Abstract
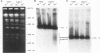
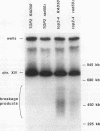
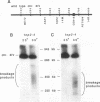
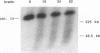
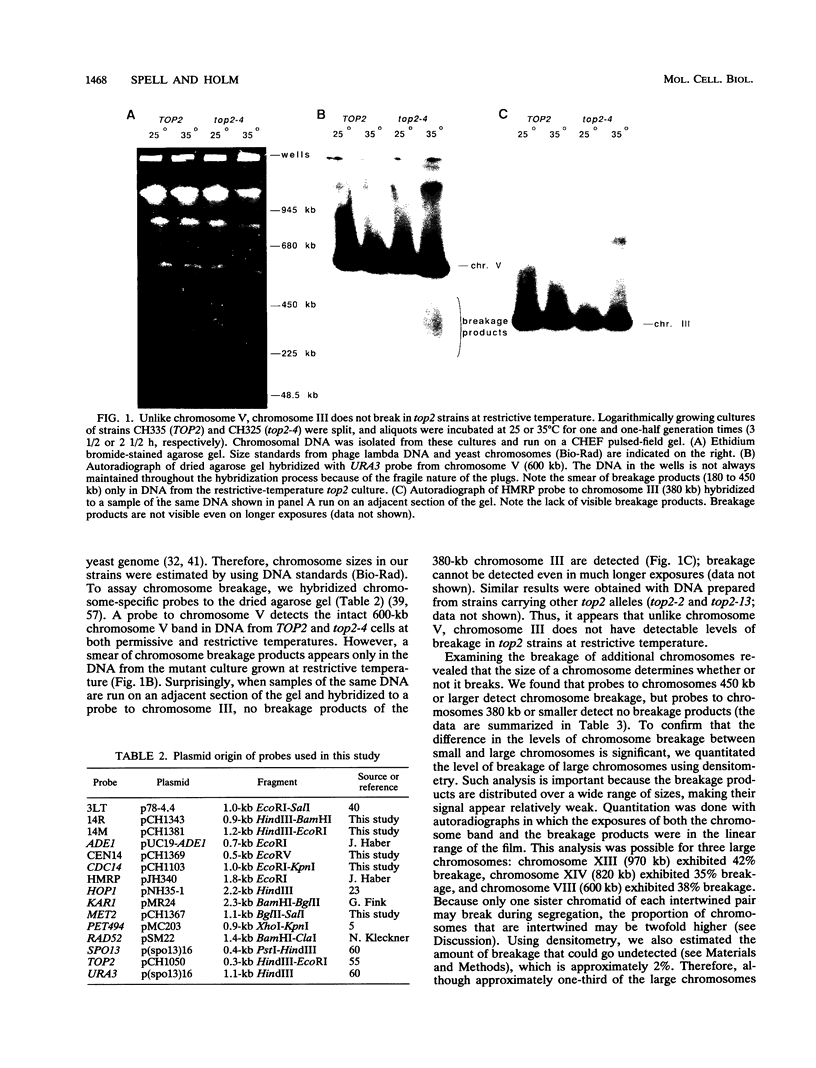
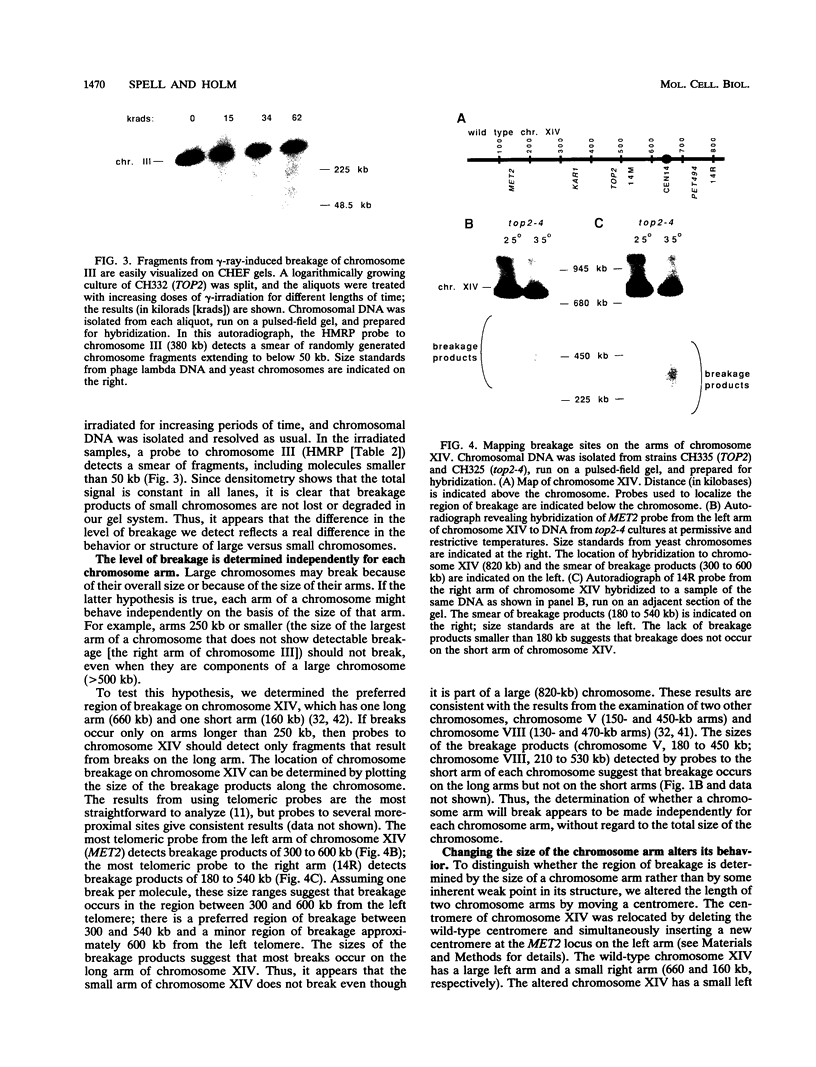
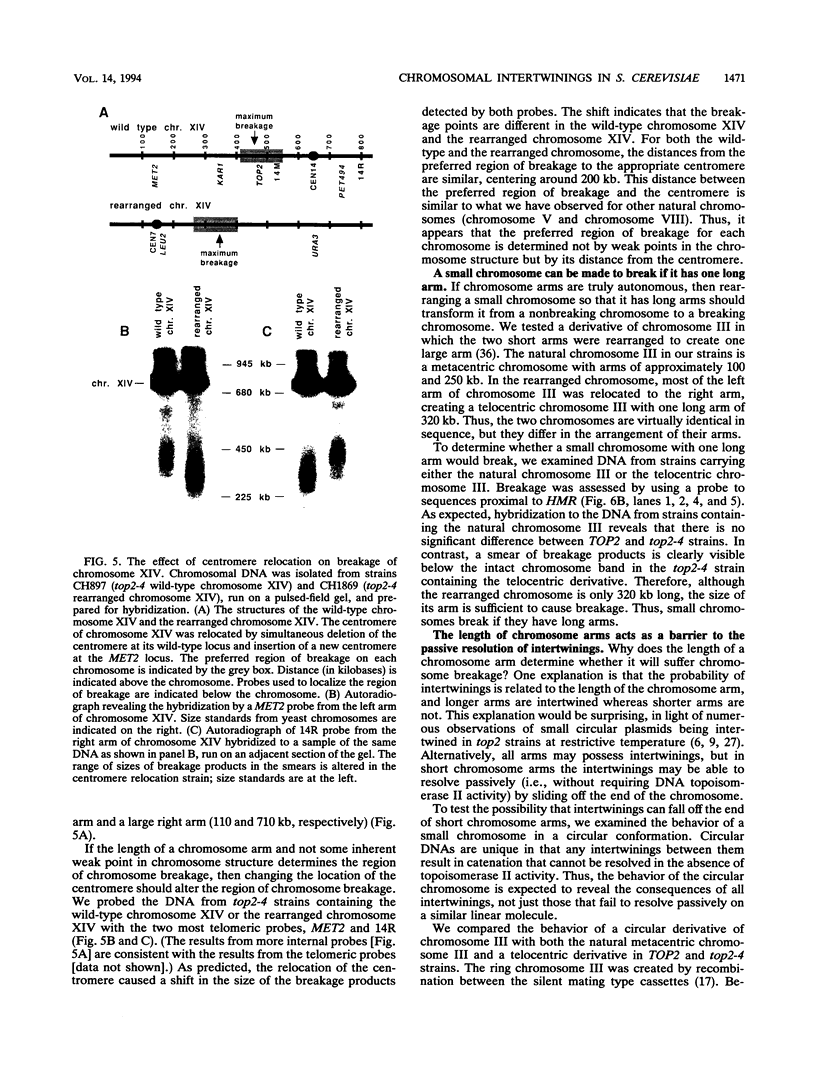
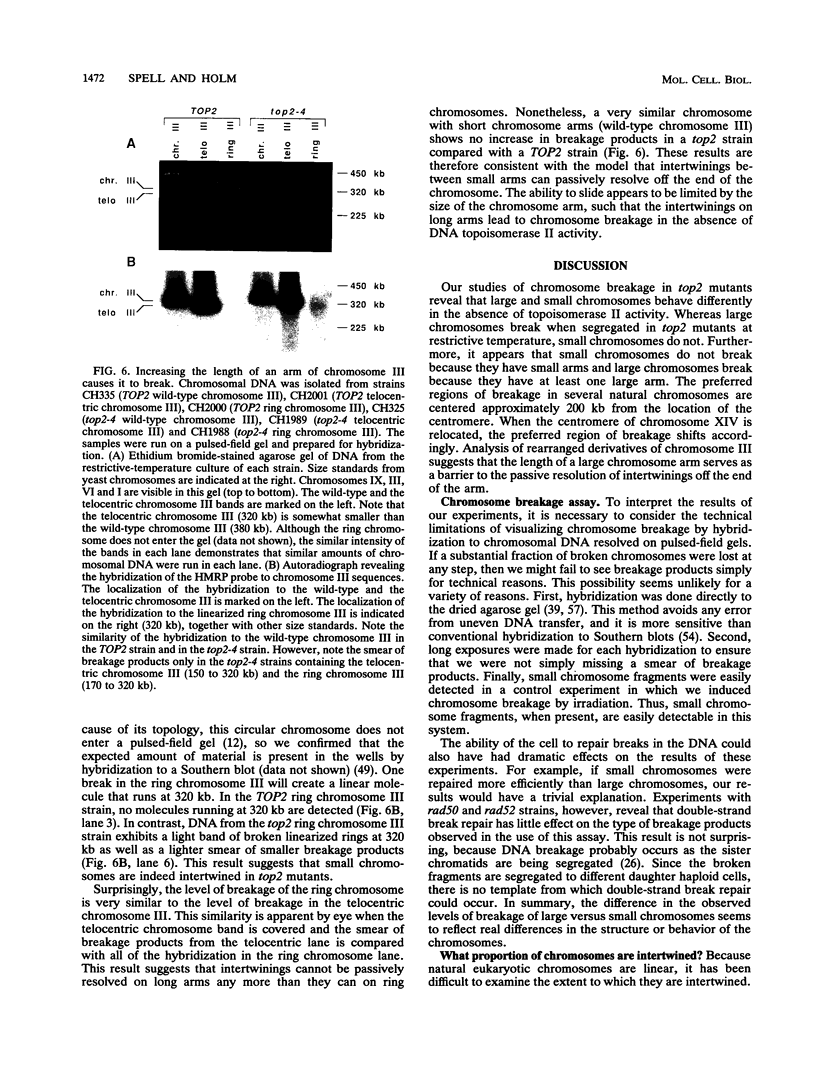
To elucidate yeast chromosome structure and behavior, we examined the breakage of entangled chromosomes in DNA topoisomerase II mutants by hybridization to chromosomal DNA resolved by pulsed-field gel electrophoresis. Our study reveals that large and small chromosomes differ in the nature and distribution of their intertwinings. Probes to large chromosomes (450 kb or larger) detect chromosome breakage, but probes to small chromosomes (380 kb or smaller) reveal no breakage products. Examination of chromosomes with one small arm and one large arm suggests that the two arms behave independently. The acrocentric chromosome XIV breaks only on the long arm, and its preferred region of breakage is approximately 200 kb from the centromere. When the centromere of chromosome XIV is relocated, the preferred region of breakage shifts accordingly. These results suggest that large chromosomes break because they have long arms and small chromosomes do not break because they have small arms. Indeed, a small metacentric chromosome can be made to break if it is rearranged to form a telocentric chromosome with one long arm or a ring with an "infinitely" long arm. These results suggest a model of chromosomal intertwining in which the length of the chromosome arm prevents intertwinings from passively resolving off the end of the arm during chromosome segregation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. O., Cozzarelli N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979 Nov 30;206(4422):1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986 Nov;6(11):3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci U S A. 1984 May;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K. Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Mar;1(3):245–253. doi: 10.1128/mcb.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields-Berry S. C., DePamphilis M. L. Sequences that promote formation of catenated intertwines during termination of DNA replication. Nucleic Acids Res. 1989 Apr 25;17(8):3261–3273. doi: 10.1093/nar/17.8.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993 Jun;121(5):961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Bell M., King J. S., Mortimer R. K. Random-breakage mapping, a rapid method for physically locating an internal sequence with respect to the ends of a DNA molecule. Nucleic Acids Res. 1990 Aug 11;18(15):4453–4461. doi: 10.1093/nar/18.15.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Sitney K. C., Cook V. E., Mortimer R. K. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989 Dec;123(4):695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S. M., Amati B. B., Cardenas M. E., Hofmann J. F. Studies on scaffold attachment sites and their relation to genome function. Int Rev Cytol. 1989;119:57–96. doi: 10.1016/s0074-7696(08)60649-x. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986 Apr 20;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Gerring S. L., Spencer F., Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990 Dec;9(13):4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder S. A., Newlon C. S. A replication map of a 61-kb circular derivative of Saccharomyces cerevisiae chromosome III. Mol Biol Cell. 1992 Sep;3(9):999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., Thorburn P. C., Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984 Feb;106(2):185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M. M., Earnshaw W. C. Topoisomerase II: A specific marker for cell proliferation. J Cell Biol. 1986 Dec;103(6 Pt 2):2569–2581. doi: 10.1083/jcb.103.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hirano T., Mitchison T. J. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993 Feb;120(3):601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989 Mar;121(3):445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway S. L., Glotzer M., King R. W., Murray A. W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993 Jul 2;73(7):1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989 Jan;9(1):159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Hartwell L. H. The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science. 1987 Dec 18;238(4834):1713–1716. doi: 10.1126/science.3317838. [DOI] [PubMed] [Google Scholar]
- Langin T., Faugeron G., Goyon C., Nicolas A., Rossignol J. L. The MET2 gene of Saccharomyces cerevisiae: molecular cloning and nucleotide sequence. Gene. 1986;49(3):283–293. doi: 10.1016/0378-1119(86)90364-1. [DOI] [PubMed] [Google Scholar]
- Liras P., McCusker J., Mascioli S., Haber J. E. Characterization of a mutation in yeast causing nonrandom chromosome loss during mitosis. Genetics. 1978 Apr;88(4 Pt 1):651–671. [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Contopoulou C. R., King J. S. Genetic and physical maps of Saccharomyces cerevisiae, Edition 11. Yeast. 1992 Oct;8(10):817–902. doi: 10.1002/yea.320081002. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Schultes N. P., Szostak J. W. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986 May 23;45(4):529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction and behavior of circularly permuted and telocentric chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Sep;6(9):3166–3172. doi: 10.1128/mcb.6.9.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction of artificial chromosomes in yeast. Nature. 1983 Sep 15;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B. Chromosome micromanipulation. II. Induced reorientation and the experimental control of segregation in meiosis. Chromosoma. 1967;21(1):17–50. doi: 10.1007/BF00330545. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Koch C. A. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol. 1969 Oct;43(1):40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purrello M., Balazs I. Direct hybridization of labeled DNA to DNA in agarose gels. Anal Biochem. 