Abstract
Heterochromatin protein 1 (HP1), a small non-histone chromosomal protein, was recently shown to form a complex in vivo with Proliferating Cell Nuclear Antigen (PCNA), a key factor in DNA replication. The complex, which requires HP1β in a form of a dimer, is engaged in DNA repair and replication. We now provide further evidence based on FRET-FLIM live cell studies confirming the association and close proximity between HP1β and PCNA in the complex. We also demonstrate using FRAP, that although HP1β-PCNA complexes are highly mobile in nonreplicating nuclei, when engaged in DNA replication, they become bound and do not exchange with the mobile pool. These observations are in agreement with a notion that a subpopulation of HP1 molecules interact with PCNA in vivo during DNA replication. Similarly, HP1β which is associated with PCNA in regions of DNA repair, is bound and does not exchange with the mobile pool, suggesting that HP1β in association with PCNA may be a component of a DNA repair complex.
Keywords: DNA repair, DNA replication, FRAP, FRET-FLIM, HP1β, PCNA, protein interactions
Introduction
Heterochromatin protein 1 (HP1) was originally described as an epigenetic regulator and a major component of heterochromatin in Drosophila melanogaster.1 In human cells HP1 exists as three isoforms, HP1α, HP1β and HP1γ, which differ in subnuclear localizations and functions. An HP1 molecule consists of N-terminal chromodomain (CD) and C-terminal chromoshadow domain (CSD) linked by a flexible hinge region.2 CD is responsible for HP1 interactions with eu- and heterochromatin, through double or triple methylated lysine 9 of histone H3.3,4 CSD allows protein homo- and hetero-dimerization5 that leads to the formation of a hydrophobic groove6 through which HP1 can interact with proteins containing a PxVxL peptide motif.
It is becoming apparent now that HP1 is a multifunctional nuclear protein.7 Its various posttranslationally modified forms are involved in a number of nuclear processes including chromatin remodeling, gene silencing, replication and repair of DNA.8-11 The list of proteins that interact with HP1 includes structural (e.g., lamin B), chromosome-associated (e.g., SP100), DNA replication and repair (e.g., CAF-1p150, Ku70, ORC1–6) and many others proteins.8 We have shown recently, using the Bimolecular Fluorescence Complementation assay (BiFC) that HP1β interacts also with PCNA, a key protein in DNA replication and repair,12,13 and that both proteins are components of complexes involved in DNA replication and repair.14
We now provide evidence supporting a notion that HP1β interacts with PCNA during DNA replication and repair. FRET-FLIM studies demonstrate that when HP1 and PCNA interact in complexes recruited to regions of DNA replication and repair, the proteins are located in close vicinity, within a few nanometers of each other. FRAP studies confirm that in nonreplicating nuclei the mobility of the complex is as high as the mobility of proteins that do not interact with any intranuclear targets, like GFP or PCNA. In S-phase, however, the complex is tightly bound at replication foci and shows no dynamic behavior, as does the individually tagged PCNA.
Although, for the sake of brevity, we use the term 'HP1-PCNA complexes' in this report, we imply protein assemblies that likely contain not only a closely spaced HP1 dimer and PCNA trimer, but other proteins as well.
Results
FRET between the FP-tagged HP1β and PCNA at DNA replication foci and DNA repair sites
Although it has been demonstrated that the HP1-PCNA complex is present at replication foci and in the regions of damaged DNA,14 one can still argue against the existence of such a complex on the grounds that the BiFC assay may lead to a false positive result. The propensity of the two parts of YFP to attach to each other could be a driving force behind the formation of the HP1-PCNA complex and the reassociated YFP might stabilize the complex beyond its natural stability. Indeed, it has been shown that some FP fragments exhibit high spontaneous association ratio.15 Even though these particular proteins were not used in our BiFC assay, such an argument of caution should still be considered. One can also argue that, even if the complexes do form in a living cell out of the untagged, endogenous proteins, the BiFC assay says little about the distance between the HP1β and PCNA molecules in the endogenous complexes. In order to address these issues and provide an independent confirmation of the existence of the HP1-PCNA complex in subnuclear regions of DNA replication and repair, we studied interaction between GFP-PCNA and mCherry-HP1β fusion proteins by measuring Förster resonance energy transfer (FRET) between their fluorescent tags. FRET was detected by means of Fluorescence Lifetime Imaging Microscopy (FLIM) (Fig. 1).
