Abstract
Oesophageal squamous cell carcinoma (OSCC) has a high prevalence in the Black and Mixed Ancestry populations of South Africa. Recently, three genome-wide association studies in Chinese populations identified five new OSCC susceptibility loci, including variants at PLCE1, C20orf54, PDE4D, RUNX1 and UNC5CL, but their contribution to disease risk in other populations is unknown. In this study, we report testing variants from these five loci for association with OSCC in the South African Black (407 cases and 849 controls) and Mixed Ancestry (257 cases and 860 controls) populations. The RUNX1 variant rs2014300, which reduced risk in the Chinese population, was associated with an increased risk of OSCC in the Mixed Ancestry population [odds ratio (OR) = 1.33, 95% confidence interval (CI) = 1.09–1.63, P = 0.0055], and none of the five loci were associated in the Black population. Since PLCE1 variants increased the risk of OSCC in all three Chinese studies, this gene was investigated further by sequencing in 46 Black South Africans. This revealed 48 variants, 10 of which resulted in amino acid substitutions, and much lower linkage disequilibrium across the PLCE1 locus than in the Chinese population. We genotyped five PLCE1 variants in cases and controls, and found association of Arg548Leu (rs17417407) with a reduced risk of OSCC (OR = 0.74, 95% CI = 0.60–0.93, P = 0.008) in the Black population. These findings indicate several differences in the genetic contribution to OSCC between the South African and Chinese populations that may be related to differences in their genetic architecture.
Introduction
Oesophageal cancer is the eighth most common cancer worldwide and the sixth most common form of death from cancer (1). The predominant subtype in developing countries is oesophageal squamous cell carcinoma (OSCC), whereas oesophageal adenocarcinoma is more common in the western world where its incidence is increasing. High-risk regions for OSCC include southern Africa, Japan, China and northern Iran. In southern Africa, oesophageal cancer is the third most common cancer in both males and females with age-adjusted incidence rates of 22.3 and 11.7 per 100 000, respectively (2); higher rates are observed in certain regions such as the Eastern Cape Province of South Africa (3). Environmental risk factors for the development of OSCC in South Africa include alcohol intake and tobacco use, nutritional deficiencies, consumption of Fusarium-(fungi) contaminated maize and infection with human papilloma virus (reviewed in ref. 4).
Candidate gene studies have analysed multiple genetic variants for association with OSCC in two indigenous South African populations, the Mixed Ancestry and the Black populations, with a high prevalence of this disease. Single nucleotide polymorphisms (SNPs) in GSTM1, GSTP1 (5), CYP3A5 (6) and CYP2E1 (7) showed some evidence of association, although sample sizes were small and data from the two populations were pooled in some analyses. In view of the differences in population structure between these two South African populations (8), recent studies by our group have analysed the Black and Mixed Ancestry populations separately, and increased the power to detect association by expansion of the sample sizes. We reported significant association of a 37kb deletion in GSTT2B (9) and of the variant ALDH2 + 82 G>A (rs886205) (10) with OSCC in the Mixed Ancestry population. However, none of the variants tested in these studies were associated with OSCC in the Black South African population, which may be related to differences in their ancestry or environmental exposures (10), or to chance.
The development of genome-wide association scans (GWAS) has had a major impact on the discovery of susceptibility genes for complex disease. The first GWAS for OSCC was published in 2009 and identified significant associations with ALDH2 Glu504Lys (rs671) on chromosome 12q24 and ADH1B Arg48His (rs1229984) on chromosome 4q23 in the Japanese population (11). These SNPs were tested for association in the South African Black and Mixed Ancestry populations; the ALDH2 Glu504Lys variant was non-polymorphic in both populations, and ADH1B Arg48His showed a suggestive association in the Mixed Ancestry population but was non-polymorphic in the Black population (10). Recently, three independent OSCC GWAS in Chinese populations have identified a total of eight SNPs in six susceptibility loci, including PLCE1 His1927Arg (rs2274223) on chromosome 10q23, which was the only locus significantly associated in all three studies (12–14). Other loci identified were C20orf54/SLC52A3 (rs13042395) on chromosome 20p13 (13), PDE4D (rs10052657) on chromosome 5q12 (14), RUNX1 (rs2014300) on chromosome 21q22.3 (14), a variant near UNC5CL (rs10484761) on chromosome 6p21.1 (14), and three SNPs at a locus on 12q24—ACAD10 (rs11066015), C12orf51 (rs2074356) and rs11066280 (14). The contribution of the new loci to the risk of OSCC in other populations is unknown.
The aim of this study was to determine whether the new loci identified in the Chinese GWAS were associated with OSCC in the South African Black and Mixed Ancestry populations, and to investigate genetic variation and the genetic architecture of the PLCE1 gene in the South African population.
Materials and methods
Study subjects
This study consisted of 407 OSCC patients and 849 controls from the South African Black population, and 257 OSCC patients and 860 controls from the South African Mixed Ancestry population. The Black patients were mainly Xhosa-speakers (98.8%) from the Eastern or Western Cape of South Africa. The Black controls were recruited from factories and outpatient clinics in the Western Cape. The proportion of Xhosa-speakers was 98.2%; they had no history of any cancer, lived in the same residential areas, and had a similar socioeconomic status to the patients. The Mixed Ancestry cases and controls were all recruited from the Western Cape. This is an admixed population with major ancestral components from the indigenous Khoisan, Bantu-speaking Africans, Europeans and Asians (8). Patients with histologically confirmed primary invasive OSCC were recruited between March 2000 and August 2011 at Groote Schuur and Tygerberg Hospitals in Cape Town. Data on alcohol consumption and tobacco use were available for both cases and controls. Smoking status was subdivided into ever-smokers (those who had smoked at some point in their lives) or never-smokers. Drinkers were defined as subjects who consumed alcohol at least once every week. Demographic and exposure data are given in Table I. Whole blood samples were collected with informed consent from all subjects and DNA was extracted at the University of Cape Town. Ethical approval for the study was obtained from the joint University of Cape Town/Groote Schuur Hospital Research Ethics Committee and the University of Stellenbosch/Tygerberg Hospital Ethics Committee.
