Abstract
Bacteria causing infections in hospitalized patients are increasingly antibiotic resistant. Classical infection control practices are only partially effective at preventing spread of antibiotic-resistant bacteria within hospitals. Because the density of intestinal colonization by the highly antibiotic-resistant bacterium vancomycin-resistant Enterococcus (VRE) can exceed 109 organisms per gram of feces, even optimally implemented hygiene protocols often fail. Decreasing the density of intestinal colonization, therefore, represents an important approach to limit VRE transmission. We demonstrate that reintroduction of a diverse intestinal microbiota to densely VRE-colonized mice eliminates VRE from the intestinal tract. While oxygen-tolerant members of the microbiota are ineffective at eliminating VRE, administration of obligate anaerobic commensal bacteria to mice results in a billionfold reduction in the density of intestinal VRE colonization. 16S rRNA gene sequence analysis of intestinal bacterial populations isolated from mice that cleared VRE following microbiota reconstitution revealed that recolonization with a microbiota that contains Barnesiella correlates with VRE elimination. Characterization of the fecal microbiota of patients undergoing allogeneic hematopoietic stem cell transplantation demonstrated that intestinal colonization with Barnesiella confers resistance to intestinal domination and bloodstream infection with VRE. Our studies indicate that obligate anaerobic bacteria belonging to the Barnesiella genus enable clearance of intestinal VRE colonization and may provide novel approaches to prevent the spread of highly antibiotic-resistant bacteria.
INTRODUCTION
The emergence and spread of highly antibiotic-resistant bacteria represent a major clinical challenge (1). In recent years, the numbers of infections caused by organisms such as methicillin-resistant Staphylococcus aureus, carbapenemase-resistant Enterobacteriaceae, vancomycin-resistant Enterococcus (VRE), and Clostridium difficile have increased markedly, and many of these strains are acquiring resistance to the few remaining active antibiotics. Most infections produced by highly antibiotic-resistant bacteria are acquired during hospitalizations, and preventing patient-to-patient transmission of these pathogens is one of the major challenges confronting hospitals and clinics. Most highly antibiotic-resistant bacterial strains belong to genera that colonize mucosal surfaces, usually at low densities. The highly complex microbiota that normally colonizes mucosal surfaces inhibits expansion of and domination by bacteria such as Enterobacteriaceae and Enterococcaceae. Destruction of the normal flora by antibiotic administration, however, disinhibits antibiotic-resistant members of these bacterial families, leading to their expansion to very high densities (2). High-density colonization by these organisms can be calamitous for the susceptible patient, resulting in bacteremia and sepsis (3). An additional problem associated with high-density colonization, however, is the difficulty of preventing transmission. One microgram of fecal material, representing a volume of one-thousandth of a microliter, can contain over 1,000 viable VRE bacteria. Given the density with which resistant bacteria colonize patients, it is not surprising that hand washing and changing of gloves and gowns between patients do not completely prevent the transfer of resistant microbes.
Although mechanisms remain to be defined, the recognition that commensal bacteria can prevent infection by pathogens is far from new. Early studies by Freter, Bohnhoff, and their colleagues roughly 50 years ago demonstrated that antibiotic administration enhanced infection with Vibrio cholerae or Salmonella enterica serovar Typhimurium by eliminating obligate anaerobic bacteria (4, 5). Subsequent studies by Thijm et al. demonstrated that antibiotic administration resulted in the expansion of oxygen-tolerant bacteria, such as enterococci and Enterobacteriaceae (6), and led to the concept of colonization resistance, i.e., the ability of commensal bacteria to prevent colonization by exogenous bacteria or marked expansion of low-frequency commensal bacteria. The association between antibiotic administration and the development of VRE infections was supported by epidemiologic studies demonstrating that administration of antibiotics that kill obligate anaerobic bacteria increases the density of VRE colonization in hospitalized patients (7).
Recent advances in DNA sequencing have enabled more-comprehensive analyses of the commensal microbiota in health and disease. Studies in mice have demonstrated that while untreated mice rapidly eliminate VRE from the gut, antibiotic-mediated disruption of the microbiota enables VRE to expand dramatically in the ileum, cecum, and colon, achieving a state of dominance in which as much as 99% of bacteria are VRE (2). Once established, VRE persists for months afterwards. Studies in patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT) also demonstrated intestinal domination by VRE, an outcome that was associated with the administration of metronidazole, an antibiotic with potent activity against obligate anaerobic bacteria (3). Although it was evident from these studies that elimination of some members of the commensal flora promoted expansion and domination by VRE, it remained unclear which bacterial taxa restrict VRE colonization and whether the reintroduction of normal preantibiotic flora can eliminate VRE from the gut.
