Abstract
Periodontitis is a highly prevalent, biofilm-mediated chronic inflammatory disease that results in the loss of the tooth-supporting tissues. It features two major clinical entities: chronic periodontitis, which is more common, and aggressive periodontitis, which usually has an early onset and a rapid progression. Natural killer (NK) cells are a distinct subgroup of lymphocytes that play a major role in the ability of the innate immune system to steer immune responses. NK cells are abundant in periodontitis lesions, and NK cell activation has been causally linked to periodontal tissue destruction. However, the exact mechanisms of their activation and their role in the pathophysiology of periodontitis are elusive. Here, we show that the predominant NK cell-activating molecule in periodontitis is CD2-like receptor activating cytotoxic cells (CRACC). We show that CRACC induction was significantly more pronounced in aggressive than chronic periodontitis and correlated positively with periodontal disease severity, subgingival levels of specific periodontal pathogens, and NK cell activation in vivo. We delineate how Aggregatibacter actinomycetemcomitans, an oral pathogen that is causally associated with aggressive periodontitis, indirectly induces CRACC on NK cells via activation of dendritic cells and subsequent interleukin 12 (IL-12) signaling. In contrast, we demonstrate that fimbriae from Porphyromonas gingivalis, a principal pathogen in chronic periodontitis, actively attenuate CRACC induction on NK cells. Our data suggest an involvement of CRACC-mediated NK cell activation in periodontal tissue destruction and point to a plausible distinction in the pathobiology of aggressive and chronic periodontitis that may help explain the accelerated tissue destruction in aggressive periodontitis.
INTRODUCTION
Periodontitis is a highly prevalent, biofilm-mediated chronic inflammatory disease that results in destruction of the tooth-supporting structures, ultimately resulting in tooth loss. Epidemiologic studies suggest that up to 60% of the population is affected by the common form of the disease, termed chronic periodontitis, while 5 to 15% may suffer from aggressive periodontitis, a clinically challenging entity characterized by an early onset and rapid disease progression (1). Although more than 700 different bacterial species have been shown to inhabit periodontal biofilms (2), a limited number of species are currently considered to be causally associated with periodontitis. These include Gram-negative species, including the anaerobic Porphyromonas gingivalis, a principal pathogen in chronic periodontitis, and the facultative Aggregatibacter actinomycetemcomitans, causally linked to aggressive periodontitis (3, 4).
From a pathophysiology perspective, both forms of periodontitis are the result of host-mediated inflammatory damage of the supporting tissues triggered in response to the microbial infection (5, 6). However, the mechanisms accounting for the difference in the course of tissue damage between chronic and aggressive periodontitis are poorly understood, and lesions cannot be distinguished by histological means (7, 8).
Here, we focused on the role of NK cells, a distinct cytotoxic lineage of lymphocytes, in the pathobiology of periodontitis. NK cells are considered to be a major component of the innate immune system and exert regulatory effects by secreting cytokines, primarily gamma interferon (IFN-γ) (9). NK cell activity is mediated by several activating or inhibitory receptors, among which the self-associating CD2-like receptor activating cytotoxic cells (CRACC/CD319/CS1/SLAMF7), which is found on NK cells, activated CD8 T cells and B cells (10, 11).
Infiltration by NK cells has been shown in inflamed gingival tissues (12–14), and limited data suggest that NK cells are activated in chronic periodontitis lesions (14). IFN-γ production by NK cells was shown to be triggered by coculture with dendritic cells (DCs) stimulated with P. gingivalis (15) or A. actinomycetemcomitans (16). A recent study reported that the periodontal pathogen Fusobacterium nucleatum is directly recognized by the NK cell-activating receptor NKp46 and that NK cell activation is associated with periodontal bone loss in vivo (17). However, the mechanisms underlying the activation of NK cells in human periodontal tissues are not fully understood.
In this study, we first examined the expression of NK cell activation molecules in the gingival tissues of a well-phenotyped cohort of patients with chronic or aggressive periodontitis and showed that the predominant NK cell-activating molecule is CRACC. Importantly, CRACC was found to be significantly overexpressed in pathological gingival tissues in aggressive periodontitis compared to tissues from chronic periodontitis lesions. Therefore, we subsequently examined in vitro mechanisms of CRACC activation in response to bacterial challenge by A. actinomycetemcomitans or P. gingivalis.
We demonstrate that CRACC induction on NK cells is mediated by DC activation and IL-12 signaling in response to infection of DCs with A. actinomycetemcomitans and, to a much lesser extent, with P. gingivalis. Finally, we show that the fimbriae of P. gingivalis contribute to active immune modulation resulting in attenuated induction of CRACC on NK cells.
MATERIALS AND METHODS
Transcriptomic analyses of periodontitis.
Whole-genome gene expression profiles were generated using Affymetrix HG-U133plus 2.0 gene arrays from 310 gingival tissue samples (69 clinically healthy and 241 diseased) from 120 nonsmoking patients (55 with aggressive and 65 with chronic periodontitis) phenotyped with respect to demographic information, clinical periodontal status, and levels of subgingival bacteria with respect to 11 species, as described previously (18, 19) (Columbia University Medical Center IRB approval number AAAB0896; GEO [Gene Expression Omnibus at the NCBI] accession number GSE16134). Data were analyzed using R/Bioconductor (limma and SPIA packages) and SAS (for population-specific expression analysis [PSEA] [20]). Mixed-effects linear regression models were used to study differential expression of mRNA in diseased versus healthy tissue. These statistical models contain both fixed and random effects. Specifically, patients were conditioned as random effect in these models to account for the within-patient correlation in gene expression, as previously described (18).
Cell culture.