Table I.
Characteristics of OSCC cases and controls in the South African Black and Mixed Ancestry populations
| Black population | Mixed Ancestry population | ||||
|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||
| n | 407 | 849 | 257 | 860 | |
| *Age, mean years (SD) | 59.8 (11.3) | 48.8 (16.7) | 60.6 (10.6) | 46.7 (16.8) | |
| *Sex, n (%) | Male | 199 (48.9%) | 335 (39.5%) | 165 (64.2%) | 309 (35.9%) |
| Female | 208 (51.1%) | 511 (60.2%) | 91 (35.4%) | 551 (64.1%) | |
| Unknown | 0 (0.0%) | 3 (0.4%) | 1 (0.4%) | 0 (0.0%) | |
| *Smoking status, n (%) | Ever-smoker | 242 (59.5%) | 333 (39.2%) | 240 (93.4%) | 597 (69.4%) |
| Never-smoker | 164 (40.3%) | 505 (59.5%) | 15 (5.8%) | 258 (30.0%) | |
| Unknown | 1 (0.2%) | 11 (1.3%) | 2 (0.8%) | 5 (0.6%) | |
| *Alcohol consumption, n (%) | Drinker | 253 (62.2%) | 452 (53.2%) | 212 (82.5%) | 419 (48.7%) |
| Non-drinker | 151 (37.1%) | 393 (46.3%) | 45 (17.5%) | 436 (50.7%) | |
| Unknown | 3 (0.7%) | 4 (0.5%) | 0 (0.0%) | 5 (0.6%) | |
*Age, sex, smoking status and alcohol consumption were significantly different between cases and controls in both populations (P < 0.01).
SNP selection and genotyping
Index SNPs (those with the strongest evidence for association with OSCC in the Chinese GWAS studies) from five of the six loci were polymorphic in the Yoruban (Nigerian) and Masai in Kinyawa (Kenyan) HapMap populations, suggesting that they were likely to be informative in the Black South African population. These five SNPs, PLCE1 His1927Arg (rs2274223), C20orf54/SLC52A3 (rs13042395), PDE4D (rs10052657), RUNX1 (rs2014300) and a variant near UNC5CL (rs10484761), were selected for genotyping. Two of the SNPs at the 12q24 locus (rs2074356 and rs11066280) were non-polymorphic in African HapMap populations. No HapMap data was available for rs11066015, but this SNP is in strong linkage disequilibrium (LD) with rs671 in the Chinese population, which we found to be absent in both the Black and Mixed Ancestry South African populations (10). Thus, these three SNPs are likely to be rare or absent in our South African populations, and we would have limited power to detect association with OSCC. The five prioritized SNPs were genotyped in 407 cases and 849 controls from the South African Black population and 257 OSCC cases and 860 controls from the Mixed Ancestry population using validated TaqMan 5′ exonuclease SNP genotyping assays (Applied Biosystems). Reactions were carried out in 2.5 µl volumes in 96-well plates. Each reaction contained 20ng DNA, Absolute QPCR ROX mix (ABgene) and TaqMan SNP assay mix (Applied Biosystems) according to assay instructions and were performed on a PTC-0225 DNA Engine (MJ Research). Fluorescent levels at the PCR endpoint were determined using a 7900HT Fast Real-Time PCR system (Applied Biosystems) and genotypes assigned using SDS 2.2.2 software (Applied Biosystems).
Sequencing of PLCE1
The 34 exons of PLCE1 were sequenced by Sanger sequencing in 46 cancer patients (38 OSCC and 8 other cancers) from the Black South African population. Exons were amplified by PCR with primers designed using Primer3 (15) and synthesized by Integrated DNA Technologies. PCR was carried out in a 10 µl reaction containing 10ng DNA, 5 µl 2× PCR mix (Promega), 0.4 pmoles of each forward and reverse primer for all exons apart from exon 1. For this exon, the reaction contained 1× Flexi reaction buffer (Promega), 1.5mM MgCl2, 0.4 pmoles of each forward and reverse primer, 1U Flexi Taq polymerase (Promega), 0.2mM dNTP and 10% dimethyl sulfoxide. All reactions were carried out on a PTC-0225 DNA Engine (MJ Research) using the following conditions: 2min at 92°C; then 30 cycles of 20 s at 92°C, 30 s at the optimized annealing temperature, and between 30 s and 3min at 72°C (depending on amplimer length); with a final 5min at 72°C. Primer sequences and PCR conditions for each amplicon are shown in Supplementary Table 1, available at Carcinogenesis Online. Subsequent ExoSAP-IT clean up (USB Europe, Staufen, Germany) followed by forward and/or reverse cycle sequencing was performed for each exon using 8 pmoles of sequencing primer (see Supplementary Table 2, available at Carcinogenesis Online) and 0.25 μl of BigDye Terminator v3.1 (Applied Biosystems) in a 5.25 μl reaction volume under recommended reaction conditions. Products were analysed on an ABI3730xl DNA sequencer (Sequence Analysis, Applied Biosystems) and aligned to the human reference genome using Staden software package (16).
