Abstract
Mycoplasma pneumoniae, the causative agent of atypical pneumonia, is one of the bacteria with the smallest genomes that are nonetheless capable of independent life. Because of their longstanding close association with their human host, the bacteria have undergone reductive evolution and lost most biosynthetic abilities. Therefore, they depend on nutrients provided by the host that have to be taken up by the cell. Indeed, M. pneumoniae has a large set of hitherto unexplored transporters and lipoproteins that may be implicated in transport processes. Together, these proteins account for about 17% of the protein complement of M. pneumoniae. In the natural habitat of M. pneumoniae, human lung epithelial surfaces, phospholipids are the major available carbon source. Thus, the uptake and utilization of glycerol and glycerophosphodiesters that are generated by the activity of lipases are important for the nutrition of M. pneumoniae in its common habitat. In this study, we have investigated the roles of several potential transport proteins and lipoproteins in the utilization of glycerol and glycerophosphodiesters. On the basis of experiments with the corresponding mutant strains, our results demonstrate that the newly identified GlpU transport protein (MPN421) is responsible for the uptake of the glycerophosphodiester glycerophosphocholine, which is then intracellularly cleaved to glycerol-3-phosphate and choline. In addition, the proteins MPN076 and MPN077 are accessory factors in glycerophosphocholine uptake. Moreover, the lipoproteins MPN133 and MPN284 are essential for the uptake of glycerol. Our data suggest that they may act as binding proteins for glycerol and deliver glycerol molecules to the glycerol facilitator GlpF.
INTRODUCTION
Mycoplasma pneumoniae belongs to a group of bacteria, the Mollicutes, that is characterized by the lack of a cell wall, a strongly reduced genome, and very limited metabolic capabilities. This genome reduction is the result of an adaptive evolution that reflects the close association of the different members of the class with their eukaryotic hosts. M. pneumoniae is a human pathogen that causes atypical pneumonia, but sometimes extrapulmonary complications such as myocarditis, encephalitis, polyradiculopathy, and otitis are observed, especially in children and older people (1–5).
M. pneumoniae thrives on lung epithelia, and the major source of carbon and energy in this habitat are phospholipids (6). Thus, these compounds are likely to be the main nutrients for M. pneumoniae in its natural environment. However, while M. pneumoniae and other mollicutes are able to utilize several carbohydrates, they are unable to use the fatty acid component of phospholipids because of the lack of the β-oxidation pathway and the citric acid cycle (7). So far, M. pneumoniae has been shown to use glucose, fructose, and glycerol as carbon sources. While glucose and fructose are transported by specific phosphotransferase systems, glycerol is thought to be taken up by the essential glycerol facilitator GlpF (8, 9). After phosphorylation, glucose and fructose are metabolized via glycolysis. Glycerol is first phosphorylated and oxidized to dihydroxyacetone phosphate, which can then also enter glycolysis. The enzyme that catalyzes the oxidation of glycerol 3-phosphate is an interesting feature of M. pneumoniae: While the glycerol 3-phosphate dehydrogenase in most organisms uses NAD as the electron acceptor, M. pneumoniae and related species have a glycerol 3-phosphate oxidase that produces hydrogen peroxide (9). In addition to the ADP-ribosylating and vacuolating CARDS toxin (10), hydrogen peroxide acts as the major virulence factor of M. pneumoniae. Thus, there is a direct link between glycerol metabolism and virulence.
Phospholipids are composed of glycerol 3-phosphate to which two long-chain fatty acids and a polar moiety are bound. In humans, lecithin, a choline-containing phospholipid, is most abundant. The degradation of phospholipids is initiated by the deacylation catalyzed by phospholipases. M. pneumoniae does not possess any phospholipases, but it is assumed that these enzymes are present in the pulmonary surfactant, where they produce free glycerophosphodiesters, mainly glycerophosphocholine (GPC) (11). This compound can be used by M. pneumoniae and many other bacteria; however, the details of its metabolism are not well understood. Transport systems for glycerophosphodiesters have so far been identified only in yeast and Escherichia coli (see reference 12 for a review). In yeast, glycerophosphodiesters are taken up by a proton symporter of the major facilitator superfamily whereas the E. coli Ugp system is an ABC transporter (13–15). In addition to glycerophosphodiesters, many bacteria are able to transport and utilize glycerol 3-phosphate. This is important when secreted or periplasmic glycerophosphodiesterases generate free extracellular glycerol 3-phosphate, as is the case in E. coli and Gram-positive bacteria such as Bacillus subtilis. In E. coli, glycerol 3-phosphate is transported by the Ugp system and the proton symporter GlpT. The latter protein is also present in B.subtilis. As described for glycerol metabolism, internal glycerol 3-phosphate becomes oxidized to dihydroxyacetone phosphate and enters the glycolytic pathway. Only a little is known about the transport of glycerophosphodiesters and glycerol 3-phosphate in M. pneumoniae. A Ugp system is annotated in the genome (16); however, M. pneumoniae is unable to utilize glycerol 3-phosphate as a carbon source (17). This is in good agreement with the weak similarity of the candidate Ugp transporter proteins (MPN134, MPN135, MPN136) to its suspected E. coli counterparts. In contrast, M. pneumoniae is able to take up and utilize glycerophosphodiesters such as GPC (17). The genome of M. pneumoniae encodes two proteins with similarity to glycerophosphodiesterases; however, only one of them, GlpQ, has this activity (17). If the M. pneumoniae glpQ gene is inactivated, the bacteria exhibit altered expression of several lipoproteins and transporters, among them the glycerol facilitator GlpF (17). In our opinion, it seemed likely that other proteins involved in the transport of glycerol and glycerophosphodiesters might be among the proteins controlled by the glycerophosphodiesterase GlpQ.