Primary human peripheral blood mononuclear cells (PBMCs) were isolated from blood samples donated by healthy, nonsmoking volunteers using gradient centrifugation, as described previously (21). NK cells were isolated from PBMCs by positive sorting using a commercial magnetic bead system (Miltenyi Biotech, Bergisch Gladbach, Germany). The purity of isolated NK cells was routinely assessed by flow cytometry, and preparations with >90% purity were used for experiments (see Fig. S2 in the supplemental material). Primary human dendritic cells were generated from bead-isolated monocytes by culture in RPMI medium supplemented with interleukin 4 (IL-4) and granulocyte-macrophage colony-stimulating factor (GM-CSF) by following current protocols (22).
Bacteria.
Aggregatibacter actinomycetemcomitans strain Y4 (23) was cultured on blood agar plates at 37°C and 5% CO2. Porphyromonas gingivalis strain FDC381 (referred to hereafter as 381) (24) was cultured under anaerobic conditions on blood agar plates. The major flagellum-deficient mutant DPG3, generated by insertional inactivation of the fimA gene in P. gingivalis 381 (25), was cultivated anaerobically on blood agar plates supplemented with erythromycin. Bacterial suspensions in RPMI medium without antibiotics were adjusted to 109 CFU/ml using a spectrophotometer at 600 nm and established growth curves.
Assessment of protein expression by flow cytometry.
For fluorescence-activated cell sorting (FACS), we used anti-CD3 and anti-CD56 (BD Biosciences, Heidelberg, Germany) and anti-CRACC and anti-2B4 (R&D Systems, Wiesbaden, Germany). Samples were analyzed on a FACS Canto II (BD Biosciences) using the BD Diva and FlowJo 7.5.5 (Treestar, Ashland, OR) software packages.
Statistical analyses.
All analyses not involving microarray data were performed using Graphpad Prism V (Graphpad, San Diego, CA). All data are presented as means ± standard deviations or as whisker plots (minimum to maximum). A P value of <0.05 was considered significant.
RESULTS
CRACC is the predominant NK cell activator in human periodontitis.
First, we evaluated the expression of NK cell-related genes in human periodontitis lesions (n = 241, from 120 patients each contributing ≥2 diseased tissue samples, with a mean probing pocket depth [PPD] of 6.58 ± 1.91 mm; range, 4 to 11 mm) compared to clinically healthy gingivae (n = 69, each patient contributing a healthy tissue sample, when available; PPD, 2.29 ± 0.52 mm; range, 1 to 4 mm) (18, 19). As summarized in Table 1, the predominant NK cell-activating gene with increased expression in periodontitis lesions versus health was the CRACC gene (3.18-fold; false discovery rate [FDR] = 5.24 × 10−30); high correlation of array data and quantitiative PCR (qPCR) findings for this gene (Spearman's r = −0.81; P = 0.005) was shown previously (26).
Table 1.
Expression of NK cell-related molecules in periodontal diseasea
| Description(s) | Fold change | FDR |
|---|---|---|
| SLAM family member 7 (CRACC) | 3.17 | 5.24 × 10−30 |
| Selectin L (lymphocyte adhesion molecule 1) | 2.76 | 6.53 × 10−34 |
| Fc fragment of IgG, low-affinity IIIa, receptor (CD16a); Fc fragment of IgG, low-affinity IIIb, receptor (CD16b) | 2.42 | 6.18 × 10−30 |
| CD48 molecule | 1.83 | 1.97 × 10−28 |
| Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 1.58 | 2.04 × 10−19 |
| SLAM family member 6 | 1.50 | 4.85 × 10−26 |
| Leukocyte-associated immunoglobulin-like receptor 1 | 1.37 | 8.49 × 10−21 |
| Killer cell lectin-like receptor subfamily G, member 1 | 1.24 | 1.15 × 10−15 |
| CD33 molecule | 1.24 | 9.74 × 10−15 |
| Killer cell lectin-like receptor subfamily C, member 1; killer cell lectin-like receptor subfamily C, member 2; killer cell lectin-like receptor subfamily C, member 3 | 1.24 | 3.31 × 10−10 |
| Sema domain, immunoglobulin domain, transmembrane domain, and short cytoplasmic domain, (semaphorin) 4D | 0.84 | 2.15 × 10−8 |
| Killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1, 2, 3, 4, 5A, 5B; three domains, long cytoplasmic tail, 3; three domains, short cytoplasmic tail, 1; killer cell immunoglobulin-like receptor 2DL4-like | 1.17 | 4.69 × 10−9 |
| Killer cell lectin-like receptor subfamily F, member 1 | 1.11 | 1.87 × 10−5 |
| CD160 molecule | 1.10 | 1.09 × 10−5 |
| Natural cytotoxicity triggering receptor 1 | 1.08 | 3.70 × 10−6 |
| CD226 molecule | 1.07 | 1.59 × 10−4 |
| Killer cell lectin-like receptor subfamily D, member 1 | 1.06 | 3.92 × 10−3 |
| Cytotoxic and regulatory T cell molecule | 1.06 | 3.86 × 10−2 |
| CD96 molecule | 1.04 | 3.65 × 10−2 |
| CD28 molecule | 1.04 | 2.34 × 10−2 |
| Natural cytotoxicity triggering receptor 3 | 1.02 | 2.37 × 10−1 |
| CD244 molecule, natural killer cell receptor 2B4 | 1.02 | 3.44 × 10−2 |
Expression of NK cell-related molecules in diseased versus healthy gingival tissues. Data stem from 310 gingival tissue samples (241 diseased and 69 clinically healthy) from 120 nonsmoking patients (55 with aggressive and 65 with chronic periodontitis) analyzed using Affymetrix HG-U133plus 2.0 gene arrays, as described previously (18, 19). Fold change describes expression levels in diseased over healthy tissues.