Genotyping of PLCE1 SNPs
Three non-synonymous SNPs in PLCE1, Arg548Leu (rs17417407), Pro1890Leu (rs58539480), and the novel variant Gly1199Ser were selected for genotyping in the Black South African population on the basis of strong evolutionary conservation of the amino acid residue, a predicted damaging effect in at least one of the two programs, Polyphen 2 (17) and SIFT (Sorting Intolerant From Tolerant) (18), and a minor allele frequency (MAF) of >0.05 in this population. The variant Ile1777Thr (rs3765524) was also selected since it was very strongly associated with OSCC in the Chinese population and common in the Black South African population. A common insertion/deletion (indel) in the 5′-untranslated region (UTR) was also selected. These five variants were genotyped in an initial sample of 323 cases and 459 controls that was available from the Black South African population. If a suggestive association with OSCC was observed (P < 0.05) then further samples that subsequently became available were genotyped in an expanded sample set from this population (a total of 407 cases and 849 controls), and in the Mixed Ancestry population (257 cases and 860 controls). The SNPs were genotyped using custom KASP By-Design assays (KBioscience) following manufacturer’s instructions. The 14bp indel (CCCGGGCTCTGCCT) in the 5′UTR of exon 1 was PCR amplified and genotyped by size separation of PCR products using 3% agarose gel electrophoresis and visualized with ethidium bromide/UV light. Primers used for amplification were as follows: forward GGGAGCGGACTGTGAACG and reverse GTGTCCCCGCTACTGTGTGT. The 10 µl PCR reaction contained 1× Flexi reaction buffer (Promega), 1.5mM MgCl2, 0.4 pmoles of each forward and reverse primer, 1U Flexi Taq polymerase (Promega), 0.2mM dNTP and 10% DMSO. Reaction conditions were as described previously, with 63°C annealing temperature and 30 s extension time.
Statistical analysis
Pearson’s chi-squared (χ2) test was used to determine whether the proportions of genotypes were consistent with Hardy–Weinberg equilibrium, using a cut-off of P < 0.05. P values were >0.05 for all SNPs tested. Genotype and allele frequencies were calculated for cases and controls and the allele frequencies compared using the Pearson’s chi-squared (χ2) test to test for association with OSCC. For the association tests of the five Chinese GWAS SNPs, a Bonferroni-corrected P value of <0.01 (0.05/5) was used as a significance threshold to account for multiple testing. This threshold was also applied to the association tests of the five PLCE1 variants genotyped after identification by sequencing. No additional correction was applied for the two populations tested. Allelic odds ratios (OR) and 95% confidence intervals (CI) were calculated using the common allele as the reference. As a secondary analysis, logistic regression was carried out adjusting for age, sex, smoking and alcohol consumption to determine whether these covariates influenced the association results determined using the Pearson’s (χ2) test; these results are reported as P (adjusted). For SNPs with suggestive evidence of allelic association (P < 0.05), the effect of alcohol and tobacco was investigated by testing for association in cases and controls stratified by smoking and drinking status. We tested for interactions by performing a case-only analysis of alleles based on smoking and drinking status, and by carrying out a gene–environment interaction test using logistic regression in a case–control analysis. The power of the study was determined using Quanto (http://hydra.usc.edu/gxe/). LD between PLCE1 variants in the South African Black population was assessed using Haploview (19). PLCE1 haplotype analysis was performed using UNPHASED (20).
Results
Case–control analysis
Five SNPs (PLCE1 rs2274223, RUNX1 rs2014300, C20orf54 rs13042395, PDE4D rs10052657 and a variant near UNC5CL rs10484761) were tested for association in the South African Black and Mixed Ancestry populations (Table II). In the South African Mixed Ancestry population, the minor ‘A’ allele of rs2014300 in RUNX1 was significantly associated with an increased risk of OSCC (OR = 1.33, 95% CI = 1.09–1.63, P = 0.0055), with minor allele frequencies of 43.8% and 37.0% in cases and controls, respectively. However, this effect is in the opposite direction to that found in the Chinese population, where the minor ‘A’ allele confers a reduced risk of OSCC (OR = 0.70) (14). The other four SNPs were not associated with OSCC in this population. In the Black South African population there was no evidence of association with OSCC for any of the variants tested. In both populations, adjusting for covariates in logistic regression produced very similar results. None of the five loci was associated with OSCC in the Black population, and ORs were similar to the unadjusted analysis. In the Mixed Ancestry population, the four loci not associated with the unadjusted analysis were also not associated with the adjusted analysis, and the RUNX1 variant rs201400 remained associated with a moderately increased effect (ORadjusted = 1.51, 95% CI = 1.19–1.92; P (adjusted) = 0.0007). There were substantial differences in allele frequencies for three of the five SNPs in the two South African populations; for rs2014300 and rs10484761, the minor allele in the Black population was the common allele in the Mixed Ancestry population, and the ‘T’ allele of rs13042395 was very rare (0.5%) in the Black population compared with the Mixed Ancestry population (6.8%) (Table II).
Table II.
