Abstract
Background
In this report we studied the genetic regulation of juvenile development of wild barley introgression lines (S42ILs) under two contrasting hydroponic nitrogen (N) supplies. Ten shoot and root related traits were examined among 42 S42ILs and the recurrent parent ‘Scarlett’. The traits included tiller number, leaf number, plant height, leaf and root length, leaf to root length ratio, shoots and root dry weight, shoot to root weight ratio, and chlorophyll content. Our aims were (1) to test the suitability of a hydroponic system for early detection of favourable S42ILs, (2) to locate quantitative trait loci (QTL) that control the examined traits, (3) to identify favourable wild barley alleles that improve trait performances in regard to N treatment and, finally, (4) to validate the identified QTL through comparison with previously reported QTL originating from the same parental cross.
Results
The phenotypic data were analysed in a mixed model association study to detect QTL. The post-hoc Dunnett test identified 28 S42ILs that revealed significant (P < 0.01) effects for at least one trait. Forty-three, 41 and 42 S42ILs revealed effects across both N treatments, under low N and under high N treatment, respectively. Due to overlapping or flanking wild barley introgressions of the S42ILs, these associations were summarised to 58 QTL. In total, 12 QTL of the hydroponic N study corresponded to QTL that were also detected in field trials with adult plants of a similar S42IL set or of the original S42 population. For instance, S42IL-135, -136 and -137, revealed increasing Hsp effects for tiller number, leaf number, leaf length, plant height and leaf to root ratio on the long arm of chromosome 7H. These QTL correspond to QTL for ears per plant and plant height that were previously detected in field trials conducted with the same S42ILs or with the S42 population.
Conclusion
Our results suggest that the QTL we identified under hydroponic N cultivation partly correspond to QTL detected in field experiments. Due to this finding, screening of plants in early developmental stages grown in a hydroponic system may be a fast and cost effective method for early QTL detection and marker-assisted allelic selection, potentially speeding up elite barley breeding programs.
Keywords: Introgression lines, Hordeum vulgare ssp. spontaneum, QTL, Nitrogen stress tolerance, Hydroponics, Juvenile plant development
Background
The domestication of barley, starting about 10,000 years ago [1], led to a multitude of genotypes, which are well adapted to farming under a broad range of conditions [2]. Besides, intensive barley breeding for over a century led to modern barley cultivars revealing a low degree of genetic diversity [3]. One aspect in innovating modern breeding programs is the introduction of exotic germplasm into new varieties [4]. The mlo powdery mildew resistance is a prominent example for the introduction of favourable wild barley alleles into elite barley varieties [5]. In this regard, Tanksley and Nelson [6] established the advanced backcross (AB) quantitative trait loci (QTL) approach in tomato, by which exotic germplasm is introduced into modern cultivars and QTL are characterised in the AB populations. A further approach to detect QTL with exotic germplasm is described by Zamir [7]: the establishment of lines that carry single chromosomal introgressions from exotic donors, so called introgression lines (ILs). In comparison to AB populations, the genetic background of ILs is fixed to the recurrent elite parent. Thus, a complete set of ILs represents the entire donor genome on the background of the recurrent parent. Compared to other mapping populations like recombinant inbred lines (RILs), ILs offer a higher statistical power and allow the detection of small QTL effects [8]. Within the last years, IL sets were developed for important crop species like maize [9], wheat [10] and rice [11].
In barley, von Korff et al. [12] crossed the German spring barley cultivar ‘Scarlett’ (Hordeum vulgare ssp. vulgare, Hv) with the Israeli wild barley accession ‘ISR42-8’ (Hordeum vulgare ssp. spontaneum, Hsp) to develop the BC2DH population S42. By further backcrossing of population S42 with ‘Scarlett’ and subsequent marker assisted selection, a set of wild barley introgression lines (S42IL) was established [13]. Different S42IL sets were used to locate Hsp QTL that regulate pathogen resistances, flowering time, agronomic performance, malting quality and drought stress tolerance [14-18]. Afterwards, the S42IL population was subjected to high-resolution (HR) genotyping with an Illumina 1536-SNP array and the threshability locus thresh1 was located on chromosome 1H by means of high-resolution mapping with segregating offspring of the original S42IL [19].
Nitrogen (N) is one of the nutrients plants need in high quantity [20]. Therefore, it is necessary to understand plant stress responses to N deficiency including changes in growth, development and differentiation. Various QTL were found in field trials and greenhouse experiments with barley under N deficiency, e.g. in recombinant inbred lines (RILs) [21]. Also wild barley has been used to identify favourable alleles under N deficiency. In population S42 Saal et al. [22] identified 14 QTL with reference to N supply. For instance, the authors mapped a QTL on the long arm of chromosome 7H that caused an effect on adult plant height across both N treatments. Later on, Schnaithmann and Pillen [23] mapped QTL for chlorophyll content and N content in the same region of chromosome 7H within the S42IL population.
The plant response to N deficiency affects plant growth and development and is a crucial component of yield [24]. One major problem in studying the interaction between plant growth and N deficiency is the uncertainty of the soil N supply in the field [25]. An alternative approach is the examination of N deficiency under controlled conditions in hydroponic systems. Although research on plants under N deficiency has been carried out in hydroponics [25-27], little is known about the direct comparison between data from single crop plants under N stress in a hydroponic system and the crop performance of the same genotype in field trials and greenhouse experiments. For instance, Beatty et al. [28] reported that barley genotypes studied under field and hydroponic conditions showed similar growth and nitrogen used efficiency characteristics. They assumed that using less-variable growth environments is another way to gain knowledge about traits and genetic targets related to N nutrition. So far, a genome wide QTL analysis with a set of wild barley ILs, focussing on these questions, has not been carried out.
In our study, juvenile wild barley ILs were grown in a hydroponic system under two contrasting N treatments. By conducting the hydroponic experiments, we aimed to (1) test the suitability of our hydroponic system for early selection of favourable S42ILs, to (2) detect QTL in the S42IL population that contribute to the control of juvenile plant growth, to (3) find favourable wild barley alleles that improve trait performances either across both N treatments or under low N supply and, finally, to (4) compare the QTL results of our experiments on juvenile barley plants with QTL found in field trials and greenhouse experiments with adult plants of the S42IL and S42 populations and other QTL studies.
Methods
Plant material
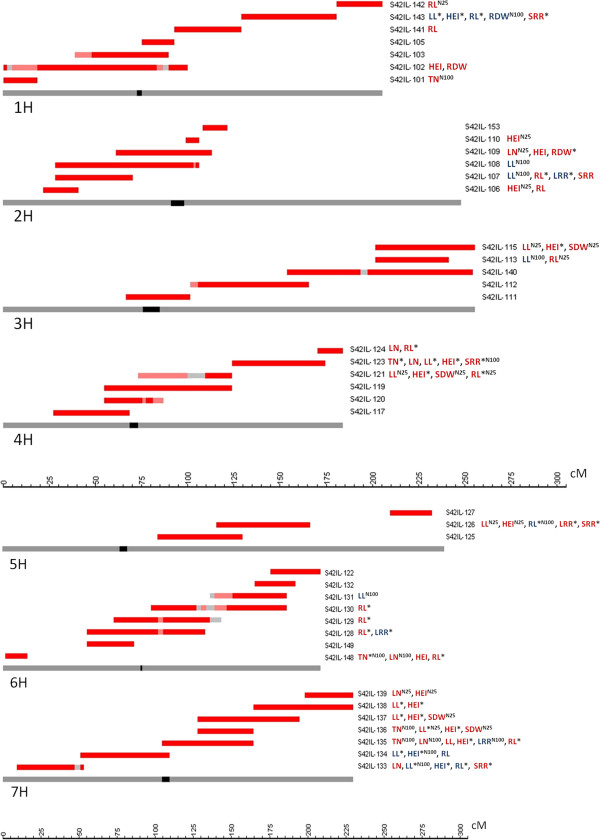
Our investigations on juvenile barley plants in a hydroponic system were carried out with introgression lines (ILs) of the population S42IL. The S42ILs are derived from a cross between the German spring barley (Hv) cultivar ‘Scarlett’ and the Israeli wild barley (Hsp) accession ‘ISR42-8’. Advanced backcrossing with ‘Scarlett’ and marker assisted selection with SSRs resulted in the development of a set of 59 S42ILs in BC3S4 generation as described in Schmalenbach et al. [13]. Based on genotypic data with 636 informative SNPs [19] we selected 42 representative S42ILs with target introgressions located on chromosomes 1H to 7H (Figure 1). The recurrent parent ‘Scarlett’ served as the control genotype in the hydroponic experiments conducted.
Figure 1.
SNP map with 58 significant (P <0.01) QTL effects among 42 S42ILs and nine traits. The chromosomes 1H to 7H are indicated as grey bars. The centromeric region is indicated as a black box within the chromosome. The cM positions of the SNP loci are indicated with the ruler at the bottom, according to Schmalenbach et al. [19]. The extent of the target Hsp introgression of each S42IL is given above the respective chromosome. Homozygous Hsp loci, heterozygous Hsp loci and loci without SNP data are presented in red, pink and light grey, respectively. The associated QTL effects are indicated right to the introgressions with abbreviations of the nine traits, i.e. TN, LN, LL, HEI, SDW, RL, RDW, LRR, SRR. Trait abbreviations are explained in Table 2. The colour of the abbreviations indicates an increasing (red) or decreasing (blue) Hsp effect. Highly significant (P < 0.001) associations are highlighted with an asterisk. Trait associations that were only detected under one treatment are labelled with N100 or N25, respectively.