Since periodontal lesions are generally characterized by increased infiltration of various types of mononuclear cells known to express CRACC, we further explored whether the high CRACC expression in the periodontitis lesions was due solely to increased numbers of infiltrating cells rather than increased CRACC transcription. To this end, we corrected mathematically for the net change in cell numbers using well-established markers for different cell populations. Indeed, when generating NK cell-specific expression profiles by correcting for increased numbers of NK cells (using PSEA [20] and NKp46 expression as an NK cell marker) in diseased periodontal lesions, we still observed significant increases (2.83-fold; FDR [27] < 0.05) in CRACC expression in periodontally diseased versus clinically healthy gingival tissues.
CRACC expression is increased in aggressive periodontitis.
A comparison of clinically diseased gingival tissue samples from subjects diagnosed with either chronic or aggressive periodontitis revealed that CRACC expression is elevated approximately 60% (Table 2) (P = 2.7 × 10−5) in aggressive periodontitis lesions. In addition, signaling pathway impact analysis of diseased tissues from aggressive versus chronic periodontitis revealed a significant induction of NK cell-mediated cytotoxicity in aggressive periodontitis lesions (FDR [27] < 0.05).
Table 2.
CRACC expression correlated with clinical features of periodontitis
| Comparison | Fold change | P value |
|---|---|---|
| Disease vs. health | 3.18 | 8.44 × 10−28 |
| Aggressive vs. chronic periodontitis | 1.6 | 2.7 × 10−5 |
| Generalized vs. localized periodontitis | 1.6 | 1.9 × 10−5 |
| 1-mm increase in periodontal probing depth | 1.24 | 2.6 × 10−29 |
CRACC expression correlates with disease severity.
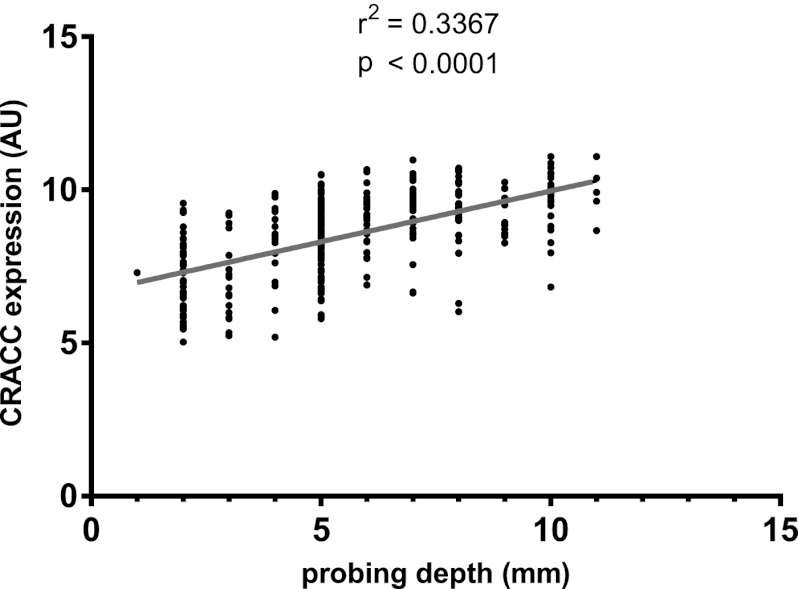
CRACC mRNA expression correlated positively with clinical (Table 2) and microbiological (Table 3) measures of periodontitis severity, such as PPD (Fig. 1) and subgingival levels of established periodontal pathogens. On the other hand, CRACC expression did not correlate with measures indicating past exposure to periodontitis, such as clinical attachment loss (CAL), or with subgingival levels of health-associated bacteria.
Table 3.
CRACC expression correlated with microbiological features of periodontitisa
| Species | Fold change/SD increase | P value |
|---|---|---|
| Aggregatibacter actinomycetemcomitans | 1.29 | 1.9 × 10−6 |
| Porphyromonas gingivalis | 1.46 | 7.9 × 10−12 |
| Treponema denticola | 1.43 | 1.8 × 10−11 |
| Tannerella forsythia | 1.48 | 3.6 × 10−13 |
| Campylobacter rectus | 1.37 | 1.0 × 10−9 |
| Eikenella corrodens | 1.26 | 0.01 |
| Fusobacterium nucleatum | 1.19 | 0.0001 |
| Parvimonas micra | 1.24 | 2 × 10−5 |
| Prevotella intermedia | 1.25 | 5 × 10−6 |
| Actinomyces naeslundii | 1.00 | 0.84 |
| Veillonella parvula | 1.14 | 0.009 |
CRACC expression fold change associated with an increase by 1 standard deviation in subgingival bacterial colonization by selected bacterial species. All results are adjusted for periodontal pocket depth and clinical attachment loss.
Fig 1.
CRACC expression in 310 clinically healthy or diseased gingival tissue biopsy specimens from 120 patients with chronic or aggressive periodontitis correlated positively with the probing depth of the periodontal pocket, a marker for periodontitis severity.
Collectively, these data suggest that CRACC expression is strongly associated with infection by specific etiologic periodontal pathogens, the severity of the periodontitis lesion, and the form of periodontitis signifying accelerated loss of periodontal tissue support (aggressive periodontitis).
CRACC expression correlates with markers of NK cell activation in vivo.
In both whole gingival tissue transcriptomes and NK cell-specific expression profiles generated from whole tissue data using PSEA algorithms (20), CRACC expression correlated positively with markers of NK cell activation, such as CD69 (in whole tissue data set, r = 0.16 and P = 0.005; in NK cell-specific data set, r = 0.16 and P = 0.003) and CD38 (in whole tissue data set, r = 0.79 and P < 0.0001; in NK cell-specific data set, r = 0.80 and P < 0.0001).