To address these issues, we used a deep-sequencing platform to identify bacterial populations that correlate with the clearance of VRE. We find that elimination of VRE from the gut of mice correlates with intestinal recolonization with bacteria belonging to the Barnesiella genus. Analysis of the fecal microbiota of allo-HSCT patients revealed that patients colonized with Barnesiella are protected from VRE domination.
MATERIALS AND METHODS
Mouse models, housing conditions, and VRE infection.
Experiments were done with 7-week-old C57BL/6J female mice purchased from Jackson Laboratory, housed with irradiated food, and provided with acidified water. Mice were individually housed to avoid contamination between mice due to coprophagia. When knockout mice were infected with VRE, each knockout mouse was cohoused with a wild-type mouse for at least 1 month before infection to minimize differences between their intestinal microbiotas. To generate mice defective in Toll-like receptor (TLR) signaling, Myd88−/− mice were bred in-house with Triflps2/lps2 mice. Triflps2/lps2 mice were provided by B. Beutler (The Scripps Research Institute, University of California, San Diego, CA), and MyD88−/− mice were obtained from S. Akira (University of Osaka, Osaka, Japan). Rag1−/− mice, which cannot develop T cells or B cells, were obtained from Jackson Laboratory. Rip2−/− mice, which are defective in NOD-like receptor signaling, were provided by R. Flavell (Yale University, New Haven, CT). All mice were backcrossed at least nine times onto the C57BL/6 background. For experimental infections with VRE, mice were treated with ampicillin (0.5 g/liter) in their drinking water, which was changed every 3 days. After 1 week of treatment, mice were infected by means of oral gavage with 108 CFU of the vancomycin-resistant Enterococcus faecium strain purchased from ATCC (stock number 700221). One day after infection, antibiotic treatment was stopped and VRE levels were determined at different time points by plating serial dilutions of fecal pellets on Enterococcosel agar plates (Difco) with vancomycin (8 μg/ml; Sigma). VRE colonies were identified by appearance and confirmed by Gram staining. In addition, as previously described (2), PCR of the vanA gene, which confers resistance to vancomycin, confirmed the presence of VRE in infected mice. In the experiment shown in Fig. 5, mice were infected 2 weeks after antibiotic treatment was stopped. All animal studies were performed in compliance with Memorial Sloan-Kettering institutional guidelines and approved by the institution's institutional animal care and use committee (IACUC).
Fecal transplantation and bacterial culture administration.
Fecal pellets from untreated mice were resuspended in phosphate-buffered saline (PBS) (1 fecal pellet/1 ml of PBS). For each experiment, several fecal pellets from different untreated mice were resuspended together in PBS. A total of 200 μl of the resuspended pool fecal material was given by oral gavage to VRE-infected mice over 3 consecutive days, starting 1 day after antibiotic treatment was stopped. For bacterial cultures, a 100-fold dilution of resuspended fecal material was grown on CDC anaerobe blood agar plates (BD) under anaerobic conditions or aerobic conditions at 37°C for 3 days. Bacterial colonies were resuspended in PBS. A total of 108 CFU from the bacterial cultures were administered, by oral gavage, to VRE-infected mice over 3 consecutive days, beginning 1 day after antibiotic treatment was stopped.
Sample collection and DNA extraction.
Fresh stool pellets were obtained before mice were euthanized. The samples were immediately frozen and stored at −80°C. DNA was extracted using a phenol-chloroform extraction technique with mechanical disruption (bead beating) as previously described (2).
16S rRNA gene amplification and 454 pyrosequencing.
For each sample, 3 replicate 25-μl PCRs were performed, with each containing 50 ng of purified DNA, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1.5 mM MgCl2, 1.25 U of Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.2 μM concentrations of the following primers, designed to amplify the V1-V3 region as previously described (8): a modified primer 8F (5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAGAGAGTTTGATCCTGGCTCAG-3′), composed of 454 Lib-L primer B (underlined) and the universal bacterial primer 8F (italics), and the modified primer 534R (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNNNATTACCGCGGCTGCTGG-3′), composed of 454 Lib-L primer A (underlined), a unique 6- or 7-base barcode (Ns), and the broad-range bacterial primer 534R (italics). Cycling conditions were 94°C for 3 min and 23 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min. Replicate PCRs were pooled, and amplicons were purified using the QIAquick PCR purification kit (Qiagen). PCR products were sequenced on a 454 GS FLX Titanium platform by following the 454 Roche recommended procedures. Sequences from allo-HSCT patients were obtained from a previously published study (3).
Sequence analysis.