Hydroponic plant cultivation and phenotyping
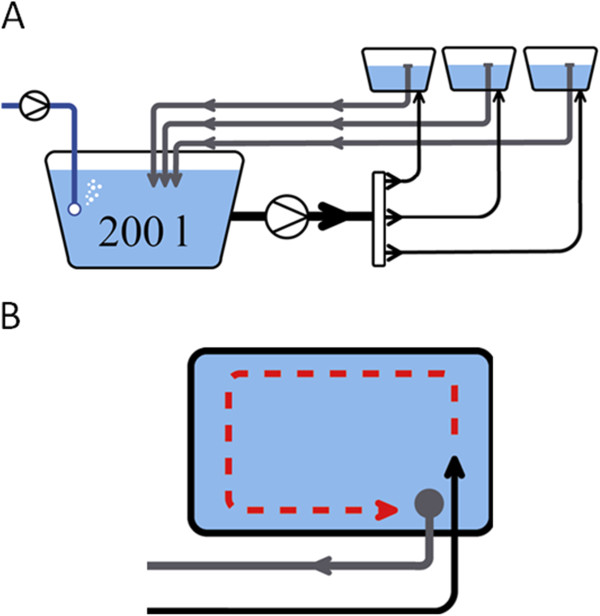
Juvenile barley plants were tested during two years in six hydroponic experiments at two locations of the University of Halle. In 2008, one and two experiments were carried out under low and high N supply, respectively, in a glasshouse at the experimental station ‘Kühnfeld’, Halle. In 2011, the same experimental set up was repeated in a second glasshouse at the experimental station ‘Heide’, Halle. In each of the six experiments, an accumulation tank for the nutrient solution was connected to three boxes for plant cultivation (Figure 2). On each of the three boxes, a perforated tray was placed containing 140 holes to hold the single test plants. The outer perimeter of the tray was filled with 44 non-measured ‘Scarlett’ plants in order to reduce border effects in the box due to inhomogeneous light and space availability. The remaining 96 holes were split into two blocks of 48 holes. ‘Scarlett’, the spring barley cultivar ‘Barke’ and the 42 S42ILs grew in four, two and one replications per block, respectively. The plants were completely randomised within each block.
Figure 2.
Hydroponic system. A. The utilised hydroponic system included an accumulation tank for 200 l of nutrient solution that was connected to three boxes for plant cultivation. B. The nutrient media circulated permanently between the boxes and the tank, ensuring that within and between boxes all plants were exposed to the same nutrient composition.
During the experiments, the nutrient solution circulated between the boxes and the accumulation tank. The composition of the modified Hoagland solution [Hoagland and Amon 1950, cited in 20] is shown in detail in Table 1. To ensure that the nutrients in the solution were available to the plants the pH and EC values of the solution were measured daily. If the pH deviated from 5.8 ± 0.25, it was adjusted by adding 4 M KOH or 4 M HCl, respectively. Plants were grown under high and low N supply solutions with 2.0 mM (i.e. 100% N = N100) and 0.5 mM (i.e. 25% N = N25) of NO3-, respectively. Cultivation of the plants lasted for 14 days in the hydroponic system with 16 h photoperiod, an irradiance of 300 μmol m-2 s-1 and day and night temperatures of 24°C and 16°C, respectively. To ensure that the roots received an ample supply of oxygen, air bubbles were constantly pressed with an air pump into the tank.
Table 1.
Composition of the nutrient media for high (100% N = N100) and low (25% N = N25) N treatments
| Component | Concentration N100(in μM) | Concentration N25(in μM) |
|---|---|---|
| Ca(NO3)2* 4H2O |
2000.00 |
500.00 |
| CaCl2 |
0.00 |
500.00 |
| MES a |
2400.00 |
2400.00 |
| K2SO4 |
700.00 |
700.00 |
| MgSO4 |
500.00 |
500.00 |
| KCl |
100.00 |
100.00 |
| KH2PO4 |
100.00 |
100.00 |
| Na-EDTA b |
50.00 |
50.00 |
| Fe(II)SO4* 7H2O |
50.00 |
50.00 |
| H3BO3 |
10.00 |
10.00 |
| MnSO4 * H2O |
0.50 |
0.50 |
| CuSO4 * 5H2O |
0.20 |
0.20 |
| ZnSO4 * 7H2O |
0.10 |
0.10 |
| (NH4)6Mo7O24 | 0.01 | 0.01 |
a 2-(N-morpholino)ethanesulfonic acid (MES) adjusted to pH 5.8.
b Na-Ethylenediaminetetraacetic acid (EDTA).
For each experiment, seeds of ‘Scarlett’, ‘Barke’ and the S42ILs were coated with the fungicide Abavit UF® (BASF, Ludwigshafen). Seed germination and plant cultivation is shown in detail in Figure 3. To study the plant growth and responses to N deficiency on juvenile barley plants, ten traits were investigated. The methods of trait measurements and trait abbreviations are given in Table 2.
Figure 3.
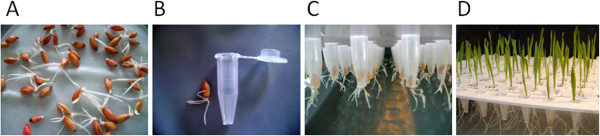
Seed germination and plant cultivation. A. Barley seeds were germinated in the dark at room temperature. B. After 72 h, seedlings that had three to five roots of 1.5 to 2.0 cm length and that had not yet started to extend the shoot were selected. Each seedling was inserted into a 1.5 ml reaction tube (Eppendorf, Hamburg) that was cropped at the bottom. C. The tubes with the seedlings were subsequently plugged into the perforated trays and placed on top of the boxes. D. Throughout the experiments the roots plunged into the nutrient solution whereas plant shoots grew on top of the tray.
Table 2.
List of ten quantitative traits investigated in the hydroponic system
| Abbr. | Trait | Method of measurement | Unit |
|---|---|---|---|
| TN |
Tiller number |
Number of tillers with first leaf unfolded after 14 days |
|
| LN |
Leaf number |
Number of leaves longer than 2 cm after 14 days |
|
| LL |
Leaf length |
Length of the youngest completely unfolded leaf after 14 days |
cm |
| HEI |
Plant height |
Length of the plant from basis to leaf tip after 14 days |
cm |
| SDW |
Shoot dry weight |
Weight of shoot mass after 2 days of drying at 80°C |
mg |
| RL |
Root length |
Length of the longest root from crown to root tip after 14 days |
cm |
| RDW |
Root dry weight |
Root weight after 2 days of drying at 80°C |
mg |
| LRR |
Leaf to root ratio |
Ratio of leaf to root length |
|
| SRR |
Shoot to root ratio |
Ratio of shoot to root dry weight |
|
| CC | Chlorophyll content | Average of 10 measurements with SPAD-502 meter a in the middle of the youngest leaf after 10 days | SPAD |
a SPAD 502 meter: Minolta Camera Co., Osaka, Japan.
Statistical analyses
Analyses of data were carried out with SAS version 9.2 (SAS Institute 2010). Coefficients of broad sense heritability (h2, [29]) for the ten traits were estimated with S42IL data across treatments (ALL) as
| (1) |
and separately for N100 and N25 treatments, respectively, as
| (2) |
where VG, VGxT, VGxE, VGxTxE and Vε are the variance components genotype, genotype by treatment, genotype by experiment, genotype by treatment by experiment and the experimental error, with t, e and r being the number of treatments (t = 2), experiments (e = 6) and replications within an experiment (r = 6), respectively. Variance components were calculated with the VARCOMP procedure in SAS.
To calculate phenotypic correlations between the ten traits within a treatment and, in addition, within a trait between the two treatments, first the least squares means (LSMeans) were estimated for each S42IL with the MIXED procedure in SAS (see below). Subsequently, Pearson correlation coefficients (r) were calculated using the SAS procedure CORR.
We conducted the detection of significant line by trait associations for every trait revealing a heritability of h2 > 10% across treatments or within the N100 and N25 treatments, respectively. In this regard, a mixed model analysis of variance was carried out with the SAS procedure MIXED, either across treatments (ALL) as
| (3) |
or separately for the N100 and N25 treatment as
| (4) |
where μ is the general mean, Gi the fixed effect of the ith genotype (42 S42ILs plus ‘Scarlett’), Tj is the fixed effect of the jth treatment (N100 and N25), Lk is the random effect of the kth location (Kühnfeld and Heide), El(Tj) is the random effect of the lth experiment (1 to 6), nested within the jth treatment, Bm the random effect of the mth box (1 to 18), Gi × Tj is the fixed interaction effect of the ith genotype with the jth treatment, Gi × Lk is the random interaction effect of the ith genotype with the kth location and εn(ijklm) and εn(iklm) are the error terms of the phenotypes Yijklmn and Yiklmn, respectively, calculated from n replications (2 to 8) per box.
Afterwards a post-hoc Dunnett test was carried out to compare the LSMeans of the S42ILs with the LSMeans of the control genotype ‘Scarlett’ [30]. A line by trait association was accepted when the LSMeans of an S42IL differed significantly (P < 0.01) from ‘Scarlett’ across treatments (Model 1) and/or under the N100 and N25 treatment (Model 2), respectively. Significant line by trait associations were summarised to one QTL if the corresponding S42ILs (1) carried overlapping and/or flanking target introgressions and (2) showed genotypic effects of the same direction. The relative performance (RP) of an S42IL compared to the control genotype ‘Scarlett’ was calculated as
| (5) |
where for each trait the LSMeans were calculated across both treatments or separately for N100 or N25, respectively.
Results
Trait performances and heritabilities of S42ILs
Table 3 indicates the trait performances of the S42IL population for ten studied traits in regard to the parameters mean, minimum, maximum, standard deviation (SD), coefficient of variation (CV) and heritability (h2). The values of each parameter are given across treatments and separately for the high and low N treatment. The high N treatment evoked an increase in the mean, minimum and maximum of the S42ILs for all traits except for root length. The highest CV was observed for root dry weight with 45.2% across both N levels. On the contrary, CVs for leaf length, plant height and chlorophyll content were below 20% across and within both N levels. Five traits showed higher heritability across treatments than per treatment, e.g. plant height with 80.8% across treatments versus 78.6% and 60.1% for the high and low N treatment, respectively. Of all investigated traits, root length showed the highest heritability (85.2% across N treatments). On the contrary, chlorophyll content was the trait that revealed the least heritability (0.0%, within N25).
Table 3.