Taken together, these findings indicate a role for CRACC in NK cell activation that is eventually linked to increased severity of periodontal destruction and to the rapid progression of disease in the entity of aggressive periodontitis. Therefore, we next proceeded to further characterize the determinants and effects of CRACC induction and NK cell activation in periodontal infections in vitro.
CRACC is induced on NK cells after infection with periodontal pathogens in PBMC culture in vitro.
First, we assessed whether infection of primary human peripheral blood mononuclear cells (PBMCs) with Aggregatibacter actinomycetemcomitans, a pathogen causally related to aggressive periodontitis, or Porphyromonas gingivalis, a principal pathogen in chronic periodontitis, would lead to increased expression of CRACC protein on NK cells.
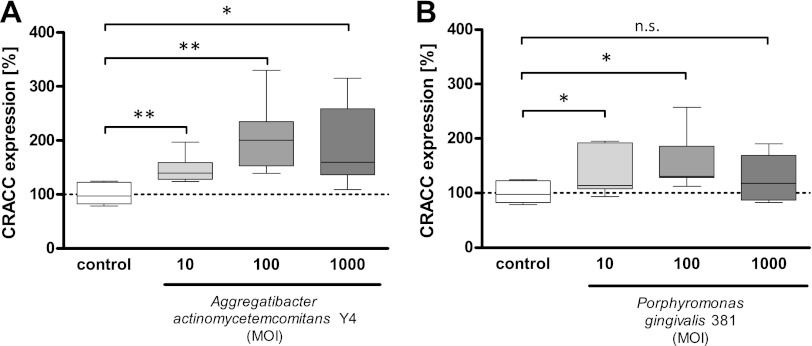
Indeed, CRACC expression on NK cells was induced by both A. actinomycetemcomitans and P. gingivalis infections in a dose-dependent manner (Fig. 2). The induction observed was significantly stronger in cells challenged with A. actinomycetemcomitans than in cells infected with P. gingivalis.
Fig 2.
CRACC expression on NK cells in PBMC culture is upregulated by A. actinomycetemcomitans or P. gingivalis in a dose-dependent manner. PBMCs were cultured for 24 h with A. actinomycetemcomitans or P. gingivalis at different multiplicities of infection (MOI) for 24 h. CRACC expression on NK cells was analyzed by flow cytometry. In A. actinomycetemcomitans coculture, CRACC upregulation was significant in comparison to control (A) (n = 8) (*, P < 0.05; **, P < 0.001), whereas in P. gingivalis-treated PBMCs, only MOIs of 10 and 100 resulted in a significantly higher CRACC expression (B) (n = 8) (*, P < 0.05; n.s., not significant).
Lack of direct recognition and CRACC induction of A. actinomycetemcomitans or P. gingivalis by NK cells.
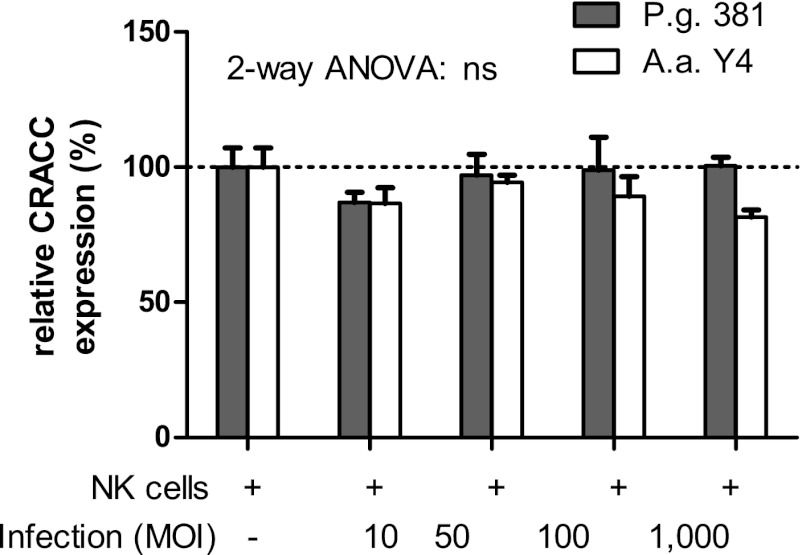
Incubation of isolated NK cells of sufficient purity (see Fig. S1 in the supplemental material) with A. actinomycetemcomitans or P. gingivalis (Fig. 3), or with Toll-like receptor 2 (TLR2) and TLR4 ligands (see Fig. S2 in the supplemental material) had no effect on CRACC expression and did not lead to activation of the NK cells (data not shown). These data suggest that CRACC induction on NK cells is not mediated by direct recognition of the periodontal pathogens.
Fig 3.
Lack of CRACC induction by NK cells after A. actinomycetemcomitans (A.a.) or P. gingivalis (P.g.) infection. Primary human NK cells were incubated with either P. gingivalis 381 or A. actinomycetemcomitans Y4 at the indicated multiplicities of infection (MOI) for 24 h. No changes in CRACC protein expression were observed after infection with either periodontal pathogen. n = 4; 2-way repeated-measures analysis of variance (ANOVA) showed that the difference was not significant (ns).
DC-mediated CRACC induction on NK cells.
Based on the finding that infection of isolated primary human NK cells with periodontal bacteria (Fig. 3) or ligation of TLR2 or TLR4 (see Fig. S1 in the supplemental material) did not result in CRACC induction, while infection of isolated PBMCs triggered CRACC on NK cells, we hypothesized that CRACC induction was likely mediated by indirect signaling via antigen-presenting cells, i.e., dendritic cells (DCs).