Sequence data were compiled and processed using mothur (9). Sequences were converted to standard FASTA format. Sequences shorter than 300 bp, containing undetermined bases or homopolymer stretches longer than 8 bp, with no exact match to the forward primer or a barcode, or that did not align with the appropriate 16S rRNA variable region were not included in the analysis. Using the 454 base quality scores, which range from 0 to 40 (0 being an ambiguous base), sequences were trimmed using a sliding-window technique such that the minimum average quality score over a window of 50 bases never dropped below 30. Sequences were trimmed from the 3′ end until this criterion was met. Sequences were aligned to the 16S rRNA gene using as the template the SILVA reference alignment (10) and the Needleman-Wunsch algorithm with the default scoring options. Potentially chimeric sequences were removed using the ChimeraSlayer program (11). To minimize the effect of pyrosequencing errors in overestimating microbial diversity (12), rare-abundance sequences that differ in 1 or 2 nucleotides from a high-abundance sequence were merged to the high-abundance sequence using the pre.cluster option in mothur. Sequences were grouped into operational taxonomic units (OTUs) using the average-neighbor algorithm. Sequences with distance-based similarity of 97% or greater were assigned to the same OTU. OTU-based microbial diversity was estimated by calculating the Shannon diversity index (13). Phylogenetic classification was performed for each sequence using the Bayesian classifier algorithm described by Wang and colleagues with the bootstrap cutoff 60% (14). In most cases, classification was able to be assigned to the genus level. Hierarchical clustering shown in Fig. 1 was performed using the option hclust of the statistical computing program R, version 2.14.0, with the default parameters.
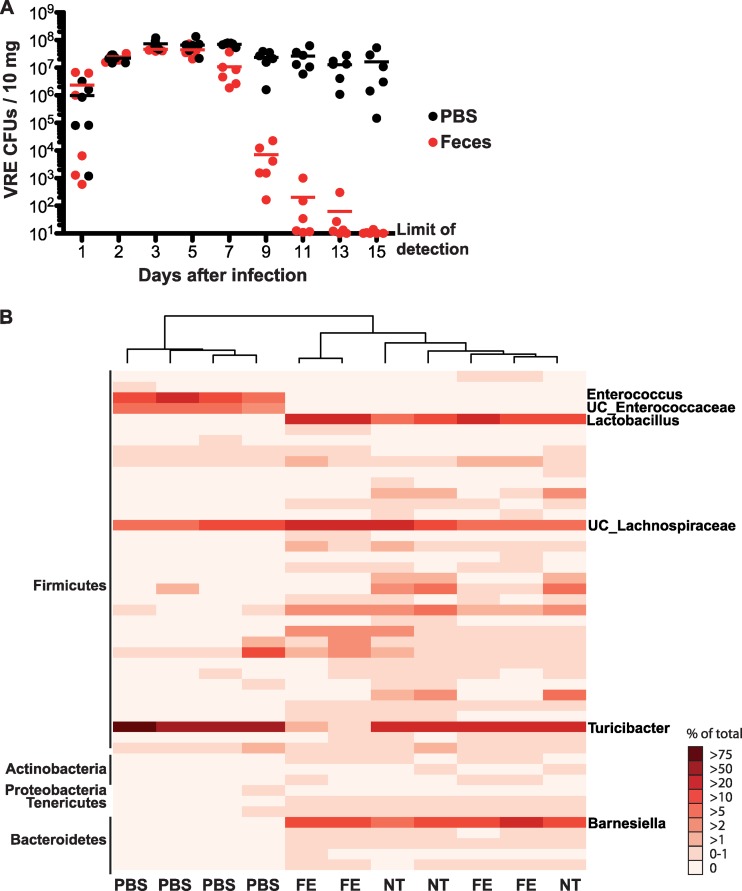
Fig 1.
Fecal transplantation clears VRE colonization in antibiotic-treated mice. Mice were infected with 108 VRE CFU after 1 week of ampicillin treatment. One day after infection, ampicillin treatment was stopped. Mice received PBS or a suspension of fecal pellets from untreated mice by oral gavage for three consecutive days, starting 1 day after antibiotic cessation. (A) Numbers of VRE CFU in the fecal pellets of infected mice were quantified every other day for two consecutive weeks (n = 6). Black dots represent PBS-treated mice and red dots represent fecal transplant-treated mice. (B) Composition of the microbiotas of four PBS and four fecal transplant (FE) mice was analyzed 15 days following infection and compared with that of the microbiotas of three untreated mice (NT). Hierarchical clustering was used to cluster samples by their microbiota composition at the genus level. Each column represents one mouse. Each row represents one genus. The most predominant phyla (left) and genera (right) are indicated.
Statistical analysis.
An unpaired Student t test was used within the GraphPad Prism program to determine if VRE levels were statistically significantly different (P < 0.05) between groups of VRE-infected mice.