Parameters describing trait performances of the S42IL population in ALL, N100 and N25, respectively
| Traita | Treatmentb | Nc | Meand | Mine | Maxe | SDf | CVg | h2 h | |
|---|---|---|---|---|---|---|---|---|---|
| TN |
|
ALL |
1379 |
1.5 |
1.0 |
4.0 |
0.6 |
39.9 |
31.1 |
| |
N100 |
922 |
1.7 |
1.0 |
4.0 |
0.6 |
35.5 |
52.9 |
|
| |
N25 |
457 |
1.1 |
1.0 |
3.0 |
0.3 |
30.5 |
11.9 |
|
| LN |
|
ALL |
1379 |
4.0 |
2.0 |
8.0 |
1.1 |
26.4 |
21.2 |
| |
N100 |
922 |
4.2 |
2.0 |
8.0 |
1.1 |
26.7 |
37.8 |
|
| |
N25 |
457 |
3.6 |
2.0 |
6.0 |
0.8 |
21.0 |
12.1 |
|
| LL |
(in cm) |
ALL |
1379 |
24.3 |
11.1 |
33.1 |
3.3 |
13.5 |
75.4 |
| |
N100 |
922 |
24.8 |
11.1 |
33.1 |
3.2 |
13.0 |
73.0 |
|
| |
N25 |
457 |
23.4 |
11.3 |
30.2 |
3.2 |
13.6 |
47.5 |
|
| HEI |
(in cm) |
ALL |
1379 |
34.6 |
17.4 |
46.1 |
4.7 |
13.5 |
80.8 |
| |
N100 |
922 |
35.4 |
17.9 |
46.1 |
4.6 |
13.1 |
78.6 |
|
| |
N25 |
457 |
33.0 |
17.4 |
43.1 |
4.3 |
13.1 |
60.1 |
|
| SDW |
(in mg) |
ALL |
1379 |
117.8 |
15.0 |
249.0 |
43.7 |
37.1 |
7.3 |
| |
N100 |
922 |
133.1 |
15.0 |
249.0 |
40.9 |
30.7 |
3.3 |
|
| |
N25 |
457 |
87.0 |
15.0 |
190.0 |
31.2 |
35.9 |
32.6 |
|
| RL |
(in cm) |
ALL |
1379 |
34.3 |
7.6 |
61.0 |
8.8 |
25.6 |
85.2 |
| |
N100 |
922 |
33.3 |
7.6 |
58.1 |
9.3 |
27.9 |
78.9 |
|
| |
N25 |
457 |
36.3 |
15.0 |
61.0 |
7.2 |
19.7 |
73.4 |
|
| RDW |
(in mg) |
ALL |
1379 |
41.5 |
4.0 |
129.0 |
18.7 |
45.2 |
32.3 |
| |
N100 |
922 |
46.0 |
4.0 |
129.0 |
19.2 |
41.7 |
39.2 |
|
| |
N25 |
457 |
32.3 |
4.0 |
75.0 |
13.8 |
42.6 |
35.9 |
|
| LRR |
|
ALL |
1379 |
0.8 |
0.4 |
2.6 |
0.2 |
31.2 |
72.4 |
| |
N100 |
922 |
0.8 |
0.4 |
2.6 |
0.3 |
32.8 |
48.8 |
|
| |
N25 |
457 |
0.7 |
0.4 |
1.1 |
0.1 |
18.7 |
78.4 |
|
| SRR |
|
ALL |
1379 |
3.0 |
1.3 |
7.3 |
0.7 |
24.6 |
80.3 |
| |
N100 |
922 |
3.1 |
1.3 |
7.3 |
0.8 |
25.6 |
73.4 |
|
| |
N25 |
457 |
2.8 |
1.4 |
6.6 |
0.6 |
20.4 |
54.0 |
|
| CC | (SPAD) |
ALL |
1377 |
36.4 |
20.8 |
51.8 |
6.5 |
17.8 |
3.1 |
| |
N100 |
922 |
37.3 |
21.3 |
51.8 |
6.2 |
16.6 |
2.0 |
|
| N25 | 455 | 34.5 | 20.8 | 48.9 | 6.6 | 19.2 | 0.0 | ||
a Trait abbreviations are given in Table 2.
b Treatment abbreviations.
ALL: across treatments, N100 and N25: high (100%) and low (25%) N supply.
c Number of observations.
d Average trait performance.
e Minimum and maximum trait performance.
f Standard deviation.
g Coefficient of variation in %.
h Heritability in %.
Trait correlations
Pearson correlations between N treatments N100 and N25 were significant (P < 0.05) for all traits (diagonal in Table 4). Highly significant (P < 0.001) correlations between treatments were observed for the length traits leaf length, plant height, root length and both ratio traits. The highest correlation coefficient between treatments was found for root length (r = 0.70), whereas the correlation between N100 and N25 within tiller number was weak (r = 0.32).
Table 4.
Pearson correlation coefficients (r) between ten quantitative traits, calculated separately for treatment N25 (bottom left triangle) and treatment N100 (upper right triangle) and within trait between treatments N100 and N25 (diagonal), based on LSMeans of 42 S42ILs
| Traita | TN | b | LN | LL | HEI | SDW | RL | RDW | LRR | SRR | CC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TN |
0.32 |
* |
0.74 |
*** |
0.55 |
*** |
0.49 |
*** |
0.61 |
*** |
0.07 |
|
0.53 |
*** |
0.23 |
|
−0.13 |
|
0.00 |
|
| LN |
0.76 |
*** |
0.38 |
* |
0.43 |
** |
0.48 |
*** |
0.66 |
*** |
0.20 |
|
0.52 |
*** |
0.03 |
|
−0.04 |
|
0.01 |
|
| LL |
0.21 |
|
−0.08 |
|
0.64 |
*** |
0.90 |
*** |
0.70 |
*** |
0.31 |
* |
0.69 |
*** |
0.03 |
|
−0.34 |
* |
0.25 |
|
| HEI |
0.08 |
|
−0.07 |
|
0.91 |
*** |
0.68 |
*** |
0.75 |
*** |
0.53 |
*** |
0.81 |
*** |
−0.02 |
|
−0.47 |
** |
0.40 |
** |
| SDW |
0.48 |
*** |
0.44 |
** |
0.55 |
*** |
0.63 |
*** |
0.44 |
** |
0.37 |
* |
0.54 |
*** |
−0.14 |
|
0.03 |
|
0.45 |
** |
| RL |
0.11 |
|
0.25 |
|
0.41 |
** |
0.43 |
** |
0.44 |
** |
0.70 |
*** |
0.60 |
*** |
−0.89 |
*** |
−0.47 |
** |
0.39 |
** |
| RDW |
0.13 |
|
0.07 |
|
0.55 |
*** |
0.59 |
*** |
0.78 |
*** |
0.43 |
** |
0.45 |
** |
−0.28 |
|
−0.79 |
*** |
0.53 |
*** |
| LRR |
−0.10 |
|
−0.29 |
|
0.16 |
|
−0.11 |
|
−0.03 |
|
−0.82 |
*** |
−0.25 |
|
0.61 |
*** |
0.34 |
* |
−0.32 |
* |
| SRR |
0.16 |
|
0.22 |
|
−0.34 |
* |
−0.34 |
* |
−0.08 |
|
−0.28 |
|
−0.66 |
*** |
0.25 |
|
0.64 |
*** |
−0.36 |
* |
| CC | 0.42 | ** | 0.40 | ** | 0.35 | * | 0.48 | ** | 0.63 | *** | 0.39 | ** | 0.58 | *** | −0.20 | −0.15 | 0.39 | ** |
a Trait abbreviations are given in Table 2.
b Significant correlation coefficients are indicated with * P < 0.05, ** P < 0.01 and *** P < 0.001.
When comparing the correlation matrices of both N treatments, pairwise correlations among traits showed a higher number of significant (P < 0.05) correlations within N100 (31, upper right triangle in Table 4) compared to N25 (24, lower left triangle in Table 4). For example the correlation between tiller number and plant height was highly significant at N100 (r = 0.49 with P < 0.001) but absent at N25 (r = 0.08 with P > 0.05). The highest correlation coefficients were found between traits that belong to the same trait complex like leaf length and plant height (r = 0.90 within N100) or root length and leaf to root ratio (r = -0.89 within N100). No correlation was found between tiller number and chlorophyll content (r = 0.00 within N100). The trait leaf to root ratio showed the lowest number of significant correlations to the other traits. Only correlations to root length and shoot to root ratio were significant within N100.
QTL detection
With regard to the trait heritabilities, QTL detection was carried out for all traits except for SDW (ALL and N100) and CC (ALL, N100 and N25), revealing trait heritabilities below 10%. In total, the post-hoc Dunnett test revealed 126 significant (P < 0.01) line by trait associations for nine traits. Fifty-six associations (44%) were highly significant (P < 0.001). As presented in Table 5, 43 associations were found across treatments (Model 1), 41 under N100 and 42 under N25 (Model 2), respectively. S42IL-143, -133 and -135, with target introgressions on chromosomes 1HL, 7HS and 7HL, respectively, revealed the highest number of trait associations (twelve, ten and ten associations, respectively).
Table 5.