Figure 1. Förster resonance energy transfer detected by FLIM. (A–D) Averaged (nmin = 8) lifetimes of GFP-PCNA (donor) in the absence and presence of acceptor measured in the nucleoplasm of nonreplicating nuclei (A, B), DNA replication foci (C) and DNA repair sites (D). Acceptor (A): mCherry, (B–D): mCherry-HP1β. Images show representative cells used for measurements, with specific regions used in FRET calculations indicated by the arrowheads. (E): Representative fluorescence decay curves and residuals obtained at replication foci. Scale bars 5 μm.
Interpretation of the measurements of FRET between PCNA and HP1β shown in Figure 1, took into account different dynamic properties of PCNA (donor) in the subsequent stages of the cell cycle. PCNA protein is highly mobile in nonreplicating nuclei (Fig. 2A and D). In S phase its large subpopulation is immobilized at replication foci (Fig. 2B and E). Similarly, a subpopulation of PCNA is bound at the sites of locally induced DNA damage (Fig. 2C and E). In areas of replicating DNA the mobility of PCNA could be neglected (Fig. 2B) and no nonspecific energy transfer was expected to occur. The FRET efficiency measured at discrete replication foci reached 9% (Fig. 1C and E). This result proves a direct interaction of a subpopulation of HP1β with PCNA in the regions of DNA replication. In the regions undergoing DNA repair (Fig. 1D), a subpopulation of PCNA binds to DNA and remains immobile, while another fraction behaves as in nonreplicating cells, i.e., moves freely. Under these conditions an energy transfer between PCNA and HP1β reached the value of 5% (Fig. 1B), which was similar to the energy transfer measured between a tagged PCNA and a freely diffusing mCherry (6%, Fig. 1A). In the regions undergoing DNA repair, fluorescence lifetime of the donor (GFP-PCNA) was shortened by 18% in the presence of the acceptor (mCherry-HP1β). To obtain a specific value of the efficiency of energy transfer we subtracted the value of nonspecific energy transfer from the value measured at the repair sites. This led to a conclusion that the efficiency of energy transfer at DNA repair sites reached 12–13%. These results are consistent with the previously reported engagement of HP1β and PCNA in a complex involved in DNA repair.14 The presence of FRET also corroborates the mutual proximity between HP1β and PCNA, which was unveiled by the BiFC studies previously.14
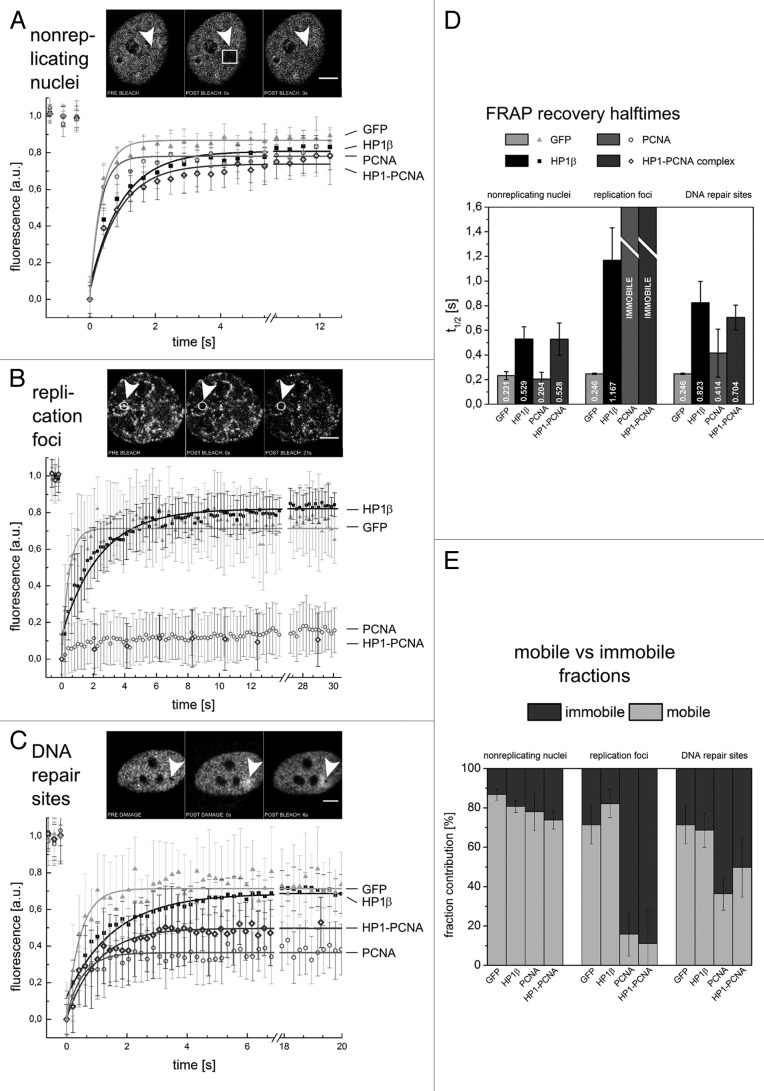
Figure 2. Dynamics of HP1β, PCNA and the HP1-PCNA complexes during DNA replication and repair. FRAP curves (A–C) and the averaged recovery halftimes (D) recorded in experiments on nonreplicating nuclei (A), at replication foci (B) and in DNA repair regions (C). Insets in A–C present images from representative experiments: arrowheads indicate FRAP ROIs (A–C) and sites of DNA damage induced by blue light (C). Mobile and immobile protein fractions (E) were calculated based on A–C; Scale bars 5 μm. GFP serves as an internal, highly mobile standard.18
Dynamics of the HP1β-PCNA complex in nonreplicating cells
In order to gain further insight into the interactions between HP1 and PCNA and the involvement of the complex in nuclear processes, we used a FRAP approach to assess the mobility and the bound fractions of HP1 and PCNA tagged individually with FPs, and compared their dynamics with that of the HP1-PCNA complex tagged with a reassociated YFP.14 The studies were performed in nonreplicating and replicating cells, as well as at DNA repair sites. The postulated interaction between HP1β and PCNA should result in the same dynamic behavior of the subpopulations of HP1β and PCNA which form the complex.