In order to determine the correlation between VRE colonization levels or Barnesiella levels and bacterial taxon recovery, the Spearman correlation test was applied using the statistical R package command cor.test. The Spearman correlation test calculates a coefficient value ranging from −1 to +1, with positive values indicating positive correlation between bacterial taxa and VRE levels, negative values indicating an inverse correlation, and 0 indicating no association. P values were computed using the asymptotic t approximation within the statistical R package command cor.test in order to test if a detected association was statistically significant.
In order to determine if the Barnesiella genus is differentially abundant in patients that develop VRE domination (VRE represents ≥30% of their fecal microbiota) than in patients that do not, we made use of a set of 16S rRNA sequences from a previous study in which the fecal microbiota composition of allo-HSCT patients was determined during the transplant course (3). In order to calculate the relative abundance of Barnesiella in each stool sample, sequences were classified using mothur as described above. For each sample, sequences that were classified as the genus Barnesiella were divided by the total number of sequences in that specific sample. The Kruskal-Wallis test was applied to analyze if the relative abundance of Barnesiella was significantly lower in all samples from VRE-dominated patients prior to domination (67 samples from 34 patients) than in all samples from patients that never developed VRE domination (251 samples from 55 patients). In a second step, we performed 10,000 iterations of first randomly selecting 34 negative patients and then comparing their samples to the predomination samples of all 34 VRE-dominated patients, again using the Kruskal-Wallis test on Barnesiella abundance. The mean and standard error over the corresponding P values were calculated (see Results).
RESULTS
Fecal transplantation eliminates VRE from the intestine.
Previous studies from our laboratory demonstrated that untreated mice rapidly and completely eliminate orally administered VRE from the intestine while mice that have been treated with ampicillin become dominated by VRE (2). Once dominated, mice continue to harbor large numbers of VRE in the colon, as determined by quantitative cultures (Fig. 1A). To determine whether administration of normal intestinal flora to dominated mice can eliminate VRE, we obtained feces from untreated mice and mixed them with PBS under anaerobic conditions for administration to VRE-dominated mice by oral gavage. Quantitative culturing of “transplanted” mice demonstrated that VRE colonization was reduced to undetectable levels within 15 days, with reduction in the density of VRE within 7 days of fecal transfer (Fig. 1A).
To determine the breadth of microbial reconstitution, we amplified bacterial 16S rRNA genes from fecal pellets isolated from untreated mice and mice that had been treated with ampicillin and colonized with VRE and that either received PBS or were reconstituted with normal fecal microbiota. Hierarchical clustering of mice on the basis of microbiota composition demonstrated that untreated and reconstituted mice were similar while VRE-dominated mice were distinct (Fig. 1B).
Obligate anaerobic bacteria clear VRE.
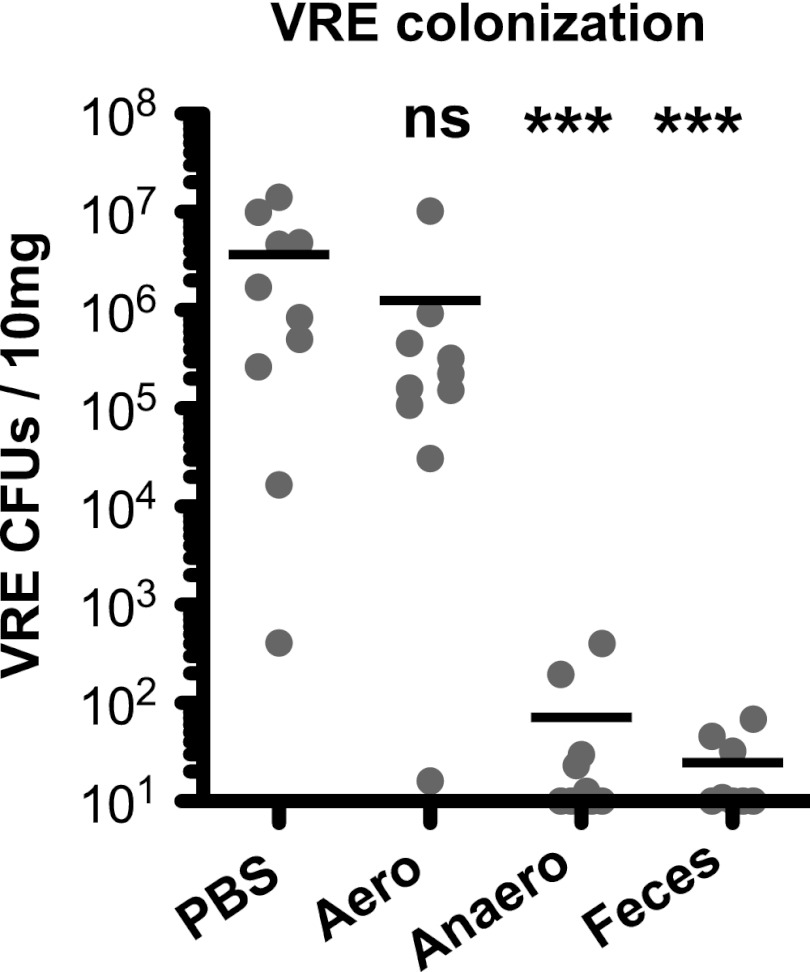
The large intestine microbiota consists of many different bacterial taxa that can be grouped into obligate anaerobes, i.e., oxygen-intolerant bacteria, and oxygen-tolerant bacteria. To determine whether VRE elimination depends on reconstitution with oxygen-tolerant or -intolerant bacteria, we cultured murine feces under strictly anaerobic conditions or under normal atmospheric conditions and reconstituted mice with bacteria harvested from these cultures. While aerobically cultured fecal pellets did not eliminate VRE, anaerobically cultured bacteria were as effective as unfractionated feces at reducing the density of VRE colonization (Fig. 2).
