant murine gene or the matrix gene of Udorn/72 (Table 2).
Table 2.
Primers used for qPCR
| Primer | Sequence (5′→3′) |
|---|---|
| GAPDH_F | AGGTCGGTGTGAACGGATTTG |
| GAPDH_R | TGTAGACCATGTAGTTGAGGTCA |
| pro IL-1β_F | CAACCAACAAGTGATATTCTCCATG |
| pro IL-1β_R | GATCCACACTCTCCAGCTGCA |
| SAA_F | AGCCTTCCATTGCCATCATTCTT |
| SAA_R | AGTATCTTTTAGGCAGGCCAGCA |
| TNF-α_F | CATCTTCTCAAAATTCGAGTGACAA |
| TNF-α_R | TGGGAGTAGACAAGGTACAACCC |
| CXCL2_F | AGTGAACTGCGCTGTCAATGC |
| CXCL2_R | AGGCAAACTTTTTGACCGCC |
| IL-1α_F | ACTCATTGGCGCTTGAGTCGGCA |
| IL-1α_R | TGTCACCCGGCTCTCCTTGAAGGT |
| Udorn/72_F | AAGACCAATCCTGTCACCTCTGA |
| Udorn/72_R | TCCTCGCTCACTGGGCA |
Microarray and analysis.
Six independent samples were used for each condition and analyzed using a NimbleGen platform (12 × 135K mouse gene expression arrays; Roche Nimblegen). Each sample consisted of the two middle ear bullas taken from one mouse. Array images were acquired with a NimbleGen MS200 scanner and processed with NimbleScan software using the RMA algorithm. Data were processed using ArrayStar (DNAStar). Differential expression tests were performed with a moderated t test implemented in ArrayStar, followed by false-discovery-rate (FDR) correction of the P values (Q-values) according to the method of Storey and Tibshirani (22). A gene was considered to be differentially expressed when an expression ratio of >4 or <0.025 relative to the control was obtained with a corresponding Q-value that was <0.05. Differentially expressed genes were mapped to Gene Ontology (GO) terms to enrich for gene class using the DAVID tool (23). DNA microarray data are available at Gene Expression Omnibus (GEO).
Histology.
Middle ears were collected and processed for histology essentially as described previously (15). Sections were scored by a pathologist blinded to the study design, based on the percentage of the middle ear cavity containing an inflammatory cell infiltrate, as previously published (15). Immunofluorescence was performed as described previously (15).
Infection of HMEECs.
HMEECs (24) were kindly donated by David Lim (House Ear Institute, Los Angeles, CA) and cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium (DMEM) (Gibco) and fully supplemented BEGM (Lonza, The Netherlands). For infection studies, confluent monolayers were washed with DMEM (Gibco) and infected with IAV at a multiplicity of infection (MOI) of 1 for 45 min at 37°C. Cells were then washed and incubated in a 1:1 mixture of DMEM (Gibco) and fully supplemented BEGM (Lonza). To evaluate the percentage of infected cells, cells were fixed with 80% (vol/vol) methanol and stained using fluorescein isothiocyanate (FITC)-conjugated anti-influenza A virus nucleoprotein (Abcam, United Kingdom). The percentage of infected cells was calculated by counting a minimum of 70 cells per well.
Cytokine production.
Levels of interleukin-6 (IL-6) were measured by an enzyme-linked immunosorbent assay (ELISA) (Sanquin, The Netherlands), while IL-8 was measured using a cytometric bead assay (CBA) Human Flex Set (BD Biosciences).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA).
RESULTS
The viral HA dictates pneumococcal replication in the middle ear.
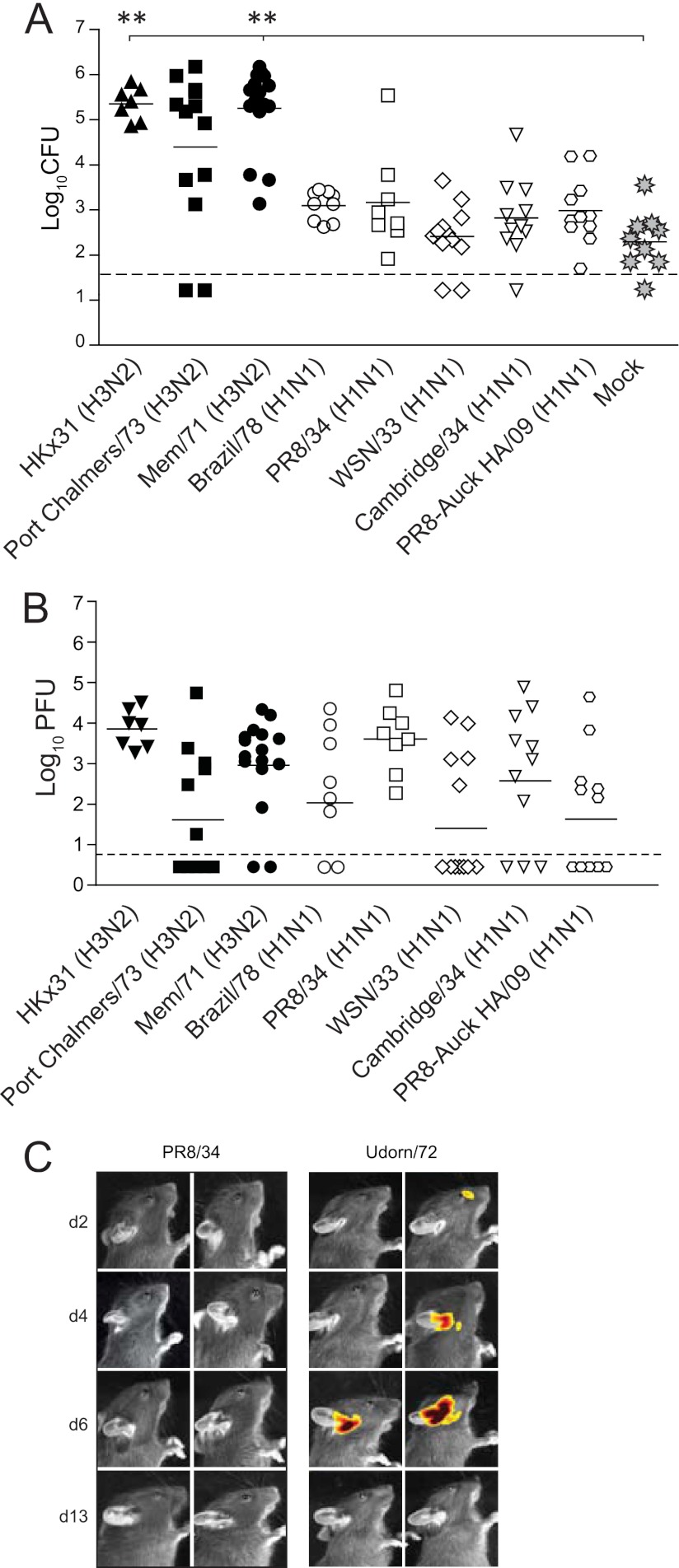