In this work, we identified M. pneumoniae proteins essential for the uptake of both glycerol and glycerophosphodiesters by analyzing the corresponding mutant strains with respect to growth, metabolism of glycerol and GPC (as judged from the production of the unique product hydrogen peroxide), and cytotoxicity. This allowed us to identify the MPN421 protein as the transporter of glycerophosphodiesters. Therefore, the MPN421 protein was renamed GlpU and the corresponding gene was renamed glpU. The two paralogous proteins MPN076 and MPN077 also contribute to GPC transport. Moreover, two additional proteins (MPN133 and MPN284) were found to be essential for the utilization of glycerol.
MATERIALS AND METHODS
Bacterial strains, oligonucleotides, and growth conditions.
All of the M. pneumoniae strains used in this study are derived from M. pneumoniae M129 (ATCC 29342) in the 32nd broth passage. All of the mutant strains used in this study are listed in Table 1. The oligonucleotides used in this study are listed in Table S1 in the supplemental material. M. pneumoniae was grown at 37°C in 150-cm2 tissue culture flasks containing 100 ml of modified Hayflick medium with glucose (1%, wt/vol) as the carbon source as described previously (8). Strains harboring transposon insertions were cultivated in the presence of 80 μg/ml gentamicin.
Table 1.
M. pneumoniae mutants isolated in this study
| Strain | Gene | Locus | Direction | Transcription unit | Function | Full protein length (aa)a | Transposon insertion (nucleotide position) | Truncated protein length in aa (additional aa) |
|---|---|---|---|---|---|---|---|---|
| glpF | MPN043 | + | Single | Glycerol uptake facilitator | 264 | Essentialb | ||
| GPM89 | MPN076 | − | Single | Uncharacterized transporter, MFSc group | 564 | After 506 | 168 (5 + stop codon) | |
| GPM90 | MPN077 | − | Single | Uncharacterized transporter, MFS group | 546 | After 685 | 228 (1 + stop codon) | |
| GPM91 | MPN133 | + | mpn133-136 | Uncharacterized lipoprotein | 301 | After 425 | 141 (5 + stop codon) | |
| ugpC | MPN134 | + | mpn133-136 | ABCd transporter ATP-binding protein | 586 | Essential | ||
| ugpA | MPN135 | + | mpn133-136 | ABC transporter permease protein | 329 | Essential | ||
| ugpE | MPN136 | + | mpn133-136 | ABC transporter permease protein | 319 | Essential | ||
| GPM86 | MPN162 | + | Single | Uncharacterized lipoprotein | 320 | After 312 | 104 (7 + stop codon) | |
| GPM87 | MPN284 | + | Single | Uncharacterized lipoprotein | 794 | After 501 | 167 (7 + stop codon) | |
| GPM25 | glpU | MPN421 | − | glpUQ | Glycerophosphodiester transporter, MFS group | 475 | After 707 | 235 (5 + stop codon) |
| cbiO | MPN433 | − | Single | Metal ion ABC transporter | 270 | Essential | ||
| GPM88 | MPN506 | + | mpn506-507 | Uncharacterized lipoprotein | 793 | After 389 | 129 (5 + stop codon) |
aa, amino acids.
Previously published in reference 9.
MFS, major facilitator superfamily.
ABC, ATP-binding cassette.
Construction of an mpn421 (glpU) complementation strain.
For complementation of the glpU mutant (GPM25), we constructed strain GPM28 as follows: The functional gene glpU, plus 985 bp upstream to include its own promoter, was amplified with primers SG17/SG18. The product was digested with EcoRI and XhoI and cloned into complementation plasmid pMTnTetM438 (18), which had been linearized with the same enzymes. The resulting plasmid, pGP692, contains the transposon with a tetracycline resistance cassette for the selection of complementing clones. Using electroporation, pGP692 was introduced into the genome of M. pneumoniae GPM25. The resulting complementation strain was designated GPM28. This strain was cultivated in the presence of gentamicin (80 μg/ml) and tetracycline (2 μg/ml).
Southern blot analysis.
For the preparation of M. pneumoniae chromosomal DNA, cells of a 100-ml culture were harvested as described previously (8). The cell pellet was resuspended in 750 μl of 50 mM Tris/HCl (pH 8.0)–25 mM EDTA, and RNase A was added to a final concentration of 25 μg/ml. After an incubation step at 37°C for 15 min, 50 μl proteinase K (25 mg/ml) and 75 μl 10% SDS were added. The mixture was incubated at 50°C until the lysate was clarified and subsequently cooled on ice. To precipitate debris, 300 μl of 5 M NaCl was added and the mixture was incubated for 20 min on ice. The precipitate was pelleted by centrifugation (25 min, 15,000 × g, 4°C), and the resulting supernatant was mixed with 500 μl isopropanol to precipitate the chromosomal DNA. The DNA pellet was washed with 70% ethanol and finally resolved in 300 μl TE buffer. Digests of chromosomal DNA were separated using 1% agarose gels and transferred onto a positively charged nylon membrane (Roche Diagnostics) and probed with digoxigenin (DIG)-labeled riboprobes obtained by in vitro transcription with T7 RNA polymerase (Roche Diagnostics) using PCR-generated fragments as the templates (for the primers used, see Table S1 in the supplemental material). The reverse primers contained a T7 RNA polymerase recognition sequence. In vitro RNA labeling, hybridization, and signal detection were carried out according to the manufacturer's instructions (DIG RNA labeling kit and detection chemicals; Roche Diagnostics).
