Abstract
The structure of the centromere-specific histone centromeric protein A (CENP-A) nucleosome has been a hot topic of debate for the past five years. Structures proposed include octamers, hexamers, homotypic and heterotypic tetramers. This controversy has led to the proposal that CENP-A nucleosomes undergo cell-cycle dependent transitions between the multiple states previously documented to exist in vivo and in vitro. In recent work from our laboratory, we sought to test this hypothesis. We discovered that CENP-A nucleosomes undergo unique oscillations in human cells, a finding mirrored in a parallel study performed in budding yeast. This review provides additional insights into the potential mechanisms for the interconversion of CENP-A nucleosomal species, and speculates on a biological role for oscillations in vivo.
Keywords: CENP-A, nucleosome, structure, oscillation, octamer, tetramer, hemisome, cell-cycle, AFM, acetylation
Introduction
CENP-A is a variant of histone H31 found at the centromere, wherein it replaces its canonical nucleosome counterpart in a significant fraction of the nucleosomes.2 However, unlike canonical nucleosomes, whose structure was firmly established in the 1970s,3 the structure of CENP-A nucleosomes has been at the center of controversy for the past five years. In vitro assembly of both yeast and human CENP-A nucleosomes yields standard octameric structures containing two copies each of CENP-A, H2A, H2B and H4 histones.4,5 Human CENP-A also produces rigidified homotypic CENP-A/H4 tetramers in vitro.6,7 These in vitro studies provide important information about the default state of CENP-A nucleosomes reconstituted in the absence of key biological processes such as mitosis, replication, or transcription, in the absence of the CENP-A chaperone HJURP, and in the absence of key binding partners, such as CENP-C and CENP-N, all of which converge upon centromeric chromatin in vivo. Indeed, a range of non-canonical structures have been documented for CENP-A nucleosomes in vivo, including unstable octamers in Drosophila,8,9 hexameric nucleosomes (homotypic tetramer bound to two copies of the chaperone Scm3) in budding yeast,10 right-handed nucleosomes in budding yeast,11 and “hemisomal” tetramers containing one each of CENP-A, H2A, H2B and H4 -in Drosophila and human cells.12,13
How a single histone variant with a conserved function can adopt so many variable structures has baffled researchers. There are several key differences that may contribute to the difference in data gathered. First, in vitro assembly of histones onto DNA performed under typical salt reconstitution conditions, in the absence of the CENP-A chaperone, Holliday junction-recognition protein (HJURP), may favor octamers over other forms. Indeed, in vitro experiments performed with the chaperone Scm3 in yeast, resulted in CENP-A nucleosomes from yeast yielding non-canonical forms.10 Second, the use of N- and C-terminal tags may alter important nucleosomal properties. Third, the use of chemical fixatives such as formaldehyde can severely impact nucleosomal structure. Fourth, it is feasible that differences in nucleosomal stability could present as non-canonical structures. Nevertheless, proposals attempting to reconcile the in vitro and in vivo data have suggested that CENP-A nucleosomes are unstable14 or adopt different structures across the cell cycle.15-17 Spanning almost four decades of chromatin research, no extant data indicates that a nucleosome can adopt different conformations within the same cell in vivo, making the latter model unprecedented and exciting. As discussed later, such proposed structural oscillations have consequences for epigenetic inheritance and for cell division.
In a recent study, we tested the hypothesis that CENP-A nucleosomes change in structure during the cell cycle.18 Native CENP-A and H3 nucleosomes were purified from various points of the human cell cycle, and their dimensions measured using Atomic Force Microscopy (AFM). Nucleosomal heights and volumes reflect conformation, since octamers containing two copies of each histone, are approximately double the height and volumes of tetramers containing only one copy of each histone.12 Unlike H3 nucleosomes, which exist as invariant octamers, our data revealed that native CENP-A nucleosomes adopt a stable tetrameric structure for the majority of the cell cycle, but alter in shape to an octameric structure at the transition from G1/S to S-phase. In G2 phase, CENP-A nucleosomes convert back to tetramers, suggesting that they are structurally flexible. Unexpectedly, the timing of CENP-A conversion from tetramers to octamers coincides with depletion18 of its chaperone, HJURP.19,20 Using an independent approach that couples Fluorescence Correlation Spectroscopy (FCS) to avalanche photodiode (APD)-confocal imaging, a parallel study reported similar oscillations for CENP-A nucleosomes and the HJURP homolog, Scm3, in S. cerevisiae and C. albicans.21 Thus, these data provide the first glimpse into oscillations that occur within the single-nucleosome context in vivo, and point to a new phenomenon in chromosome biology, which appears to be conserved from yeast to humans, despite the fact that the proteins involved share minimal homology.