To understand the role of IAV in bacterial OM, we tested the ability of a panel of different IAV strains to induce bacterial OM in mice colonized with the bioluminescent S. pneumoniae strain EF3030lux. Bacterial OM was monitored by enumeration of bacterial load in the middle ear 6 days post-IAV infection. This time point was selected as day 6 is the peak of OM in infant mice (15). Infection with HKx31 (H3N2), Port Chalmers/73 (H3N2), and Mem/71(H3N2) resulted in approximately 105 bacteria in the middle ear (Fig. 1A). These findings are consistent with our previous observation of bacterial OM in mice coinfected with S. pneumoniae and Udorn/72 (H3N2), and we have demonstrated that this amount of bacteria in the ears is associated with increased middle ear inflammation and a 20-dB hearing loss (15). In contrast, infection with H1N1 strains resulted in lower middle ear bacterial titers (a mean of approximately 103 bacteria), which were equivalent to the titers seen with mock-infected mice (Fig. 1A). We have previously demonstrated that this level of pneumococci in the middle ear does not result in OM (as defined by middle ear inflammation and hearing loss) (15). The inability of the tested H1N1 strains to induce bacterial outgrowth in the middle ear was in spite of the fact that these strains replicated efficiently in the nasal cavity (Fig. 1B). These results were also not due to the lower infectious doses used for the more virulent H1N1 IAV strains (such as PR8/34), as mice infected with only 20 PFU of HKx31 still developed bacterial OM (data not shown). Likewise, the differences could not be explained by differences in the kinetics of disease development, as mice infected with PR8/34 showed no bacterial OM by in vivo imaging on day 2, day 4, or day 13 post-IAV infection (Fig. 1C). Thus, of the IAV strains tested, H3N2 strains induced a higher rate of bacterial OM in mice.
Fig 1.
H3N2 viruses facilitate pneumococcal otitis media. (A) Titers of S. pneumoniae EF3030lux in the middle ears of mice 6 days after i.n. infection with different IAV strains. Bacterial counts are represented as the averages of titers derived from the left and right ears of each mouse. Statistical significance was determined using a Mann-Whitney U test and Bonferroni posttest and was analyzed only relative to mock-infected mice (rather than comparing all the different permutations of the relative treatment groups). Statistical significance is denoted by two asterisks (P < 0.01). Data are pooled from a minimum of two independent experiments. The dashed line indicates the detection limit of the assay. (B) Replication of different IAV strains in the nasal cavity. Mice were coinfected with S. pneumoniae EF3030lux and IAV, and IAV titers were assessed 6 days post-viral infection. Statistical significance was determined using a one-way analysis of variance (ANOVA) and Bonferroni posttest. Data are pooled from a minimum of two independent experiments. The dashed line indicates the detection limit of the assay. (C) Representative in vivo images of S. pneumoniae EF3030lux in the middle ears of mice at various days (d) after intranasal (i.n.) infection with IAV. A luminescent signal indicates the presence of live, actively replicating pneumococci at that site. The detection limit of in vivo imaging in the middle ear is approximately 103 CFU (16).