Case–control analysis of five loci from Chinese GWAS in South African Black and Mixed Ancestry OSCC
| Black population | Mixed Ancestry population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | P value | Cases | Controls | OR (95% CI) | P value | |||
| PLCE1 rs2274223 | His/His | 140 (33.5%) | 302 (35.5%) | His/His | 78 (30.7%) | 310 (36.2%) | ||||
| His/Arg | 208 (49.8%) | 411 (48.4%) | His/Arg | 130 (51.2%) | 408 (47.6%) | |||||
| Arg/Arg | 70 (16.7%) | 137 (16.1%) | Arg/Arg | 46 (18.1%) | 139 (16.2%) | |||||
| His | 488 (58.4%) | 1015 (59.7%) | Reference | — | His | 286 (56.3%) | 1028 (60.0%) | Reference | — | |
| Arg | 348 (41.6%) | 685 (40.3%) | 1.06 (0.89–1.25) | 0.5208 | Arg | 222 (43.7%) | 686 (40.0%) | 1.16 (0.95–1.42) | 0.1386 | |
| C20orf54 rs13042395 | C/C | 402 (99.5%) | 837 (98.9%) | C / C | 221 (87.0%) | 742 (87.2%) | ||||
| C/T | 2 (0.5%) | 9 (1.1%) | C / T | 32 (12.6%) | 103 (12.1%) | |||||
| T/T | 0 (0.0%) | 0 (0.0%) | T / T | 1 (0.4%) | 6 (0.7%) | |||||
| C | 806 (99.8%) | 1683 (99.5%) | Reference | — | C | 474 (93.3%) | 1587 (93.2%) | Reference | — | |
| T | 2 (0.2%) | 9 (0.5%) | 0.46 (0.10–2.15) | 0.3150 | T | 34 (6.7%) | 115 (6.8%) | 0.99 (0.67–1.47) | 1.0000 | |
| near UNC5CL rs10484761 | G/G | 107 (26.9%) | 225 (26.6%) | A / A | 105 (41.5%) | 398 (47.2%) | ||||
| G/A | 210 (52.8%) | 435 (51.4%) | G / A | 117 (46.2%) | 362 (42.9%) | |||||
| A/A | 81 (20.4%) | 186 (22.0%) | G / G | 31 (12.3%) | 84 (10.0%) | |||||
| G | 424 (53.3%) | 885 (52.3%) | Reference | — | A | 327 (64.6%) | 1158 (68.6%) | Reference | — | |
| A | 372 (46.7%) | 807 (47.7%) | 0.96 (0.81–1.14) | 0.6542 | G | 179 (35.4%) | 530 (31.4%) | 1.20 (0.97–1.47) | 0.0933 | |
| PDE4D rs10052657 | C/C | 300 (74.6%) | 642 (76.0%) | C / C | 171 (67.1%) | 613 (71.6%) | ||||
| C/A | 94 (23.4%) | 190 (22.5%) | C / A | 79 (31.0%) | 220 (25.7%) | |||||
| A/A | 8 (2.0%) | 13 (1.5%) | A / A | 5 (2.0%) | 23 (2.7%) | |||||
| C | 694 (86.3%) | 1474 (87.2%) | Reference | — | C | 421 (82.5%) | 1446 (84.5%) | Reference | — | |
| A | 110 (13.7%) | 216 (12.8%) | 1.08 (0.85–1.38) | 0.5329 | A | 89 (17.5%) | 266 (15.5%) | 1.15 (0.88–1.50) | 0.3005 | |
| RUNX1 rs2014300 | A/A | 152 (37.7%) | 311 (37.1%) | G / G | 81 (32.1%) | 346 (40.9%) | ||||
| A/G | 197 (48.9%) | 378 (45.1%) | A / G | 121 (48.0%) | 374 (44.2%) | |||||
| G/G | 54 (13.4%) | 149 (17.8%) | A / A | 50 (19.8%) | 126 (14.9%) | |||||
| A | 501 (62.2%) | 1000 (59.7%) | Reference | — | G | 283 (56.2%) | 1066 (63.0%) | Reference | — | |
| G | 305 (37.8%) | 676 (40.3%) | 0.90 (0.76–1.07) | 0.2342 | A | 221 (43.8%) | 626 (37.0%) | 1.33 (1.09–1.63) | 0.0055 | |
The association tests in the South African populations had good power to detect the ORs reported in the Chinese GWAS studies for four out of the five loci tested, if it is assumed that the associated variant and the causal variant are either the same or are in complete LD. The ORs reported in the Chinese studies were 1.34–1.43 (PLCE1), 1.33 (UNC5CL), 0.67 (PDE4D), 0.70 (RUNX1) and 0.66 (C20orf54) (12–14). Using the allele frequencies determined in the Black controls, power in the Black population was >80% to detect the effects seen in PLCE1, UNC5CL, PDE4D and RUNX1, and there was 80% power to detect ORs of 1.27, 1.28, 1.49 and 1.28 for these four loci, respectively. In the Mixed Ancestry population, power was somewhat lower but still adequate at >75% for these four SNPs, with 80% power to detect ORs of 1.33, 1.35, 1.54 and 1.35, respectively. The low frequency of the C20orf54 SNP rs13042395 in both populations (0.5% and 6.8% in Black and Mixed Ancestry controls, respectively) did not provide sufficient power to detect the association reported in the Chinese population.
Sequencing and genotyping of PLCE1
The strong association of the SNP rs2274223 (His1927Arg) in PLCE1 with OSCC in the Chinese population (12–14) was not replicated in either of the South African populations tested. Since genetic variability is highest in African populations and LD is generally much lower, we investigated the PLCE1 locus in the Black South African population by sequencing all 34 exons of PLCE1 and adjacent splice sites in 46 individuals from this population to examine LD and to identify potential functional sequence variants. A total of 48 polymorphic variants were detected, including 26 known SNPs, 11 novel variants and one known 14bp insertion/deletion in the 5′-UTR. Ten of these 48 variants produce amino acid substitutions in the encoded PLCE1 protein and could therefore be considered as candidate OSCC causal variants; their locations relative to the putative protein domains of PLCE1 are shown in Figure 1. The potential functional consequences of these variants were assessed by examination of evolutionary conservation across multiple species and use of the predictive programs Polyphen2 and SIFT (see Materials and methods). Six of the ten SNPs were predicted to have probable or possible damaging effects by one or more of these methods (Table III).