This Extra View article provides insights into potential kinetochore and epigenetic components that facilitate structural transitions, and speculates on the interplay between biological processes and CENP-A nucleosomes in vivo.
Single Molecule Microscopy can Distinguish Between CENP-A Octamers and Tetramers in vitro and in vivo
Our recent study utilized AFM to measure the heights of purified native CENP-A nucleosomes18 without artifacts resulting from chemical fixatives, dehydration and cryo-freezing typically associated with Electron Microscopy (EM) and cryo-EM. Thus, AFM is advantageous because it can be performed under gentle conditions, and yet yield a detailed single molecule perspective of native chromatin structures at high resolution.
We first confirmed the default “ground” state of CENP-A nucleosomes, reconstituted in vitro in the absence of any biological processes or chaperones, is octameric. Following standard salt dialysis of histones purified from human cells22 combined with plasmid DNA containing α-satellite arrays, we observed that bulk histones form chromatin arrays packaged with nucleosomes whose sizes were consistent with those of canonical octamers (Fig. 1 and Table 1). Next, to mimic conditions typically used for in vivo purifications, we gently digested these bulk in vitro arrays with Micrococcal Nuclease (MNase) and immuno-precipitated (IP’ed) CENP-A-containing nucleosomes from the reconstituted mix. Analysis of such in vitro reconstituted CENP-A particles indicated that their sizes corresponded closely to those of canonical octameric nucleosomes (Fig. 1 and Table 1). These in vitro data are consistent with previous reports suggesting that the octamer is the default structure for CENP-A nucleosomes assembled by salt dialysis.4,5 This experiment also suggests that there is no inherent instability within octameric CENP-A nucleosomes, at least when subjected to experimental conditions such as nuclease digestion, IP or AFM analyses. The logical conclusion from these results is that alternative species observed for native CENP-A nucleosomes in vivo are not likely to be generated from technical artifacts arising during purification or analysis.
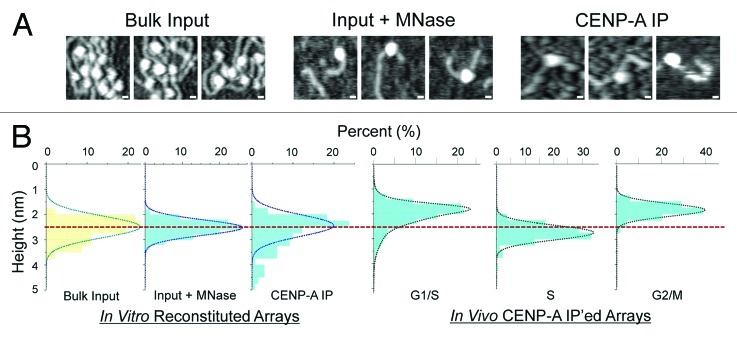
Figure 1. Comparing in vitro reconstituted CENP-A nucleosomes to CENP-A nucleosome purified from human cells using AFM. (A) AFM micrographs showing in vitro reconstituted bulk nucleosomes onto plasmids containing α-satellite DNA derived from human centromeres (left), in vitro reconstituted chromatin digested for 1 min with MNase (middle), and IP’ed in vitro reconstituted CENP-A chromatin digested with MNase (right). Scale bar = 10 nm. (B) Graphical representation of AFM measurements comparing in vitro (listed above and Table 1) and in vivo samples from Bui et al., 2012. Dotted red line designates nucleosomes of mean octameric heights.
Table 1. AFM measurements of in vitro and in vivo CENPA nucleosomes.
| In Vitro Reconstituted Nuclesomes | In Vivo Purified CENP-A Nucleosomes | |||||
|---|---|---|---|---|---|---|
| |
Bulk Input (n) |
Input + MNase (n) |
CENP-A IP (n) |
G1/S-phase (n) |
S-phase (n) |
G2/M-phase (n) |
|
Height (nm) |
2.5 ± 0.5 (44) |
2.5 ± 0.4 (533) |
2.45 ± 0.5 (102) |
1.9 ± 0.28 (1372) |
2.7 ± 0.34 (188) |
1.8 ± 0.27 (479) |
|
Volume (nm3) |
290 ± 75 (44) |
275 ± 65 (533) |
257 ± 75 (102) |
168 ± 33 (1372) |
273 ± 53 (188) |
198 ± 72 (479) |
| Diameter (nm) |
14.6 ± 1.9 (47) |
13.2 ± 1.3 (1063) |
13.3 ± 1.1 (144) |
12.7 ± 0.8 (288) |
13.3 ± 1.9 (246) |
14.6 ± 2.2 (584) |
AFM measurements of in vitro reconstituted bulk chromatin (before and after MNase digestion), IP’ed in vitro reconstituted CENP-A chromatin, and native CENP-A chromatin purified from G1/S, S and G2-phase cells.18 Numbers reported are mean values, standard deviations and sample size (n).