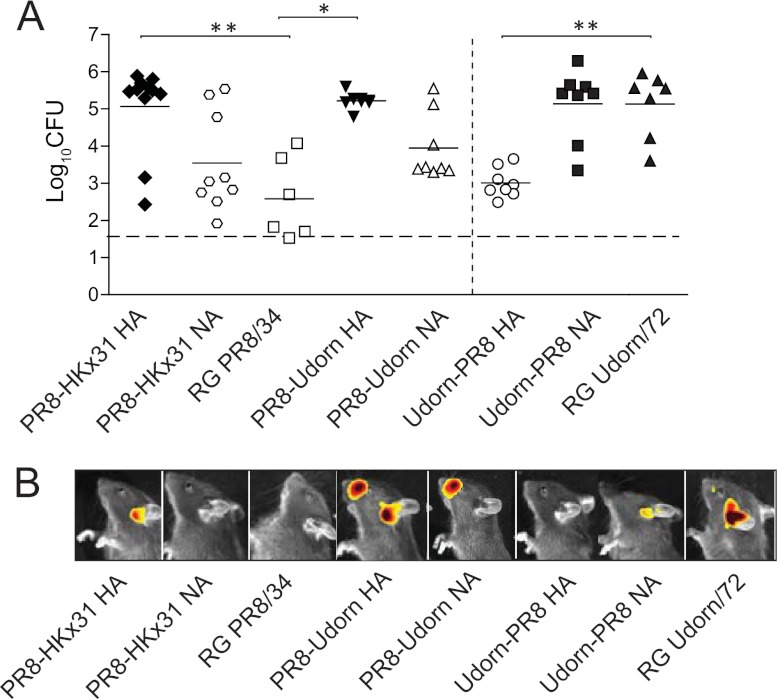
The HKx31 reassortant virus, associated with bacterial OM, has all the internal genes of PR8/34 (H1N1) but the HA and NA of A/Aichi/2/68 (H3N2). As PR8/34 did not promote OM, this implicates the NA and/or HA as the key viral determinant(s) of OM in this mouse model of disease. Reverse genetics (RG) was thus used to create two isogenic PR8/34 strains bearing either the HA or the NA from HKx31 (PR8-HKx31 HA or PR8-HKx31 NA, respectively). Mice infected with PR8-HKx31 NA showed no significant increase in pneumococcal titers (P > 0.05; Kruskal-Wallis test and Dunn's posttest; Fig. 2A) or luminescence in the middle ear (Fig. 2B) compared to mice infected with the reverse-engineered parental virus RG PR8/34. Conversely, mice infected with PR8-HKx31 HA had significantly higher pneumococcal titers (P < 0.05; Kruskal-Wallis test and Dunn's posttest; Fig. 2A) and luminescence (Fig. 2B) in the middle ear. These data suggest that it is the HA of A/Aichi/2/1968 that confers susceptibility to pneumococcal OM.
Fig 2.
The H3 HA facilitates pneumococcal otitis media. (A) Titers of S. pneumoniae EF3030lux in the middle ears of mice 6 days after i.n. infection with different IAV strains. Bacterial counts are represented as the averages of titers derived from the left and right ears of each mouse. Statistical significance was determined using a Kruskal-Wallis test and Dunn's posttest relative to the relevant reverse genetics (RG) parental virus. Statistical significance is denoted by two asterisks (P < 0.01) or one asterisk (P < 0.05). Data are pooled from a minimum of two independent experiments. The dashed line indicates the detection limit, while solid lines indicate the mean values of the data. H3 viruses are shown in black, while H1 viruses are shown in white. (B) Representative in vivo images of S. pneumoniae EF3030lux in the middle ears of mice 6 days after intranasal (i.n.) infection with IAV. The detection limit of in vivo imaging in the middle ear is approximately 103 CFU (16).
To confirm that the HA was responsible for promoting the OM observed in our previous study in Udorn/72 (H3N2)-infected mice (15), we used reverse genetics to generate two additional viruses, Udorn-PR8 HA and Udorn-PR8 NA, which bear the internal genes of Udorn/72 with the HA and NA, respectively, of PR8/34. The transfer of the PR8/34 HA to a Udorn/72 backbone was sufficient to significantly reduce pneumococcal titers compared to mice infected with RG Udorn/72 (P < 0.05; Kruskal-Wallis test and Dunn's posttest; Fig. 2A). Similarly, the transfer of the Udorn/72 HA to a PR8/34 backbone increased pneumococcal titers in the middle ear, while the transfer of the Udorn/72 NA did not (P > 0.05; Kruskal-Wallis test and Dunn's posttest; Fig. 2A). Taken together, these data show that, among these viruses, the viral HAs of Udorn/72 and HKx31 confer enhanced susceptibility to pneumococcal OM while that of PR8/34 does not.
The viral HA determines middle ear inflammation.
To gain further insight into the mechanism facilitating pneumococcal OM in vivo, we focused on the differences between infection with Udorn/72 and Udorn-PR8 HA. Transcriptional profiling was performed on the middle ears of mice 6 days postinfection with one of the two viruses. We identified 78 upregulated and 6 downregulated probe sets in Udorn/72-infected mice relative to Udorn-PR8 HA-infected mice (Fig. 3A). Gene expression analysis demonstrated that Udorn/72 induced a much stronger inflammatory response than Udorn-PR8 HA, with 16 probe sets assigned to the inflammatory response (GO:0006954, Fig. 3B). Of note, there were significantly higher levels of pro-IL-1β, IL-1α, and CXCL2 in the middle ears of Udorn/72-infected mice (see Table S1 and S2 in the supplemental material). The expression of key proinflammatory genes was subsequently confirmed by qPCR (Fig. 3C). To verify that these findings were not restricted to a single time point, qPCR was used to assess the expression of key proinflammatory cytokines 3 days post-IAV infection. Similar to day 6 results, pro-IL-1β, CXCL2, serum amyloid A (SAA), and IL-1α were upregulated ≥2-fold in Udorn/72-infected mice (Fig. 3C). The ability of a broader panel of virus strains to induce middle ear inflammation was then examined histologically. Figure 3D clearly shows differential abilities of H3 and H1 IAV strains to induce middle ear inflammation (for the statistical analysis of these data, see Table S3 in the supplemental material). Virus strains that caused middle ear inflammation in the absence of a S. pneumoniae infection (i.e., H3 strains) were those that facilitated bacterial outgrowth in the middle ear in a coinfection. This virus-induced inflammation was characterized by an influx of inflammatory cells (the overwhelming majority of which were neutrophils) into the middle ear cavity (results not shown). Viruses that did not cause middle ear inflammation in a single infection (i.e., H1 strains) were also those that did not facilitate bacterial outgrowth in the middle ear in a coinfection (Fig. 3D). Although these results show a clear correlation between viral inflammation and pneumococcal replication, viral suppression of the immune response or virus-induced damage to the epithelium may also contribute to enhanced pneumococcal replication. Thus, we investigated the pneumococcal response in the context of nonviral inflammation. Instead of a viral infection, LPS or PBS was administered directly into the middle ear cavity of mice colonized with S. pneumoniae. Transtympanic injection of LPS, and not PBS, induced middle ear inflammation, which was characterized by submucosal edema and a neutrophilic infiltrate in the lumen of the middle ear cavity (Fig. 3E) akin to what is observed following IAV infection. LPS, and not PBS, also induced pneumococcal outgrowth in the middle ear cavity (Fig. 3E). These findings demonstrate that the simple induction of middle ear inflammation is sufficient to induce bacterial OM and suggest that the ability of the IAV to induce middle ear inflammation may facilitate secondary bacterial OM.