List of 58 significant (P <0.01) QTL effects among nine traits and 42 S42ILs
| Traita | QTL name | Line | Chr.b | Target intro.b(cM) | Effect | LSMeans S42ILd | LSMeans Scarlettd | Diffe | RP(IL)f(in %) | QTL effect in S42 or S42ILg | Candidate geneh |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TN |
QTn.S42IL-1H |
S42IL-101 |
1H |
001.1 - 013.5 |
N100 |
2.0 |
1.6 |
0.4 |
22.9 |
|
|
| |
QTn.S42IL-4H |
S42IL-123 |
4H |
128.9 - 172.3 |
ALL+N100 |
1.7 |
1.4 |
0.4 |
26.4* |
I, V |
|
| |
QTn.S42IL-6H |
S42IL-148 |
6H |
003.3 - 010.7 |
N100 |
2.1 |
1.6 |
0.5 |
30.7* |
|
|
| |
QTn.S42IL-7H |
S42IL-135 |
7H |
101.2 - 152.3 |
N100 |
2.1 |
1.6 |
0.4 |
25.5 |
|
|
| |
|
S42IL-136 |
7H |
134.4 - 152.3 |
N100 |
2.0 |
1.6 |
0.4 |
22.7 |
|
|
| LN |
QLn.S42IL-2H |
S42IL-109 |
2H |
064.0 - 110.8 |
N25 |
4.2 |
3.2 |
0.9 |
28.3 |
|
Eam61, Flt-2L2 |
| |
QLn.S42IL-4H |
S42IL-123 |
4H |
128.9 - 172.3 |
ALL |
4.3 |
3.6 |
0.8 |
21.3 |
|
|
| |
|
S42IL-124 |
4H |
171.3 - 183.5 |
ALL+N100 |
4.2 |
3.6 |
0.7 |
19.1 |
|
VRN-H23 |
| |
QLn.S42IL-6H |
S42IL-148 |
6H |
003.3 - 010.7 |
N100 |
4.7 |
3.9 |
0.8 |
21.0 |
|
|
| |
QLn.S42IL-7H.a |
S42IL-133 |
7H |
017.3 - 051.9 |
ALL |
4.3 |
3.6 |
0.7 |
19.6 |
|
VRN-H34 |
| |
QLn.S42IL-7H.b |
S42IL-135 |
7H |
101.2 - 152.3 |
N100 |
4.6 |
3.9 |
0.7 |
18.8 |
|
|
| |
QLn.S42IL-7H.c |
S42IL-139 |
7H |
198.7 - 229.7 |
N25 |
4.1 |
3.2 |
0.9 |
27.5 |
|
|
| LL |
QLl.S42IL-1H |
S42IL-143 |
1H |
130.7 - 173.5 |
ALL+N100+N25 |
20.3 |
23.7 |
−3.4 |
−14.3* |
|
HvFT35 |
| |
QLl.S42IL-2H |
S42IL-107 |
2H |
034.3 - 066.8 |
N100 |
22.9 |
25.3 |
−2.4 |
−9.3 |
|
|
| |
|
S42IL-108 |
2H |
034.3 - 104.8 |
N100 |
22.8 |
25.3 |
−2.5 |
−9.7 |
|
|
| |
QLl.S42IL-3H.a |
S42IL-113 |
3H |
204.5 - 239.7 |
N100 |
23.0 |
25.3 |
−2.3 |
−9.1 |
|
sdw16 |
| |
QLl.S42IL-3H.b |
S42IL-115 |
3H |
204.5 - 255.1 |
N25 |
25.4 |
22.1 |
3.3 |
15.0 |
|
sdw16 |
| |
QLl.S42IL-4H |
S42IL-121 |
4H |
074.1 - 119.1 |
N25 |
24.7 |
22.1 |
2.6 |
11.8 |
|
|
| |
|
S42IL-123 |
4H |
128.9 - 172.3 |
ALL+N25 |
26.0 |
23.7 |
2.3 |
9.7* |
|
|
| |
QLl.S42IL-5H |
S42IL-126 |
5H |
145.6 - 200.1 |
N25 |
25.1 |
22.1 |
3.1 |
13.8 |
|
ari-e.GP7 |
| |
QLl.S42IL-6H |
S42IL-131 |
6H |
140.0 - 180.7 |
N100 |
22.9 |
25.3 |
−2.4 |
−9.5 |
|
cul28 |
| |
QLl.S42IL-7H.a |
S42IL-133 |
7H |
017.3 - 051.9 |
N100 |
22.6 |
25.3 |
−2.6 |
−10.4* |
|
brh19 |
| |
|
S42IL-134 |
7H |
051.9 - 107.4 |
ALL+N100 |
21.6 |
23.7 |
−2.0 |
−8.6* |
|
brh19 |
| |
QLl.S42IL-7H.b |
S42IL-135 |
7H |
101.2 - 152.3 |
ALL+N25 |
25.6 |
23.7 |
1.9 |
8.0 |
|
|
| |
|
S42IL-136 |
7H |
134.4 - 152.3 |
N25 |
26.3 |
22.1 |
4.2 |
18.9* |
|
|
| |
|
S42IL-137 |
7H |
134.4 - 193.9 |
ALL+N25 |
26.1 |
23.7 |
2.5 |
10.4* |
|
|
| |
QLl.S42IL-7H.c |
S42IL-138 |
7H |
176.4 - 229.7 |
ALL+N25 |
25.6 |
23.7 |
1.9 |
8.1* |
|
|
| HEI |
QHei.S42IL-1H.a |
S42IL-102 |
1H |
001.1 - 098.2 |
ALL |
35.3 |
33.0 |
2.3 |
7.0 |
IV |
|
| |
QHei.S42IL-1H.b |
S42IL-143 |
1H |
130.7 - 173.5 |
ALL+N100+N25 |
27.0 |
33.0 |
−6.0 |
−18.1* |
I, III, V |
HvFT35 |
| |
QHei.S42IL-2H.a |
S42IL-106 |
2H |
022.4 - 034.3 |
N25 |
34.5 |
30.6 |
3.9 |
12.9 |
V |
|
| |
QHei.S42IL-2H.b |
S42IL-109 |
2H |
064.0 - 110.8 |
ALL+N25 |
35.3 |
33.0 |
2.3 |
6.9 |
I - V |
Eam61, Flt-2L2 |
| |
|
S42IL-110 |
2H |
102.7 - 104.8 |
N25 |
34.3 |
30.6 |
3.7 |
12.1 |
I, III, IV, V |
Flt-2L2 |
| |
QHei.S42IL-3H |
S42IL-115 |
3H |
204.5 - 255.1 |
ALL+N25 |
36.6 |
33.0 |
3.6 |
10.9* |
I, IV, V |
sdw16 |
| |
QHei.S42IL-4H |
S42IL-121 |
4H |
074.1 - 119.1 |
ALL+N25 |
35.4 |
33.0 |
2.4 |
7.3* |
II, V |
|
| |
|
S42IL-123 |
4H |
128.9 - 172.3 |
ALL+N25 |
35.5 |
33.0 |
2.6 |
7.8* |
V |
|
| |
QHei.S42IL-5H |
S42IL-126 |
5H |
145.6 - 200.1 |
N25 |
34.6 |
30.6 |
4.1 |
13.3 |
|
ari-e.GP7 |
| |
QHei.S42IL-6H |
S42IL-148 |
6H |
003.3 - 010.7 |
ALL |
35.4 |
33.0 |
2.5 |
7.5 |
|
|
| |
QHei.S42IL-7H.a |
S42IL-133 |
7H |
017.3 - 051.9 |
ALL+N100 |
30.0 |
33.0 |
−3.0 |
−9.1* |
|
brh19 |
| |
|
S42IL-134 |
7H |
051.9 - 107.4 |
N100 |
30.9 |
35.4 |
−4.5 |
−12.8* |
I - V |
brh19 |
| |
QHei.S42IL-7H.b |
S42IL-135 |
7H |
101.2 - 152.3 |
ALL+N25 |
36.1 |
33.0 |
3.2 |
9.6* |
I, IV, V |
|
| |
|
S42IL-136 |
7H |
134.4 - 152.3 |
ALL+N25 |
35.7 |
33.0 |
2.8 |
8.4* |
I, V |
|
| |
|
S42IL-137 |
7H |
134.4 - 193.9 |
ALL+N25 |
37.2 |
33.0 |
4.2 |
12.8* |
I, II, V |
|
| |
QHei.S42IL-7H.c |
S42IL-138 |
7H |
176.4 - 229.7 |
ALL+N25 |
35.9 |
33.0 |
2.9 |
8.9* |
|
|
| |
|
S42IL-139 |
7H |
198.7 - 229.7 |
N25 |
34.5 |
30.6 |
3.9 |
12.9 |
|
|
| SDW |
QSdw.S42IL-3H |
S42IL-115 |
3H |
204.5 - 255.1 |
N25 |
104.1 |
65.8 |
38.3 |
58.2 |
I |
|
| |
QSdw.S42IL-4H |
S42IL-121 |
4H |
074.1 - 119.1 |
N25 |
100.7 |
65.8 |
34.9 |
53.0 |
I, VI |
|
| |
QSdw.S42IL-7H |
S42IL-136 |
7H |
134.4 - 152.3 |
N25 |
101.1 |
65.8 |
35.3 |
53.6 |
VI |
|
| |
|
S42IL-137 |
7H |
134.4 - 193.9 |
N25 |
102.0 |
65.8 |
36.2 |
55.0 |
VI |
|
| RL |
QRl.S42IL-1H.a |
S42IL-141 |
1H |
094.9 - 127.7 |
ALL+N100 |
37.3 |
33.0 |
4.3 |
13.0 |
|
|
| |
QRl.S42IL-1H.b |
S42IL-143 |
1H |
130.7 - 173.5 |
ALL+N100 |
27.9 |
33.0 |
−5.1 |
−15.5* |
|
|
| |
QRl.S42IL-1H.c |
S42IL-142 |
1H |
188.5 - 205.1 |
N25 |
40.0 |
33.8 |
6.2 |
18.2 |
VII |
|
| |
QRl.S42IL-2H |
S42IL-106 |
2H |
022.4 - 034.3 |
ALL+N100 |
37.8 |
33.0 |
4.8 |
14.6 |
|
|
| |
|
S42IL-107 |
2H |
034.3 - 066.8 |
ALL+N100+N25 |
39.3 |
33.0 |
6.3 |
19.0* |
|
|
| |
QRl.S42IL-3H |
S42IL-113 |
3H |
204.5 - 239.7 |
N25 |
40.2 |
33.8 |
6.3 |
18.7 |
|
sdw16 |
| |
QRl.S42IL-4H.a |
S42IL-121 |
4H |
074.1 - 119.1 |
N25 |
41.1 |
33.8 |
7.2 |
21.4* |
|
|
| |
QRl.S42IL-4H.b |
S42IL-124 |
4H |
171.3 - 183.5 |
ALL+N100+N25 |
38.7 |
33.0 |
5.6 |
17.0* |
|
|
| |
QRl.S42IL-5H |
S42IL-126 |
5H |
145.6 - 200.1 |
N100 |
25.5 |
32.2 |
−6.7 |
−20.8* |
|
ari-e.GP7 |
| |
QRl.S42IL-6H.a |
S42IL-148 |
6H |
003.3 - 010.7 |
ALL+N25 |
38.1 |
33.0 |
5.0 |
15.2* |
|
|
| |
QRl.S42IL-6H.b |
S42IL-128 |
6H |
071.4 - 132.2 |
ALL+N100+N25 |
39.9 |
33.0 |
6.9 |
20.9* |
|
|
| |
|
S42IL-129 |
6H |
073.9 - 133.5 |
ALL+N100+N25 |
38.4 |
33.0 |
5.4 |
16.4* |
|
|
| |
|
S42IL-130 |
6H |
098.7 - 180.7 |
ALL+N100+N25 |
39.5 |
33.0 |
6.5 |
19.7* |
|
|
| |
QRl.S42IL-7H.a |
S42IL-133 |
7H |
017.3 - 051.9 |
ALL+N100+N25 |
27.2 |
33.0 |
−5.8 |
−17.7* |
|
brh19 |
| |
|
S42IL-134 |
7H |
051.9 - 107.4 |
ALL |
28.1 |
33.0 |
−5.0 |
−15.1 |
|
brh19 |
| |
QRl.S42IL-7H.b |
S42IL-135 |
7H |
101.2 - 152.3 |
ALL+N100+N25 |
41.5 |
33.0 |
8.5 |
25.7* |
|
|
| RDW |
QRdw.S42IL-1H.a |
S42IL-102 |
1H |
001.1 - 098.2 |
ALL+N100 |
45.1 |
33.9 |
11.2 |
33.0 |
|
|
| |
QRdw.S42IL-1H.b |
S42IL-143 |
1H |
130.7 - 173.5 |
N100 |
31.2 |
43.5 |
−12.3 |
−28.4 |
|
|
| |
QRdw.S42IL-2H |
S42IL-109 |
2H |
064.0 - 110.8 |
ALL+N100+N25 |
48.1 |
33.9 |
14.2 |
41.9* |
|
|
| LRR |
QLrr.S42IL-2H |
S42IL-107 |
2H |
034.3 - 066.8 |
ALL+N100 |
0.6 |
0.8 |
−0.2 |
−23.7* |
|
|
| |
QLrr.S42IL-5H |
S42IL-126 |
5H |
145.6 - 200.1 |
ALL+N100 |
0.9 |
0.8 |
0.2 |
24.1* |
|
ari-e.GP7 |
| |
QLrr.S42IL-6H |
S42IL-128 |
6H |
071.4 - 132.2 |
ALL+N100 |
0.6 |
0.8 |
−0.2 |
−20.9* |
|
|
| |
QLrr.S42IL-7H |
S42IL-135 |
7H |
101.2 - 152.3 |
N100 |
0.7 |
0.8 |
−0.2 |
−21.0 |
|
|
| SRR |
QSrr.S42IL-1H |