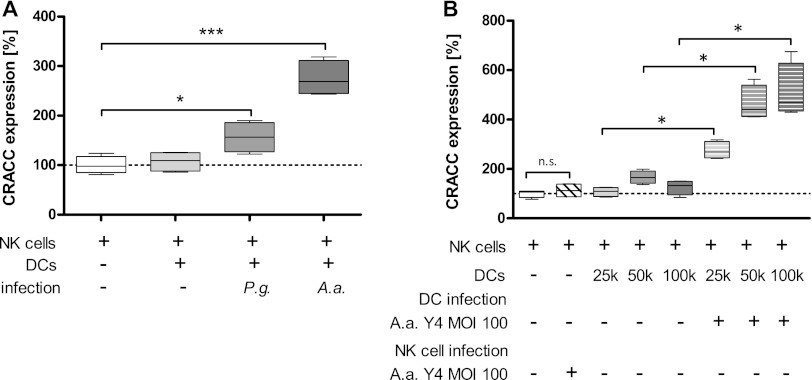
We therefore tested whether infection of primary human monocyte-derived DCs with either A. actinomycetemcomitans or P. gingivalis and subsequent incubation of the washed DCs with primary NK cells was sufficient to induce CRACC protein expression on the surface of NK cells. Indeed, direct interaction of preinfected DCs with periodontal pathogens with NK cells resulted in significant induction of CRACC on NK cells (Fig. 4A). This interaction showed a clear dose dependency (Fig. 4B).
Fig 4.
Induction of CRACC protein on NK cell surfaces is mediated by DCs. (A) Primary human NK cells were incubated (24 h) with or without washed primary human DCs previously challenged with either A. actinomycetemcomitans (A.a.) or P. gingivalis (P.g.) or left uninfected (multiplicity of infection [MOI], 100). Coculture of NK cells with infected DCs resulted in induction of CRACC protein on NK cell surfaces. However, the induction was far more pronounced in response to A. actinomycetemcomitans than P. gingivalis challenge. (B) Dose-dependent induction of CRACC in response to coculture with different numbers of DCs previously infected with A. actinomycetemcomitans. NK cells were incubated with or without A. actinomycetemcomitans at an MOI of 100 and in coculture with or without DC in different cell ratios. n = 4; 1-way repeated-measures analysis of variance with post hoc Dunnett test. *, P < 0.05; ***, P < 0.001.
Soluble factors mediate CRACC upregulation on NK cells.
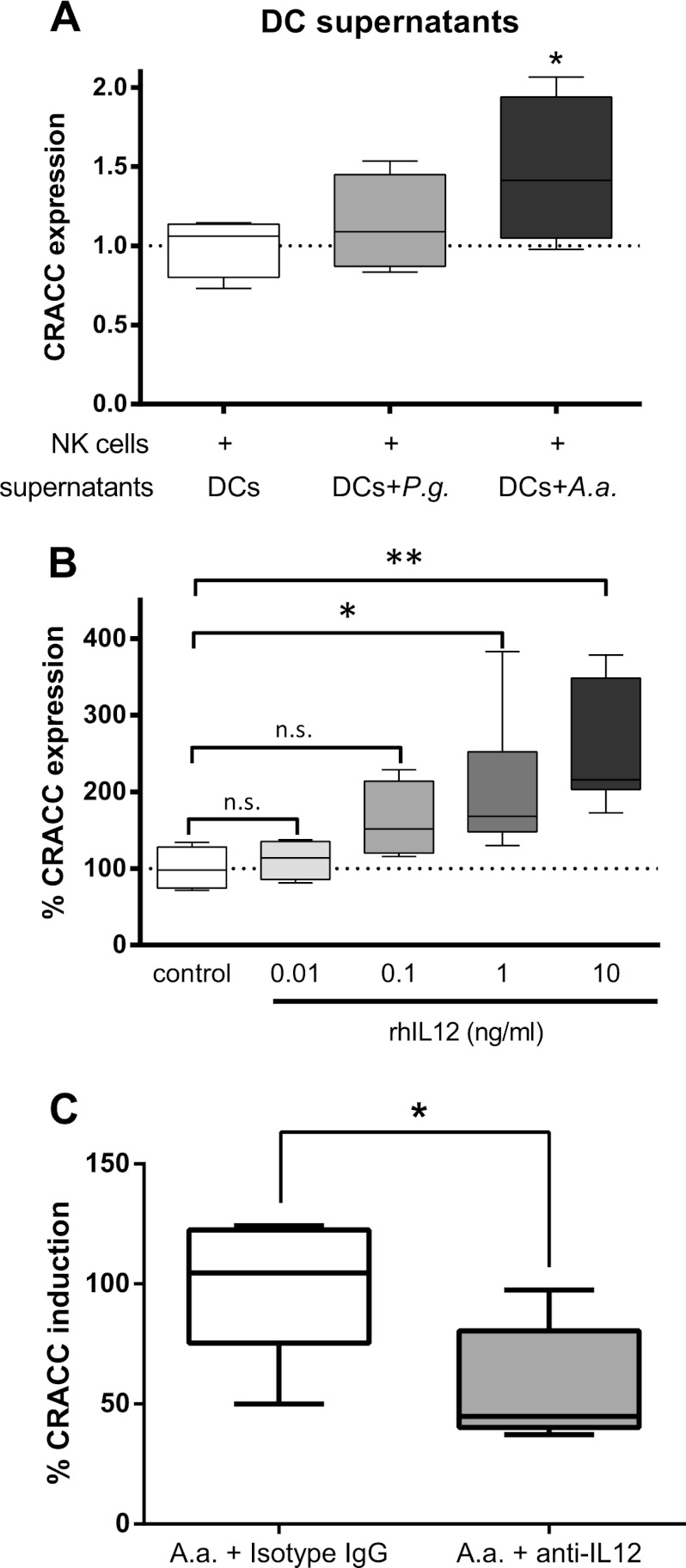
Next, we evaluated mechanisms responsible for the DC-mediated CRACC induction on NK cell surfaces. To this end, we incubated primary human NK cells with filtered, cell- and bacterium-free culture supernatants of DCs previously infected with either A. actinomycetemcomitans or P. gingivalis. Supernatants from A. actinomycetemcomitans-infected DCs were able to significantly induce CRACC expression on NK cell surfaces, while a trend toward higher CRACC expression was observed when using supernatants from P. gingivalis-infected DCs, although it did not reach statistical significance (Fig. 5A). This suggests that soluble factors secreted by infected DCs are sufficient to induce CRACC induction.