Fig 3.
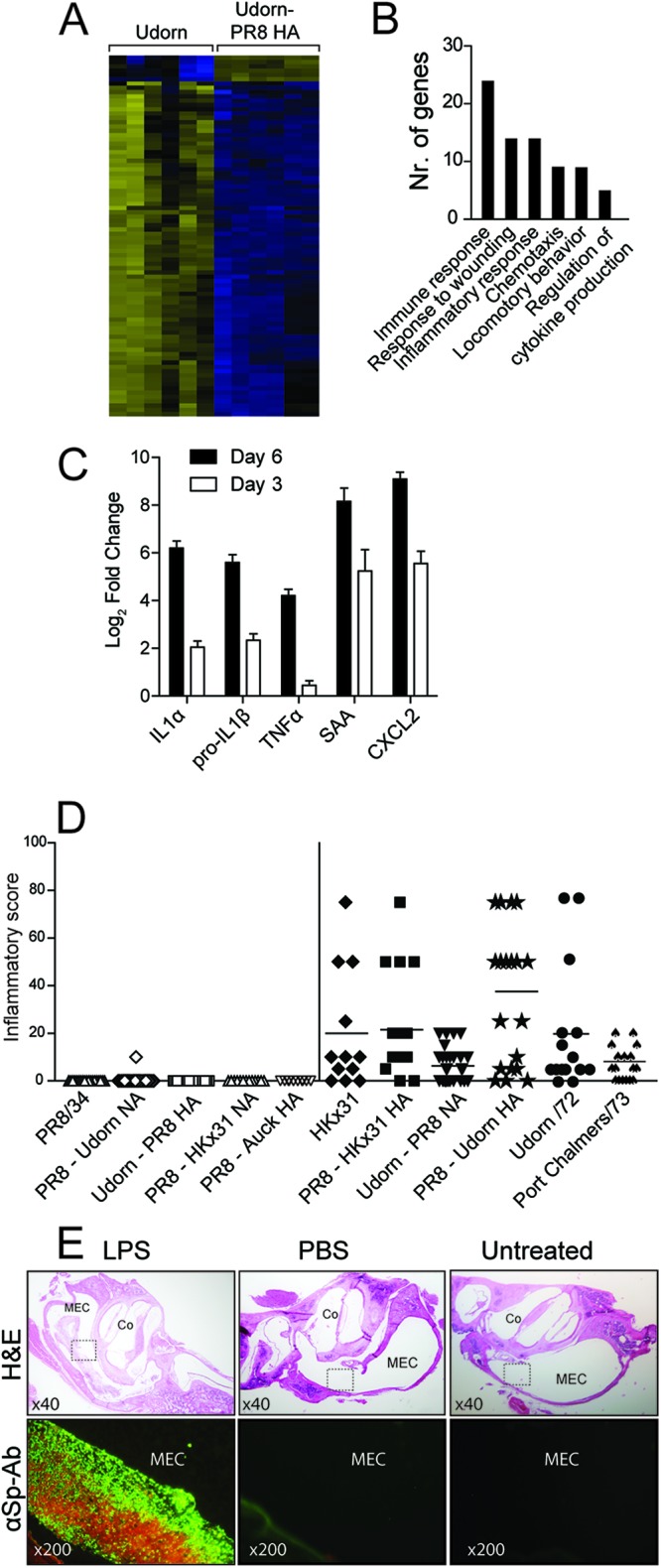
Inflammation facilitates bacterial OM. (A) Heat map demonstrating differential gene expression in the middle ears of mice 6 days postinfection with Udorn/72 or Udorn PR8-HA. Upregulated genes are represented in yellow, downregulated genes are shown in blue, and black represents genes that were not differentially expressed. (B) Functional classification of genes upregulated in the middle ear of Udorn/72-infected mice relative to that of Udorn-PR8 HA-infected mice 6 days postinfection. (C) Fold increase in the expression of selected genes in the middle ear of Udorn/72-infected mice relative to that of Udorn-PR8 HA-infected mice 6 days postinfection by qPCR. Means ± standard errors of the means (SEM) are shown, where each bar represents a minimum of four data points. Data are normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. SAA: serum amyloid A. (D) Middle ear inflammation observed by histology 6 days after infection with IAV. Inflammatory score data represent the percentage of the middle ear cavity with an inflammatory cell infiltrate, as previously published (15). Sections were scored by a pathologist blinded to the experimental design, and each data point represents a single ear from an infected mouse. Data are pooled from a minimum of two independent experiments. Data points in black indicate H3-bearing viruses, while data points in white indicate H1-bearing viruses. (E) Immunofluorescence using a FITC-labeled anti-S. pneumoniae antibody (αSp-Ab) on middle ear sections from mice colonized with S. pneumoniae and challenged transtympanically with LPS/PBS or left untreated. Hematoxolin and eosin (H&E) staining of the relevant sections is shown (upper panels). Boxes indicate the regions examined by immunofluorescence in the lower panels. Relevant image magnifications are shown. MEC, middle ear cavity; Co, cochlea.
