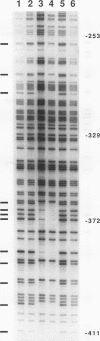
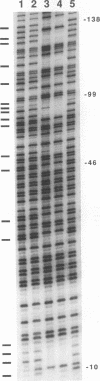
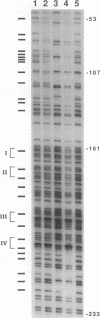
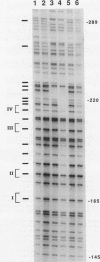
Abstract
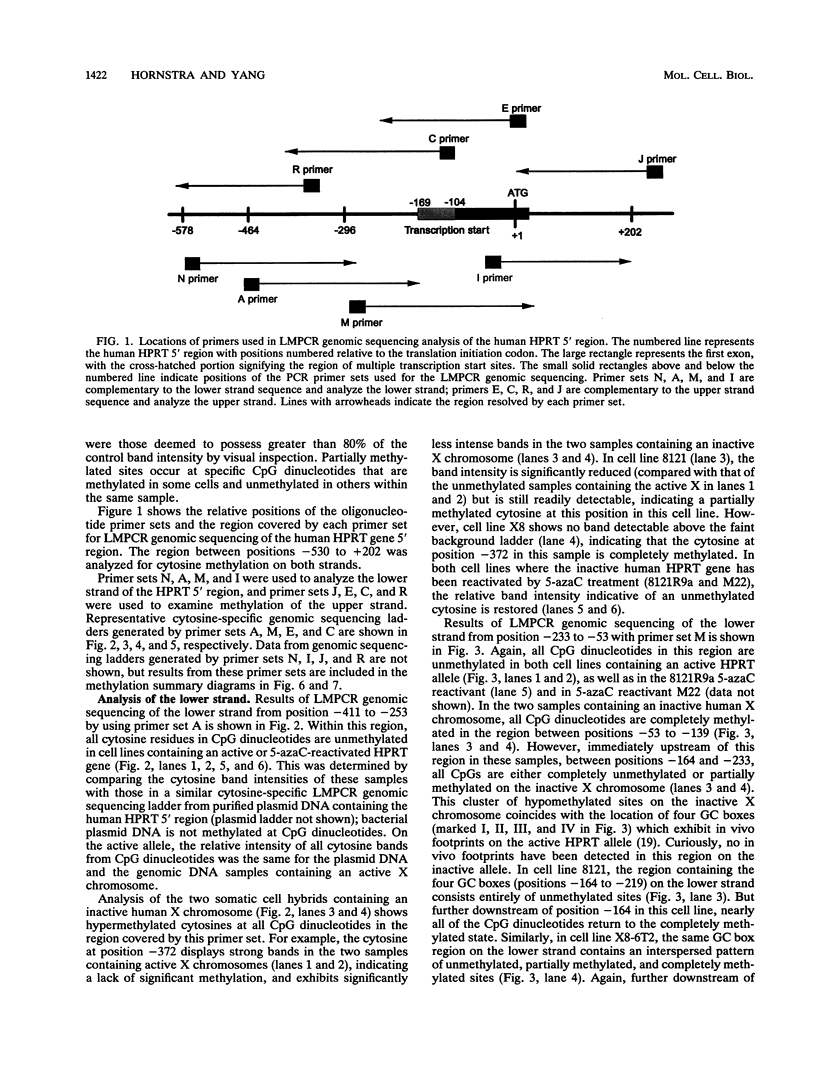
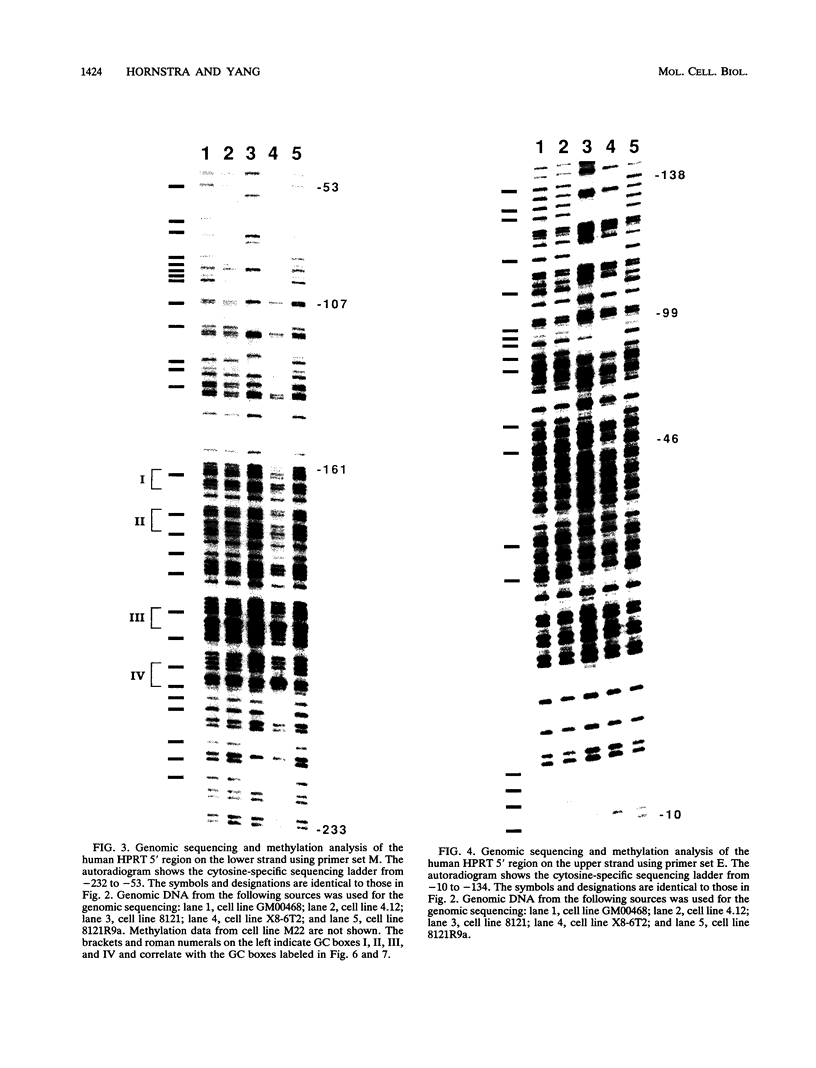
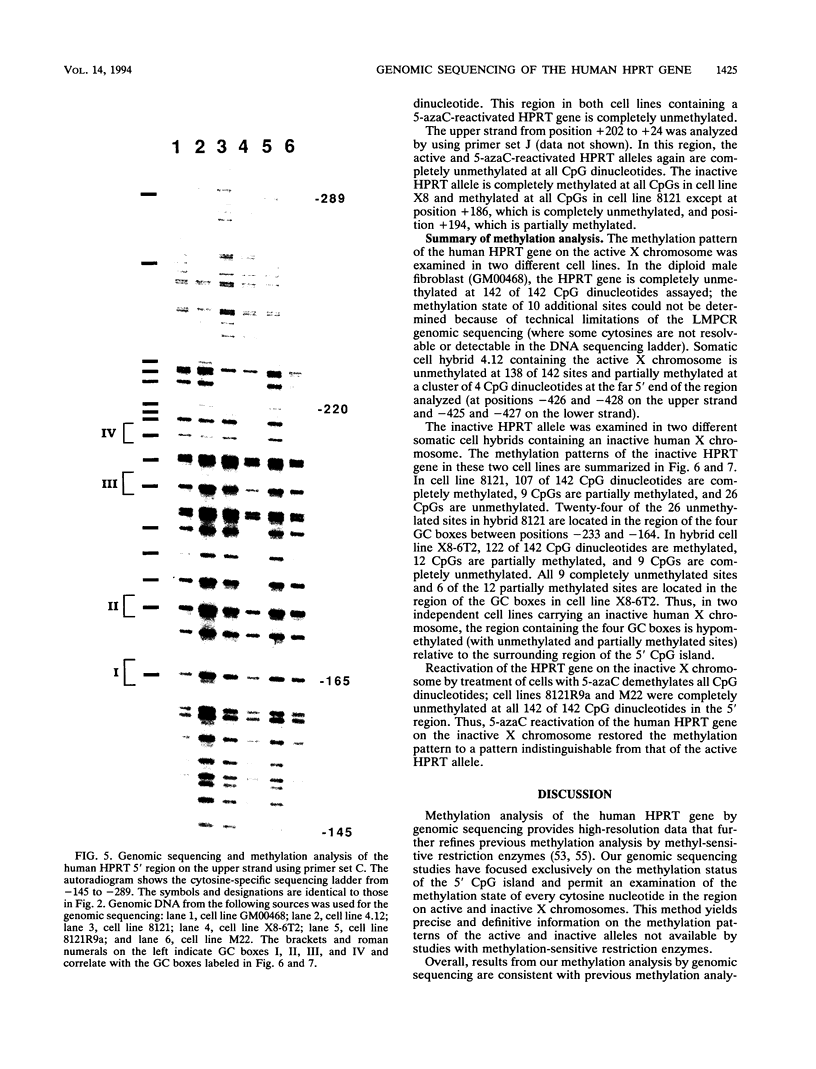
DNA methylation within GC-rich promoters of constitutively expressed X-linked genes is correlated with transcriptional silencing on the inactive X chromosome in female mammals. For most X-linked genes, X chromosome inactivation results in transcriptionally active and inactive alleles occupying each female nucleus. To examine mechanisms responsible for maintaining this unique system of differential gene expression, we have analyzed the methylation of individual cytosine residues in the 5' CpG island of the human hypoxanthine phosphoribosyltransferase (HPRT) gene on the active and inactive X chromosomes. Methylation analysis of 142 CpG dinucleotides by genomic sequencing was carried out on purified DNA using the cytosine-specific Maxam and Gilbert DNA sequencing reaction in conjunction with ligation-mediated PCR. These studies demonstrate the 5' CpG islands of active and 5-azacytidine-reactivated alleles are essentially unmethylated while the inactive allele is hypermethylated. The inactive allele is completely methylated at nearly all CpG dinucleotides except in a 68-bp region containing four adjacent GC boxes where most CpG dinucleotides are either unmethylated or partially methylated. Curiously, these GC boxes exhibit in vivo footprints only on the active X chromosome, not on the inactive X. The methylation pattern of the inactive HPRT gene is strikingly different from that reported for the inactive X-linked human phosphoglycerate kinase gene which exhibits methylation at all CpG sites in the 5' CpG island. These results suggest that the position of methylated CpG dinucleotides, the density of methylated CpGs, the length of methylated regions, and/or chromatin structure associated with methylated DNA may have a role in repressing the activity of housekeeping promoters on the inactive X chromosome. The pattern of DNA methylation on the inactive human HPRT gene may also provide insight into the process of inactivating the gene early in female embryogenesis.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Hattar J., Beard P., Jiricny J. Cytosine methylation in CTF and Sp1 recognition sites of an HSV tk promoter: effects on transcription in vivo and on factor binding in vitro. Nucleic Acids Res. 1989 Dec 25;17(24):10179–10190. doi: 10.1093/nar/17.