1983 Feb 1;128(2):393–397. doi: 10.1016/0003-2697(83)90391-3. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., McCarroll R. M., Newlon C. S., Fangman W. L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989 Oct;9(10):4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D., Thomas W., Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990 Mar 23;60(6):1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Scherthan H., Loidl J., Schuster T., Schweizer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma. 1992 Oct;101(10):590–595. doi: 10.1007/BF00360535. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Surosky R. T., Newlon C. S., Tye B. K. The mitotic stability of deletion derivatives of chromosome III in yeast. Proc Natl Acad Sci U S A. 1986 Jan;83(2):414–418. doi: 10.1073/pnas.83.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky R. T., Tye B. K. Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics. 1985 Jul;110(3):397–419. doi: 10.1093/genetics/110.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J. R., Sedat J. W., Agard D. A. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three-dimensional wide-field microscopy. Cell. 1993 Apr 9;73(1):97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- Thomas W., Spell R. M., Ming M. E., Holm C. Genetic analysis of the gyrase A-like domain of DNA topoisomerase II of Saccharomyces cerevisiae. Genetics. 1991 Aug;128(4):703–716. doi: 10.1093/genetics/128.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987 Sep 11;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Wang H. T., Frackman S., Kowalisyn J., Esposito R. E., Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987 Apr;7(4):1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D. T., Fields-Berry S. C., DePamphilis M. L. The termination region for SV40 DNA replication directs the mode of separation for the two sibling molecules. Cell. 1985 Jun;41(2):565–575. doi: 10.1016/s0092-8674(85)80029-5. [DOI] [PubMed] [Google Scholar]