Analysis of mRNA amounts.
Preparation of total M. pneumoniae RNA was done as described previously (8). For slot blot analysis, serial 2-fold dilutions of the RNA extract (2 to 0.25 μg) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were blotted onto a positively charged nylon membrane using a PR 648 Slot Blot Manifold (Amersham Biosciences). Equal amounts of yeast tRNA (Roche) and M. pneumoniae chromosomal DNA served as controls. DIG-labeled riboprobes were obtained by in vitro transcription from PCR products that cover open reading frame internal sequences using T7 RNA polymerase (Roche). The reverse primers used to generate the PCR products contained a T7 promoter sequence (see Table S1 in the supplemental material).
Determination of in vivo hydrogen peroxide production.
The hydrogen peroxide production of M. pneumoniae was determined using the Merckoquant peroxide test (Merck, Darmstadt, Germany) as previously described (9). Briefly, growing cells were resuspended in assay buffer and after incubation for 1 h at 37°C, glucose, glycerol, or glycerophosphodiesters (final concentration, 100 μM) were added to one aliquot. An aliquot without any added carbon source served as the control. The test strips were dipped into the suspensions for 1 s and subsequently read.
HeLa cell cytotoxicity assay.
Infection of HeLa cell cultures with M. pneumoniae cells was done as described previously (9, 19). At 4 days after infection, the HeLa cell cultures were fixed with formalin buffered with 10% phosphate-buffered saline (pH 7.4) for 30 min at room temperature. The fixative solution was discarded, and the cells were stained with 0.1% aqueous crystal violet solution for 30 min with gentle shaking. After incubation, the cell cultures were rinsed with water and photographed. To dissolve the dye for quantification, 0.5% SDS in water was added to each well and the culture plates were incubated for 30 min while shaking. Finally, 100 μl of the dissolved staining solution was used to measure the optical density at 595 nm. The cytotoxicity of M. pneumoniae cells was quantified by calculating the percentage of viable HeLa cells compared to that of a noninfected HeLa cell control. The cytotoxicity assays were performed in triplicate.
RESULTS
Identification of genes tentatively involved in the transport of glycerol and GPC.
In the genome of M. pneumoniae, the glpF gene was suggested to encode the glycerol facilitator. This gene is essential in M. pneumoniae, and it was suggested that the encoded protein is also required for water uptake (9). The proteins encoded in the mpn133-136 operon were annotated as lipoprotein (MPN133) and as a Ugp glycerol 3-phosphate ABC transport system (MPN134, MPN135, MPN136). Since M. pneumoniae is unable to utilize glycerol 3-phosphate (17), this system may have different substrate specificity. The proteome analysis of a glpQ mutant defective in glycerophosphodiesterase revealed overexpression of GlpF, the putative metal ion ABC transporter CbiO, and the uncharacterized lipoprotein MPN162 in the mutant. Moreover, the expression of the uncharacterized lipoproteins MPN284 and MPN506 was reduced in the glpQ mutant (17). The impact of GlpQ on the regulation of these genes suggests that the corresponding proteins might be involved in functions related to glycerol and/or GPC metabolism. The glpQ gene is located downstream of a gene encoding a potential permease of unknown function (MPN421). Thus, this protein might be implicated in the transport of GPC. The analysis of protein-protein interactions in M. pneumoniae revealed an interaction between MPN284 and the uncharacterized transporter MPN076 (20). Finally, the proteins MPN077 and MPN506 are highly similar (75 to 80% identity) to MPN076 and MPN284, respectively. Therefore, we considered all of these proteins as potential candidates for the uptake of GPC.
Isolation of M. pneumoniae mutants.
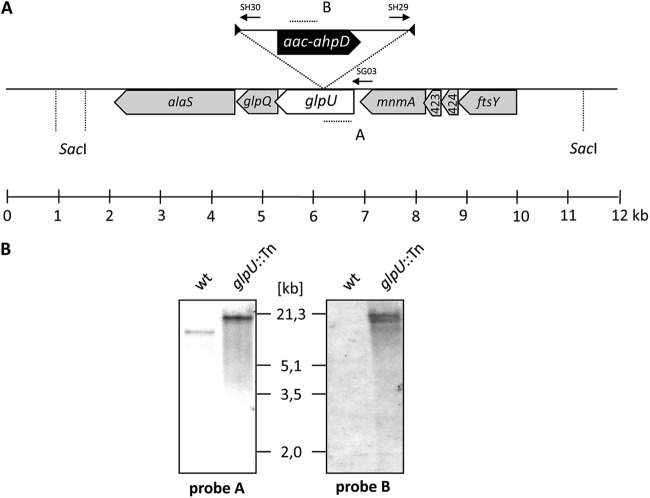
To get more insights into the function of the candidate permeases, we attempted to isolate corresponding mutants. This was done by using “haystack mutagenesis” (21). This strategy is based on an ordered collection of pooled random transposon insertion mutants that can be screened for junctions between the transposon and the gene of interest due to transposon insertion. Sixty-four pools containing 2,976 individual mutants (21) were used in a PCR to detect junctions between the gene of interest and the minitransposon. For mpn421 (glpU), this was done by using oligonucleotides SG03 and SH29 for glpU and the minitransposon, respectively (Fig. 1A). From one pool that gave a positive signal, a colony PCR with the 50 individual mutants resulted in the identification of one glpU mutant. The presence of the transposon insertion in glpU was verified by Southern blotting using a glpU-specific probe (Fig. 1B). To test whether this strain contained only a unique transposon insertion, we did another Southern blot assay using a probe specific for the aac-aphD resistance gene present on the minitransposon. As shown in Fig. 1B, a band with the same size as the SacI fragment hybridizing to the glpU probe was detected (Fig. 1B). The isolated glpU mutant strain was designated GPM25. The position of the transposon insertion in the glpU gene of M. pneumoniae GPM25 was determined by DNA sequencing. The glpU gene was disrupted between nucleotides 707 and 708, resulting in a truncated protein of 235 amino acids with five additional amino acids and the following stop codon encoded by the inserted minitransposon. The transposon library was screened the same way for all desired mutants. No mutants were obtained for mpn134, mpn135, mpn136, and cbiO, suggesting that these genes are essential. The isolation and initial characterization of mutants affected in mpn076, mpn077, mpn133, mpn162, mpn284, glpU, and mpn506 are summarized in Table 1 (see also Fig. S1 in the supplemental material). To make sure that all of the biological effects of the transposon insertion are due to the intended gene disruptions and not to possible polar effects on the expression of downstream genes, we analyzed their transcription organization. With the exception of glpU, mpn133, and mpn506, all of the disrupted genes are expressed as monocistronic transcripts, excluding the possibility of polar effects. For the respective glpU, mpn133, and mpn506 mutants, regular expression of the downstream genes was demonstrated by slot blot analysis (see Fig. S2 in the supplemental material). However, since the glpQ gene is located immediately downstream of glpU (Fig. 1A), we wanted to ascertain that the phenotypes observed for the glpU mutant were indeed the result of the disruption of the glpU gene. For this purpose, we introduced a functional copy of glpU into the genome of glpU mutant strain GPM25 (for details, see below).
