Abstract
Background
Visceral leishmaniasis (VL), caused by infection with Leishmania donovani complex, remains a major public health problem in endemic regions of South Asia, East Africa, and Brazil. If untreated, symptomatic VL is usually fatal. Rapid field diagnosis relies principally on demonstration of anti-Leishmania antibodies in clinically suspect cases. The rK39 immunochromatographic rapid diagnostic test (RDT) is based on rK39, encoded by a fragment of a kinesin-related gene derived from a Brazilian L. chagasi, now recognised as L. infantum, originating from Europe. Despite its reliability in South Asia, the rK39 test is reported to have lower sensitivity in East Africa. A reason for this differential response may reside in the molecular diversity of the rK39 homologous sequences among East African L. donovani strains.
Methodology/Principal Findings
Coding sequences of rK39 homologues from East African L. donovani strains were amplified from genomic DNA, analysed for diversity from the rK39 sequence, and compared to South Asian sequences. East African sequences were revealed to display significant diversity from rK39. Most coding changes in the 5′ half of repeats were non-conservative, with multiple substitutions involving charge changes, whereas amino acid substitutions in the 3′ half of repeats were conservative. Specific polymorphisms were found between South Asian and East African strains. Diversity of HASPB1 and HASPB2 gene repeat sequences, used to flank sequences of a kinesin homologue in the synthetic antigen rK28 designed to reduce variable RDT performance, was also investigated. Non-canonical combination repeat arrangements were revealed for HASPB1 and HASPB2 gene products in strains producing unpredicted size amplicons.
Conclusions/Significance
We demonstrate that there is extensive kinesin genetic diversity among strains in East Africa and between East Africa and South Asia, with ample scope for influencing performance of rK39 diagnostic assays. We also show the importance of targeted comparative genomics in guiding optimisation of recombinant/synthetic diagnostic antigens.
Author Summary
Visceral leishmaniasis (VL) is caused by infection with parasites of the Leishmania donovani complex, spread by the bite of blood-sucking sandflies, especially in South Asia, East Africa, and Brazil. If untreated, symptomatic VL leads to systemic pathologies and is usually fatal. Up to approximately 400,000 new cases are estimated to occur annually, and regional epidemics have been devastatingly severe. Diagnosis of clinically suspect cases in the field, and thus appropriate treatment, relies principally on a rapid diagnostic test (RDT) based on detection of antibodies against a Leishmania antigen known as rK39. Although this test is reliable in South Asia, it has shown less success in East Africa. One reason may reside in diversity of the rK39 homologue among East African L. donovani, with consequent limitations in the binding of diagnostic antibodies. Here, we investigate the sequences of rK39 homologues in a panel of East African L. donovani strains, and show that there is significant diversity compared to rK39 and to South Asian sequences. Additionally, we examine diversity in another diagnostic antigen known as HASPB1/2. Our results indicate that an improved RDT may need to encompass East-African-specific antigen diversity to provide high performance field diagnosis for this region.
Introduction
Visceral leishmaniasis (VL) remains a major public health concern in many parts of the tropical world, with the great majority of the estimated 200,000 to 400,000 annual new cases found in South Asia, East Africa, and Brazil [1], [2]. VL is caused by kinetoplastid protozoa of the Leishmania donovani complex. These are: L. donovani in South Asia and East Africa; L. infantum, mainly in Europe, the wider Mediterranean region, and in Latin America, where it was historically also known as L. chagasi but is now demonstrated to be synonymous with L. infantum originating from Europe [3].
Leishmania promastigotes, transmitted during bloodmeal feeding by female sandflies (Phlebotomus and Lutzomyia spp., in the Old and New World respectively), are internalised by local dermal macrophages and dendritic cells. Within these host cells, flagella are lost, and transformation into proliferative amastigote forms is followed by cell lysis, re-invasion of other cells, and parasite dissemination by the lymphatic and vascular systems, which can lead to infiltration of bone marrow and hepatosplenomegaly. In symptomatic cases, VL is fatal if untreated [4].
Parasitological diagnosis, by demonstration of amastigotes in spleen aspirates, approximates to a gold standard for VL diagnosis but is applied cautiously due to associated risk. Serological (anti-Leishmania antibody) tests include enzyme-linked immunosorbent assay (ELISA), indirect fluorescent antibody test (IFAT), and direct agglutination test (DAT) [5]. However, after drug treatment and cure current serological tests may still give positive results and therefore cannot readily diagnose relapse. Furthermore, such tests can also detect anti-leishmanial antibodies in asymptomatic individuals living in endemic areas, but with no VL history or subsequent progression to VL [6].
Burns et al [7] identified a kinesin-related gene product, LcKin, as a candidate diagnostic antigen by screening a Brazilian L. infantum (L. chagasi) genomic library with serum of an L. donovani patient. A part of the coding sequence, comprising a 46aa region followed by 6.5×39aa repeats, forms the recombinant diagnostic protein rK39. In recent multicentre evaluations, the use of the rK39 in a lateral-flow immunochromatographic, rapid test format reported less success in East Africa than in the Indian subcontinent for point-of-care diagnosis of VL [8], [9]. Underlying explanatory factors may reside in molecular divergence between East African L. donovani kinesin gene homologues and the Brazilian L. infantum (L. chagasi)-derived rK39 sequence, and/or may be due to differential immunocompetence and antibody levels produced among African and Asian human populations.
Studies on South Asian L. donovani strains using PCR primers based on LcKin have identified rK39 homologous sequences [10]–[12]. Gerald et al [13] reported the first East African (Sudanese) L. donovani kinesin homologue, LdK39. The first two of the 39-aa repeats of LdK39, flanked by sequences of the L. donovani antigens HASPB1 and HASPB2 [14], comprise rK28, a novel recombinant protein for diagnosis of VL, designed to be an improvement over rK39 [15]–[16]. HASPB proteins are expressed on the surface of infective promastigote and amastigote Leishmania life cycle stages [14]. The first 3×14aa repeats of HASPB1 are incorporated into rK28, along with the complete ORF of HASPB2, which includes three imperfect consecutive repeats, 2×14aa and 1×13aa. HASPB1 and HASPB2 correspond with K26 and K9, respectively [17], which were originally identified in L. infantum (L. chagasi) at the same time as the HASPBs, but were given different nomenclature. A recent comparison of rK26 and rK9 showed significantly lower diagnostic efficacy than rK28 [15].