In nonreplicating HeLa cells the half-time of GFP-PCNA fluorescence recovery (t1/2) was similar to the t1/2 measured for GFP alone (Fig. 2A and D). GFP served as an internal standard, since it does not bind to any targets and diffuses rapidly.16-18 As anticipated, the recovery of GFP fluorescence was rapid (Fig. 2A), almost too fast in comparison with the time resolution of the recording method used. The mobility of PCNA, a trimeric protein tagged with GFP, which is at least 4 times larger than GFP alone, was also very fast. This result indicates a lack of detectable binding of PCNA to any cellular targets in nonreplicating nuclei.19 The difference between half-time recoveries of PCNA and GFP was not resolved, since the fluorescence of both tagged proteins recovered too fast for the t1/2 values to be measured with required accuracy.
HP1β mobility in nonreplicating cells was significantly lower than that of PCNA or GFP (Fig. 2A). A low mobility of HP1 was expected, considering that HP1 binds dynamically to histone tails.4,20
In nonreplicating nuclei the HP1-PCNA complex was as dynamic as GFP-HP1β, and far less dynamic than GFP-PCNA or GFP alone. This difference in mobility may be a consequence of differences in protein sizes or their binding properties. It is reasonable to expect that even in nonreplicating nuclei the complex contains not only an HP1 dimer and a PCNA trimer, but other proteins as well. Therefore, if the complex does not bind to any targets, the diffusion rates of the complex should be lower than the rates measured for GFP-PCNA fusion protein (see Discussion). The differences in binding properties cannot be ruled out, either. It is possible that even in nonreplicating nuclei the HP1-PCNA complex does bind transiently to some intranuclear targets, while PCNA trimers, which presumably also exist in cytoplasm and are not engaged in the complex, do not exhibit any binding interactions. Such a putative binding of the complex would be expected to result in a longer fluorescence recovery times than PCNA alone—as we have observed.
In addition to measuring the dynamic properties of the HP1-PCNA complex, we estimated the immobile fractions of the individual proteins and the complex. The FRAP curve of GFP reached plateau at the level of only 87% of the initial fluorescence value. This low level of fluorescence recovery could be interpreted as an evidence of the existence of an immobile fraction or a result of a loss of a part of the overall pool of the fluorescently tagged protein during the bleaching insult. Since GFP is known to exhibit no specific binding interactions,18 this difference between the initial level of fluorescence and the final plateau of the FRAP curve cannot be explained by the existence of an immobile fraction. Considering that in these experiments a relatively large volume of the nucleus (3 μm × 3 μm area in the confocal plane) was bleached out, and GFP is very mobile, the final level of GFP fluorescence recovery must reflect the loss of a part of the fluorophore population, which occurred during the bleaching phase of the experiment. A loss of a measurable part of the entire pool of the HP1-PCNA complex is expected as well. The plateaus of FRAP curves representing PCNA, HP1β and the HP1-PCNA complex reached only 78%, 81% and 74% of the initial fluorescence level, respectively. By analogy to GFP, we ascribe these incomplete fluorescence recoveries to bleaching out parts of the entire pools of these proteins during the bleach phase. In conclusion, the FRAP curves revealed no measurable immobile fractions of HP1β, PCNA or the HP1-PCNA complex in nonreplicating nuclei, and suggest that HP1-PCNA complex may transiently bind to some target in nonreplicating cells.