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Buschhausen G., Wittig B., Graessmann M., Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C. R., Hubberman P., Kaslow D. C., Migeon B. R. Comparison of factor IX methylation on human active and inactive X chromosomes: implications for X inactivation and transcription of tissue-specific genes. EMBO J. 1986 Sep;5(9):2223–2229. doi: 10.1002/j.1460-2075.1986.tb04488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracopoli N. C., Rettig W. J., Albino A. P., Esposito D., Archidiacono N., Rocchi M., Siniscalco M., Old L. J. Genes controlling gp25/30 cell-surface molecules map to chromosomes X and Y and escape X-inactivation. Am J Hum Genet. 1985 Jan;37(1):199–207. [PMC free article] [PubMed] [Google Scholar]
- Ellis N., Keitges E., Gartler S. M., Rocchi M. High-frequency reactivation of X-linked genes in Chinese hamster X human hybrid cells. Somat Cell Mol Genet. 1987 May;13(3):191–204. doi: 10.1007/BF01535202. [DOI] [PubMed] [Google Scholar]
- Fuscoe J. C., Fenwick R. G., Jr, Ledbetter D. H., Caskey C. T. Deletion and amplification of the HGPRT locus in Chinese hamster cells. Mol Cell Biol. 1983 Jun;3(6):1086–1096. doi: 10.1128/mcb.3.6.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARTLER S. M., BURT B. REPLICATION PATTERNS OF BOVINE SEX CHROMOSOMES IN CELL CULTURE. Cytogenetics. 1964;3:135–142. doi: 10.1159/000129804. [DOI] [PubMed] [Google Scholar]
- Garrity P. A., Wold B. J. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler S. M., Riggs A. D. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Chapman V. M. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Ellis N. A., Gartler S. M. Demethylation of specific sites in the 5' region of the inactive X-linked human phosphoglycerate kinase gene correlates with the appearance of nuclease sensitivity and gene expression. Mol Cell Biol. 1988 Nov;8(11):4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5' CpG island. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington M. A., Jones P. A., Imagawa M., Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstra I. K., Yang T. P. In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal Biochem. 1993 Sep;213(2):179–193. doi: 10.1006/abio.1993.1407. [DOI] [PubMed] [Google Scholar]
- Hornstra I. K., Yang T. P. Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5' region: implications for X chromosome inactivation. Mol Cell Biol. 1992 Dec;12(12):5345–5354. doi: 10.1128/mcb.12.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Keith D. H., Singer-Sam J., Riggs A. D. Active X chromosome DNA is unmethylated at eight CCGG sites clustered in a guanine-plus-cytosine-rich island at the 5' end of the gene for phosphoglycerate kinase. Mol Cell Biol. 1986 Nov;6(11):4122–4125. doi: 10.1128/mcb.6.