Fig 5.
DC-dependent induction of CRACC protein on NK cell surfaces is mediated by soluble factors. (A) Primary human NK cells were incubated (24 h) with filtered, cell-free supernatants from DCs previously infected with either P. gingivalis (P.g.) or A. actinomycetemcomitans (A.a.) or left uninfected (24 h; multiplicity of infection, 100). Culture supernatants from A. actinomycetemcomitans-infected DCs triggered a significant induction of CRACC protein on NK cells. (B) CRACC protein expression on primary human NK cells is induced by recombinant human IL-12 (rhIL12) in a dose-dependent manner. n = 4; 1-way repeated-measures analysis of variance with post hoc Dunnett tests. *, P < 0.05. (C) IL-12-blocking antibodies lead to significantly reduced A. actinomycetemcomitans infection-mediated CRACC induction on NK cells. n = 4; paired t tests. *, P < 0.05.
Interleukin 12 (IL-12) is one of the factors known to mediate DC-NK cell cross talk and was previously reported to be secreted by DCs infected with either A. actinomycetemcomitans or P. gingivalis (15, 16). Indeed, IL-12 was able to trigger CRACC induction on primary human NK cells in a dose-dependent manner (Fig. 5B). Conversely, concomitant incubation of A. actinomycetemcomitans-challenged DCs with IL-12-blocking antibodies led to a strong decrease in A. actinomycetemcomitans-mediated CRACC induction (Fig. 5C), suggesting that IL-12 blockade in fact prevented the transfer of the ability to induce CRACC on NK cells in conditioned medium supernatants from DCs infected with periodontal pathogens. This indicates that the DC-mediated upregulation of CRACC on NK cell surfaces is mediated by IL-12 signaling.
Fimbriated P. gingivalis attenuates CRACC induction.
Our data show strong differences in CRACC induction in response to A. actinomycetemcomitans or P. gingivalis infection of DCs, with P. gingivalis infection resulting in significantly attenuated induction of CRACC expression on NK cells.
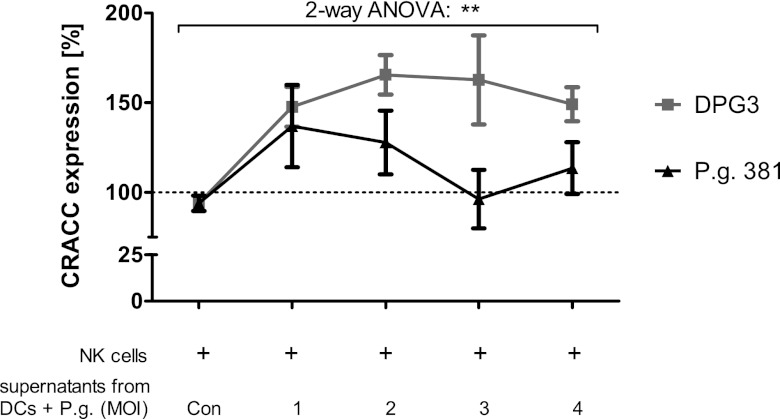
Interestingly, a fimbria-deficient mutant (DPG3) engineered from P. gingivalis 381 by deletion of the fimA gene (25) triggered significantly higher levels of CRACC protein expression on primary human NK cells than wild-type P. gingivalis 381 used for all other experiments (Fig. 6).
Fig 6.
CRACC activation in response to P. gingivalis (P.g.) infection is attenuated by fimbriae. Primary human NK cells were incubated with cell culture supernatants from DCs previously challenged with fimbriated P. gingivalis 381 or major-fimbria-deficient DPG3 at the indicated multiplicity of infection (MOI) for 24 h, or from uninfected DCs (Con). The graph shows significantly stronger induction of CRACC protein expression on NK cell surfaces in response to the fimbria-deficient mutant. n = 4; two-way repeated-measures analysis of variance (ANOVA). **, P < 0.01.
These data indicate that the apparently attenuated response to P. gingivalis infection is due to the organism's fimbriae.
DISCUSSION
Our present work shows that the predominant NK cell-activating receptor in human periodontitis is CRACC. A strong induction of CRACC was observed not only in the gingival tissue transcriptomic profiles in a large, well-phenotyped cohort of patients with periodontitis but also in cell population-specific expression profiles generated using the PSEA algorithm (20). Our data clearly show a significant induction of CRACC on NK cells in states of periodontitis compared to gingival health in vivo.
Furthermore, CRACC expression in periodontal tissues correlated positively with clinically important markers of periodontitis, such as probing pocket depth (PPD) or levels of subgingival colonization by specific periodontal pathogens. This is relevant, since the composition of the subgingival microbiome is known to trigger specific expression profiles in the gingival tissues (26, 28).
Importantly, we show an increased expression of CRACC in pathological gingival tissues from subjects diagnosed with aggressive periodontitis, compared to chronic periodontitis, pointing to an increased activation of NK cells in this clinically challenging form of periodontitis that is characterized by an accelerated rate of destruction. It must, however, be realized that these data are cross-sectional in nature and therefore cannot be used to infer causality. Still, in the absence of established animal models for aggressive periodontitis, our data from a large and well-controlled human cohort provide valuable insights into the potential molecular mechanisms of aggressive periodontitis in vivo.