Mobility of the HP1-PCNA complex at replication foci
In areas of DNA replication almost no recovery of fluorescence of the HP1-PCNA complex was detected, indicating that the complex was bound during replication, as was the individually tagged PCNA (Fig. 2B). PCNA engaged in replication factory slides on DNA;21,22 it is not dynamic and cannot exchange with a mobile pool, since its detachment requires ATP and RFC complex support.23 The engaged PCNA is expected to detach from DNA and exchange with a mobile pool only after the replication of a given stretch of DNA has been completed.24 It can be anticipated that only approximately 30% of the PCNA pool is engaged in replication at any given time, while the rest remains mobile.25 Thus, in order to specifically measure the mobility of the PCNA engaged in replication, it was necessary to minimize the influence of its mobile pool on the observed fluorescence recovery. We achieved this goal by bleaching out and measuring the fluorescence recovery in the smallest possible volume embracing an individual replication focus. This task was somewhat complicated by small sizes of replication foci and by movements of chromatin - the latter necessitated precise tracking of each replication focus. The averaged FRAP curve that represents PCNA in Figure 2B shows the expected lack of protein mobility at replicating regions (while the mobility of the GFP standard was the same as in nonreplicating nuclei). The HP1-PCNA complex investigated at replication foci by FRAP showed a lack of mobility as well. This observation confirms that the PCNA which is engaged in the complex, behaves just as the individually tagged PCNA. Taken together, these results are consistent with the notion that PCNA molecules, which are components of the HP1-PCNA complex, are involved in replication. In addition they strongly suggest that HP1β, which is also a component of this complex, has some role to play in DNA replication.
The mobility of individually tagged HP1β at replication foci was lower than in nonreplicating cells, but still higher than the mobility of the HP1-PCNA complex (Fig. 2D). This observation may seem to contradict the notion that HP1β is a component of the complex which is bound in the regions of replicating DNA. However, we note that the recovery of GFP-HP1β fluorescence is a convolution of two curves representing at least two HP1β subpopulations, because the bleaching spot (volume), centered onto the replication focus, unavoidably embraces not only replicating but also nonreplicating DNA. Thus, HP1β FRAP curves represent not only the putative immobile HP1β subpopulation, which is bound within HP1-PCNA complex and engaged in replication, but also the abundant, dynamic HP1β interacting with histone tails.26 The latter class of HP1β is not associated with PCNA and likely to represent the majority of the total pool of HP1β (see Discussion).
The relatively fast initial recovery of GFP-HP1β fluorescence compared with no recovery of CAG-PCNA and the HP1-PCNA comples (Fig. 2B) indicates, that most of HP1β molecules interact dynamically with some nuclear targets, while only a small proportion of HP1β is engaged with PCNA in a process of replication. It is therefore reasonable to postulate that, at replication foci, all PCNA molecules have an HP1 dimer as a partner in the complex they form, but only a subpopulation of HP1β (most likely posttranslationally modified) interacts with PCNA and is involved in replication. These observations confirm our previous finding, which implicated only a subpopulation of HP1β in interaction with the PCNA engaged in replication.14
Mobility of an HP1-PCNA complex at DNA repair sites
HP1 recruited to DNA damage sites11,27 remains in a complex with PCNA.14 It is known that in DNA damage response (DDR) processes ubiquitinylated PCNA trimer is involved in RAD6 repair pathway.28 However, the role of HP1 in DNA damage signaling or repair is unknown. In order to establish if HP1 is a stable or an exchanging, dynamic component of the putative repair complex with PCNA, we compared the mobilities of HP1β, PCNA and the HP1-PCNA complex in the regions undergoing DNA repair. It is important to emphasize that, in our approach, local DNA damage was inflicted without the use of exogenous photosensitizers, i.e., in a way which is different from many experiments described in literature previously.11,29,30 The absence of a photosensitizer is a precondition for undisturbed continuation of repair processes and noninvasive imaging after the induction of local damage.31 This also applies to the HP1-PCNA dynamic behavior we describe here.