Here we investigate whether molecular divergence in rK39 kinesin sequence homologues of East African L. donovani may contribute to lower rK39 diagnostic test success rates in East Africa. We analyse the rK39 homologues in a panel of East African L. donovani strains and compare their diversity against published South Asian sequences. In addition, we compare sequence diversity of HASPB1 and HASPB2 in East African and South Asian strains.
Methods
L. donovani strains
Table 1 lists the East African L. donovani strains used in this study. Strains were selected to represent genetic groups within the L. donovani complex in East Africa (Baleela et al, unpublished data; identified by multilocus sequence typing (MLST) and microsatellite analysis (MLMT)). Strains were cultured in αMEM medium supplemented by foetal calf serum (Sigma, UK), and genomic DNA was extracted from uncloned cultures using Gentra Puregene Tissue Core Kit A (Qiagen, UK). Uncloned cultures were specifically used here because the intention was to capture the diversity present within natural L. donovani populations. Table 2 lists the GenBank sequences derived in other studies [7], [10]–[14], [18]–[19] and used here for comparisons.
Table 1. East African L. donovani strains for which sequences were determined, with GenBank accession numbers.
| Strain | Origin | MON/LON a | Kinesin | HASPB amplicon | ||
| HASPB1 | HASPB2 | Unpredicted | ||||
| MHOM/ET/67/HU3 (LV9) | Ethiopia | MON18/LON46 | KC342866 | KC342849 | KC342855 | - |
| MHOM/ET/00/HUSSEN | Ethiopia | MON31/LON42 | KC342867 | - | - | KC342861 |
| MHOM/ET/72/GEBRE1 | Ethiopia | MON82/LON50 | - | KC342850 | - | - |
| MHOM/SD/87/UGX-MARROW | Sudan | MON31 | KC342868 | - | - | KC342862 |
| MHOM/SD/82/GILANI | Sudan | MON30/LON48 | KC342869 | KC342851 | KC342856 | - |
| MHOM/SD/98/LEM3582 | Sudan | MON18 | KC342870 | KC342852 | KC342857 | - |
| MHOM/SD/XX/SUDAN1 | Sudan | MON18 | KC342871 | - | KC342858 | - |
| MCAN/SD/98/LEM3556 | Sudan | MON82 | - | KC342853 | KC342859 | - |
| MHOM/SD/97/LEM3458 | Sudan | MON18 | - | KC342854 | KC342860 | - |
| IMAR/KE/62/LRC-L57 | Kenya | MON37/LON44 | KC342872 | - | - | KC342863 |
| MHOM/KE/67/MRC(L)3 | Kenya | MON37/LON44 | - | - | - | KC342864 |
| MCAN/IQ/81/SUKKAR 2 | Iraq | LON43 | - | - | - | KC342865 |
MON and LON reference numbers refer to multilocus sequence enzyme electrophoresis (MLEE) profiles.
Table 2. GenBank sequences used in comparisons.
| Strain | Origin | Gene product | GenBank | Reference |
| MHOM/BR/82/BA-2,C1 | Brazil | Kinesin LcKin/rK39 | L07879 | [7] |
| MHOM/IN/KE16/1998 | India | Kinesin/Ld-rKE16 | AY615886 | [10] |
| Morenaa | India | Kinesin | DQ648599 | [11] |
| MHOM/IN/80/DD8 | b | Kinesin/rKRP42 | AB256033 | [12] |
| MHOM/SD/62/1S-CL2D | Sudan | Kinesin/LdK39 | DQ831678 | [13] |
| MCAN/ES/98/LLM-877 (JPCM5)c | Spain | Kinesin | XM_00146426e | [18] |
| MHOM/NP/2003/BPK282/0cl4d | Nepal | Kinesin | FR799601e | [19] |
| MHOM/ET/67/L28 (LV9) | Ethiopia | HASPB1 | AJ011810 | [14] |
| MHOM/ET/67/L28 (LV9) | Ethiopia | HASPB2 | AJ011809 | [14] |
| MHOM/NP/2003/BPK282/0cl4d | Nepal | HASPB | FR799601e | [19] |
Described in [11] as clinical isolate, not laboratory strain, so WHO code not given here.
Described in [12] as isolated from a Bangladeshi patient, but MHOM/IN/80/DD8 is also reported as being from India [6].
L. infantum reference genome.
L. donovani reference genome.
GenBank numbers refer to locations on the entire chromosome 23 sequence, to which both the kinesin and HASPB BLAST searches map.