Fig 2.
Commensal anaerobic bacteria suppress VRE colonization in antibiotic-treated mice. Mice were infected with 108 VRE CFU after 1 week of ampicillin treatment. One day after infection, ampicillin treatment was stopped. Mice were orally gavaged for three consecutive days, starting 1 day after antibiotic cessation, with PBS, a suspension of fecal pellets from untreated mice (feces), or an aerobic (aero) or anaerobic (anaero) culture of fecal microbiota from untreated mice. Numbers of VRE CFU in the fecal pellets of infected mice were analyzed 5 weeks after infection (n = 8 to 10). Limit of detection, 10 CFU/10 mg. ***, significantly different (P < 0.001) from the PBS group; ns, not significant.
The mechanism by which anaerobic bacteria prevent the expansion of oxygen-tolerant bacteria, such as Enterococcus, in the gut is unclear. A possible mechanism that limits VRE colonization of the gut is stimulation of innate immune effector mechanisms by commensal bacteria and their products. For example, intestinal lipopolysaccharide (LPS) or systemic flagellin can stimulate TLRs and induce epithelial cell expression of Reg3γ, a bactericidal C-type lectin that kills VRE (15, 16). To determine the potential role of TLRs, Nod1 or Nod2, or T cell- or antibody-mediated mechanisms in VRE elimination by fecal transfer, we used MyD88/Trif, Rip2, and Rag1 knockout mice for colonization by VRE, followed by reconstitution with normal fecal flora (see Fig. S1 in the supplemental material). Fecal reconstitution resulted in equivalent reductions of VRE colonization in wild-type and knockout mice, indicating that adaptive immune mechanisms and TLR or Nod1/2 signaling are not required for VRE elimination by normal colonic microbes.
Microbiota reconstitution and VRE clearance vary after fecal transplantation.
Although reconstitution with fecal flora markedly reduced VRE density in the colon, the magnitude of reduction varied between experiments and also between mice within the same experiment. We reasoned that variability in VRE elimination reflected differences in reconstitution with distinct bacterial taxa following transfer and that comparing the microbiota of mice that eliminated VRE with that of mice that only partially cleared VRE might enable us to correlate the presence of specific anaerobic bacteria with VRE clearance. Figure 3 shows the relative abundance of different bacterial taxa and VRE density in colon contents isolated from experimental mice described in Fig. 2 (Fig. 3A and B). These results demonstrate that reconstitution of mice varies and that the magnitude of VRE elimination also varies within specific groups by a factor of up to 1,000 (Fig. 3B, PBS group).
Fig 3.
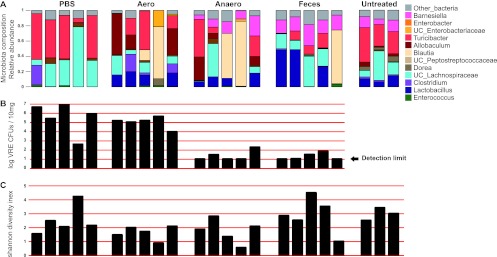
Microbiota reconstitution and VRE clearance vary after fecal transplantation. Mice were infected with 108 VRE CFUs after a week of ampicillin treatment. One day after infection, ampicillin treatment was stopped. Mice were orally gavaged for three consecutive days, starting 1 day after antibiotic cessation, with PBS, a suspension of fecal pellets from untreated mice (feces), or an aerobic or anaerobic culture of fecal microbiota from untreated mice. (A) Genus level composition. (B) VRE CFU numbers. (C) Shannon diversity index of murine fecal samples obtained 5 weeks after infection. Each column represents one mouse. For the microbiota composition, the most abundant bacterial taxa are indicated with different colors.