S42IL-143 |
1H |
130.7 - 173.5 |
ALL+N100+N25 |
3.9 |
2.9 |
1.0 |
35.1* |
|
|
| |
QSrr.S42IL-2H |
S42IL-107 |
2H |
034.3 - 066.8 |
ALL+N100 |
3.4 |
2.9 |
0.5 |
16.3 |
|
|
| |
QSrr.S42IL-4H |
S42IL-123 |
4H |
128.9 - 172.3 |
N100 |
3.6 |
3.0 |
0.6 |
18.4* |
|
|
| |
QSrr.S42IL-5H |
S42IL-126 |
5H |
145.6 - 200.1 |
ALL+N100 |
3.3 |
2.9 |
0.4 |
13.5* |
|
ari-e.GP7 |
| QSrr.S42IL-7H | S42IL-133 | 7H | 017.3 - 051.9 | ALL+N100+N25 | 3.8 | 2.9 | 0.9 | 30.2* |
a Trait abbreviations are given in Table 2.
b Chromosomal location and extent (in centiMorgan) of the target introgression according to Schmalenbach et al. [19].
c Significant (P < 0.01) line effect across treatments (ALL), under high N treatment (N100) or under low N treatment (N25), determined by the Dunnett test.
d Least squares means of the S42IL and 'Scarlett', respectively. If the S42IL showed a significant association across treatments, the LSMeans across treatments (ALL) is given. If the S42IL showed a significant association solely in N100 or in N100 and N25, respectively, the LSMeans of N100 is given. If the S42IL showed a significant effect solely under N25, the LSMeans of N25 is given.
e Difference between the LSMeans of the S42IL and the LSMeans of 'Scarlett'.
f Relative trait performance of the S42IL compared to 'Scarlett', calculated as RP(IL) = 100 × [LSMeans(S42IL) – LSMeans(‘Scarlett’)]/LSMeans(‘Scarlett’). Highly significant (P < 0.001) trait associations of a QTL are labelled with an asterisk.
g Corresponding QTL in S42 or S42IL published in: I von Korff et al. [45], II Schmalenbach et al. [14], III Wang et al. [18], IV von Korff et al. [44], V Saal et al. [22], VI Schnaithmann and Pillen [23], VII Naz et al. [16].
h Candidate genes published in: 1 Franckowiak and Konishi 1996, cited in [46], 2 Chen et al. [47], 3 Laurie et al. [49], 4 Yan et al. [48], 5 Faure et al. [58], 6 Laurie et al. [54], 7 Chloupek et al. [56], 8 von Wettstein 1993, cited in [61], 9 Li et al. [57].
With regard to the position and extent of the corresponding target introgressions, all associations were summarised to 58 QTL. Among the nine traits, root length, plant height and leaf length showed the highest number of QTL effects (twelve, eleven and ten QTL, respectively). On the contrary, root and shoot dry weight were the traits that revealed only three and two QTL effects, respectively. For 43 QTL (74%), the Hsp allele showed an increasing trait effect compared to ‘Scarlett’. In the following paragraphs, the results shown in Table 5 are described in detail for each of the nine traits.
Shoot traits
A total of five S42ILs showed significant line by trait associations for tiller number (TN). The associations were summed up to four QTL, located on chromosomes 1H, 4H, 6H and 7H. At QTn.S42IL-4H the line effects were detected across treatments whereas the other QTL showed only effects under N100. All QTL were due to a higher trait performance of the S42IL compared to ‘Scarlett’, with a maximum increase of 0.5 more tillers per plant (30.7%) by S42IL-123 across treatments (QTn.S42IL-6H).
For leaf number (LN), seven S42ILs significantly differed from ‘Scarlett’. The effects were summarised to six QTL, which were located on chromosomes 2H, 4H, 6H and 7H. Two QTL each were detected across treatments, under N100 and under N25, respectively. At all QTL the S42ILs showed an increase in LN. S42IL-109 showed the maximum difference to ‘Scarlett’ at QLn.S42IL-2H with 0.9 (28.3%) more leaves per plant under N25.
There were 15 S42ILs that were significantly associated to leaf length (LL). The associations were subsequently summarised to 10 QTL. Target introgressions of the corresponding S42ILs were located on all seven barley chromosomes. Two and three QTL showed significantly different trait performances of the S42IL solely under N25 and N100, respectively. Five QTL included lines with relatively shorter leaves than Scarlett. At QLl.S42IL-1H.b, S42IL-143 revealed the maximum decreasing effect compared to ‘Scarlett’ under N100 with -3.4 cm (-14.3%). The Hsp introgression of S42IL-136 caused the maximum elongation of the youngest leaf with 4.2 cm (18.9%) at QLl.S42IL-7H.b.
The Dunnett test revealed 17 lines that were significantly associated with plant height (HEI). Altogether eleven QTL from all seven barley chromosome were defined. One QTL effect was only found under N25. Two QTL were due to relatively lower plant height of the corresponding S42ILs. As shown for LL, S42IL-143 revealed the maximum decreasing effect compared to ‘Scarlett’ across treatments with -6.0 cm (-18.1%, QHei.S42IL-1H.b). At QHei.S42IL-7H.c, S42IL-126 had the maximum increasing effect compared to ‘Scarlett’ under N25 with 4.1 cm (13.3%).
Due to low heritabilities with h2 < 10% (see Table 3), QTL detection for shoot dry weight (SDW) was only conducted within the N25 treatment. There, four S42ILs differed significantly from ‘Scarlett’. The effects were summarised to three QTL, which were located on chromosomes 3H, 4H and 7H. The QTL showed an increase in trait performance of the S42ILs revealing an up to 58.2% higher SDW.
Root traits
A total of 16 lines showed significantly different root lengths (RL) compared to ‘Scarlett’. The associations were summed up to twelve QTL, located on all barley chromosomes. Three QTL effects, on chromosomes 1H, 2H and 3H, were only found under N25. At QRl.S42IL-5H the strongest decreasing QTL effect was found for S42IL-126 under N100 with -6.7 cm (-20.8%). S42IL-121 showed the strongest increasing QTL effect at QRl.S42IL-4H.a with a 7.2 cm (21.4%) longer root compared to ‘Scarlett’.
Three S42ILs showed significant line by trait associations for root dry weight (RDW). According to this, three QTL were defined, located on chromosomes 1H and 2H. At QRdw.S42IL-1H.b the line effect was solely present under N100. This QTL caused a lower trait performance of S42IL-143 compared to ‘Scarlett’, with a decrease of -12.3 mg (-28.4%). The maximum increase was found at QRdw.S42IL-2H with a plus of 14.2 mg (41.9%).
Shoot to root trait ratios
For leaf to root ratio (LRR), four S42ILs differed significantly from ‘Scarlett’. These effects corresponded to four QTL, located on chromosomes 2H, 5H, 6H and 7H. QLrr.S42IL-7H was only detected under N100. The QTL on chromosome 5H, QLrr.S42IL-5H, was the only one with an increase in the trait performance by 0.2 (24.1%). S42IL-107 showed the maximum decline compared to ‘Scarlett’ at QLrr.S42IL-2H with -0.2 (-23.7%).
A total of five S42ILs showed a significantly different trait performance from ‘Scarlett’ for shoot to root ratio (SRR). The effects corresponded to five QTL, located on chromosomes 1H, 2H, 4H, 5H and 7H. QSrr.S42IL-4H was only detected under N100. All QTL revealed an increase in trait performance of the S42ILs. S42IL-143 showed the maximum increase compared to ‘Scarlett’ at QSrr.S42IL-1H with 1.0 (35.1%).
Chlorophyll content
Due to low heritabilities with h2 < 10% (see Table 3), no QTL detection was carried out for chlorophyll content (CC).