We next examined CENP-A purified from human cells at early G1, G1/S, S, and G2 phases. In contrast to the single form of CENP-A nucleosomes observed in vitro, the in vivo data demonstrate that while CENP-A nucleosomes are octamers from late G1/S to late S-phase, they present a tetrameric conformation at all other points of the cell cycle18 (recapitulated in Figure 1B and Table 1). Further evidence supporting the altered shape includes the DNA length associated with CENP-A nucleosomes elongating from ~110bp to the canonical 150bp at S-phase, and increases in intra-nucleosomal interactions between tagged and endogenous copies of CENP-A histones during early S-phase. Immunofluorescence (IF) performed on centromeric chromatin fibers demonstrates a strong correlation between HJURP depletion and the formation of stable CENP-A octamers in the G1/S to S-phase transition, suggesting a provocative causal link between HJURP loss and CENP-A octamer formation. G1/S also marks a transition within CENP-A histones by the appearance of acetylation at K124 in its Histone Fold Domain (HFD).18 Furthermore, inter-nucleosomal Forster Resonance Energy Transfer (FRET) measurements indicate that chromatin fibers at centromeres convert from a closed to opened state at the G1/S to S transition.18 In sum, the G1/S to S, and S to G2 phase transitions appear to be critical for CENP-A structural conversions. Three questions emerge from this study: (1) how do CENP-A nucleosomes interconvert between two (or more) conformations in vivo? (2) which cell cycle related factors are responsible for the interconversion specifically at the G1/S to S, and S to G2 transitions? and (3) do CENP-A nucleosomal oscillations serve a biological purpose?
Factors that Control Interconversion between Different Conformations of the CENP-A Nucleosome in vivo
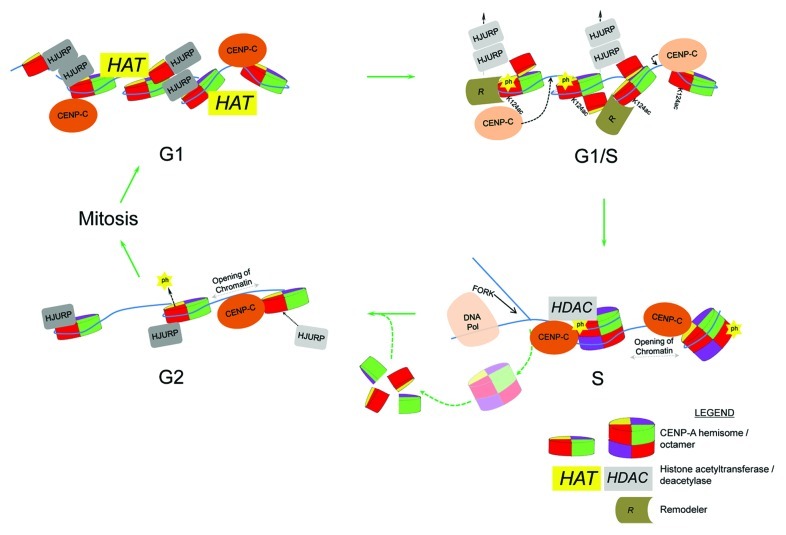
There are several mechanisms by which tetramers and octamers can be envisioned to interconvert in vivo. The simplest hypothesis is that CENP-A undergoes incomplete assembly as a hemisomal tetramer in early G1, whereas stable octamer formation requires a subsequent “maturation” step during G1/S. This explanation would appear to be the most parsimonious based on the following results. The presence of HJURP/Scm3 at centromeric chromatin coincides with the presence of CENP-A hemisomes.18,21 The implication of this result is that either HJURP stabilizes the tetramer, or that HJURP must be evicted in order for CENP-A octamers to form, because HJURP competes for the four-helix bundle, presenting a steric challenge for the second copy of CENP-A in the octamer. The most likely candidate to enforce eviction is CENP-C, because this protein has dual binding abilities: it binds the linker DNA between centromeric nucleosomes23 and also directly interacts with CENP-A’s C-terminus.24 Thus, repositioning of CENP-C to the C-terminus of CENP-A might displace HJURP from CENP-A and from linker DNA (Fig. 2). CENP-N interacts with CENP-A independent of CENP-C25 and stably associates with kinetochores in S and G2-phase.26 Thus, synergistic action between these two key interacting partners of CENP-A, serving as a “cap and tail” complex, may promote CENP-A octamer stability. We have observed that HJURP is depleted from centromeres at the G1/S to S-phase transition, and remains depleted until the next G2 phase. How is HJURP sequestered away from centromeres until replication is complete? One plausible explanation relates to the finding that a residue in the α1 helix of CENP-A, Serine 68 (S68), specifically interacts with HJURP.27 When CENP-A S68 is replaced by glutamine (S68Q), the bulky residue creates a steric clash in the hydrophobic pocket of HJURP, disrupting the interaction between CENP-A and HJURP.27 A contrasting study, however, concluded that HJURP recognition is solely dependent on CENP-A’s centromere-targeting domain (CATD).28 Nevertheless, a speculative mechanism whereby HJURP/CENP-A interactions may remain disrupted is if CENP-A’s S68 is phosphorylated from G1/S through early G2, thus inhibiting HJURP binding. Although human CENP-A nucleosomes do coalesce into octamers at early S phase,18 CENP-A octamers in yeast are susceptible to pulling forces in vitro.8 Thus, the force applied by advancing DNA polymerases or gyrases during replication may be sufficient to disrupt the four-helix-bundle within the CENP-A octamer. We speculate that concomitant displacement of HJURP,18 potentially by phosphorylating S68, repositioning of CENP-C and CENP-N,24 along with S/G2-phase-dependent complexes containing Cdc2029 and Cdk230 stabilize CENP-A octamers during S-phase, whereas replication-fork mediated splitting of CENP-A nucleosomes subsequently allows HJURP to recycle “old” CENP-A back as hemisomes onto newly replicated daughter strands at early G2.
Figure 2. A model depicting dynamic CENP-A-kinetochore protein interactions, CENP-A histone modifications and nucleosomal interconversion across the cell cycle. Lightened HJURP and CENP-C proteins signify eviction and repositioning, respectively. K124ac = CENP-A acetylated at K124 and ph = potential phosphorylation of CENP-A S68.