Fig 1.
Isolation of an M. pneumoniae glpU transposon insertion mutant. (A) Schematic representation of the genomic region surrounding the glpU gene of M. pneumoniae and the site of the transposon insertion in glpU knockout strain GPM25. The annealing sites of the oligonucleotides used for determination of the transposon insertion site are indicated by arrows. Probes hybridizing to internal fragments of the glpU and aac-ahpD genes are depicted as dotted lines. (B) Southern blot analysis to confirm the single insertion of the minitransposon into the glpU gene of strain GPM25. Chromosomal DNA of the wild type (wt) and that of strain GPM25 were digested using SacI. Blots were hybridized with the glpU-specific probe (left blot) and a probe hybridizing to the aac-ahpD gene of the minitransposon (right blot).
Implication of the putative transporters in growth on glucose and glycerol.
For an initial characterization of the isolated mutants, we observed the growth of these strains in modified Hayflick medium (Fig. 2 and 3). A comparison of the growth of the wild-type and mutant strains revealed that the glpU mutant, as well as the mpn076 and mpn077 mutants, were impaired in the utilization of glucose, whereas the other mutants grew at a rate similar to that of the wild-type strain (Fig. 2A and 3A). In the presence of glycerol, the mpn133 and mpn284 mutants exhibited reduced growth; specifically, they did not reach the same yield as the wild type and the other mutant strains (Fig. 3B). This suggests that MPN133 and MPN284 may be involved in the utilization of glycerol. In the absence of any added carbohydrate, the bacteria can use the serum phospholipids present in Hayflick medium. Under these conditions, all of the mutants have a very low yield; however, the glpU and mpn076 mutants seemed to be impaired in phospholipid utilization (Fig. 2C and 3C).
Fig 2.
Growth of M. pneumoniae in modified Hayflick medium containing different carbon sources. One hundred milliliters of medium was inoculated with 2.5 mg of the relevant M. pneumoniae cells and incubated for up to 6 days at 37°C in 150-cm2 cell culture flasks. The following strains were used: the wild type (wt), a glpU::Tn mutant, and a glpU::Tn + glpU mutant. Glucose (A) and glycerol (B) were added to a final concentration of 1% (wt/vol). Moreover, cells were grown without the addition of any carbon source (C), allowing the serum phospholipids in the modified Hayflick medium to serve as the only growth factor. Attached cells were collected by scraping, and growth was monitored by determination of the wet weight of the cell pellets. All measurements were done three times. Results are from a representative experiment.
Fig 3.
Growth of different M. pneumoniae strains in medium containing different carbon sources. Growth assays were performed as described in the legend to Fig. 2. The following strains were analyzed concerning their growth in modified Hayflick medium containing glucose (A), glycerol (B), or no additional carbon source (C): the wild type (wt), an mpn076::Tn mutant, an mpn077::Tn mutant, an mpn133::Tn mutant, an mpn162::Tn mutant, an mpn284::Tn mutant, and an mpn506::Tn mutant.
The roles of the disrupted transporters in the production of hydrogen peroxide.