Given our incomplete understanding of the molecular differences between chronic and aggressive periodontitis (8), we then proceeded to characterize the determinants and effects of CRACC induction in an in vitro model of periodontal infection.
Using primary human NK cells and DCs isolated from healthy donors, and the periodontal pathogens A. actinomycetemcomitans, which has been etiologically linked to aggressive periodontitis, and P. gingivalis, a principal pathogen in chronic periodontitis, we replicated the induction of CRACC by periodontal infections in vitro. Importantly, in line with the finding of increased CRACC expression in aggressive periodontitis lesions, we consistently observed significantly stronger CRACC induction in response to infection with A. actinomycetemcomitans than with P. gingivalis.
In this study, we exclusively used primary cells isolated from healthy donors. It is, however, conceivable that cells from patients suffering from periodontitis could exhibit differing activation patterns due to inherent or acquired characteristics. However, the design of our in vitro experiments was based upon in vivo data that stem solely from periodontitis patients, and their results corroborate the in vivo findings. Therefore, it is reasonable to assume that the specific responses in primary cells from both healthy and diseased subjects are likely similar. Nevertheless, further experimentation is needed to definitely address this issue.
We demonstrate that the mechanism by which periodontal pathogens induce CRACC expression is indirect and is mediated by DC-secreted IL-12 triggering increased expression of the NK cell-activating receptor. Isolated NK cells infected with either A. actinomycetemcomitans or P. gingivalis did not display increased CRACC expression (Fig. 3), nor did they show other markers of activation (data not shown). This is important, since most recently, a direct recognition of another periodontal species, Fusobacterium nucleatum, by the activating NK cell receptor NKp46 was shown (17), and it may indicate a level of specificity in the ability of different periodontal bacteria to mobilize components of the immune system.
Furthermore, our data indicate substantial differences in the ability of established periodontal pathogens to induce CRACC expression on NK cells via DC infection, with A. actinomycetemcomitans consistently triggering significantly stronger induction of CRACC expression than P. gingivalis. Interestingly, infection of DCs with major-fimbria-deficient P. gingivalis 381 mutant DPG3 (25) resulted in a significantly increased induction of CRACC on cocultured NK cells. These data indicate that the presence of fimbriae is critical for the suppression of NK cell activation in response to P. gingivalis, in line with observations that this fimbria-deficient mutant, but not the wild-type strain, induced periodontal bone loss in mice (29). Furthermore, these data appear to corroborate findings by Hajishengallis and coworkers demonstrating the ability of P. gingivalis to actively evade the immune system. These investigators demonstrated that P. gingivalis utilizes its surface fimbriae to bind CXCR4 on human monocytes, trigger cyclic AMP (cAMP)-dependent protein kinase A signaling, and lead to inhibition of TLR2-mediated proinflammatory and antimicrobial responses to the infection (30).
On the other hand, these data show that CRACC induction on NK cells by P. gingivalis is not mediated through active invasion of host cells but is, rather, due to its inability to circumvent TLR-dependent detection by antigen-presenting cells. In contrast, invasion of host cells, a critical element of P. gingivalis's virulence in the context of atherogenesis (31), is a property requiring fimbriae, specifically, the FimA protein that DPG3 lacks (32).
In this context, however, it needs to be noted that the observed differences in the abilities of DPG3 and its parent strain, P. gingivalis 381, to induce CRACC expression on NK cell surfaces via DCs cannot exclusively be attributed to the fimA deletion in DPG3 but may be partly due to unaccounted-for concomitant alterations of the bacterial cells. Nevertheless, use of the wild-type strain and its fimbria-deficient mutant is a well-established experimental model for the study of the specific effects of fimA.
Importantly, a pattern of CRACC expression similar to the one shown here in the context of periodontal infections was reported for patients with systemic lupus erythematosus (SLE), a disease with a number of pathobiological similarities to periodontitis (33). Specifically, the frequency of CRACC+ CD56bright NK cells was found to be higher in patients with SLE than in controls. In vitro, stimulation of PBMCs with SLE immune complexes induced CRACC expression on NK cell surfaces (34).
Taken together, our data suggest an active involvement of CRACC-mediated NK cell activation in periodontal tissue destruction. Furthermore, they point to a conceivable distinction in the pathophysiology of the two major forms of periodontitis that may account for the accelerated course of destruction in aggressive periodontitis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wenzel Vogel for expert technical assistance.
This study was supported by grants from the NIH (number DE-015649 and number DE-021820 to P.N.P. and number DE-018739 to R.T.D.), Colgate-Palmolive USA (to P.N.P.), the German Research Foundation (KFO208, TP6, and TP9 to M.K.), the German Society for Oral and Maxillofacial Sciences (to M.K.), the German Society for Periodontology (to M.K.), and “Neue Gruppe” Scientific Fund (to M.K.).
Footnotes
Published ahead of print 17 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00895-12.