A second plausible mechanism relates to histone stability. Native CENP-A ChIP studies have previously isolated HDAC1, a histone deacetylase.31 Given our recent report of acetylations within the HFD of CENP-A (K124ac) and histone H4 (K79ac) at G1/S,18 it is possible that centromere-bound HDAC1 removes the acetylation in subsequent phases of the cell cycle (Fig. 2). About half of the CENP-A residues within the HFD are shared with canonical histone H3 (~43% identity). CENP-A K124 is found within the α3 helix, and this residue is conserved with canonical histone H3K122. H3K122 is located at the histone-DNA dyad, is also acetylated, and reported to destabilize DNA-histone interactions which can affect nucleosome stability.32 It is possible that acetylation within the HFD of CENP-A plays a structural role, modulating the stability of the CENP-A octamer. With the availability of more advanced proteomic and genetic tools, there is no doubt that more modifications will be discovered and their functions investigated within the context of CENP-A dynamics.
A third mechanism driving oscillations might rely on centromeric DNA transcription. The α-thalassaemia mental retardation X-linked protein (ATRX) is a chromatin remodeler and, in conjunction with Death domain associated protein (DAXX), serves as a chaperone for the transcription-coupled histone variant H3.3.33,34 ATRX is capable of forming large complexes to regulate transcription35,36 and to facilitate heterochromatin formation at centromeres.37 H3.3 is loaded to centromeres during S phase, as a potential “gap holder” for CENP-A.38 ATRX/DAXX-mediated H3.3 assembly at centromeres presumably also relies on transcription, without which H3.3 would be unable to compete with replication-coupled deposition of H3.1. Chromatin remodelers also likely participate in CENP-A dynamics in vivo. For example, the remodeling and spacing factor 1 (RSF1) is present at centromeres during mid-G1, and its activity is required for CENP-A inheritance through the following S phase.39 Chromatin remodeling, or transcription, in G2-phase may play a role in evicting the gap-holder H3.3 to allow HJURP to re-deposit CENP-A in advance of the next mitosis. Additional chromatin remodelers that may contribute to centromeric remodeling include nucleophosmin1 (NPM1), which associates with CENP-A,20 and has dual functions as a remodeler and as a histone chaperone.40
Biological Significance of CENP-A Nucleosomal Oscillations
How do histones get inherited through replication? Semi-conservative chromatin inheritance, similar to that seen for DNA after replication, was proposed nearly 30 years ago.41 This model was ingenious since it provided a mechanism by which the chromatin state that defined a particular locus could be inherited by both daughter strands, thus preserving epigenetic information into the next generation. This model was controversial since it required splitting of the four-helix bundle at the heart of octamers, and has since proven incorrect for the vast majority of replication-coupled H3.1 nucleosomes. However, a recent discovery has resurrected it for histone variants. SILAC-tracking demonstrates that a quarter of transcriptionally-coupled histone variant H3.3 nucleosomes follow a semi-conservative inheritance pattern, presumably arising from H3.3 nucleosomes splitting at the replication fork, so that each daughter strand receives one copy of “old” H3.3.42
It is unknown if other histone variants also mark chromatin in a semi-conservative fashion after replication. In the case of CENP-A, it is not known how it is re-deposited after the replication fork passes through centromeric chromatin, because its chaperone HJURP does not appear to return until G2.18 CENP-A re-deposition either relies on other assembly factors more intimately tied to the replication fork (such as CAF-1), or, as noted above, H3.3 serves as a place-holder until “old” CENP-A can be recycled to the centromere before the next mitosis.38 H3.3 is transcription coupled, making its assembly at centromeres during replication intriguing in the absence of RNA polymerase activity. How HJURP is able to distinguish H3.3-diluted centromeric chromatin from H3.3 chromatin elsewhere in the genome also remains mysterious. However, if CENP-A nucleosomes split at the replication fork, then a plausible biological role for hemisomes is to segregate one copy of “old” CENP-A to each daughter strand, thus re-establishing centromeric domains. The transition back to CENP-A hemisomes in G2 may also allow better accessibility to previously reported mitotic interacting partners which require access to the internal domains of CENP-A.19,20,43,44
Conclusions