IL-1β is detected in the middle ear during clinical cases of OM (25), and in mice, IL-1β in the middle ear triggers otitis media with effusion (25). Given that pro-IL-1β was upregulated in the middle ear following Udorn/72 (H3N2) infection, we reasoned that IL-1β contributed to the observed outgrowth of bacteria in the middle ear. However, B6.ICE−/− mice (which are unable to produce the caspase-1 necessary for the production of active IL-1β) still displayed pneumococcal OM and middle ear inflammation (data not shown). Similarly, coinfected B6.IL1R−/− mice (which are unable to respond to IL-1β or IL-1α) and B6.MyD88−/− mice (which are defective in their immune response to selective bacterial/viral ligands) still developed high bacterial titers in the middle ear (data not shown). Thus, although inflammation facilitates pneumococcal outgrowth in the middle ear, there is a large degree of redundancy in the inflammatory cascade in the middle ear.
Middle ear inflammation is associated with viral replication in the middle ear.
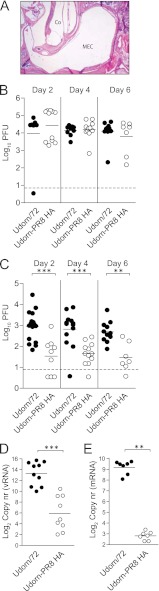
We then sought to assess if virus-induced middle ear inflammation was dependent on active viral replication. Udorn/72 was thus inactivated by β-propiolactone (BPL) treatment, and a pressure cabin system (20) was used to deliver the virus from the nasal cavity to the middle ear of 14-day-old mice. Preliminary experiments showed that a pressure increase of 40 kPa delivered an intranasally administered inoculum to the middle ear (see Fig. S1 in the supplemental material), consistent with previous studies (20). The administration of inactivated Udorn/72 to the middle ear did not induce inflammation (Fig. 4A). Thus, we assessed the replication of live Udorn/72 and Udorn-PR8 HA over time. There was no significant difference between the levels of replication of the two viruses in the nasal cavity at any time point (P > 0.05; Mann-Whitney U test; Fig. 4B). However, in the middle ear Udorn/72 replicated to significantly higher titers than Udorn-PR8 HA at all time points (P < 0.01; Mann-Whitney U test; Fig. 4C). Consistent with these data, we detected significantly more viral particles and viral mRNA in the middle ears of Udorn/72-infected mice than in mice infected with Udorn-PR8 HA (P < 0.01; Mann-Whitney U test; Fig. 4D and E). To confirm these data, we then investigated viral replication in the middle ear across a broader panel of IAV strains. HKx31, PR8-Udorn HA, and Port Chalmers/73 (which induced middle ear inflammation and pneumococcal outgrowth) replicated efficiently in the middle ear, while PR8/34 did not (see Fig. S2 in the supplemental material). Thus, these data would suggest that IAV-induced middle ear inflammation in mice is associated with increased viral replication in this organ.
Fig 4.
Induction of middle ear inflammation by IAV reflects increased viral replication. (A) Representative image of middle ear sections stained with H&E (magnification, ×40). Samples were taken 4 days after administration of BPL-inactivated Udorn/72 to the middle ear. MEC, middle ear cavity; Co, cochlea. (B) Replication of Udorn/72 and Udorn-PR8 HA in the nasal cavity of mice various days post-IAV infection. (C) Replication of Udorn/72 and Udorn-PR8 HA strains in the middle ears of mice various days post-IAV infection. Viral titers are represented as the averages of titers derived from the left and right ears of each mouse. (D) Genomic viral RNA (vRNA) detected by qPCR in the middle ear 6 days post-IAV infection. Copy number (nr) is expressed per 3.5 μg of RNA. (E) Viral mRNA detected by qPCR in the middle ear 6 days post-IAV infection. Copy number is expressed per 5 μg of RNA. Statistical significance was determined using a Mann-Whitney U test and is denoted by three asterisks (P < 0.001) or two asterisks (P < 0.01). Data are pooled from a minimum of two independent experiments, and a dashed line indicates the detection limit of the assay.
The HA determines infection and inflammation in HMEECs.