Kinesin homologues
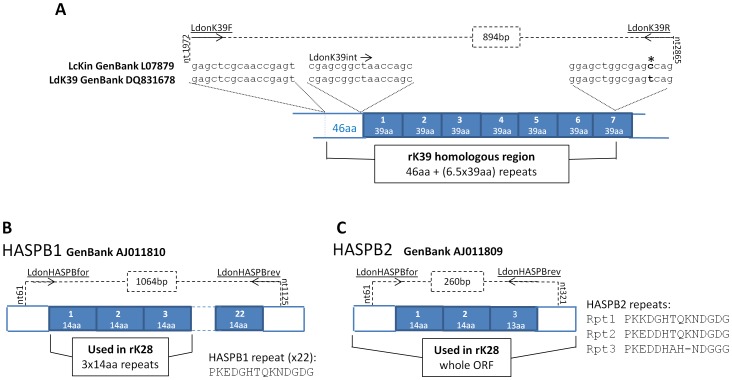
PCR primers LdonK39F (gagctcgcaaccgagt) and LdonK39R (ctgrctcgccagctcc) were designed for this study based on a comparison of LcKin and LdK39 coding sequences (Figure 1A), and were targeted to amplify the 894 bp region of the L. donovani kinesin gene that is incorporated into the diagnostic antigen rK39. The level of conservation in the sequences encoding the 39-aa repeats predicts that primer LdonK39R would be expected to anneal to multiple sites within the kinesin gene template, generating multiple amplicons. Amplification reactions were performed in a total volume of 20 ul, and comprised of 1× NH4 Reaction buffer supplemented with 1.5 mM MgCl2 (Bioline, UK), 200 µM dNTPs (New England Biolabs, UK), 10 pmol of each primer, and 1 U BioTaq DNA polymerase (Bioline). Amplification conditions were: 1 cycle of 94°C, 2 mins; 25 cycles of 94°C for 30 secs, 55°C for 30 secs, 72°C for 1 min; 1 cycle of 72°C for 5 mins. PCR products were separated by electrophoresis on 1.5% agarose gels (Bioline). Amplicons corresponding to rK39 homologues were excised and purified from gels using QIAquick Gel Extraction Kit (Qiagen), cloned into pGEM-T easy vector (Promega, UK), and transformed into XL1-blue E. coli on blue-white selection. Primers Sp6/T7 and M13for/rev were used to sequence from the vector (between 2–5 colonies per strain were taken forward for sequencing, except UGX-MARROW and SUDAN1, where due to difficulty in cloning the corresponding inserts only one clone was sequenced); in addition, LdonK39int (cgagcggctaaccagc), which binds to the 3′ end of the non-repeat region immediately upstream of the repeats, was used as an internal sequencing primer (Figure 1A). Sequences were analysed using BioEdit [20].
Figure 1. Leishmania rK39 and HASPB antigen repeats and the PCR primer target sequences.
Repetitive coding regions depicted as filled boxes, PCR primers underlined, and 5′ and 3′ binding positions with amplicons are indicated by dashed lines. [A] Kinesin gene comparison for primer design (* = non-conserved nucleotide). [B] HASPB1 GenBank sequence displays 22× perfect 14aa repeats. [C] HASPB2 GenBank sequence displays 3 imperfect repeats.
HASPB
PCR primers LdonHASPBfor (cataaaaccactgaggc) and LdonHASPBrev (atcttcgttcttctcctg) were designed for this study to flank the repeat regions of the HASPB1 ORF, amplifying a 1064 bp product (Figure 1B). Due to the identity between HASPB1 and HASPB2 at the primer binding sites, a 260 bp product would also be predicted to be amplified from HASPB2 by these primers (Figure 1C). Composition of PCR mix was as described above for kinesin, except that 40 µM dNTPs were used. Amplification conditions were: 1 cycle of 94°C, 2 mins; 25 cycles of 94°C for 30 secs, 50°C for 30 secs, 72°C for 1 min; 1 cycle of 72°C for 10 mins. PCR products were separated by electrophoresis on 1% agarose gels (Bioline). Amplicons were excised and purified from gels using QIAquick Gel Extraction Kit, and sequenced directly using the amplification primers. Sequences were analysed using BioEdit.
Accession numbers
Kinesin nucleotide sequences derived in this manuscript are available under GenBank accession numbers KC342866- KC342872. HASPB Nucleotide sequences derived in this manuscript are available under GenBank accession numbers KC342849- KC342865.
Results
East African L. donovani kinesin diversity
Multiple kinesin amplicons were produced using a combination of primers LdonK39F, which binds to the non-repeat region and LdonK39R, because the latter primer binds to nucleotide sequence that is conserved across the repeats. An example is shown in Figure 2. Sequencing of cloned amplicons containing the rK39 homologous sequences into plasmid vectors revealed the presence of nucleotide and predicted amino acid diversity among East African L. donovani strains. Table 3 shows the amino acid polymorphisms that were found among the East African strains, together with their divergence from L. infantum (L. chagasi) derived LcKin rK39, and alongside the GenBank sequence for L. donovani derived kinesin LdK39 used in rK28. Substitutions between a non-charged and a charged residue (D−, E−, H+, K+, R+) in comparison with the LcKin rK39 sequence are shown underlined.
Figure 2. Multiple amplicons corresponding to kinesin tandem repeats are produced by PCR primers LdonK39F and LdonK39R.
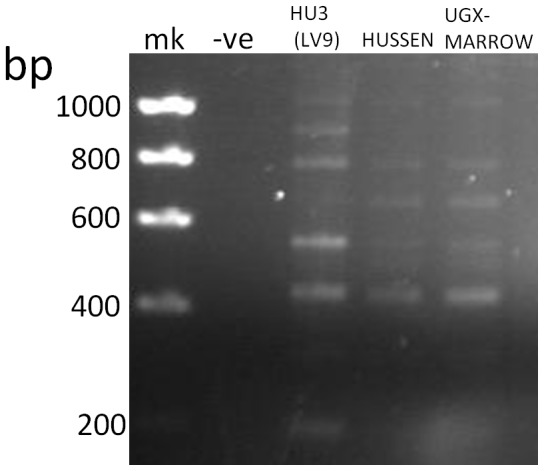
Amplifications from strains HU3 (LV9), Hussen, and UGX-MARROW, are depicted. Major amplicon sizes differ by 117 bp, the size of the nucleotide sequence encoding the 39aa repeat in the kinesin gene; mk = Hyperladder I (Bioline).