REFERENCES
- 1. Demmer RT, Papapanou PN. 2010. Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol. 2000 53:28–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Darveau R. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490 [DOI] [PubMed] [Google Scholar]
- 4. Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. 2008. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet 371:237–242 [DOI] [PubMed] [Google Scholar]
- 5. Liu YC, Lerner UH, Teng YT. 2010. Cytokine responses against periodontal infection: protective and destructive roles. Periodontol. 2000 52:163–206 [DOI] [PubMed] [Google Scholar]
- 6. Graves D. 2008. Cytokines that promote periodontal tissue destruction. J. Periodontol. 79:1585–1591 [DOI] [PubMed] [Google Scholar]
- 7. Smith M, Seymour GJ, Cullinan MP. 2010. Histopathological features of chronic and aggressive periodontitis. Periodontol. 2000 53:45–54 [DOI] [PubMed] [Google Scholar]
- 8. Ford PJ, Gamonal J, Seymour GJ. 2010. Immunological differences and similarities between chronic periodontitis and aggressive periodontitis. Periodontol. 2000 53:111–123 [DOI] [PubMed] [Google Scholar]
- 9. Caligiuri MA. 2008. Human natural killer cells. Blood 112:461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Claus M, Meinke S, Bhat R, Watzl C. 2008. Regulation of NK cell activity by 2B4, NTB-A and CRACC. Front. Biosci. 13:956–965 [DOI] [PubMed] [Google Scholar]
- 11. Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. 2009. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat. Immunol. 10:297–305 [DOI] [PubMed] [Google Scholar]
- 12. Stelin S, Ramakrishan H, Talwar A, Arun KV, Kumar TS. 2009. Immunohistological analysis of CD1a Langerhans cells and CD57 natural killer cells in healthy and diseased human gingival tissue: a comparative study. J. Indian Soc. Periodontol. 13:150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujita S, Takahashi H, Okabe H, Ozaki Y, Hara Y, Kato I. 1992. Distribution of natural killer cells in periodontal diseases: an immunohistochemical study. J. Periodontol. 63:686–689 [DOI] [PubMed] [Google Scholar]
- 14. Muthukuru M. 2012. Technical advance: decreased helper T cells and increased natural killer cells in chronic periodontitis analyzed by a novel method for isolating resident lymphocytes. J. Leukoc. Biol. 92:683–692 [DOI] [PubMed] [Google Scholar]
- 15. Kikuchi T, Willis DL, Liu M, Purkall DB, Sukumar S, Barbour SE, Schenkein HA, Tew JG. 2005. Dendritic-NK cell interactions in P. gingivalis-specific responses. J. Dent. Res. 84:858–862 [DOI] [PubMed] [Google Scholar]
- 16. Kikuchi T, Hahn CL, Tanaka S, Barbour SE, Schenkein HA, Tew JG. 2004. Dendritic cells stimulated with Actinobacillus actinomycetemcomitans elicit rapid gamma interferon responses by natural killer cells. Infect. Immun. 72:5089–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaushu S, Wilensky A, Gur C, Shapira L, Elboim M, Halftek G, Polak D, Achdout H, Bachrach G, Mandelboim O. 2012. Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog. 8:e1002601 doi:10.1371/journal.ppat.1002601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, Pavlidis P, Papapanou PN. 2008. Transcriptomes in healthy and diseased gingival tissues. J. Periodontol. 79:2112–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kebschull M, Papapanou PN. 2010. The use of gene arrays in deciphering the pathobiology of periodontal diseases. Methods Mol. Biol. 666:385–393 [DOI] [PubMed] [Google Scholar]
- 20. Kuhn A, Thu D, Waldvogel HJ, Faull RL, Luthi-Carter R. 2011. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat. Methods 8:945–947 [DOI] [PubMed] [Google Scholar]
- 21. Fuss IJ, Kanof ME, Smith PD, Zola H. 2009. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. 85:7.1.1–7.1.8 doi:10.1002/0471142735.im0701s85 [DOI] [PubMed] [Google Scholar]
- 22. O'Neill DW, Bhardwaj N. 2005. Differentiation of peripheral blood monocytes into dendritic cells. Curr. Protoc. Immunol. 67:22F.4.1–22F.4.9 doi:10.1002/0471142735.im22f04s67 [DOI] [PubMed] [Google Scholar]
- 23. Kiley P, Holt SC. 1980. Characterization of the lipopolysaccharide from Actinobacillus actinomycetemcomitans Y4 and N27. Infect. Immun. 30:862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ono M, Okuda K, Takazoe I. 1987. Purification and characterization of a thiol-protease from Bacteroides gingivalis strain 381. Oral Microbiol. Immunol. 2:77–81 [DOI] [PubMed] [Google Scholar]
- 25. Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, Cho MI, Genco RJ, Evans RT, Dyer DW. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papapanou PN, Behle JH, Kebschull M, Celenti R, Wolf DL, Handfield M, Pavlidis P, Demmer RT. 2009. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kebschull M, Papapanou PN. 2011. Periodontal microbial complexes associated with specific cell and tissue responses. J. Clin. Periodontol. 38(Suppl 11):17–27 [DOI] [PubMed] [Google Scholar]
- 29. Baker PJ, Dixon M, Evans RT, Roopenian DC. 2000. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol. Immunol. 15:27–32 [DOI] [PubMed] [Google Scholar]
- 30. Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. 2008. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc. Natl. Acad. Sci. U. S. A. 105:13532–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pollreisz A, Huang Y, Roth G, Cheng B, Kebschull M, Papapanou P, Schmidt A, Lalla E. 2010. Enhanced monocyte migration and pro-inflammatory cytokine production by Porphyromonas gingivalis infection. J. Periodontal Res. 45:239–245 [DOI] [PubMed] [Google Scholar]
- 32. Deshpande RG, Khan MB, Genco CA. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kobayashi T, Ito S, Yasuda K, Kuroda T, Yamamoto K, Sugita N, Tai H, Narita I, Gejyo F, Yoshie H. 2007. The combined genotypes of stimulatory and inhibitory Fc gamma receptors associated with systemic lupus erythematosus and periodontitis in Japanese adults. J. Periodontol. 78:467–474 [DOI] [PubMed] [Google Scholar]
- 34. Hagberg N, Theorell J, Alm GV, Eloranta ML, Bryceson Y, Rönnblom L. 2012. SLE immune complexes upregulate the expression of slamf7 (cd319) on plasmacytoid dendritic cells. Ann. Rheum. Dis. 71:A3 doi:10.1136/annrheumdis-2011-201230.6 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.