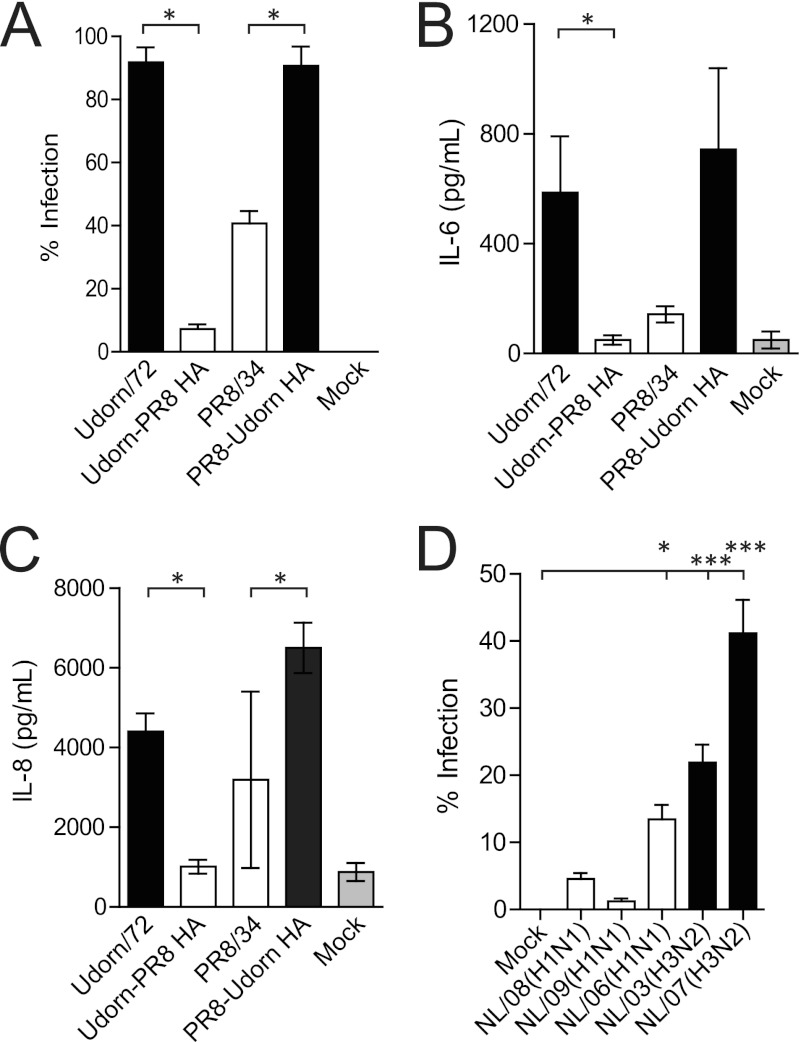
To assess the relevance of these findings to human cells in vitro, HMEECs were infected with Udorn/72, Udorn-PR8 HA, PR8/34, and PR8-Udorn HA. The percentage of viral infection and the production of IL-6 and IL-8 were then measured. These cytokines were selected to reflect those upregulated in the murine middle ear following IAV infection (IL-6) and those important in the subsequent recruitment of neutrophils into the middle ear (IL-8). Udorn/72 and PR8-Udorn HA infected a significantly higher percentage of HMEECs than Udorn-PR8 HA and PR8/34, respectively (P < 0.05; Mann-Whitney U test; Fig. 5A), and this resulted in the production of significantly higher levels of IL-6 and IL-8 (P < 0.05; Mann-Whitney U test; Fig. 5B and C). We then used a broader panel of human IAV strains to investigate whether or not more recent H3N2 strains still displayed an increased infection rate of HMEECs relative to H1N1 strains. While these more recent virus strains were less efficient at infecting HMEECs (compared to, for example, Udorn/72), a similar trend was observed: H3N2 strains had a higher rate of infection than H1N1 strains (Fig. 5D).
Fig 5.
The HA mediates infection and inflammation in HMEECs. (A) Percentages of HMEECs infected by different IAV strains 8 h postinfection at an MOI of 1. (B) IL-6 levels in the cell supernatant of HMEECs 20 h postinfection with IAV (MOI of 1) or post-mock infection. (C) IL-8 levels in the cell supernatant of HMEECs 20 h postinfection with IAV (MOI of 1) or post-infection. Statistical significance was determined using a Mann-Whitney U test, and significance is denoted by one asterisk (P < 0.05). Data are pooled from three independent experiments, and data represent the means ± SEM. (D) Percentages of HMEECs infected by recently circulating clinical IAV strains 8 h postinfection at an MOI of 1. Statistical significance was determined using a one-way ANOVA and Dunnett's posttest and was analyzed relative to mock-treated cells. Data are pooled from three independent experiments, and data represent the means ± SEM.
DISCUSSION
OM affects more than 80% of children under the age of 3 years. Although typically a self-limiting disease, OM can lead to meningitis, hearing loss, and learning difficulties. OM frequently arises following coinfection with S. pneumoniae and IAV. However, the role of IAV in the pathogenesis of pneumococcal-influenza virus OM remains unclear.
Here, we provide the first evidence that the viral HA plays a role in the development of secondary pneumococcal OM. Specifically, we noted an increased incidence of pneumococcal OM in mice following infection with H3N2 (rather than H1N1) IAV strains. This strain-dependent difference in disease development is congruent with previous studies in ferrets (26) as well as previous studies on secondary pneumococcal pneumonia (4, 27). Such studies have suggested that these differences reflect the differential NA activities of these viruses (4). Although it was speculated that the viral NA may also be important in the development of pneumococcal OM (14, 26), our data would suggest that the viral factors important in secondary bacterial disease are specific for each disease phenotype. Interestingly, our data contrast those of McCullers and colleagues (28), who showed that mice coinfected with PR8/34 and S. pneumoniae develop bacterial OM. However, the differences in the animal models used between studies may preclude direct comparisons (28).