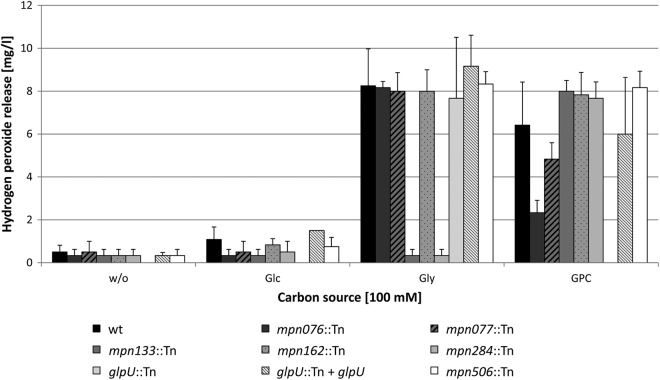
The oxidation of glycerol 3-phosphate results in the generation of hydrogen peroxide, the major cytotoxic product of M. pneumoniae. We asked, therefore, whether the disruption of the putative transporters would affect hydrogen peroxide formation. Hydrogen peroxide formation was assayed in M. pneumoniae cultures that contained no carbon source, glucose, glycerol, or GPC. In the absence of an added carbon source or in the presence of glucose, none of the mutants formed substantial amounts of hydrogen peroxide (Fig. 4). When glycerol or GPC was available, maximal hydrogen peroxide formation (6 to 9 μg/ml) was observed in the wild-type strain. This is in good agreement with previous reports of increased hydrogen peroxide generation in the presence of these substrates (17, 22). It is important to note that the glycerol concentration used here (100 μM) corresponds to the physiological concentration in blood serum (23). The mutant strains defective in mpn162 and mpn506 were indistinguishable from the wild-type strain with respect to hydrogen peroxide formation under all of the conditions studied here. This is in good agreement with the absence of any particular phenotypes in the growth tests (Fig. 3). In contrast, the mpn133 and mpn284 mutants were unable to form hydrogen peroxide in the presence of glycerol while they behaved similarly to the wild type when GPC was present in the medium. This demonstrates that the principal ability to synthesize hydrogen peroxide is not affected by the mutations. In conclusion, these mutants seem to be unable to accumulate glycerol. This idea is in excellent agreement with the observed inability of the two mutants to utilize glycerol as a carbon source (Fig. 3B). The mpn076, mpn077, and glpU mutants produced large amounts of hydrogen peroxide in the presence of glycerol. However, the mpn076 and mpn077 mutants exhibited reduced hydrogen peroxide formation in the presence of GPC, whereas the glpU mutant did not produce any hydrogen peroxide with GPC as the substrate. In agreement with the growth test (Fig. 2C), this result suggests that the putative transporter MPN421 (GlpU) is essential for GPC transport. Moreover, the two homologous proteins MPN076 and MPN077 are involved in GPC utilization.
Fig 4.
Examination of M. pneumoniae hydrogen peroxide release. The hydrogen peroxide production of M. pneumoniae mutant strains in the presence of different carbon sources (100 μM) was measured after 2 h. The following strains were used: the wild type (wt), an mpn076::Tn mutant, an mpn077::Tn mutant, an mpn133::Tn mutant, an mpn162::Tn mutant, an mpn284::Tn mutant, a glpU::Tn mutant, a glpU::Tn + glpU mutant, and an mpn506::Tn mutant. Error bars indicate standard deviations (based on three independent experiments). Glc, glucose; Gly, glycerol; w/o, without the addition of any carbon source.
Impact of potential transporters on the cytotoxicity of M. pneumoniae.
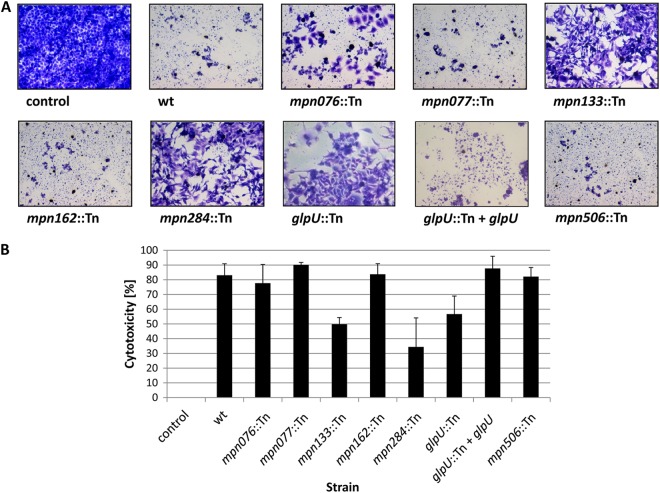
The formation of hydrogen peroxide is one of the major virulence factors of M. pneumoniae that contribute to cytotoxicity. To assess the cytotoxicity of the transporter mutants, we infected confluently grown HeLa cell cultures with cells of the wild-type and mutant strains of M. pneumoniae (multiplicity of infection, 2). As shown in Fig. 5, the HeLa cells had undergone nearly complete lysis after 4 days upon infection with wild-type M. pneumoniae (cytotoxicity of 83%). Similar results were observed with strains GPM86 and GPM88, which are affected in mpn162 and mpn506, respectively. Thus, these proteins are not involved in any of the phenotypes addressed in this study. In contrast, a large portion of viable cells was observed after infection of the cell culture with mpn133 and mpn284 mutants GPM91 and GPM87, respectively. Thus, the lack of glycerol utilization and hydrogen peroxide formation in the presence of glycerol correlates perfectly with the impaired cytotoxicity of these mutants. Interestingly, cytotoxicity was also severely impaired in glpU mutant GPM25. This observation suggests that the transport and subsequent metabolism of both glycerol and GPC contribute to the host cell damage seen in our assay. No significant effects were detectable for the mpn076 and mpn077 mutant strains. These results suggest that the residual hydrogen peroxide formation observed for these mutants is still sufficient to cause damage to host cells. These results perfectly match the minor roles of these proteins in GPC utilization observed in the growth tests and the hydrogen peroxide formation assay (see above).
Fig 5.