Discussion
Suitability of the hydroponic system for early selection of favourable S42ILs
In 2008 and 2011, a set of 42 introgression lines was tested in six hydroponic experiments under two different N treatments. The fixation of the plants in Eppendorf tubes during growth in the hydroponic system allowed us to analyse shoots and roots simultaneously. Huang et al. [31], who used a similar plant fixation in their hydroponic system, also highlighted the flexibility of those systems. Also other studies reported that plant responses to low N availability vary among genotypes, developmental stages and plant organs [32]. Regarding the N treatments, we did not observe any symptoms of starvation on the plants under full supply condition, which indicates that the macro and micro nutrients were sufficiently supplied throughout the experiment [33]. In contrast, the reduction of N supply caused a severe reaction in most traits studied. For example, root length was increased while plant height was decreased under N deficiency (Table 3).
Taking the parameter heritability as a quality measure of genetic to phenotypic variation, we found that the phenotypic data collected in our hydroponic system predominantly revealed strong and medium heritabilities with h2 > 70% for five traits and 70% >h2 > 30% for two traits, respectively. Shoot dry weight (ALL and N100) and chlorophyll content (ALL, N100 and N25) have to be considered as not heritable with h2 > 10% (Table 3). In this regard, the mixed model analysis of variance was not carried out for the respective treatments of those traits.
For the other traits, the phenotypic data collected from the hydroponic experiments were subsequently analysed in a line by trait association study. In total, 12 out of 58 QTL effects (21%) that we found with juvenile S42ILs in the hydroponic system corresponded to QTL for comparable traits that were detected in previous field and greenhouse studies with adult plants of a similar S42IL set or with the original S42 population (Table 5). The corresponding QTL predominantly controlled the traits plant height (8 x) and, to a lesser extent, tiller number (1 x) and root length (1 x). Both findings, the medium to strong heritabilities observed and the high amount of corresponding QTL, support the idea to use the hydroponic system for early mapping of QTL and early selection of the QTL-bearing genotypes in plant physiology experiments and in plant breeding.
In the following paragraphs, we discuss the detection of juvenile plant growth QTL in wild barley ILs in general and in regard to N deficiency. Finally, we will draw connections between our QTL results, candidate genes and previously described QTL.
Detection of juvenile plant development QTL
The major advantage of growing juvenile barley plants is the fast collection of phenotypic data, for instance within 14 days in our hydroponic system. This time-, cost- and effort-reducing aspects are of major importance for plant breeding where the reduction of time for phenotypic assessment can substantially improve the breeding process because new varieties may be introduced earlier and at lower selection costs to the market. Furthermore, the hydroponic system allows to simultaneously study the development of barley root and shoot traits [32]. Mattsson et al. [34], for instance, could show that in barley root relative growth rate increased while root dry weight decreased at the same time. In addition, the reaction of nutrient deficiencies on barley growth has often been carried out with juvenile plants [24,27,35]. We also conducted a juvenile hydroponic experiment to map QTL that control barley root and shoot traits in regard to N supply.
With 126 significant (P < 0.01) line by trait associations for nine traits under study, the number of QTL detected in the hydroponics experiment was higher than in any other study that was carried out with adult plants of the S42IL population. For example, Schmalenbach et al. [14] found 65 associations for seven field agronomical traits in the S42IL population. The increase in the number of associations may be attributed to the different screening methods (hydroponic vs. field testing), the different developmental stages (juvenile vs. adult plants), different traits (shoot and root vs. agronomic traits) and the varying N levels, as already reported for the S42 population by Saal et al. [22].
The highest number of QTL was found for the length parameters leaf length, plant height and root length. The three traits showed high heritabilities of 75.4%, 80.8% and 85.2% across both treatments, respectively. Trait heritability is known to influence the number and the probability of detected QTL. For traits with heritabilities of h2 < 10%, Ellis et al. [35] did not detect any QTL.
The QTL we mapped were scattered across the whole genome (Figure 1). We detected the highest number of QTL effects on chromosome arm 7HL. Here, QTL were found for all traits except root dry weight and shoot to root ratio (Figure 1). In the same chromosomal region QTL effects for plant height and shoot dry weight where already described in studies of the S42IL and S42 population (Table 5). In this regard, genes located on chromosome arm 7HL may be particularly important for regulation of plant growth.
At 75% of all detected QTL, the S42ILs showed increasing Hsp effects (Table 5). In contrast, when studying agronomic traits with S42ILs in the field, only 47% of the detected associations showed increasing Hsp effects [14]. Since most of the traits measured are related to a fast growth during the juvenile phase, this finding was expected. It may be explained by the fact that the donor of the S42ILs, the Hsp accession ‘ISR42-8’, is well adapted to survival under low nutrient supply. Those S42ILs that reveal QTL effects compared to the elite barley cultivar ‘Scarlett’ may have, thus, contributed exotic Hsp alleles that speed up or increase juvenile growth. Under high N treatment an increase of the traits we evaluated is generally desirable.
A fast juvenile plant development is of great importance, especially when plants are grown under field conditions. Baethgen et al. [24] reported that the application of a high dosage of N early in the growing season stimulated tiller formation of malting barley. However, many of these tillers did not develop fertile ears and, thus, the extra dosage of N fertilizer was wasted. In addition, for malting barley one has to consider that a high N supply during grain filling may be conflicting with grain quality [25]. On the other hand, if plants are cultivated under low N supply, many tillers may also be undesirable since plants may not be able to produce ears and fill the grains for all of the tillers. Multivariate analyse methods like the principle component analysis, are important statistical tools for primary assessment of data structure and, in addition, may support to identify the most important traits for phenotypic selection. However, our primary aim was to locate QTL early on under control and N starvation conditions in a hydroponic system. Thus, the difference in trait performance of the S42ILs compared to ‘Scarlett’ was of greatest interest to us. Due to space and time limitations, a thorough multivariate analysis will be delayed to a follow up study where the most promising traits and, in addition, favourable QTL alleles, will be used for genotype selection. Subsequently, the potential selection gain will be measured within the selected offspring generation.
QTL and N deficiency
Twenty-five of the 42 associations we found under N25 (Model 2) were simultaneously present across treatments (Model 1, see Table 5). Jana and Wilen [36] recommended that it is optimal, when lines that are good under non stress conditions, also perform well under stress conditions. This may be of great importance for breeders, who do not have to conduct two parallel breeding programs, and for farmers, who do not have to select different varieties based on expected N stress conditions. On the other hand, selection of breeding lines in stress environments may result in genetic gains by using adapted germplasm, especially in regard to originally low-input crops like malting barley [37]. However, our results indicated that a number of QTL effects were not in common between both N treatments. Sixteen and 17 QTL were solely detected under N100 or N25, respectively. In one case, the direction of the exotic QTL effect was actually opposed, decreasing leaf length under high N supply while increasing leaf length under low N supply (see Table 5, QLl.S42IL-3H.a and QLl.S42IL-3H.b, present in S42IL-113 and-115, respectively). We, thus, conclude that it may be worth to select barley cultivars for N stress tolerance separately from experiments under low N and high N fertilization.
Associations to tiller number and leaf number were detected across treatments, under N100 and under N25. Similar to our results, Andersen [38] also found an increase in the number of leaves under low N treatment in juvenile and adult plants. For leaf length, two QTL where found in the same region of chromosome 3H. The decreasing effect at QLl.S42IL-3H.a and the increasing effect at QLl.S42IL-3H.b were detected under N100 and N25, respectively. Our results suggest that the gene effect on reducing leaf length is reversed under N deficiency. Shoot dry weight of the S42ILs was increased by up to 58.2% under N25 (Table 3), which was already observed by Marshall and Ellis [25] (Table 5).
Regarding plant development, root characteristics are of great importance, which is especially shown when plants are grown under stress conditions [39]. Most root length QTL showed an increase of trait performance under N25, which may indicate that the S42ILs try to react to N starvation by increasing their capacity to take up N from the solution. In contrast, root dry weight showed a mean reduction of approximately 30% compared to the control treatment, which was also shown in various other studies [28,40], and only two increasing QTL were found for this trait. In addition, Karley et al. [27] demonstrated in barley that trait differences between the N treatments increased with advancing growth stages, except for root parameters. The authors reported a lack of genotype by N supply interaction for root traits and assumed a limited potential for exploiting genetic variation to improve barley root performance [27]. However, this finding is in contrast to our study, where we detected a substantial number of root-related QTL effects. Also Naz et al. [16] reported a severe Hsp QTL effect on chromosome 5H that is present in a S42IL and caused an substantial increase in root biomass, both, under drought and controlled water conditions. Unfortunately, this particular line, S42IL-176, was not included in the set of S42ILs that we studied under hydroponics.
We did not detect leaf to root ratio or shoot to root ratio QTL solely under low N supply. Also Bahrman et al. [32] did not detect variety by N level effects in the shoot to root ratio of eight week old wheat plants. We conclude that in order to detect a change in shoot to root ratio, plants may have to be under stress for a longer time [41].
Because of the missing heritability for chlorophyll content we did not conduct the analysis of variance for this trait. In our experiments genetic effects on chlorophyll content were, thus, not stable across or within treatments. This finding is in accordance with Cartelat et al. [42], who reported that the change in chlorophyll content was reflecting the N nutrition status, but showed no direct association to genotype, growth stage or environment, respectively.
Comparison of our results with known candidate genes and QTL
As highlighted before, we revealed more QTL than in any other QTL study conducted with the S42IL or S42 populations, indicating that new genes may have been expressed in our hydroponic study. Furthermore, comparing trait performances of coinciding S42ILs, analysed simultaneously in our study and in field and greenhouse studies [14,23], revealed low correlations, for example between juvenile and adult plant height (r < 0.08, data not shown) as well as between juvenile and adult number of tillers and ears, respectively (r < 0.12, data not shown). However, in Table 5 altogether 12 QTL for tiller number, plant height, shoot dry weight and root length under hydroponics corresponded to previously detected field and greenhouse QTL for number of ears, plant height, biomass and root length [14,16,18,22,23,43,44]. It remains open if the same genes in the respective S42ILs caused the QTL effects under hydroponic and field or greenhouse cultivation. Alternatively, it may also be possible that linked Hsp alleles that are present on the same introgression may have caused the QTL effects in the independent studies. To further elaborate on this question, interesting QTL effects should be narrowed down and validated in high-resolution offspring lines that are derived from the original S42ILs and segregate for the detected QTL effects [19].