Other studies have suggested that the recovery of microbial diversity, lost during antibiotic administration, may be important for elimination of antibiotic-resistant pathogens (17). Analysis of the overall microbial diversity using the Shannon diversity index showed that some mice that were transplanted with the anaerobic culture of the fecal microbiota did not recover their overall microbial diversity but were able to suppress VRE colonization (Fig. 3C). This result suggests that the recovery of key members of the microbiota, rather than the recovery of a complex microbiota, is important for VRE elimination.
Reconstitution with Barnesiella correlates with VRE clearance.
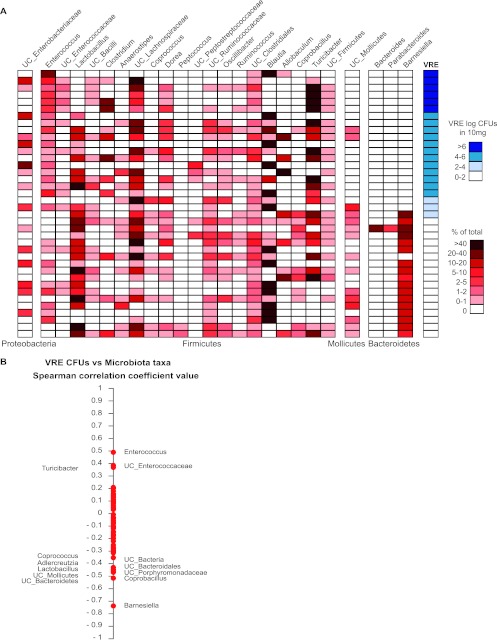
In order to identify key members of the microbiota that suppress VRE colonization and to facilitate the analysis of these complex data, we stratified experimental mice according to VRE density in the colon and plotted the densities of different bacterial taxa on a heat map (Fig. 4A). While reconstitution of mice with bacterial taxa varied from mouse to mouse irrespective of VRE density, clearance of VRE was markedly enhanced in mice recolonized with bacteria belonging to the Barnesiella genus (Fig. 4A). The Spearman rank correlation test demonstrates that recolonization with Barnesiella negatively correlates with VRE colonization (Fig. 4B) (P = 1.32−7). These results suggest that the Barnesiella genus contributes to the clearance of VRE colonization. It is possible, however, that additional bacterial genera corecover with Barnesiella and contribute to VRE elimination. To identify genera that positively correlate with recovery of Barnesiella, we used the Spearman correlation test (see Fig. S2 in the supplemental material) to compare the abundance of Barnesiella with the relative abundance of other bacterial genera. The Barnesiella genus belongs to the family Porphyromonadaceae, within the phylum Bacteroidetes. While unclassified sequences belonging to the Porphyromonadaceae family correlated positively with the prevalence of Barnesiella, we also found a positive correlation with the genera Coprobacillus and Adlercreutzia. To determine if these two genera contribute to VRE clearance, we analyzed their prevalence and the presence of Barnesiella in mice with reduced prevalence of VRE (<100 VRE CFU/10 mg feces). While 16 out of 17 mice with low VRE levels were colonized with Barnesiella, only 11 out of 17 mice were colonized with Coprobacillus or Adlercreutzia, demonstrating a greater correlation between Barnesiella colonization and suppression of VRE colonization.
Fig 4.
Colonization with the Barnesiella genus correlates with VRE elimination. (A) Genus level composition of the fecal microbiota (red heat map) and VRE CFU levels (blue heat map) for the different mice described in Fig. 2 5 weeks after VRE infection. Only the microbial taxa representing at least 1% of the microbiota are shown. Each heat map row represents one mouse, and mice are sorted by VRE colonization levels in descending order. (B) Spearman correlation analysis of the different bacterial genera present in the fecal microbiota of mice with the VRE CFU levels 5 weeks after infection. Spearman correlation coefficient values range from +1 (maximum positive correlation value) to −1 (maximum inverse correlation value). Each point represents one genus. To analyze the statistical significance of a given coefficient Spearman value, P values were computed using the asymptotic t approximation. Spearman coefficient values with a P value of <0.05 are indicated.
To determine whether Barnesiella can prevent intestinal colonization with VRE, mice that had been treated with ampicillin were reconstituted with commensal bacterial cultures prior to VRE infection. As shown in Fig. 5, mice that were gavaged with PBS or the aerobically cultured fecal microbiota became densely VRE colonized. However, most mice that received anaerobically cultured fecal microbiota became resistant to VRE colonization. One mouse that received anaerobically cultured microbiota lacked Barnesiella and was densely colonized with VRE.
Fig 5.