The viral HA was found to be important in controlling the development of secondary pneumococcal OM in mice, as it helped dictate the replication efficiency of IAV in the middle ear. Our data suggested that, of the virus strains tested (Udorn/72, PR8/34, HKx31, and PC/73 and recombinants thereof), those that were able to replicate efficiently in the middle ear were able to induce an inflammatory response in this organ and that it was this inflammatory response that was important for pneumococcal outgrowth. The role of inflammation in pneumococcal colonization and pneumonia has previously been established (8–10, 29). However, this report provides, to the best of our knowledge, the first evidence that middle ear inflammation may mediate pneumococcal OM. Previous studies have suggested that inflammation facilitates bacterial disease by upregulating pneumococcal receptors on epithelial cells (10). However, in this study, as well as previously (15), we have demonstrated that pneumococci in the middle ear localize to the lumen, rather than to the middle ear epithelium. This would suggest that increased pneumococcal-epithelial cell adherence is unlikely to be relevant to bacterial OM. Previous studies in the chinchilla suggest that pneumococci persist in the middle ear by forming biofilms consisting of bacterial components and host components derived from the influx of neutrophils (30). Given that in the present study (i) neutrophils were the predominant cellular infiltrate in the middle ear and (ii) pneumococci colocalized with the neutrophils in the middle ear, a similar mechanism of disease may be occurring in our murine model. This is supported by our observation that LPS injected into the middle ear (instead of IAV) also induced both a neutrophillic infiltrate and pneumococcal outgrowth. Our attempts to use antibody treatment to deplete neutrophils in the middle ear have thus far been unsuccessful. However, a clearer understanding of the role of neutrophils in OM remains a key goal of future studies. In this respect, it is important to note that we have used naïve mice without immunological memory against IAV and S. pneumoniae. This may partially explain why recruited neutrophils are unable to clear the bacteria.
In the present study, we used a panel of different IAV strains of which some (such as PR8/34) were mouse adapted while others (such as Udorn/72) were largely unaltered from the available sequences of relevant human strain. However, the limitation of using the murine model of disease is that post-1975 H3N2 strains are unable to replicate efficiently in mice without extensive adaptation (due to the presence of additional glycosylation sites on the head of the HA) (31); thus, their ability to induce pneumococcal OM cannot be tested. Moreover, studies from a murine model of disease cannot necessarily be extrapolated to human infections. To determine the relevance of our findings to human cells in vitro, and across a broader panel of IAV strains, we assessed viral infection in HMEECs. While we have found that these cells produce limited infectious virus particles (data not shown), these cells are a well-established model for examining the interactions between IAV and human middle ear cells (32, 33). Consistent with what was observed in our in vivo murine model, Udorn/72 and PR8-Udorn HA mediated increased viral replication and inflammation relative to Udorn-PR8 HA and PR8/34. Infection of HMEECs with a broader panel of more recent human IAV strains supported these findings. That is, there was a trend toward increased infection of HMEECs following infection with an H3N2, rather than H1N1, IAV strain. These data are consistent with observations that in human populations, bacterial OM is associated more with H3N2 infections than with H1N1 infections (7). However, the difference in the infection rates of HMEECs observed in this study between NL/06 (H1N1) and NL/03(H3N2) was not substantial. Thus, while H3N2 strains may have an increased tendency to replicate in the middle ear and cause bacterial OM, the difference between H3N2 and H1N1 strains in this regard is not unequivocal. Our data emphasize the need for future clinical studies into the association between infection with an H1 or H3 IAV strain and the development of pneumococcal OM in children. Should our mouse and in vitro models accurately mimic disease development in the human population, our data would suggest that that limiting viral replication and inflammation in the middle ear may be key in preventing secondary pneumococcal OM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kelly Rogers for help with the in vivo imaging and David Lim for his kind donation of the HMEECs. We also thank the Molecular Virology Section at the University Medical Centre Groningen for their assistance in the preparation of the BPL-inactivated virus, Saskia van Selm and Fred van Opzeeland for assistance with the pressure cabin experiments, Thomas Gebhardt for technical assistance, Elles Simonetti for assistance with the CBA, and Richard Strugnell and Deborah Middleton for helping guide the study. We also thank Ron Fouchier for his kind donation of virus strains and Andrew Giraud for supply of IL1R−/− mice.
K.R.S. is supported by a GlaxoSmithKline postgraduate support grant and the Elizabeth and Vernon Puzey postgraduate research scholarship. D.A.D. is supported by the 7th Framework Programme of the European Commission (ETB grant 08010). O.L.W. is supported by a Career Development Fellowship (RD Wright Fellowships) from the Australian National Health and Medical Research Council. The Melbourne World Health Organization Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing. Part of this work was supported by a University of Melbourne Overseas Research Scholarship, a Boehringer-Ingelheim Fonds Travel grant, and an EMBO Short-Term Research fellowship.
Footnotes
Published ahead of print 14 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01278-12.