Cytotoxicity of M. pneumoniae for HeLa cell cultures. (A) Infection assay to verify cytotoxic effects of M. pneumoniae mutant strains. HeLa cells were infected with M. pneumoniae wild-type (wt), mpn076::Tn mutant, mpn077::Tn mutant, mpn133::Tn mutant, mpn162::Tn mutant, mpn284::Tn mutant, glpU::Tn mutant, glpU::Tn + glpU mutant, and mpn506::Tn mutant cells. A HeLa cell culture without the addition of any M. pneumoniae cells served as a control. After 4 days, HeLa cell cultures were stained with crystal violet and photographed. All pictures are shown at the same magnification. (B) Quantification of cytotoxicity caused by M. pneumoniae in HeLa cell cultures. HeLa cells were infected with different M. pneumoniae strains or left uninfected (negative control). The amount of crystal violet bound to viable HeLa cell cultures at 4 days postinfection was used as an index of cytotoxicity. Cytotoxicity was calculated as the percentage of dead HeLa cells in comparison to that in the uninfected HeLa cell control. Error bars indicate standard deviations (based on three independent experiments).
Complementation analysis of the glpU mutant.
Inactivation of the GPC transporter gene glpU resulted in phenotypes of the corresponding mutant strain, GPM25, that were indistinguishable from those reported for the glycerophosphodiesterase gene glpQ (mpn420) (17). The glpQ gene is located immediately downstream of glpU (Fig. 1A), thus leaving the possibility of indirect polar effects of glpU disruption on glpQ expression. To test whether the observed effects of the glpU mutation were indeed caused by the disruption of the transporter gene or rather due to the potential polar effect, we performed two sets of experiments. (i) RNA slot blot analyses revealed that the glpQ gene is expressed in glpU mutant GPM25, and in fact, this expression is even stronger than that observed in the wild type (see Fig. S2 in the supplemental material). Thus, a polar effect on glpQ transcription can be excluded. (ii) We complemented the mutant by introducing a functional copy of glpU into the genome of glpU mutant strain GPM25, resulting in strain GPM28. This resulted in complementation of the growth defects with glucose and phospholipids (Fig. 2A and C). Similarly, the phenotypes of the glpU mutant with respect to hydrogen peroxide production and cytotoxicity were completely complemented by ectopic expression of glpU in GPM28 (Fig. 4 and 5). Thus, all of the phenotypes observed with the glpU mutant were complemented, demonstrating that it was the lack of the GPC transporter GlpU itself that was responsible for these phenotypes.
DISCUSSION
Of the genes and proteins of M. pneumoniae, only a small fraction has been studied with respect to their function. The transporters and lipoproteins are a notable example of this statement. The genome of M. pneumoniae encodes 53 membrane-spanning transporters and 67 lipoproteins. These lipoproteins are members of six distinct families (24). This accounts for about 17% of the protein-coding genes that are devoted to transport processes. In this work, we propose functions for three of these transporters, as well as for two lipoproteins the function of which was previously unknown. Because of the limited metabolic biosynthetic potential of M. pneumoniae and its relatives, it is obvious that these bacteria depend on the provision of many metabolites and intermediates by the host, and this requires a large set of transporters. However, while many transporters are vaguely annotated on the basis of membrane segments or similarities to established classes of transporters (such as ABC transporters or members of the major facilitator superfamily) (25), little is known about their actual substrates. This is particularly striking for the lipoproteins.
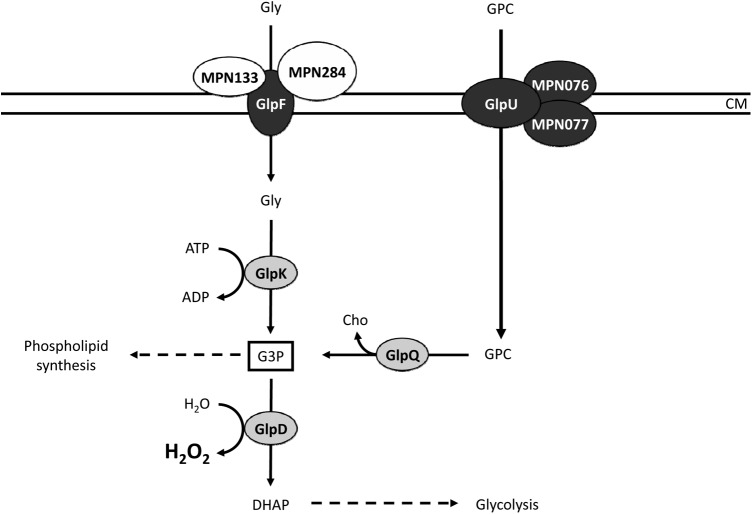
Phospholipids and derived metabolites such as glycerol are the major sources of carbon and energy for M. pneumoniae on lung epithelia (6). Moreover, the oxidation of the common intermediate glycerol 3-phosphate gives rise to the production of hydrogen peroxide, a major factor in M. pneumoniae virulence (9). With the data presented in this work, we have completed the elucidation of the catabolic pathways for glycerol and GPC (Fig. 6). In this study, we present the discovery of the GPC transporters GlpU (MPN421), MPN076, and MPN077. Our results clearly demonstrate that GlpU is the major transporter of GPC. Importantly, the gene encoding this GPC permease is located upstream from the glpQ gene encoding the glycerophosphodiesterase that degrades GPC once it has been taken up into the cell (17). The complete loss of hydrogen peroxide formation in the presence of GPC in the glpU mutant raises the question of which role the additional transporters MPN076 and MPN077 might play in the uptake of GPC. On the basis of the fact that these transporters are encoded by adjacent genes and given their high similarity (76% identical amino acids), it is tempting to speculate that these accessory transporters are the result of a gene duplication. This gene duplication is specific for M. pneumoniae and may reflect the selective pressure that acts on this bacterium on the lung epithelia: inactivation of either gene results in reduced uptake of GPC (as judged from the hydrogen peroxide formation assay). Thus, the presence of the duplicated accessory GPC transporters may help M. pneumoniae to acquire GPC more efficiently. This explains why this duplication does not occur in other mollicutes and may be a reason for the ability of M. pneumoniae but not of related species such as M. genitalium to colonize lung tissue. Of the two GPC transporters, MPN076 seems to be somewhat more efficient; thus, mutations in the corresponding gene have more pronounced effects. Interestingly, the presence of two candidate genes for the potential glycerophosphodiesterase that catalyzes the first intracellular step in GPC metabolism was also detected. However, of the two potential glycerophosphodiesterases, GlpQ and MPN566, the latter has lost important amino acid residues and its enzymatic activity toward GPC (17). Thus, in the course of evolution, the duplication products may have different fates. While one product (MPN076 and GlpQ) is optimized for its activity, the second may lose the primary activity. MPN077 is still involved in GPC transport, whereas in the case of MPN566, the original activity has already been lost. Such proteins might acquire completely novel functions in the further course of evolution; alternatively, the corresponding genes may become obsolete.