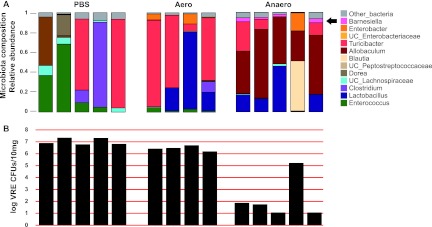
Commensal anaerobic bacteria prevent VRE intestinal colonization. Mice were treated with ampicillin and, after antibiotic treatment was stopped, were reconstituted with either PBS or an aerobic or anaerobic culture of the fecal microbiota. (A) Two weeks after reconstitution, the fecal microbiota composition of the mice was determined and mice were infected orally with 108 VRE CFU. (B) The level of VRE colonization was determined 1 week after infection. Each column represents one mouse.
The Barnesiella genus is associated with protection against VRE domination in transplant patients.
Patients undergoing allo-HSCT have a high incidence of intestinal domination and associated bacteremia with VRE (3). In general, patients have a diverse microbiota prior to allo-HSCT and then develop intestinal domination with different oxygen-tolerant bacterial species, with VRE the most prevalent and persistent. We prospectively collected fecal samples before transplantation and throughout hospitalization from 94 patients undergoing allo-HSCT and determined the microbiota composition. Thus, when patients developed VRE domination (VRE relative abundance, ≥30% of the microbiota), we were able to characterize the microbiota that preceded the development of VRE domination (Fig. 6A). This analysis demonstrated that most patients had a diverse flora prior to VRE domination (Fig. 6B), similar to that of patients who did not develop VRE domination (3). The majority of allo-HSCT patients do not develop VRE domination, a finding which may be explained by decreased exposure to VRE and/or antibiotics. We postulated that Barnesiella may play a role in protecting patients from VRE domination. Figure 6C shows the relative abundance of Barnesiella in fecal samples from patients who did not develop VRE domination compared to that in samples from patients who went on to develop VRE domination. Samples from patients who did not develop VRE domination contained higher levels of Barnesiella (average, 2.5 × 10−3 parts per unit) than samples from patients who progressed to VRE domination (average, 4.5 × 10−5 parts per unit). Results obtained using the Kruskal-Wallis test indicate that the difference observed in Barnesiella abundance between the groups of samples was statistically significant (P < 0.02). In order to ensure that the difference between these two sample sets is not attributable to a small number of outliers, we performed 10,000 iterations of randomly selecting 34 negative patients and compared their samples to the predomination samples of the 34 dominated patients. This step confirmed the results (Kruskal-Wallis P < 0.04; standard error [SE], 0.0004). These results suggest that patients harboring Barnesiella in their colonic microbiota are protected from VRE domination while the absence of this genus renders patients more vulnerable to VRE domination.
Fig 6.
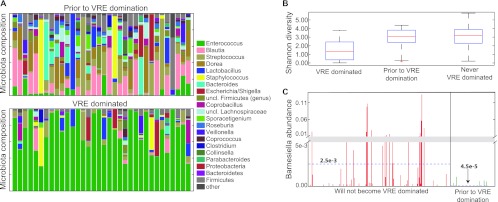
Barnesiella diminishes the risk of VRE domination in allo-HSCT patients. (A) Composition of the intestinal microbiota of allo-HSCT patients before and during VRE domination. (B) Shannon diversity index of samples from patients during VRE domination and prior to VRE domination and from patients that never developed VRE domination. (C) Relative abundance (parts per unit; total microbiota = 1) of the Barnesiella genus in samples from patients that did not develop VRE domination compared to samples from patients that developed VRE domination, taken before domination occurred. The y axis scale is divided into two sections (from 0 to 0.005 and from 0.01 to 0.11 parts per unit). Blue dashed lines indicate the means in each group of samples.
DISCUSSION
Although clinical studies indicate that antibiotics that kill obligate anaerobic bacteria result in dense colonization of the intestine with VRE (3, 7), it has remained unclear whether a specific subset of anaerobic bacteria inhibits VRE colonization. Our studies in mice and humans demonstrate that a microbiota containing bacteria that belong to the Barnesiella genus restricts colonization of the intestinal tract by VRE. Further studies will be required to determine which species and strains within the Barnesiella genus suppress VRE, the density of Barnesiella spp. required for VRE suppression, and whether Barnesiella is sufficient or whether other bacterial taxa are required to eliminate VRE.