REFERENCES
- 1.Adegbola R, Obaro S, Biney E, Greenwood B. 2001. Evaluation of Binax NOW Streptococcus pneumoniae urinary antigen test in children in a community with a high carriage rate of pneumococcus. Pediatr. Infect. Dis. J. 20:718–719 [DOI] [PubMed] [Google Scholar]
- 2.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301 [DOI] [PubMed] [Google Scholar]
- 3.Coates D, Sweet C, Smith H. 1986. Severity of fever in influenza: differential pyrogenicity in ferrets exhibited by H1N1 and H3N2 strains of differing virulence. J. Gen. Virol. 67:419–425 [DOI] [PubMed] [Google Scholar]
- 4.Peltola VT, McCullers JA. 2004. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr. Infect. Dis. J. 23:S87–S97 [DOI] [PubMed] [Google Scholar]
- 5.Peltola VT, Murti KG, McCullers JA. 2005. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 192:249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Haber M, Ray S, Farley M, Panozzo C, Klugman K. 2012. Invasive pneumococcal pneumonia and respiratory virus co-infections. Emerg. Infect. Dis. 18:294–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright PF, Thompson J, Karzon DT. 1980. Differing virulence of H1N1 and H3N2 influenza strains. Am. J. Epidemiol. 112:814–819 [DOI] [PubMed] [Google Scholar]
- 8.Shivshankar P, Boyd A, Le Saux C, Yeh I, Orihuela C. 2011. Cellular senescence increases expression of bacterial ligands in the lungs and is positively correlated with increased susceptibility to pneumococcal pneumonia. Aging Cell 10:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCullers JA, Rehg JE. 2002. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186:341–350 [DOI] [PubMed] [Google Scholar]
- 11.McNamee LA, Harmsen AG. 2006. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infect. Immun. 74:6707–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cripps A, Otczyk D, Kyd J. 2005. Bacterial otitis media: a vaccine preventable disease? Vaccine 23:2304–2310 [DOI] [PubMed] [Google Scholar]
- 13.Abramson JS, Giebink GS, Quie PG. 1982. Influenza A virus-induced polymorphonuclear leukocyte dysfunction in the pathogenesis of experimental pneumococcal otitis media. Infect. Immun. 36:289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong HH, Grants I, Liu X, DeMaria TF. 2002. Comparison of alteration of cell surface carbohydrates of the chinchilla tubotympanum and colonial opacity phenotype of Streptococcus pneumoniae during experimental pneumococcal otitis media with or without an antecedent influenza A virus infection. Infect. Immun. 70:4292–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Short KR, Diavatopoulos DA, Thorton R, Pedersen J, Strugnell RA, Wise AK, Reading PC, Wijburg OL. 2011. Influenza virus induces bacterial and non-bacterial otitis media. J. Infect. Dis. 204:1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diavatopoulos D, Short K, Price J, Wilksch J, Brown L, Briles D, Strugnell R, Wijburg O. 2010. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 24:1789–1798 [DOI] [PubMed] [Google Scholar]
- 17.Short KR, Diavatopoulos DA, Reading PC, Brown LE, Rogers KL, Strugnell RA, Wijburg OLC. 2011. Using bioluminescent imaging to investigate synergism between Streptococcus pneumoniae and influenza A virus in infant mice. J. Vis Exp. 2011:e2357 doi:10.3791/2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Sobrido L, García-Sastre A. 2010. Generation of recombinant influenza virus from plasmid DNA. J. Vis. Exp. 2011:e2057 doi:10.3791/2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stol K, van Selm S, van den Berg S, Bootsma HJ, Blokx WAM, Graamans K, Tonnaer ELGM, Hermans PWM. 2009. Development of a non-invasive murine infection model for acute otitis media. Microbiology 155(Pt 12):4135–4144 [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann E, Stech J, Guan Y, Webster R, Perez D. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3 doi:10.1186/gb-2003-4-5-p3. [PubMed] [Google Scholar]
- 24.Chun YM, Moon SK, Lee HY, Webster P, Brackmann DE, Rhim JS, Lim DJ. 2002. Immortalization of normal adult human middle ear epithelial cells using a retrovirus containing the E6/E7 genes of human papillomavirus type 16. Ann. Otol. Rhinol. Laryngol. 111:507–517 [DOI] [PubMed] [Google Scholar]
- 25.Smirnova MG, Kiselev SL, Gnuchev NV, Birchall JP, Pearson JP. 2002. Role of the pro-inflammatory cytokines tumor necrosis factor-alpha, interleukin-1beta, interleukin-6 and interleukin-8 in the pathogenesis of the otitis media with effusion. Eur. Cytokine Netw. 13:161–172 [PubMed] [Google Scholar]
- 26.Peltola VT, Boyd KL, McAuley JL, Rehg JE, McCullers JA. 2006. Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect. Immun. 74:2562–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullers JA, Bartmess KC. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187:1000–1009 [DOI] [PubMed] [Google Scholar]
- 28.McCullers JA, Karlström Å, Iverson AR, Loeffler JM, Fischetti VA. 2007. Novel strategy to prevent otitis media caused by colonizing Streptococcus pneumoniae. PLoS Pathog. 3:e28 doi:10.1371/journal.ppat.0030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura S, Davis KM, Weiser JN. 2011. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121:3657–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid SD, Hong W, Dew KE, Winn DR, Pang B, Watt J, Glover DT, Hollingshead SK, Swords WE. 2009. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J. Infect. Dis. 199:786–794 [DOI] [PubMed] [Google Scholar]
- 31.Reading PC, Morey LS, Crouch EC, Anders EM. 1997. Collectin-mediated antiviral host defense of the lung: evidence from influenza virus infection of mice. J. Virol. 71:8204–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong HH, Long JP, Li D, DeMaria TF. 2004. Alteration of gene expression in human middle ear epithelial cells induced by influenza A virus and its implication for the pathogenesis of otitis media. Microb. Pathog. 37:193–204 [DOI] [PubMed] [Google Scholar]
- 33.Tong H, Long J, Shannon P, DeMaria T. 2003. Expression of cytokine and chemokine genes by human middle ear epithelial cells induced by influenza A virus and Streptococcus pneumoniae opacity variants. Infect. Immun. 71:4289–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.