Table 3. rK39 polymorphism among East African L. donovani strains and in comparison with the LcKin diagnostic rK39 sequence.
| P c | Rpt1 | Rpt2 | Rpt3 | |||||||||||||||||||
| Amino Acid | 41 | 4 | 21 | 23 | 27 | 39 | 6 | 7 | 10 | 21 | 23 | 24 | 27 | 39 | 7 | 8 | 10 | 21 | 23 | 27 | 35 | 39 |
| LcKin(used in rK39) a | C | Q | A | A | S | T | R | E | A | A | A | A | M | T | D | S | E | S | T | M | S | T |
| LdK39(used in rK28) b | S | L | S | T | M | T | R | D | E | A | A | A | S | A | D | S | E | S | T | T | N | T |
| MHOM/ET/67/HU3(LV9) | S | L | S | T | M | T | R | D | E | A | A | A | S | A | D | SP | E | S | T | T | N | T |
| MHOM/ET/00/HUSSEN | S | Q | A | A | S | A | R | E | E | A | A | A | SP | A | E | S | A | SA | TA | MS | N | T |
| MHOM/SD/87/UGX-MAR | S | Q | A | A | S | A | R | E | E | A | A | A | S | A | E | S | A | S | T | M | N | T |
| MHOM/SD/82/GILANI | S | L | SA | TA | MS | T | R | D | E | SA | TA | A | MS | TA | D | S | E | AS | AT | ST | N | AT |
| MHOM/SD/98/LEM3582 | S | L | S | T | M | T | R | D | E | A | A | VA | S | A | D | S | E | S | T | T | N | T |
| MHOM/SD/XX/SUDAN1 | S | L | S | T | M | T | R | D | E | A | A | A | S | A | D | S | E | S | T | T | N | T |
| IMAR/KE/62/LRC-L57 | S | Q | A | A | S | T | L | E | E | S | T | A | M | A | E | S | E | A | A | S | N | T |
LcKin is derived from L. infantum (L. chagasi).
LdK39 is derived from L. donovani.
P = pre-repeat region.
– = not determined here.
When the seven rK39 homologous repeats of all the East African strains were compared with the LcKin rK39 amino acid sequence, residues 2, 6, 10, 16 and 18 were each affected three or four times by substitutions involving charge changes. In contrast, although there were multiple substitutions in the latter half of the East African rK39 tandem repeats, especially affecting residues 21, 23, 27 and 39, none of these substitutions involved charge changes. Two tracts of the rK39 repeat, residues 11 to 15 and 28 to 34 were perfectly conserved between LcKin39 across all the East African strains examined and across all seven rK39 repeats within those strains.
Sequencing of multiple plasmid clones revealed the presence of alternative residues at single sites within single strains. This was notably widespread for the two strains Gilani and Hussen, in the case of Gilani, including several alternatives within the first and second repeats.
Comparison of L. donovani kinesin diversity between East Africa and South Asia
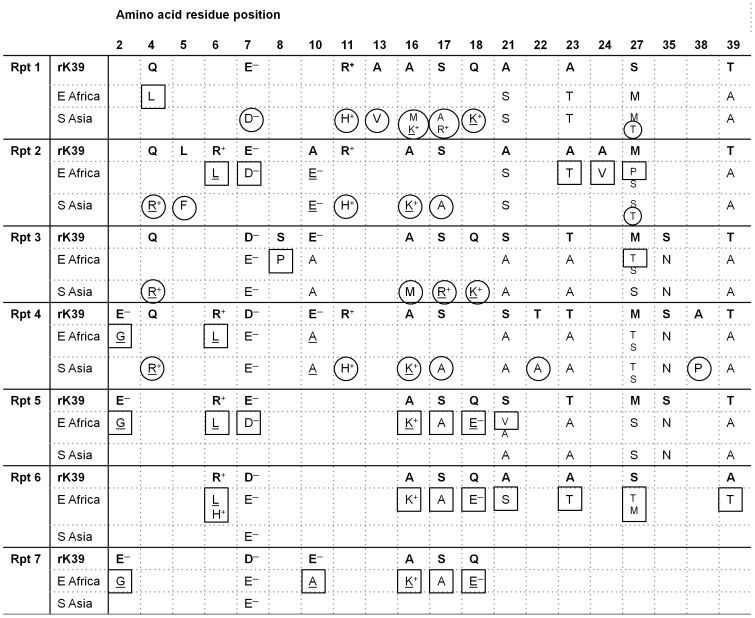
Figure 3 depicts the composite L. donovani kinesin polymorphisms for East Africa and South Asia and the divergence between the two geographic regions, in comparison with the LcKin rK39 derived from L. infantum (L. chagasi). The East African polymorphisms in Figure 3 incorporate the data obtained here together with the published LdKin (rK28) sequence. Substitutions between non-charged and charged residues (D−, E−, H+, K+, R+) compared to the LcKin rK39 sequence are shown underlined. East Africa-specific polymorphisms are boxed; South Asia-specific polymorphisms are circled.
Figure 3. Polymorphisms among seven rK39 repeats of East African and South Asian strains show region-specific substitutions.
Changes between charged and non-charged residues are underlined. No residue entered at a particular site indicates conservation of that residue with the corresponding residue of diagnostic rK39. Where two alternative residues are indicated in smaller text these are not necessarily always within the same strain, for clarification compare with Table 3. Region-specific polymorphisms in each repeat are boxed (East Africa) or circled (South Asia).
In the 46aa non-repeat region the only divergence from L. infantum (L. chagasi) diagnostic LcKin rK39 was Cys→Ser41, found among both South Asian and East African strains (Table 3). Throughout the 6.5×39aa repeats, there were polymorphisms divergent from the diagnostic LcKin rK39 that were unique to each region and other polymorphisms that were common to East Africa and South Asia. For example, Glu→Gly2 (Repeats 4, 5, 7) and Arg→His11 (Repeats 1, 2, 4) are found only in East Africa and South Asia respectively, whereas in Repeat 1, Ala→Ser21, Ala→Thr23, Ser→Met27, Thr→Ala39, which are all conservative changes found in the latter half of the repeats, were found in both regions. Indeed, as for the East African strains (Table 3) all such polymorphisms identified in the latter half of the repeats among South Asian strains are conservative, i.e., a non-charged residue is replaced by another non-charged residue; conversely, non-conservative substitutions between non-charged and charged residues are found in the first half of the repeats. Among the published South Asian sequences charges diverged from those in diagnostic LcKin rK39 at residue 4 (Gln→Arg; Repeats 2, 3, 4) and at residues 16, 17 and 18, for example uncharged to positive at these three sites in repeat 1 (Figure 3).