The majority of intestinal obligate anaerobes belong to the Firmicutes and Bacteroidetes phyla. Our analyses of murine microbiotas of reconstituted mice did not identify taxa belonging to the Firmicutes phylum that restrict VRE colonization. On the other hand, our studies demonstrate that when species of Barnesiella, a genus within the Porphyromonadaceae family of the Bacteroidetes phylum, are part of the intestinal microbiota, the density of VRE decreases until it is ultimately cleared. In addition to the family Porphyromonadaceae, the Bacteroidetes phylum includes the Bacteroidaceae and Prevotellaceae families. Bacteria belonging to the Bacteroidaceae family include Bacteroides fragilis, which produces polysaccharides that stimulate innate and adaptive immune development in the gut (18). Recent studies have implicated bacteria belonging to the Prevotellaceae family with bowel inflammation in mice treated with dextran sulfate sodium (DSS) and with the development of steatohepatitis in mice treated with a methionine-choline-deficient diet (19, 20). In contrast, the Porphyromonadaceae family, including the genus Barnesiella, has not been associated with immune development or inflammatory diseases in the intestine. Consistent with our findings, studies with antibiotic-treated mice correlated colonization with Porphyromonadaceae with resistance to infection by Salmonella enterica serovar Typhimurium and Citrobacter rodentium; however, in those studies, the protective bacterial genera were not identified (21, 22). Factors influencing the density of Porphyromonadaceae in the colon include dietary fat intake (23), exposure to stress (24), and possibly major histocompatibility complex (MHC) haplotype (25).
Previous studies demonstrated that intestinal colonization by VRE can be suppressed by MyD88-dependent and microbiota-mediated induction of RegIIIγ expression by epithelial cells (15). In this study, we find that elimination of VRE from the intestine by fecal transplantation does not depend on TLRs, NOD receptors, B cells, or T cells. Thus, commensal flora-mediated inhibition of dense intestinal VRE colonization can be indirect, by innate immune induction, or direct. The mechanisms of direct VRE inhibition by the anaerobic flora remain undefined.
Oxygen-intolerant bacteria have limited options on the earth's surface, and the metazoan colon represents an essential, and for many anaerobic species, sole, sanctuary. Despite their density in the colon and their proximity to the bloodstream, obligate anaerobes rarely cause human disease and their survival depends on their host's survival. Obligate anaerobes of the gut promote their host's survival by limiting the expansion of oxygen-tolerant bacteria, which, by and large, are the subset containing most of the intestinal pathogens. Our results suggest that Barnesiella spp., by restricting the growth of VRE, regulate the composition of the microbiota and optimize host survival. Although some studies demonstrate that obligate anaerobes of the gut can trigger and/or promote inflammatory diseases, the ability of other anaerobes to restrict the growth of intestinal pathogens suggests that these bacteria may be exploited to limit colonization with highly antibiotic-resistant bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (RO1-AI42135, RO1-AI95706, and PO1-CA23766 to E.G.P.), from the Tow Foundation (to E.G.P.), from the Spanish MICINN (SAF2011-29458 to C.U.), and from the Marie-Curie Career Integration Grant (PCIG09-GA-2011-293894 to C.U.) and by a fellowship from the Cancer Research Institute to C.U.
Footnotes
Published ahead of print 14 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01197-12.
REFERENCES
- 1. Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Comparative Sequencing Program NISC. Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4: 148ra116 doi:10.1126/scitranslmed.3004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, Van Den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M- A, Jenq RR, van den Brink MRM, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55(7): 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freter R. 1955. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J. Infect. Dis. 97: 57–65 [DOI] [PubMed] [Google Scholar]
- 5. Bohnhoff M, Miller C. 1964. Resistance of the mouse's intestinal tract to experimental Salmonella infection. J. Exp. Med. 120: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thijm HA, van der Waaij D. 1979. The effect of three frequently applied antibiotics on the colonization resistance of the digestive tract of mice. J. Hyg. (Lond.) 82: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N. Engl. J. Med. 343: 1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80: 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, Desantis TZ, Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12: 1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Magurran AE. 2004. Measuring biological diversity. Afr. J. Aquat. Sci. 29: 285–286 [Google Scholar]
- 14. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455: 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. 2010. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 201: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197: 435–438 [DOI] [PubMed] [Google Scholar]
- 18. Mazmanian S, Liu C, Tzianabos A, Kasper D. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118 [DOI] [PubMed] [Google Scholar]
- 19. Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez J-P, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482: 179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferreira RBR, Gill N, Willing BP, Antunes LCM, Russell SL, Croxen MA, Finlay BB. 2011. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6: e20338 doi:10.1371/journal.pone.0020338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu T, Hougen H, Vollmer AC, Hiebert SM. 2012. Gut bacteria profiles of Mus musculus at the phylum and family levels are influenced by saturation of dietary fatty acids. Anaerobe 18: 331–337 [DOI] [PubMed] [Google Scholar]
- 24. Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. 2011. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav. Immun. 25: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, White BA, Taneja V. 2012. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One 7: e36095 doi:10.1371/journal.pone.0036095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.