Although we do not yet know the mechanism whereby CENP-A nucleosomes change shape or alter stability across the cell cycle, it is likely that structural conversions rely on a number of factors, such as protein modifiers, chromatin remodelers, transcriptional regulators, and chaperones. Here, we propose three plausible models that may facilitate the interconversion between CENP-A tetrameric and octameric states: (1) depletion of HJURP and repositioning of CENP-C and CENP-N “capping and tailing” CENP-A octamers during S-phase, (2) post-translational histone modifications that destabilize the interaction at the DNA-histone dyad interface in the octamer and 3) cell-cycle coupled centromeric chromatin remodelers which facilitate structural changes (Fig. 2). Whether any, or all, of these mechanisms are responsible for the flexibility seen within CENP-A nucleosomes will be exciting to investigate. The split personality of CENP-A45 suggests that it is cyclically torn between its traditional and unconventional forms, both of which are crucial for its dual role as a seminal player in mitosis, and as a marker for epigenetic inheritance of centromeres.
Acknowledgments
M.B., M.W. and Y.D. are supported by the CCR/NCI Intramural Research Program, E.K.D. is supported by the NIBIB Intramural Research Program. We thank Delphine Quenet for comments on the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23588
References
- 1.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–92. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–15. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–8. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 4.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–5. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 5.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr., Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–82. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 7.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–51. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–9. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–64. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–13. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A. 2010;107:20317–22. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–37. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 16.Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol. 2010;22:392–402. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–56. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, Giebe S, et al. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–26. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–97. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 20.Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–84. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–16. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalal Y, Fleury TJ, Cioffi A, Stein A. Long-range oscillation in a periodic DNA sequence motif may influence nucleosome array formation. Nucleic Acids Res. 2005;33:934–45. doi: 10.1093/nar/gki224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–41. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–8. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–55. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellwig D, Emmerth S, Ulbricht T, Döring V, Hoischen C, Martin R, et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci. 2011;124:3871–83. doi: 10.1242/jcs.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–6. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, et al. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–62. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez M, He H, Sun S, Li C, Li F. Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proc Natl Acad Sci USA. 2013;110:606–11. doi: 10.1073/pnas.1214874110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–84. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 32.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, et al. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J Biol Chem. 2009;284:23312–21. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–80. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci. 2004;117:3807–20. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 36.Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Bassett AR, Cooper SE, Ragab A, Travers AA. The chromatin remodelling factor dATRX is involved in heterochromatin formation. PLoS One. 2008;3:e2099. doi: 10.1371/journal.pone.0002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase. Nucleus. 2011;2:146–57. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindström MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weintraub H, Worcel A, Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976;9:409–17. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- 42.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–8. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 43.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–54. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westhorpe FG, Straight AF. The split personality of CENP-A nucleosomes. Cell. 2012;150:245–7. doi: 10.1016/j.cell.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]