Comparison with L. donovani complex reference genomes
After the sequencing of the L. infantum reference genome [18], Downing et al [19] sequenced the Nepalese L. donovani strain BPK282/0cl4 as the reference genome for L. donovani. The sequence coding for the non-repeat 46-aa region of rK39 was submitted to NCBI BLAST against the reference genomes. For the L. infantum reference genome (Spanish, canine-isolated), there were no differences across the entire rK39 sequence, in accordance with this being the postulated source of L. infantum (L. chagasi) in South America [3]. For the genome sequence of BPK282, due to the repeat nature of the downstream region, only sequences of the first two repeats could be unequivocally aligned and these were incorporated into Figure 3 for comparisons with the individual repeat sequences generated here. From the unequivocal BLAST alignment information, BPK282 repeat 1 is the same as for rK39 except Thr→Ala39; in BPK repeat 2, where only the first 6 residues are unambiguously assembled, Gln→Arg4 is present.
L. donovani HASPB
PCR primers LdonHASPBfor and LdonHASPBrev were designed to bind unique sequences flanking the 22×14aa-repeat coding region of HASPB1 to produce a 1064 bp amplicon. However, with some of the strains studied here in addition to the 1064 bp product the smaller 260 bp amplicon corresponding to HASPB2 (Figure 1C) could also be seen: an example is shown for HU3 (LV9) in Figure 4. However, these primers unexpectedly gave amplicons of ∼400–500 bp for some of the strains (Hussen, UGX-marrow, LRC-L57, MRC(L)3, SUKKAR 2) (Figure 4).
Figure 4. Predicted (1064 bp, 260 bp) amplicons and unpredicted (∼400–500 bp) amplicons with HASPB PCR primers LdonHASPBfor and LdonHASPBrev.
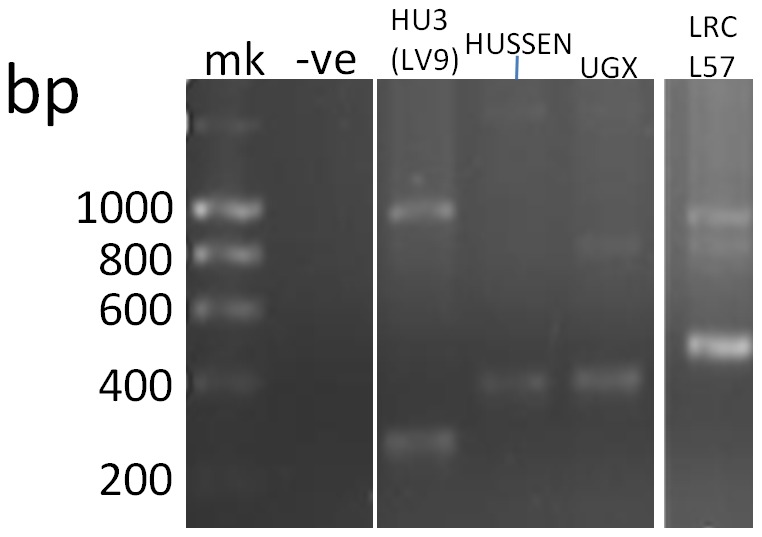
Amplifications from strains HU3 (LV9), Hussen, UGX-MARROW, and LRC-L57, are depicted; mk = Hyperladder I.
Comparisons with rK28 HASPB sequences
Strains HU3 (LV9), Gebre1, Gilani, LEM3582, LEM3556, and LEM3458, which gave 1064 bp-amplicons, had identical sequences in their first 3×14aa repeats as the HASPB1 used in diagnostic rK28. However, subsequent repeats, not incorporated into rK28, were imperfect with polymorphic residues at certain sites, such as His→Arg6 (repeats 15, 18, 22) and Gly→Ala14 (repeats 5, 14, 17) for strains LEM3582, LEM3556, and LV9.
For those strains producing the predicted 260 bp amplicon of the HASPB2 homologue, (Figure 1) a comparison with the rK28 sequence identified a Gly residue inserted between Ala32 and Val33 preceding the imperfectly repeated 14/13aa region and a Pro→Gln96 substitution present after the 14/13aa region. The only polymorphism within the imperfectly repeated 14/13aa region, compared to rK28, was Lys→Glu49, at the third residue of first 14aa region, with a consequent charge change.
Interestingly, the strains producing unexpected ∼400–500 bp amplicons (Hussen, UGX-marrow, LRC-57, SUKKAR 2, MRC(L)3) revealed on sequence analysis a HASPB1/HASPB2-like combination composition across their repeats, due to the presence of the central motifs that were HASPB1-like (HTQKN) or HASPB2-like (HAH-N). The HTQKN and HAH-N motifs were not consistently present in the same repeat numbers across these five strains and the HASPB1-like and HASPB2-like sections of the tandem repeat were not necessarily aggregated (Table 4). In the case of LRC/L57, both the 1064 bp (predicted HASPB1-homologue) and ∼500 bp amplicons contained the HASPB2-like motif HAH-N. The Lys→Glu49 substitution at the third residue of the first 14aa region, described above, was also present in those strains giving the ∼400–500 bp amplicons.