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem B. S., Goitein R., Richler C., Marcus M., Cedar H. In situ nick-translation distinguishes between active and inactive X chromosomes. Nature. 1983 Jul 7;304(5921):88–90. doi: 10.1038/304088a0. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kovesdi I., Reichel R., Nevins J. R. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Cantoni G. L., Razin A. Methylation in the preinitiation domain suppresses gene transcription by an indirect mechanism. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10119–10123. doi: 10.1073/pnas.89.21.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J., Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991 Jul 22;285(2):155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Liskay R. M., Evans R. J. Inactive X chromosome DNA does not function in DNA-mediated cell transformation for the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4895–4898. doi: 10.1073/pnas.77.8.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock L. F., Melton D. W., Caskey C. T., Martin G. R. Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol Cell Biol. 1986 Mar;6(3):914–924. doi: 10.1128/mcb.6.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock L. F., Takagi N., Martin G. R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987 Jan 16;48(1):39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McBurney M. W. X chromosome inactivation: a hypothesis. Bioessays. 1988 Aug-Sep;9(2-3):85–88. doi: 10.1002/bies.950090211. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., Bird A. P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992 Oct 11;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes R. S., Shapiro L. J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981 Jan 23;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Monaco A. P., Kunkel L. M. Cloning of the Duchenne/Becker muscular dystrophy locus. Adv Hum Genet. 1988;17:61–98. doi: 10.1007/978-1-4613-0987-1_3. [DOI] [PubMed] [Google Scholar]
- Monk M. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):299–312. doi: 10.1098/rstb.1990.0013. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- OHNO S., KAPLAN W. D., KINOSITA R. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp Cell Res. 1959 Oct;18:415–418. doi: 10.1016/0014-4827(59)90031-x. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Hansen R. S., Gartler S. M., Riggs A. D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Tanguay R. L., Steigerwald S. D., Riggs A. D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990 Aug;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989 Sep 8;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rommens J. M., Iannuzzi M. C., Kerem B., Drumm M. L., Melmer G., Dean M., Rozmahel R., Cole J. L., Kennedy D., Hidaka N. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989 Sep 8;245(4922):1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Hansen R. S., Gartler S. M. Hemimethylation and hypersensitivity are early events in transcriptional reactivation of human inactive X-linked genes in a hamster x human somatic cell hybrid. Mol Cell Biol. 1992 Sep;12(9):3819–3826. doi: 10.1128/mcb.12.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J., Grant M., LeBon J. M., Okuyama K., Chapman V., Monk M., Riggs A. D. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990 Sep;10(9):4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J. T., Caskey C. T. HPRT: gene structure, expression, and mutation. Annu Rev Genet. 1985;19:127–148. doi: 10.1146/annurev.ge.19.120185.001015. [DOI] [PubMed] [Google Scholar]
- Toniolo D., Martini G., Migeon B. R., Dono R. Expression of the G6PD locus on the human X chromosome is associated with demethylation of three CpG islands within 100 kb of DNA. EMBO J. 1988 Feb;7(2):401–406. doi: 10.1002/j.1460-2075.1988.tb02827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Gartler S. M. Comparison of transformation efficiency of human active and inactive X-chromosomal DNA. Nature. 1983 Mar 3;302(5903):82–83. doi: 10.1038/302082a0. [DOI] [PubMed] [Google Scholar]
- Venolia L., Gartler S. M., Wassman E. R., Yen P., Mohandas T., Shapiro L. J. Transformation with DNA from 5-azacytidine-reactivated X chromosomes. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2352–2354. doi: 10.1073/pnas.79.7.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988 Sep;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Jolly D. J., Lunnen K. D., Friedmann T., Migeon B. R. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc Natl Acad Sci U S A. 1984 May;81(9):2806–2810. doi: 10.1073/pnas.81.9.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen P. H., Mohandas T., Shapiro L. J. Stability of DNA methylation of the human hypoxanthine phosphoribosyltransferase gene. Somat Cell Mol Genet. 1986 Mar;12(2):153–161. doi: 10.1007/BF01560662. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Patel P., Chinault A. C., Mohandas T., Shapiro L. J. Differential methylation of hypoxanthine phosphoribosyltransferase genes on active and inactive human X chromosomes. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]