Fig. 1.
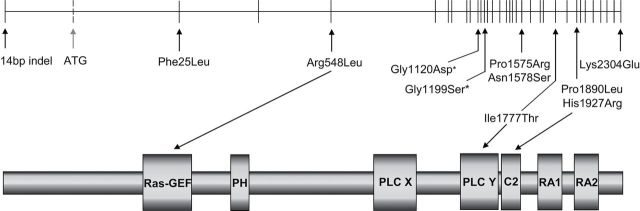
Potential functional variants detected in the PLCE1 gene. Top shows exon/intron structure with vertical lines representing exons for NM_016341.3 (dotted vertical line reflects exon present only in NM_001165979.1). Locations of non-synonymous SNPs and 5′-UTR insertion/deletion identified by sequencing in the South African Black population are indicated with vertical arrows (* indicates novel variants). Bottom shows the predicted functional domains of the PLCE1 protein and the position of variants which are located within a domain.
Table III.
PLCE1 non-synonymous SNPs in the South African Black population
| SNP identifier | Chr location (build 37), | Amino acid | MAFa | Amino acid conservationb | Polyphen 2 (score) | SIFT (score) |
|---|---|---|---|---|---|---|
| major > minor allele | change | |||||
| rs115135156 | 10:95848924, T>C | Phe25Leu | 0.043 | Rhc, Moc, Doc, Elc, Opc | Benign (0.013) | Damaging (0)d |
| rs17417407 | 10:95931087, G>T | Arg548Leu | 0.178 | Rh, Mo, Do, El, Op, Ch, X_t, Ze | Probably damaging (0.981) | Tolerated (0.12) |
| Novel | 10:96014026, G>A | Gly1120Asp | 0.011 | Rh, Mo, Do, El, Op, Ch, X_t | Probably damaging (0.968) | Damaging (0.04) |
| Novel | 10:96018597, G>A | Gly1199Ser | 0.058 | Rh, Mo, Do, El, Op, Ch, X_t | Probably damaging (0.984) | Tolerated (0.12) |
| rs2274224 | 10:96039597, C>G | Pro1575Arg | 0.318 | Not conserved | Benign (0) | Tolerated (0.53) |
| rs61732525 | 10:96039606, A>G | Asn1578Ser | 0.045 | Rh, Do, El, X_t, Ze | Benign (0.002) | Tolerated (0.42) |
| rs3765524 | 10:96058298, T>C | Ile1777Thr | 0.386 | Not conserved | Benign (0.002) | Tolerated (0.85) |
| rs58539480 | 10:96066230, C>T | Pro1890Leu | 0.087 | Rh, Mo, Do, El, Op, X_t, Ze | Probably damaging (0.999) | Tolerated (0.33) |
| rs2274223 | 10:96066341, A>G | His1927Arg | 0.489 | Ze | Benign (0) | Tolerated (0.83) |
| rs3203713 | 10:96087728, A>G | Lys2304Glu | 0.141 | Rhc, Doc, Elc | Unknown | Damaging (0.01) d |
aMAF calculated from the number of individuals genotyped successfully (n = 43–46).
bAmino acid conservation is shown for Rhesus Macaque (Rh), Mouse (Mo), Dog (Do), Elephant (El), Opossum (Op), Chicken (Ch), Xenopus Tropicalis (X_t) and Zebrafish (Ze).
cConservation only available for nucleotide sequence.
dLow confidence SIFT score indicating that the protein alignment does not have enough sequence diversity and an amino acid may incorrectly be predicted to be damaging.
Pair-wise analysis of all 48 variants detected by sequencing in the 46 Black South African individuals showed that there is very low LD across the PLCE1 gene in this population (Supplementary Figure 1, available at Carcinogenesis Online). Sixteen of these variants have also been genotyped in the HapMap project, allowing a comparison of the LD structure between the South African Black population and HapMap populations such as the Han Chinese from Beijing (CHB) and Yoruba in Ibadan, Nigeria (YRI) (Figure 2; Supplementary Figure 2, available at Carcinogenesis Online). This shows that the South African Black population has the lowest level of LD across PLCE1 among these three populations. The index SNP rs2274223 is in very strong LD with multiple other SNPs in the CHB population, but is in low to moderate LD with other SNPs in the South Africans. The variants Arg548Leu (rs17417407), Ile1777Thr (rs3765524), Pro1890Leu (rs58539480) and the novel Gly1199Ser SNP, together with the 5′-UTR 14bp indel, were selected for genotyping (see Materials and methods) and tested for association with OSCC in the Black South African population. Results for allelic association tests are shown in Table IV. Only one variant, PLCE1 Arg548Leu (rs17417407), showed suggestive evidence for association with OSCC (P = 0.035) and was therefore genotyped in an additional set of cases and controls. Combined analysis of 407 cases and 849 controls revealed a MAF (T, 548Leu) of 16.6% in cases and 21.1% in controls, giving an OR = 0.74 (95% CI = 0.60–0.93, P = 0.008). Similar results were obtained for these five PLCE1 variants when adjusting for age, sex, smoking and drinking status, except that the rs1741707 association was slightly less significant (ORadjusted = 0.75, 95% CI = 0.59–0.95, P (adjusted) = 0.019). Haplotype analysis of the five SNPs and indel of PLCE1 show that none of the haplotypes were associated with OSCC in the Black South African population (P = 0.977 for overall association, data not shown). The Arg548Leu SNP (rs17417407) was also genotyped in the Mixed Ancestry population, but no evidence of association was detected, with MAF for the Leu allele of 17.4% and 18.0% in cases and controls, respectively (OR = 0.96, 95% CI = 0.74–1.25, P = 0.764).