Table 4. Repeat sequences of unexpected amplicons.
| HUSSEN | UGX-Marrow | LRC-L57 | MRC(L)3 | SUKKAR 2 | |
| Rpt 1 | LKEDGHTQKNDGDG | LKEDGHTQKNDGDG | PKEDGHTQKNDGDG | PKEDGHTQKNDGGG | LKEDGHTQKNDGDG |
| Rpt 2 | PKEDGHTQKNDGGA | PKEDGHTQKNDGGA | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDGHTQKNDGDG |
| Rpt 3 | PKEDGHTQKNDGDG | PKEDGHTQKNDGDG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGDG |
| Rpt 4 | PKEDDHAH-NDGDG | PKEDDHAH-NDGDG | PKEDGHTQKNDGDG | PKEDGHTQKNDGDG | PKEDDHAH-NDGDG |
| Rpt 5 | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG |
| Rpt 6 | PKEDDHAH-NDGGA | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG | PKEDDHAH-NDGGG |
| Rpt 7 | PKEDDHAH-NDGGG | PKEDGHTQKNDGGV | |||
| Rpt 8 | PKEDGHTQKNDGGV |
Central motifs:
HTQKN = HASPB1-like.
HAH-N = HASPB2 like.
Comparison with L. donovani reference genome
The sequence coding for the pre-repeat region of HASPB1 (not used in rK28, Figure 1A) was submitted to NCBI BLAST against the Nepalese L. donovani reference genome, to facilitate correct alignment. Due to the repeat nature of the downstream region, only the first two repeats of the reference genome could be unequivocally aligned for comparisons with the individual sequences generated here. The first two repeats of the reference genome showed the combination composition of central motifs HTQKN in repeat 1 (HASPB1-like) and HAH-N in repeat 2 (HASPB2-like). Thus, the South Asian reference genome has a HASPB1/2 combination structure like the four East African strains Hussen, UGX-marrow, LRC-57, MRC(L)3, and the Iraqi SUKKAR 2, and with the same organisation of repeats 1 and 2 seen in the Kenyan strains LRC-57 and MRC(L)3.
Discussion
For VL there is a particular need for rapid diagnostic tests (RDTs) that can be unequivocally applied in endemic regions at primary care level and with only basic technical training. Of the available options for serological diagnosis of VL only the immunochromatographic rK39 assay can be considered a point of care test for field application. The IFAT requires a fluorescence microscope. Use of the DAT is constrained by the readout delay, the relative sophistication of the procedure and limited access to lyophilised antigen. In agreement with other studies, Boelaert et al [8] recommend the use of rK39 in South Asia, but report less satisfactory results from East Africa. One factor influencing regional differences in diagnostic performance may be divergence in kinesin gene homologues in East African L. donovani from the L. infantum (L. chagasi)-derived LcKin rK39 sequence. In an attempt to overcome the geographical limitations of the rK39 test a modified recombinant antigen, rK28, has been devised, incorporating segments of the HASPB antigen.
Analyses of L. donovani complex isolates with multiple molecular markers have revealed extensive genetic diversity with at least six distinct lineages, and both inter- and intra-lineage diversity. Furthermore, genotyping of isolates from endemic regions has shown an association between genotype and geographical origin [21]–[22]. Kuhls et al [22] used multilocus microsatellite typing (MLMT) for high resolution comparison of the six global L. donovani genetic lineages, and showed that allelic diversity in Africa was greater than in India; a subsequent MLMT study also described comparative homogeneity among South Asian L. donovani [23]. Thus, the relative homogeneity of South Asian L. donovani and greater diversity in East Africa, if mirrored in diversity of the kinesin gene, is one potential explanation of the more limited efficacy of diagnostic rK39 in East Africa.
Here we determined across a panel of East African strains the diversity of the kinesin repeat region homologous to that used in both the rK39 diagnostic antigen (repeats 1 to 6.5) and the rK28 recombinant antigen (repeats 1 and 2). We then compared this diversity to that among predetermined South Asian sequences.
Surprisingly, in view of the proven value of the rK39 RDT in South Asia, there is considerable amino acid diversity among kinesin repeats 1 to 7 of South Asian strains as well as those from East Africa (Figure 3). Alternative residues at multiple sites suggested that Gilani and Hussen are heterozygous, although sequences were not obtained from DNA clones and therefore multiclonality rather than heterozygosity cannot be excluded. Across all strains the diversity was particularly notable in the first half of each rK39 repeat, because it involved charge changes at residues 2, 6, 10, 16, and 18, with only a short stretch of consistently conserved residues (11 to 15). Such charge changes among the South Asian strains are very likely to disrupt antigenic epitopes, if present. As far as we are aware epitopes within or across adjacent rK39 repeats have not yet been precisely mapped. However, we would predict that the diagnostic epitopes lie in the latter half of the rK39 repeat, where, although there is extensive diversity, the changes are conservative, none involving charge, and there is one stretch of residues (28 to 34) that is entirely conserved across all isolates and across all seven repeats. This is consistent with the recent work of Costa et al [24] in which antigen prediction software led to the synthesis of a 22mer peptide (ESTTAAKMSAEQDRESTRATLE) encompassing the region of rK39 from residue 20 of repeat 3 to residue of 2 of repeat 4, a sequence which is also found in the next two repeats. In that study, the peptide was recognised in ELISA by Brazilian sera from symptomatic and asymptomatic canine VL and symptomatic human VL. However, our study shows that at least 5 of the residues within that 22mer are polymorphic in East Africa.
Kinesin repeat divergence from the rK39 diagnostic recombinant is complex, and some divergence is common to both South Asia and East Africa, for example at residues 7, 10, 21, 23 (Figure 3). Nevertheless, there were polymorphisms unique to each region, for example at residues 4, 16, and 18, consistent with the expectation that differential sensitivities of rK39 assays may be partially attributable to positional and physiochemical polymorphisms of the kinesin gene. The identification and confirmation of region-specific polymorphisms may be limited by the amount of sequence data so far available. Also diversity beyond the seven sequenced repeats of the kinesin tandem array may present a wider range of epitopes for immunological recognition.