In the following paragraphs, we attempt to draw connections between 12 significant Hsp effects and already described candidate genes and QTL effects.
Andersen [38] reported that juvenile tiller number is highly correlated with adult plant ear number, which is also an indicator for yield potential. We mapped one tiller number QTL on chromosome 4H that was consistent with two previously detected QTL for number of ears in population S42 [22,45]. At QTn.S42IL-7H, S42IL-135 revealed a strong increase in tiller number. Ellis et al. [35] also found a strong increasing effect on tiller number in the same region of 7HL with barley seedlings, too.
The number of leaves influences the photosynthetic capacity of the plant. Thus, growth, development and yield may be co-regulated through the control of leaf number. Cuesta-Marcos et al. [46] estimated the number of leaves until heading in barley lines and found a significant QTL effect nearby the earliness per se locus Eam6 [Franckowiak and Konishi 1995, cited in 46]. We also found a QTL in this chromosomal region with S42IL-109 under N25 at QLn.S42IL-2H. In addition, the Flt2L locus, controlling flowering time and plant height, maps to the same region [47] and may be present in S42IL-110. Furthermore, VRNH2 and VRNH3, two major genes determining the requirement for vernalisation, map to the same chromosomal regions where we found leaf number QTL in the S42IL population [18,48-50]. Their influences on growth related traits [18,51] may be due to primary effects on juvenile leaf number as it was already shown for the final leaf number in wheat [52].
We detected one decreasing and one increasing Hsp effect for leaf length on chromosome 3H under N100 and N25, respectively. Similar to this, Gregory et al. [53] described a reduction in barley shoot length under control conditions and associated the effect with the candidate gene for dwarfism on chromosome 3HL: sdw1[54,55]. Saal et al. [22] also detected two Hsp effects on total plant height under low and high N treatment on 3HL, but there was no difference between the directions of both effects. Thus, we conclude that sdw1 controlled juvenile shoot development in our study; however, plant reaction depends on the status of N supply. On chromosomes 5H and 7H two additional dwarfing genes are located: arie.GP and brh1. At both loci, we also detected QTL for leaf length. Chloupek et al. [56] already described that arie.GP has pleiotropic effects on plant growth in general, including shoot development. However, the arie.GP allele is a very rare allele in the elite barley gene pool, making it unlikely that ‘Scarlett’ carries this dwarfing gene. In addition, brh1[57] was associated with strong effects on plant height and leaf length in a barley backcross population [51]. S42IL-133 and -134, probably carrying brh1, showed Hsp effects of the same direction on leaf length.
We verified 33 QTL, previously described for adult plant height in studies with the S42 or S42IL population, with twelve lines associated to juvenile plant height (Table 5). In addition, we found candidate genes influencing barley shoot development, which may explain the Hsp effects. On chromosome 1H for example, S42IL-143 caused the strongest decreasing effect on plant height. QTL of the same direction were described with the S42 population [22,45] and the decreasing Hsp effect on adult plant height at the HvFT3 locus [58] on chromosome 1HL was already described [18]. Besides, the three dwarfing genes on chromosomes 3H, 5H and 7H revealed strong effects on plant height in our experiments. The effects are similar to those described for leaf length and are discussed in detail in the previous paragraph. Additionally, for each of the three dwarfing genes we refer to Hsp effects described in previous studies carried out with adult plants of the S42IL or S42 population.
On chromosome 4H, an increasing Hsp effect on straw weight was described by Schnaithmann and Pillen [23] which is consistent with our effect QSdw.S42IL-4H. Von Korff et al. [45] also detected a biomass effect on chromosome 4H.
Semi-dwarf genes are known to have a major impact on aerial plant organs [54,55] but their effects on root traits was also affirmed in the literature [56]. In this regard, Wojciechowski et al. [59] found differences in wheat root length due to the dwarfing-gene Rht. With our data, we found two contrasting root length QTL in the chromosomal regions of the barley semi-dwarfing genes sdw1 and arie.GP on chromosomes 3H and 5H, respectively. At both loci, contrasting effects for several shoot and root parameters like stabile isotope discrimination for C and N were reported [35,56]. We assume that the Hsp alleles at the dwarfing-gene loci may also have caused the root length QTL.
The number of QTL effects detected for the leaf to root ratio was relatively low compared to the traits it was derived from. Wang et al. [60] demonstrated that QTL for mathematically derived traits are harder to detect than the component traits they are derived from.
Conclusion
In general, the verification of a QTL is interpreted as the stability of its effect across genotypes or environments. As our data suggest, the verification of a QTL may also be interpreted as the stability of its effect across developmental stages. In total, 12 QTL effects we reported for juvenile S42ILs in the hydroponic system corresponded to QTL previously described in field and greenhouse studies conducted with the S42ILs or the parent population S42. S42IL-135, -136 and -137 revealed multiple increasing Hsp effects on the long arm of chromosome 7H. So far, we could not associate the QTL with any candidate gene. We therefore aim for further studies with high-resolution populations derived from those three lines, to fine-map the QTL affecting juvenile plant development as demonstrated in Schmalenbach et al. [19]. Additionally, there were several interesting QTL solely detected under low N treatment, for example, close to the dwarfing gene arie.GP on chromosome 5H. To conclude, our data indicate that the field performance of an adult barley genotype may be predictable by examining its juvenile development under hydroponic conditions. Thus, hydroponic studies, in particular to select genotypes tolerant to abiotic stresses, have a high potential to support modern plant breeding programs. In this regard, future fine mapping of the detected QTL in a hydroponic system, by means of phenotyping the already existing HR populations, which are derived from the original S42ILs, may be of great benefit both to breeders and plant physiologists in order to speed up selection of target phenotypes. The aforementioned three S42ILs on chromosome 7H may be excellent starting points for this endeavour.
Abbreviations
ALL: Across treatments; HR: High-resolution; Hsp: Hordeum vulgare ssp. spontaneum; Hv: Hordeum vulgare ssp. vulgare; LSMeans: Least squares means; N: Nitrogen; N100: 100% N, i.e. control treatment; N25: 25% N, i.e. low N treatment; RP: Relative performance of an S42IL compared to 'Scarlett'; S42: BC2DH population derived from the cross 'Scarlett' x ISR42-8; S42IL: Introgression line derived from S42.
Competing interests
The work was funded by the federal state of Saxony-Anhalt (FKZ: 3651A/0808, PI: Klaus Pillen).
Authors' contributions
AH carried out the experiments in 2008, performed the joint data analysis and contributed substantially to writing the manuscript. AM carried out the experiments in 2011 and performed the data analysis in 2011. KP conceived the idea, supervised the project, contributed to data analysis and to writing the manuscript. All authors read and approved the final manuscript.
Contributor Information
Astrid Hoffmann, Email: astrid.hoffmann@landw.uni-halle.de.
Andreas Maurer, Email: andreas.maurer@landw.uni-halle.de.
Klaus Pillen, Email: klaus.pillen@landw.uni-halle.de.
Acknowledgements
We would like to express our gratitude to Andrea Lossow, Max-Planck-Institute for Plant Breeding Research, Cologne, for introducing the hydroponic system to Astrid Hoffmann, to Dr. Wilfried Ilse for assistance in setting up the hydroponic system in Halle and to Florian Schnaithmann for assistance in running the hydroponic experiments.