Fig. 2.
HapMap LD structure for 16 SNPs across PLCE1 gene region. Plots show r 2 values for pairwise LD between variants in the South African Black population and two HapMap populations, Yoruban from Nigeria and Han Chinese from Beijing. Colour schemes and labelling are according to Haploview as follows: white (r2 = 0), black (r2 = 1) or shades of grey (0 < r2 < 1); numbers indicate r2 values (×100).
Table IV.
Case–control analysis of five potential functional PLCE1 variants for OSCC in the South African Black population
| Cases | Controls | OR (95% CI) | P value | ||
|---|---|---|---|---|---|
| 5′-UTR 14bp indel | Ins/Ins | 185 (57.6%) | 260 (57.0%) | ||
| Ins/Del | 122 (38.0) | 171 (37.5%) | |||
| Del/Del | 14 (4.4%) | 25 (5.5%) | |||
| Ins | 492 (76.6%) | 691 (75.8%) | Reference | — | |
| Del | 150 (23.4%) | 221 (24.2%) | 0.95 | 0.693 | |
| (0.75–1.21) | |||||
| rs17417407 Arg548Leu | Arg/Arg | 226 (70.4%) | 271 (61.5%) | ||
| Arg/Leu | 81 (25.2%) | 152 (34.5%) | |||
| Leu/Leu | 14 (4.4) | 18 (4.1%) | |||
| Arg | 533 (83.0%) | 694 (78.7%) | Reference | — | |
| Leu | 109 (17.0%) | 188 (21.3%) | 0.75 | 0.035 | |
| (0.58–0.98) | |||||
| Novel Gly1199Ser | Gly/Gly | 289 (90.0%) | 410 (91.3%) | ||
| Gly/Ser | 30 (9.3%) | 38 (8.5%) | |||
| Ser/Ser | 2 (0.6%) | 1 (0.2%) | |||
| Gly | 608 (94.7%) | 858 (95.5%) | Reference | — | |
| Ser | 34 (5.3%) | 40 (4.5%) | 1.20 | 0.446 | |
| (0.75–1.92) | |||||
| rs3765525 Ile1777Thr | Ile/Ile | 86 (27.2%) | 126 (27.9%) | ||
| Ile/Thr | 162 (51.3%) | 233 (51.5%) | |||
| Thr/Thr | 68 (21.5%) | 93 (20.6%) | |||
| Ile | 334 (52.8%) | 485 (53.7%) | Reference | — | |
| Thr | 298 (47.2%) | 419 (46.3%) | 1.03 | 0.756 | |
| (0.84–1.27) | |||||
| rs58539480 Pro1890Leu | Pro/Pro | 262 (85.3%) | 375 (87.4%) | ||
| Pro/Leu | 45 (14.7%) | 53 (12.4%) | |||
| Leu/Leu | 0 (0.0%) | 1 (0.2%) | |||
| Pro | 569 (92.7%) | 803 (93.6%) | Reference | — | |
| Leu | 45 (7.3%) | 55 (6.4%) | 1.15 | 0.490 | |
| (0.77–1.74) |
Alcohol and smoking analysis
Gene–environment interactions between OSCC and smoking and alcohol drinking habits were tested for variants that showed an association with OSCC (P < 0.05). In the Mixed Ancestry population, RUNX1 rs2014300 showed no significant associations in case–control analysis for either drinkers or non-drinkers, or for a case-only analysis of drinkers versus non-drinkers (see Supplementary Table 3, available at Carcinogenesis Online). The low number of non-smokers (n = 15) in the Mixed Ancestry population prevented analysis by smoking status. The PLCE1 Arg548Leu SNP (rs17417407) showed no interaction with alcohol use in the Black population. When stratifying by smoking status in cases and controls, an association was observed in a case–control analysis of ever-smokers, OR = 0.64 (P = 0.005), with weakened association in never-smokers (OR = 0.87). However, the case-only analysis of ever- versus never-smokers was not significant (P = 0.16). Logistic regression analysis showed no evidence for an interaction with smoking or alcohol with either RUNX1 rs2014300 or PLCE1 Arg548Leu (rs17417407).
Discussion
The aim of this study was to determine whether five new loci reported to be associated with susceptibility to OSCC in the Chinese population (12–14) also contribute to susceptibility in two South African populations. In the South African Mixed Ancestry population, only one SNP, RUNX1 rs2014300, was associated with OSCC, with the minor ‘A’ allele conferring an increased risk of disease (OR = 1.33, 95% CI = 1.09–1.63). However, the association is in the opposite direction to that found in the Chinese population, where allele ‘A’ is also the minor allele but is protective (OR = 0.70) (14). Since it is unlikely that the same allele of this SNP would have opposite effects on susceptibility to the same disease in these two populations, the finding in the Mixed Ancestry population may be a false positive, despite remaining significant after correction for multiple testing. Further analysis of this SNP in other populations would help to establish whether this association is specific to OSCC in the Chinese population.