Fig 6.
Schematic illustration of glycerol metabolism by M. pneumoniae. The glycerol uptake facilitator GlpF (MPN043) may form a complex together with MPN133 and MPN284 that is required for general glycerol import. The glycerol kinase GlpK (MPN050) and the glycerol 3-phosphate oxidase GlpD (MPN051) metabolize glycerol to the glycolytic intermediate dihydroxyacetone phosphate. Hydrogen peroxide formation by GlpD is crucial for the cytotoxic effects of M. pneumoniae. GlpQ (MPN420) encodes a glycerophosphodiesterase that is able to metabolize GPC to glycerol-3-phosphate and choline. The GPC uptake system is based on the permease GlpU (MPN421) and the two accessory proteins MPN076 and MPN077, of which MPN076 seems to be the more efficient. CM, cell membrane; DHAP, dihydroxyacetone phosphate; G3P, glycerol 3-phosphate; Gly, glycerol; Cho, choline. White, lipoproteins; light gray, cytosolic enzymes; dark gray, transmembrane transport proteins.
Two lipoproteins, MPN133 and MPN284, are required for the uptake of glycerol. The precise function of the potential glycerol facilitator GlpF could not be characterized because the corresponding gene is essential (9). From our data, we conclude that the lipoproteins may bind free glycerol in the medium and deliver it to GlpF, which catalyzes the ultimate uptake. This scenario leaves the question of why GlpF is essential while MPN133 and MPN284 are not. It is well established that glycerol transporters are similar to aquaporins that are necessary for the uptake of water (26). M. pneumoniae does not possess a specific gene encoding an aquaporin. Thus, GlpF might perform both functions, and the aquaporin activity would certainly not depend on a binding protein. Our results demonstrate that both MPN133 and MPN284 are essential for the formation of hydrogen peroxide when the cells grow in the presence of glycerol. Moreover, both proteins are essential for full cytotoxicity. The latter phenotype is easily explained by the absence of the virulence factor hydrogen peroxide in the mutants. It should, however, be noted that both proteins were identified in previous studies as virulence factors. Indeed, MPN133 seems to have a dual function: in addition to its role in glycerol transport, it also has a nuclease activity that was suggested to be important for the acquisition of nucleotides. Similar results were obtained with the homolog from M. genitalium, MG186 (27, 28). Similarly, the MPN284 homolog from Mycoplasma gallisepticum, MslA, was identified as an immunogenic virulence factor (29). It is interesting that dual functions of a lipoprotein in transport and virulence were also suggested for P37 of Mycoplasma hyorhinis (30, 31). This protein enhances the invasiveness of certain cancer cells, such as melanoma and prostate carcinoma cells (32). Given the small number of proteins in M. pneumoniae and other mollicutes, proteins with multiple functions (i.e., moonlighting proteins) may help the bacteria to fulfill all of the requirements of the cell, as well as those in the interaction with the host. Moonlighting of M. pneumoniae proteins has also been described for the glycolytic kinases that are also active in nucleotide metabolism (33) and for the glycerophosphodiesterase GlpQ, which also controls the expression of a set of transporter-encoding genes (17).
Given the importance of metabolite uptake for the host-associated lifestyle of M. pneumoniae, the investigation of transport processes remains an important task in the analysis of these bacteria. Moreover, the fact that many families of lipoproteins are specific for the genus Mycoplasma and their implication in virulence (34) make them attractive targets for the isolation of antimycoplasmal compounds.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Fonds der Chemischen Industrie. S.R.S. was supported by a personal stipend from the Studienstiftung des Deutschen Volkes.
Footnotes
Published ahead of print 7 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01212-12.