The differential immunocompetence and antibody titres of East African and South Asian human populations in the context of VL remains to be thoroughly explored, as a major alternative or contributing explanation of variable rK39 test sensitivities. Interestingly a recent WHO/TDR evaluation [9] of a rapid diagnostic test based on rKE16 antigen, a LcKin homologue derived from an Indian L. donovani [10], reported markedly lower sensitivities against Brazilian and East African sera, compared to South Asian. The same evaluation [9] also reported a lower sensitivity of rK39-based tests with Brazilian sera. However, the rK39 antigen is derived from a Brazilian L. infantum (L. chagasi) strain [7], which originated from Iberia in Europe and is considered to be relatively homogeneous, based on microsatellite typing [3]. Thus, kinesin diversity among Brazilian L. infantum should be explored further and comparative immune response levels among Brazilian and South American populations.
In the rK28 recombinant diagnostic antigen, designed to improve on performance of the rK39 test, highly conserved HASPB repetitive amino acid sequences flank the first two repeats of Ldk39, derived from an East African strain. Alce et al [14], following the work of McKean et al [25], amplified two homologues of the L. major Gene B protein from L. donovani, naming the ORF of the larger HASPB1 and that of the smaller HASPB2. Recombinant HASPB1 has been used in animal vaccine models [26]–[27], and HASPB has a role in L. major differentiation within the sandfly vector P. papatasi [28]. The PCR primers used in the current study, like those used by Alce et al [14], anneal to regions flanking HASPB repeat regions and generate two PCR products of predicted size corresponding to HASPB1 and HASPB2, compatible with the presence of two distinct HASPB1 and HASPB2 loci in the genome, as concluded for the L. major genome [29]. However, several strains produced unexpected sized amplicons with a mix of HASPB1 and HASPB2 motifs within these PCR products, indicating a structural reorganisation of HASPB in these strains. Similarly, Haralambous et al [30] in K26, a homologue of HASPB, also found unexpected amplicons for the Hussen strain as compared to Gebre1 and Gilani. Nevertheless, despite this structural reorganisation, there was limited HASPB amino acid diversity across the strains. There was only one polymorphism compared to rK28 that involved a charge change, Lys→Glu49 in HASPB2, potentially affecting antigenicity. A recent study [31] reports HASPB repeat sequence diversity in Indian L. donovani strains; a comparison with sequences derived here shows that among those East African strains producing unexpected sized amplicons (Table 4), there is the presence of repeat sequence types that are also predominant among Indian strains (PKEDDHAHNDGGG, PKEDGHTQKNDGDG, PKEDDHAHNDGDG). In addition we find repeat types not reported from Indian strains, for example repeats beginning with Lys in place of Pro (Hussen, UGX-Marrow, SUKKAR 2), or ending in Val (LRC-L57, MRC(L)3).
We did not have patient samples from which to attempt direct amplification of kinesin and HASPB and confirm diversity in situ. Nevertheless the polymorphisms described here do not occur stochastically across strains but at sites of reported kinesin diversity, or in the case of the HASPB2, are consistent among strains (Ala32-Gly-Val33 or Pro→Gln96). Furthermore, the strains were cryopreserved, not subjected to prolonged passage in vitro, and sequences were determined bi-directionally and/or repeated to confirm their validity.
We have undertaken the most comprehensive analysis of diagnostic kinesin and HASPB antigen diversity in East African strains to date. We show that there is extensive kinesin genetic diversity among strains and between East Africa and South Asia, with ample scope for influencing performance of rK39 diagnostic assays. Future research should both widen and focus the genomic comparisons between strains and also compare immune competence profiles among East African, Brazilian and South Asian populations as an alternative or contributory factor to variable RDT performance. There are broader implications from these findings. Firstly, we see the crucial importance of sustaining accessible collections of Leishmania strains representative of genetic lineages and global diversity. Secondly, due to present limitations in the capacity of whole genome sequencing to assemble complex antigen gene families, especially those comprised of repeated sequences, targeted analysis of individual strains is also necessary for these comparisons of genetic diversity. Thirdly, it is evident that comparative genomics has a vital role in guiding the optimisation of recombinant or synthetic diagnostic antigens. Not only are even better diagnostic tests needed for diagnosis of VL but biomarkers are urgently required to distinguish symptomatic cases, asymptomatic infections at risk or not of progression to VL, and post-treatment outcome (relapse versus cure).
Acknowledgments
We thank Isabel Mauricio and Rania Baleela for guidance on choice of strains, Matthew Yeo for contributing to parasite culture, and for their constructive comments on the manuscript. We also thank François Chappuis for his comments. The authors are also grateful to anonymous reviewers for their improvements to the manuscript.