In the South African Black population, none of the five SNPs showed evidence of association with OSCC. This is consistent with our previous study, in which none of the 13 variants associated with OSCC in other populations was associated in South African Black OSCC (10). There are several possible reasons for the lack of association of the five SNPs in the South African populations. One is that the GWAS findings are false positives. This is very unlikely for PLCE1 since the association has been observed in three independent studies (12–14). The associations at PDE4D and RUNX1 were convincingly replicated in the original study (14), with combined P values of 10–19 and 10–21, respectively, so these are also probably to be robust findings. Replication of the UNC5CL locus was less convincing (14), and the C20orf54 association was only observed in one of the three GWAS (13). A meta-analysis of all three Chinese GWAS would help to resolve the status of some of these loci. An alternative explanation for the lack of association in South African OSCC is insufficient power. We had high power to detect the effect observed in the Chinese studies for four of the five SNPs, with the other SNP (C20orf54 rs13042395) being very rare in the Black population. However, if the effect size at these loci is much smaller in South Africans than in Chinese, the power of this study would be reduced. A further possible reason is that the SNPs genotyped in the Chinese GWAS studies may not be the actual causal SNPs that are driving the association, but merely markers that tag them with high LD. Since LD is generally lower in African populations (21), our studies may not be able to detect an association if the causal SNP is not genotyped directly.
The importance of the PLCE1 locus in OSCC susceptibility in the Chinese population prompted us to investigate sequence variation in this gene and its genetic architecture in the South African Black population in more detail, since substantial differences in sequence variation and LD structure could exist between these two populations. Sequencing of the entire coding region in 46 individuals revealed that LD across PLCE1 was much weaker in the South African Black population compared with the Chinese. This suggests that very high density SNP analysis across its entire genomic structure would be required for a complete interrogation of the contribution of PLCE1 to OSCC in this population. We tested five coding sequence changes that were conserved and/or predicted to alter protein function and an insertion/deletion polymorphism in the 5′-UTR as these were sufficiently common to allow detection of association with OSCC in the South African Black population. Only the Arg548Leu variant (rs17417407) was associated with OSCC. The Arg548 allele is conserved across species (Rhesus, Mouse, Dog, Elephant, Opossum, Chicken, Xenopus Tropicalis and Zebrafish) and is predicted to be ‘probably damaging’ by Polyphen2 but tolerated by SIFT. This SNP was not included in the GWAS SNP micorarrays used by the three Chinese studies. However, it is in complete LD (r 2 = 1) with rs2689700 in the combined Chinese/Japanese (CHB/JPT) HapMap population, which is an intronic SNP that was present on the GWAS chips used in all three GWAS studies and is not listed as associated with OSCC. In both the Chinese/Japanese and South African Black populations, Arg548Leu rs17417407 is in very low LD with His1927Arg rs2274223 (r 2 = 0.023 and 0.03, respectively), the Chinese OSCC index risk variant. This suggests that two independent variants in PLCE1 may contribute to OSCC susceptibility loci in the Chinese and South African Black populations.
An important question regarding the association at the PLCE1 locus is whether variants within PLCE1 itself are driving this association, since the GWAS signal appears to include at least one other adjacent gene, NOC3L (12–14). The protein encoded by PLCE1, phospholipase C epsilon 1, is responsible for the hydrolysis of phosphatidyl-inositol 4,5-biphosphate to generate diacylglycerol and inositol 1,4,5-triphosphate, which causes the release of calcium and activation of protein kinase C. PLCE1 can also act as a guanine-exchange factor, activating Ras, which is unique to this class of phospholipase C enzymes; PLCE1 is itself activated by Ras and Rho family GTPases, and thus could be affected by the oncogenic properties of Ras in cancer cells, although this has not yet been shown (reviewed in ref. 22). Studies of PLCE1 expression and protein levels in tumours have produced conflicting results, with mRNA shown to be reduced in OSCC tissue (23), but with unchanged (23) or increased levels (13) of the protein being reported in tumour tissues compared with normal tissue. Activation of PLCE1 has also been linked to tumour cell migration in head and neck squamous cell carcinoma (24). However, sequence variation in PLCE1 has also been associated with non-cancer phenotypes. The Ile1777Thr variant (rs3765524), which is strongly associated with OSCC (12), is also associated with dengue shock syndrome (25), and biallelic mutations in PLCE1 are an important cause of nephrotic syndrome (26). The adjacent gene, NOC3L (also known as FAD24) has been shown to regulate DNA replication during adipogenesis (27). It has also been reported to have a role in repression of NF-κB activity and H-Ras-mediated transformation (28), and silencing of the gene produces sensitivity to Tamoxifen, a drug that inhibits estrogen receptor α signalling in breast cancer (29). Fine mapping of the region of association with a dense panel of SNPs in parallel with functional are needed to confirm the identity of the causal gene.
Two of the SNPs showing the strongest association at the PLCE1 locus in the Chinese population are the non-synonymous variants Ile1777Thr (rs3765524) and His1927Arg (rs2274223), which raises the question of whether one or other of them might be the causal variant at this locus. His1927Arg is located within the protein calcium-dependent lipid-binding C2 domain of PLCE1, but our bioinformatic analysis of this variant (Table III) shows that it is not conserved and is scored as benign or tolerated by the functional prediction programs PolyPhen and SIFT. Ile1777Thr is located in the PI-PLC Y-box domain that is important for catalytic activity, but is not evolutionary conserved and is also scored as benign or tolerated by the prediction programs. Thus, neither variant has strong credentials as the causal variant. Interestingly, the Arg548Leu variant rs17417407, which showed modest association with OSCC in the South African Black population, is located in a Ras-guanine-exchange factor domain, and may therefore affects its capacity to activate the Ras protein. It does show strong evolutionary conservation and is predicted to be damaging by one of the two prediction programs. It should also be noted that non-coding variants and synonymous SNPs may have important functional effects (30,31), and multiple associations with complex disease have been detected in gene deserts, which do not contain any known coding genes (32). These classes of variants will need to be taken into account when attempting to define causal variants at complex disease loci.