References
- Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F. On the origin and domestication history of barley (Hordeum vulgare) Mol Biol Evol. 2000;17(4):499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- von Bothmer R. In: Barley: Genetics, Biochemistry, Molecular Biology, and Biotechnology. Shewry P, editor. Wallingford: CAB Internationa; 1992. The wild species of Hordeum: relationships and potential use for improvement of cultivated barley; pp. 3–18. [Google Scholar]
- Ellis RP, Forster BP, Robinson D, Handley LL, Gordon DC, Russell JR, Powell W. Wild barley: a source of genes for crop improvement in the 21st century? J Exp Bot. 2000;51(342):9–17. doi: 10.1093/jexbot/51.342.9. [DOI] [PubMed] [Google Scholar]
- Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004;2(10):1610–1615. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63(1–2):141–152. [Google Scholar]
- Tanksley SD, Nelson JC. Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet. 1996;92(2):191–203. doi: 10.1007/BF00223376. [DOI] [PubMed] [Google Scholar]
- Zamir D. Improving plant breeding with exotic genetic libraries. Nat Rev Genet. 2001;2(12):983–989. doi: 10.1038/35103589. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Vries HBD, Effgen S, Vreugdenhil D, Koornneef M. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175(2):891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalma SJ, Hostert BM, LeDeaux JR, Stuber CW, Holland JB. QTL mapping with near-isogenic lines in maize. Theor Appl Genet. 2007;114(7):1211–1228. doi: 10.1007/s00122-007-0512-6. [DOI] [PubMed] [Google Scholar]
- Liu SB, Zhou RG, Dong YC, Li P, Jia JZ. Development, utilization of introgression lines using a synthetic wheat as donor. Theor Appl Genet. 2006;112(7):1360–1373. doi: 10.1007/s00122-006-0238-x. [DOI] [PubMed] [Google Scholar]
- Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Ying CS, Luo LJ. QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativaL.) Theor Appl Genet. 2006;112(4):648–656. doi: 10.1007/s00122-005-0167-0. [DOI] [PubMed] [Google Scholar]
- von Korff M, Wang H, L on J, Pillen K. Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor Appl Genet. 2004;109(8):1736–1745. doi: 10.1007/s00122-004-1818-2. [DOI] [PubMed] [Google Scholar]
- Schmalenbach I, Körber N, Pillen K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet. 2008;117(7):1093–1106. doi: 10.1007/s00122-008-0847-7. [DOI] [PubMed] [Google Scholar]
- Schmalenbach I, L on J, Pillen K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theor Appl Genet. 2009;118(3):483–497. doi: 10.1007/s00122-008-0915-z. [DOI] [PubMed] [Google Scholar]
- Sayed M, Schumann H, Pillen K, Naz A, Leon J. AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgareL.) BMC Genet. 2012;13(1):61. doi: 10.1186/1471-2156-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz AA, Ehl A, Pillen K, L on J. Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgareL.) Plant Breeding. 2012;3:392–398. [Google Scholar]
- Schmalenbach I, Pillen K. Detection and verification of malting quality QTLs using wild barley introgression lines. Theor Appl Genet. 2009;118(8):1411–1427. doi: 10.1007/s00122-009-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Schmalenbach I, von Korff M, Leon J, Kilian B, Rode J, Pillen K. Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet. 2010;120(8):1559–1574. doi: 10.1007/s00122-010-1276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I, March TJ, Bringezu T, Waugh R, Pillen K. High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the Illumina GoldenGate assay. G3: Genes, Genomes, Genetics. 2011;1(3):187–196. doi: 10.1534/g3.111.000182. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E, Bloom A. Mineral nutrition of plants 2. Principles and Perspectives. Sunderland: Sinauer; 2005. [Google Scholar]
- Mickelson S, See D, Meyer FD, Garner JP, Foster CR, Blake TK, Fischer AM. Mapping of QTL associated with nitrogen storage and remobilization in barley (Hordeum vulgare L.) leaves. J Exp Bot. 2003;54(383):801–812. doi: 10.1093/jxb/erg084. [DOI] [PubMed] [Google Scholar]
- Saal B, von Korff M, L on J, Pillen K. Advanced-backcross QTL analysis in spring barley: IV. Localization of QTL × nitrogen interaction effects for yield-related traits. Euphytica. 2011;177(2):223–239. doi: 10.1007/s10681-010-0252-6. [DOI] [Google Scholar]
- Schnaithmann F, Pillen K. Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines. Euphytica. 2012. [DOI]
- Baethgen WE, Christianson CB, Lamothe AG. Nitrogen fertilizer effects on growth, grain yield, and yield components of malting barley. Field Crop Res. 1995;43(2–3):87–99. [Google Scholar]
- Marshall B, Ellis RP. Growth, yield and grain quality of barley (Hordeum vulgare L.) in response to nitrogen uptake I. A low cost, controlled nutrient supply system. J Exp Bot. 1998;49(323):1049–1057. [Google Scholar]
- Ellis RP, Marshall B. Growth, yield and grain quality of barley (Hordeum vulgare L.) in response to nitrogen uptake - II. Plant development and rate of germination. J Exp Bot. 1998;49(323):1021–1029. [Google Scholar]
- Karley AJ, Valentine TA, Squire GR. Dwarf alleles differentially affect barley root traits influencing nitrogen acquisition under low nutrient supply. J Exp Bot. 2011;62(11):3917–3927. doi: 10.1093/jxb/err089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty PH, Anbessa Y, Juskiw P, Carroll RT, Wang JA, Good AG. Nitrogen use efficiencies of spring barley grown under varying nitrogen conditions in the field and growth chamber. Ann Bot. 2010;105(7):1171–1182. doi: 10.1093/aob/mcq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker H. Pflanzenzüchtung. Stuttgart: UTB; 2011. Volume 2. [Google Scholar]
- Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50(272):1096–1121. doi: 10.1080/01621459.1955.10501294. [DOI] [Google Scholar]
- Huang YZ, Zhang GP, Wu FB, Chen JX, Xiao YP. Interaction of salinity and cadmium stresses on antioxidant enzymes, sodium, and cadmium accumulation in four barley genotypes. J Plant Nutr. 2006;29(12):2215–2225. doi: 10.1080/01904160600974155. [DOI] [Google Scholar]
- Bahrman N, Gouy A, Devienne-Barret F, Hirel B, Vedele F, Le Gouis J. Differential change in root protein patterns of two wheat varieties under high and low nitrogen nutrition levels. Plant Sci. 2005;168(1):81–87. doi: 10.1016/j.plantsci.2004.07.035. [DOI] [Google Scholar]
- Le Bot J, Adamowicz S, Robin P. Modelling plant nutrition of horticultural crops: a review. Sci Hort. 1998;74(1–2):47–82. [Google Scholar]
- Mattsson M, Johansson E, Lundborg T, Larsson M, Larsson CM. Nitrogen-utilization in N-limited barley during vegetative and generative growth .1. Growth and nitrate uptake kinetics in vegetative cultures grown at different relative addition rates of nitrate-N. J Exp Bot. 1991;42(235):197–205. [Google Scholar]
- Ellis RP, Forster BP, Gordon DC, Handley LL, Keith RP, Lawrence P, Meyer R, Powell W, Robinson D, Scrimgeour CM. et al. Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J Exp Bot. 2002;53(371):1163–1176. doi: 10.1093/jexbot/53.371.1163. [DOI] [PubMed] [Google Scholar]
- Jana S, Wilen RW. In: Abiotic stress: plant resistance through breeding and molecular approaches. Ashraf M, Harris PJC, editor. Binghampton, NY: CRC; 2005. Breeding for abiotic stress tolerance in barley; pp. 491–511. [Google Scholar]
- Ceccarelli S. Adaptation to low/high input cultivation. Euphytica. 1996;92(1–2):203–214. [Google Scholar]
- Andersen S. Relation between leaf number and ear development in spring-sown barley and oats. Physiol Plantarum. 1955;8:404–417. doi: 10.1111/j.1399-3054.1955.tb08984.x. [DOI] [Google Scholar]
- Dunbabin V, Diggle A, Rengel Z. Is there an optimal root architecture for nitrate capture in leaching environments? Plant Cell Environ. 2003;26(6):835–844. doi: 10.1046/j.1365-3040.2003.01015.x. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Pettersson R, Paustian K. Shoot and root production and nitrogen uptake in barley, with and without nitrogen fertilization. J Agron Crop Sci. 1987;158(3):163–171. doi: 10.1111/j.1439-037X.1987.tb00258.x. [DOI] [Google Scholar]
- Chapin FS, Walter HS, Clarkson DT. Growth response of barley and tomato to nitrogen stress and its control by abscisic acid, water relations and photosythesis. Planta. 1988;173:352–366. doi: 10.1007/BF00401022. [DOI] [PubMed] [Google Scholar]
- Cartelat A, Cerovic ZG, Goulas Y, Meyer S, Lelarge C, Prioul JL, Barbottin A, Jeuffroy MH, Gate P, Agati G. et al. Optically assessed contents of leaf polyphenolics and chlorophyll as indicators of nitrogen deficiency in wheat (Triticum aestivum L.) Field Crop Res. 2005;91(1):35–49. doi: 10.1016/j.fcr.2004.05.002. [DOI] [Google Scholar]
- von Korff M, Wang H, Leon J, Pillen K. AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor Appl Genet. 2005;111(3):583–590. doi: 10.1007/s00122-005-2049-x. [DOI] [PubMed] [Google Scholar]
- von Korff M, L on J, Pillen K. Detection of epistatic interactions between exotic alleles introgressed from wild barley (Hordeum vulgare ssp. spontaneum) Theor Appl Genet. 2010;121(8):1455–1464. doi: 10.1007/s00122-010-1401-y. [DOI] [PubMed] [Google Scholar]
- von Korff M, Wang H, L on J, Pillen K. AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum) Theor Appl Genet. 2006;112(7):1221–1231. doi: 10.1007/s00122-006-0223-4. [DOI] [PubMed] [Google Scholar]
- Cuesta-Marcos A, Casas AM, Yahiaoui S, Gracia MP, Lasa JM, Igartua E. Joint analysis for heading date QTL in small interconnected barley populations. Mol Breed. 2008;21(3):383–399. doi: 10.1007/s11032-007-9139-1. [DOI] [Google Scholar]
- Chen A, Baumann U, Fincher GB, Collins NC. Flt-2L, a locus in barley controlling flowering time, spike density, and plant height. Funct Integr Genomic. 2009;9(2):243–254. doi: 10.1007/s10142-009-0114-2. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103(51):19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. RFLP mapping of 5 major genes and 8 quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome. 1995;38(3):575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Curr Opin Plant Biol. 2009;12(2):178–184. doi: 10.1016/j.pbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Dahleen LS, Vander Wal LJ, Franckowiak JD. Characterization and molecular mapping of genes determining semidwarfism in barley. J Hered. 2005;96(6):654–662. doi: 10.1093/jhered/esi125. [DOI] [PubMed] [Google Scholar]
- Wang SY, Ward RW, Ritchie JT, Fischer RA, Schulthess U. Vernalization in wheat II. Genetic variability for the interchangeability of plant age and vernalization duration. Field Crop Res. 1995;44(2–3):67–72. [Google Scholar]
- Gregory PJ, Bengough AG, Grinev D, Schmidt S, Thomas WTB, Wojciechowski T, Young IM. Root phenomics of crops: opportunities and challenges. Funct Plant Biol. 2009;36(10–11):922–929. doi: 10.1071/FP09150. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Romero C, Simpson E, Snape JW. Assignment of the denso dwarfing gene to the long arm of chromosome 3(3H) of barley by use of RFLP Markers. Plant Breed. 1993;111(3):198–203. doi: 10.1111/j.1439-0523.1993.tb00630.x. [DOI] [Google Scholar]
- Hellewell KB, Rasmusson DC, Gallo-Meagher M. Enhancing yield of semidwarf barley. Crop Sci. 2000;40(2):352–358. doi: 10.2135/cropsci2000.402352x. [DOI] [Google Scholar]
- Chloupek O, Forster BP, Thomas WTB. The effect of semi-dwarf genes on root system size in field-grown barley. Theor Appl Genet. 2006;112(5):779–786. doi: 10.1007/s00122-005-0147-4. [DOI] [PubMed] [Google Scholar]
- Li M, Kudrna D, Kleinhofs A. Fine mapping of a semi-dwarf gene brachytic1 in barley. Barley Genet Newsl. 2000;32:14–17. [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176(1):599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski T, Gooding MJ, Ramsay L, Gregory PJ. The effects of dwarfing genes on seedling root growth of wheat. J Exp Bot. 2009;60(9):2565–2573. doi: 10.1093/jxb/erp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li H, Zhang L, Lü W, Wang J. On the use of mathematically-derived traits in QTL mapping. Mol Breed. 2012;29(3):661–673. doi: 10.1007/s11032-011-9580-z. [DOI] [Google Scholar]
- Babb S, Muehlbauer GJ. Genetic and morphological characterization of the barley uniculm2 (cul2) mutant. Theor Appl Genet. 2003;106(5):846–857. doi: 10.1007/s00122-002-1104-0. [DOI] [PubMed] [Google Scholar]