Funding Statement
This work is part of the NIDIAG network research partnership supported by the European Commission under the Health Cooperation Work Programme of the 7th Framework Programme (Grant agreement no. 260260, http://cordis.europa.eu/fp7/home_en.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2010). First WHO report on neglected tropical diseases: Working to overcome the global impact of neglected tropical diseases. WHO, Geneva. http://www.who.int/neglected_diseases/2010report/en/index.html. [PubMed]
- 2. Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, et al. (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuhls K, Alam MZ, Cupolillo E, Ferreira GEM, Mauricio IL, et al. (2011) Comparative microsatellite typing of New World Leishmania infantum reveals low heterogeneity among populations and its recent Old World origin. PLoS Negl Trop Dis 5: e1155 doi:10.1371/journal.pntd.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S, et al. (2007) Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 5: 873–882. [DOI] [PubMed] [Google Scholar]
- 5. Srivastava P, Dayama A, Mehrotra S, Sundar S (2011) Diagnosis of visceral leishmaniasis. Trans R Soc Trop Med Hyg 105: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO (2010). WHO Technical Report Series 949. Control of the Leishmaniases. WHO, Geneva. http://apps.who.int/iris/bitstream/10665/44412/1/WHO_TRS_949_eng.pdf. [PubMed]
- 7. Burns JM, Shreffler WG, Benson DR, Ghalib HW, Badaro R, et al. (1993) Molecular characterization of a kinesin-related antigen of Leishmania chagasi that detects specific antibody in African and American visceral leishmaniasis. Proc Natl Acad Sci USA 90: 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boelaert M, El-Safi S, Hailu A, Mukhtar M, Rijal S, et al. (2008) Diagnostic tests for kala-azar: a multi-centre study of the freeze-dried DAT, rK39 strip test and KAtex in East Africa and the Indian subcontinent. Trans R Soc Trop Med Hyg 102: 32–40. [DOI] [PubMed] [Google Scholar]
- 9. Cunningham J, Hasker E, Das P, El Safi S, Goto H, et al. (2012) A global comparative evaluation of commercial immunochromatographic rapid diagnostic tests for visceral leishmaniasis. Clin Infect Dis 55: 1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sivakumar R, Sharma P, Chang K-P, Singh S (2006) Cloning, expression, and purification of a novel recombinant antigen from Leishmania donovani . Protein Expr Purif 46: 156–165. [DOI] [PubMed] [Google Scholar]
- 11. Dey A, Sharma U, Singh S (2007) First case of indigenous visceral leishmaniasis from central India. Am J Trop Med Hyg 77: 95–8. [PubMed] [Google Scholar]
- 12. Takagi H, Islam MZ, Itoh M, Islam AU, Saifuddin Ekram AR, et al. (2007) Production of recombinant kinesin-related protein of Leishmania donovani and its application in the serodiagnosis of visceral leishmaniasis. Am J Trop Med Hyg 76: 902–5. [PubMed] [Google Scholar]
- 13. Gerald NJ, Coppens I, Dwyer DN (2007) Molecular dissection and expression of the LdK39 kinesin in the human pathogen, Leishmania donovani . Mol Microbiol 63: 962–979. [DOI] [PubMed] [Google Scholar]
- 14. Alce TM, Gokool S, McGhie D, Stager S, Smith DF (1999) Expression of hydrophilic surface proteins in infective stages of Leishmania donovani . Mol Biochem Parasitol 102: 191–196. [DOI] [PubMed] [Google Scholar]
- 15. Pattabhi S, Whittle J, Mohamath R, El-Safi S, Moulton GG, et al. (2010) Design, development and evaluation of rK28-based point-of-care tests for improving rapid diagnosis of visceral leishmaniasis. PLoS Negl Trop Dis 4: e822 doi:10.1371/journal.pntd.0000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaish M, Bhatia A, Reed SG, Chakravarty J, Sundar S (2012) Evaluation of rK28 antigen for serodiagnosis of visceral Leishmaniasis in India. Clin Microbiol Infect 18: 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhatia A, Daifalla NS, Jen S, Badaro R, Reed SG, et al. (1999) Cloning, characterization and serological evaluation of K9 and K26: two related hydrophilic antigens of Leishmania chagasi . Mol Biochem Parasitol 102: 249–261. [DOI] [PubMed] [Google Scholar]
- 18. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, et al. (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21: 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall TA (1999) Bioedit: a user friendly biological sequence alignment edit and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 21. Lukeš J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, et al. (2007) Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci USA 104: 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhls K, Keilonat L, Ochsenreither S, Schaar M, Schweynoch C, et al. (2007) Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral leishmaniasis. Microbes Infect 9: 334–343. [DOI] [PubMed] [Google Scholar]
- 23. Alam MZ, Kuhls K, Schweynoch C, Sundar S, Rijal S, et al. (2009) Multilocus microsatellite typing (MLMT) reveals genetic homogeneity of Leishmania donovani strains in the Indian subcontinent. Infect Genet Evol 9: 24–31. [DOI] [PubMed] [Google Scholar]
- 24. Costa MM, Penido M, dos Santos MS, Doro D, de Freitas E, et al. (2012) Improved Canine and Human Visceral Leishmaniasis Immunodiagnosis Using Combinations of Synthetic Peptides in Enzyme-Linked Immunosorbent Assay. PLoS Negl Trop Dis 6: e1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKean PG, Trenholme KR, Rangarajan D, Keen JK, Smith DF (1997) Diversity in repeat-containing surface proteins of Leishmania major . Mol Biochem Parasitol 86: 225–235. [DOI] [PubMed] [Google Scholar]
- 26. Stäger S, Smith DF, Kaye PM (2000) Immunization with a recombinant stage-regulated surface protein from Leishmania donovani induces protection against visceral leishmaniasis. J Immunol 165: 7064–7071. [DOI] [PubMed] [Google Scholar]
- 27. Moreno J, Nieto J, Masina S, Cañavate C, Cruz I, et al. (2007) Immunization with H1, HASPB1 and MML Leishmania proteins in a vaccine trial against experimental canine leishmaniasis. Vaccine 25: 5290–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sádlová J, Price HP, Smith BA, Votýpka J, Volf P, et al. (2010) The stage-regulated HASPB and SHERP proteins are essential for differentiation of the protozoan parasite Leishmania major in its sand fly vector, Phlebotomus papatasi . Cell Microbiol 12: 1765–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Depledge DP, MacLean LM, Hodgkinson MR, Smith BA, Jackson AP, et al. (2010) Leishmania-Specific Surface Antigens Show Sub-Genus Sequence Variation and Immune Recognition. PLoS Negl Trop Dis 4: e829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haralambous C, Antoniou M, Pratlong F, Dedet J-P, Soteriadou K (2008) Development of a molecular assay specific for the Leishmania donovani complex that discriminates L. donovani/Leishmania infantum zymodemes: a useful tool for typing MON-1. Diagn Microbiol Infect Dis 60: 33–42. [DOI] [PubMed] [Google Scholar]
- 31. Maroof A, Brown N, Smith B, Hodgkinson MR, Maxwell A, et al. (2012) Therapeutic vaccination with recombinant adenovirus reduces splenic parasite burden in experimental visceral leishmaniasis. J Infect Dis 205: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]