In conclusion, this study has examined a series of new genetic associations with OSCC, which have emerged from GWAS in the Chinese population, in two indigenous populations from South Africa. None of these associations was replicated in either of the two indigenous South African populations studied. However, although none of the variants in PLCE1 that conferred an increased risk of OSCC in Chinese populations were associated in the South African populations, we found that the Arg548Leu variant rs17417407 is associated with a reduced risk of OSCC in the Black South African population. As discussed previously (10), the reasons for variation in genetic associations between populations may include differences in both genetic architecture and in environmental risk factors that interact with genetic factors to promote development of the disease. The emerging differences that we have observed between the genetic factors contributing to the development of OSCC in African versus Asian populations (9,10) suggest that well-powered GWAS in non-Asian populations are needed to define the genetic basis of this cancer in Africa and elsewhere.
Supplementary material
Supplementary Tables 1–3 and Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
Association for International Cancer Research (09-0625), the Medical Research Council UK, The Generation Trust, the National Institutes of Health Research Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London to H.B., N.J.P., C.M.L., C.G.M., The South African Research Chairs Initiative of the Department of Science and Technology and the National Research Foundation, the International Centre for Genetic Engineering and Biotechnology (ICGEB), the South African Medical Research Council and the University of Cape Town to M.I.P.
Supplementary Material
Acknowledgements
We thank Antoinette Olivier, Zenaria Abbas and Amy Salkinder for assisting with the sample collection and processing, and the patients and healthy controls for their participation in this study.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CI
confidence interval
- GWAS
genome-wide association scans
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
- OSCC
oesophageal squamous cell carcinoma
- SNP
single nucleotide polymorphisms;
- UTR
untranslated region.
References
- 1. Ferlay J., et al. (2010). Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer, 127, 2893––2917 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., et al. (2012). Cancer burden in Africa and opportunities for prevention. Cancer, doi: 10.1002/cncr.27410. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 3. Somdyala N.I., et al. (2010). Cancer incidence in a rural population of South Africa, 1998–2002. Int. J. Cancer, 127, 2420––2429 [DOI] [PubMed] [Google Scholar]
- 4. Hendricks D., et al. (2002). Oesophageal cancer in Africa. IUBMB Life, 53, 263––268 [DOI] [PubMed] [Google Scholar]
- 5. Li D., et al. (2010). The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet., 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dandara C., et al. (2005). CYP3A5 genotypes and risk of oesophageal cancer in two South African populations. Cancer Lett., 225, 275––282 [DOI] [PubMed] [Google Scholar]
- 7. Li D., et al. (2005). Association of cytochrome P450 2E1 genetic polymorphisms with squamous cell carcinoma of the oesophagus. Clin. Chem. Lab. Med., 43, 370––375 [DOI] [PubMed] [Google Scholar]
- 8. de Wit E., et al. (2010). Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum. Genet., 128, 145––153 [DOI] [PubMed] [Google Scholar]
- 9. Matejcic M., et al. (2011). Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case-control study. PLoS ONE, 6, e29366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bye H., et al. (2011). Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis, 32, 1855––1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui R., et al. (2009). Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology, 137, 1768––1775 [DOI] [PubMed] [Google Scholar]
- 12. Abnet C.C., et al. (2010). A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet., 42, 764––767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang L.D., et al. (2010). Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet., 42, 759––763 [DOI] [PubMed] [Google Scholar]
- 14. Wu C., et al. (2011). Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet., 43, 679––684 [DOI] [PubMed] [Google Scholar]
- 15. Rozen S., et al. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365––386 [DOI] [PubMed] [Google Scholar]
- 16. Bonfield J.K., et al. (1995). A new DNA sequence assembly program. Nucleic Acids Res., 23, 4992––4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adzhubei I.A., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248––249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar P., et al. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073––1081 [DOI] [PubMed] [Google Scholar]
- 19. Barrett J.C., et al. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263––265 [DOI] [PubMed] [Google Scholar]
- 20. Dudbridge F. (2008). Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum. Hered., 66, 87––98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teo Y.Y., et al. (2010). Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet., 11, 149––160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bunney T.D., et al. (2010). Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer, 10, 342––352 [DOI] [PubMed] [Google Scholar]
- 23. Hu H., et al. (2012). Putatively functional PLCE1 variants and susceptibility to esophageal squamous cell carcinoma (ESCC): a case-control study in Eastern Chinese populations. Ann. Surg. Oncol., 19, 2403––2410 [DOI] [PubMed] [Google Scholar]
- 24. Bourguignon L.Y., et al. (2006). Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J. Biol. Chem., 281, 14026––14040 [DOI] [PubMed] [Google Scholar]
- 25. Khor C.C., et al. (2011). Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet., 43, 1139––1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hinkes B., et al. (2006). Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat. Genet., 38, 1397––1405 [DOI] [PubMed] [Google Scholar]
- 27. Johmura Y., et al. (2008). FAD24, a regulator of adipogenesis, is required for the regulation of DNA replication in cell proliferation. Biol. Pharm. Bull., 31, 1092––1095 [DOI] [PubMed] [Google Scholar]
- 28. Johmura Y., et al. (2008). FAD24, a regulator of adipogenesis and DNA replication, inhibits H-RAS-mediated transformation by repressing NF-kappaB activity. Biochem. Biophys. Res. Commun., 369, 464––470 [DOI] [PubMed] [Google Scholar]
- 29. Mendes-Pereira A.M., et al. (2012). Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc. Natl. Acad. Sci. U.S.A., 109, 2730––2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sauna Z.E., et al. (2007). Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res., 67, 9609––9612 [DOI] [PubMed] [Google Scholar]
- 31. Cooper G.M., et al. (2011). Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet., 12, 628––640 [DOI] [PubMed] [Google Scholar]
- 32. Mathew C.G. (2008). New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet., 9, 9––14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