REFERENCES
- 1.Atkinson TP, Balish MF, Waites KB. 2008. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol. Rev. 32:956–973 [DOI] [PubMed] [Google Scholar]
- 2.Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsiodras S, Kelesidis I, Kelesidis T, Stamboulis E, Giamarellou H. 2005. Central nervous system manifestations of Mycoplasma pneumoniae. J. Infect. 51:343–354 [DOI] [PubMed] [Google Scholar]
- 4.Narita M. 2009. Pathogenesis of neurological manifestations of Mycoplasma pneumoniae infection. Pediatr. Neurol. 41:159–166 [DOI] [PubMed] [Google Scholar]
- 5.Narita M. 2010. Pathogenesis of extrapulmonary manifestations of Mycoplasma pneumoniae infection with special reference to pneumonia. J. Infect. Chemother. 16:162–169 [DOI] [PubMed] [Google Scholar]
- 6.Veldhuizen R, Nag K, Orgeig S, Possmayer F. 1998. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta 1408:90–108 [DOI] [PubMed] [Google Scholar]
- 7.Halbedel S, Hames C, Stülke J. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol. 12:147–154 [DOI] [PubMed] [Google Scholar]
- 8.Halbedel S, Hames C, Stülke J. 2004. In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae. J. Bacteriol. 186:7936–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hames C, Halbedel S, Hoppert M, Frey J, Stülke J. 2009. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan TR, Baseman JB. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 103:6724–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackerman SJ, Kwatia MA, Doyle CB, Enhorning G. 2003. Hydrolysis of surfactant phospholipids catalyzed by phospholipase A2 and eosinophil lysophospholipases causes surfactant dysfunction: a mechanism for small airway closure in asthma. Chest 123:355S doi:10.1378/chest.123.3_suppl.355S [DOI] [PubMed] [Google Scholar]
- 12.Patton-Vogt J. 2007. Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim. Biophys. Acta 1771:337–342 [DOI] [PubMed] [Google Scholar]
- 13.Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patton-Vogt J, Henry SA. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinosotol in Saccharomyces cerevisiae. Genetics 149:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brzoska P, Boos W. 1988. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the UGP transport system of Escherichia coli. J. Bacteriol. 170:4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, Yuan YP, Herrmann R, Bork P. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidl SR, Otto A, Lluch-Senar M, Pinol J, Busse J, Becher D, Stülke J. 2011. A trigger enzyme in Mycoplasma pneumoniae: impact of the glycerophosphodiesterase GlpQ on virulence and gene expression. PLoS Pathog. 7:e1002263 doi:10.1371/journal.ppat.1002263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lluch-Senar M, Vallmitjana M, Querol E, Piñol J. 2007. A new promoterless reporter vector reveals antisense transcription in Mycoplasma genitalium. Microbiology 153:2743–2752 [DOI] [PubMed] [Google Scholar]
- 19.Schmidl SR, Gronau K, Hames C, Busse J, Becher D, Hecker M, Stülke J. 2010. The stability of cytadherence proteins in Mycoplasma pneumoniae requires activity of the protein kinase PrkC. Infect. Immun. 78:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castaño-Diez D, Chen WH, Devos D, Güell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Böttcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC. 2009. Proteome organization of a genome-reduced bacterium. Science 326:1235–1240 [DOI] [PubMed] [Google Scholar]
- 21.Halbedel S, Busse J, Schmidl SR, Stülke J. 2006. Regulatory protein phosphorylation in Mycoplasma pneumoniae: a PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J. Biol. Chem. 281:26253–26259 [DOI] [PubMed] [Google Scholar]
- 22.Low IE. 1971. Effect of medium on H2O2 levels and peroxidase-like activity by Mycoplasma pneumoniae. Infect. Immun. 3:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilo P, Vilei EM, Peterhans E, Bonvin-Klotz L, Stoffel MH, Dobbelaere D, Frey J. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J. Bacteriol. 187:6824–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallamaa KM, G. F Browning, S. L Tang. 2006. Lipoprotein multigene families in Mycoplasma pneumoniae. J. Bacteriol. 188:5393–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JS, Reddy V, Chen JH, Shlykov MA, Zheng WH, Cho J, Yen MR, Saier MH., Jr 2011. Phylogenetic characterization of transport protein superfamilies: superiority of Superfamily Tree programs over those based on multiple alignments. J. Mol. Microbiol. Biotechnol. 21:83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroud RM, Miercke LJ, O'Connell J, Khademi S, Lee JK, Remis J, Harries W, Robles Y, Akhavan D. 2003. Glycerol facilitator GlpF and the associated aquaporin family of channels. Curr. Opin. Struct. Biol. 13:424–431 [DOI] [PubMed] [Google Scholar]
- 27.Li L, Krishnan M, Baseman JB, Kannan TR. 2010. Molecular cloning, expression, and characterization of a Ca2+-dependent, membrane-associated nuclease of Mycoplasma genitalium. J. Bacteriol. 192:4876–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somarajan SR, Kannan TR, Baseman JB. 2010. Mycoplasma pneumoniae Mpn133 is a cytotoxic nuclease with a glutamic acid-, lysine- and serine-rich region essential for binding and internalization but not enzymatic activity. Cell. Microbiol. 12:1821–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szczepanek SM, Frasca S, Jr, Schumacher VL, Liao X, Padula M, Djordjevic SP, Geary SJ. 2010. Identification of lipoprotein MslA as a neoteric virulence factor of Mycoplasma gallisepticum. Infect. Immun. 78:3475–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilson E, Alloing G, Schmidt T, Claverys JP, Dudler R, Hofnung M. 1988. Evidence for high affinity binding-protein dependent transport systems in Gram-positive bacteria and in Mycoplasma. EMBO J. 7:3971–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudler R, Schmidhauser C, Parish RW, Wettenhall RE, Schmidt T. 1988. A Mycoplasma high-affinity transport system and the in vitro invasiveness of mouse sarcoma cells. EMBO J. 7:3963–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ketcham CM, Anai S, Reutzel R, Sheng S, Schuster SM, Brenes RB, Agbandje-McKenna M, McKenna R, Rosso CJ, Boehlein SK. 2005. p37 induces tumor invasiveness. Mol. Cancer Ther. 4:1031–1038 [DOI] [PubMed] [Google Scholar]
- 33.Pollack JD, Myers MA, Dandekar T, Herrmann R. 2002. Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS 6:247–258 [DOI] [PubMed] [Google Scholar]
- 34.Chambaud I, Wróblewski H, Blanchard A. 1999. Interactions between Mycoplasma lipoproteins and the host immune system. Trends Microbiol. 7:493–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.