The plateaus of FRAP curves revealed that, in the regions of DNA repair, PCNA and the HP1-PCNA complex were represented by large immobile, bound fractions (64 ± 8% and 50 ± 15%, respectively, Figure 2C). These PCNA and HP1-PCNA complex subpopulations represent the molecules recruited and bound in the DNA repair region.
Despite the presence of large immobile fractions, the initial rates of fluorescence recovery were high for the individually tagged PCNA and the HP1-PCNA complex. This suggests that, in addition to the recruited and bound subpopulations of PCNA and the complex, their highly mobile fractions were present in ROIs as well. These mobile subpopulations most likely represent the molecules that were not associated with the damaged DNA. The presence of both fractions is a consequence of the fact that the volume, which is interrogated in FRAP, embraces both the damaged and the undamaged DNA. Moreover, the number and density of the DNA lesions created by our approach is low.31 Under such conditions the damaged region of the nucleus is expected to contain only a relatively low number of HP1-PCNA complexes and PCNA proteins bound in the region of the damaged DNA and a large number of the mobile molecules not engaged in response to DNA damage.
The FRAP curve representing HP1β also shows a rapid initial recovery, suggesting that the bulk of HP1β is mobile (Fig. 2C). It exchanges and moves at a rate similar to the one observed in nonreplicating nuclei. However, HP1β would be expected to have a reduced mobility and some immobile fraction in the region of DNA repair as well, since the HP1-PCNA complex is bound there, as discussed above. The presence of such an immobile fraction of HP1β is not clearly detectable in the plateau level of the HP1β FRAP curve, considering that the plateau of GFP, which has no bound fraction, reaches a similar value. Note that the slight difference between GFP plateau levels in the regions of DNA repair and in nonreplicating nuclei is merely a consequence of different FRAP protocols used in these two types of experiments. At replication foci the bleached volume was much smaller than the one in nonreplicating nuclei, in which all of the studied proteins move rapidly. Thus, the sizes of the parts of the total fluorescent protein populations, which are bleached out in the initial stage of these two types of FRAP experiments, are not the same. We conclude that the immobile fraction of HP1β, which is engaged in a complex with PCNA bound to damaged DNA, constitutes only a minor subpopulation of the entire pool of HP1β in the nucleus. Hence, the bound subpopulation is almost undetectable against the abundant mobile population of HP1β which interacts with chromatin via histone tails.
The size of the immobile pool of PCNA in the DNA repairing region (64 ± 8%) is very similar to the one of the HP1-PCNA complex (50 ± 15%) (Fig. 2E). The fact that these two values are not significantly different indicates that most likely all PCNA which is bound in the region of the damaged chromatin has an HP1 as a partner in the complex. The same situation was observed at replication foci.
Discussion
HP1β and PCNA are closely spaced at the replication and DNA damage sites
PCNA is known to be bound to DNA in replication sites.25 If HP1β is also tightly bound to PCNA in replication foci, a question arises as to the position and role of this protein in the replication process. Since HP1β and PCNA are detected as a complex even in nonreplicating nuclei, it is likely that they are recruited to replication sites as one entity. Not knowing if other proteins accompany HP1β and PCNA in this complex, it is impossible to envisage the position of HP1β in relation to DNA and PCNA sliding over the DNA strand. The fact that Förster resonant transfer of energy between the fluorescent tags attached to HP1β and PCNA can be detected justifies a notion that the distance between the two proteins engaged in the complex does not exceed 10 nanometers. This observation is in agreement with our previously published data that demonstrated, using the BiFC assay, that HP1β and PCNA are in close proximity in a replication complex.14
HP1β appears to be closely associated with PCNA also in areas, where local DNA damage has been inflicted. In our experiments the damage had to be sublethal to allow normal repair processes, and sufficiently severe at the same time to evoke a detectable recruitment of repair proteins. Thus, a window of experimental conditions required to induce damage and allow undisturbed recruitment of the HP1-PCNA complex was very narrow. A 12–13% Förster resonant transfer detected under these conditions constitutes convincing evidence in favor of the notion that both proteins are in close proximity in the recruited HP1-PCNA complex. The FRAP data reinforce a notion that the complex is bound to DNA at the DNA repair sites.
The donor lifetimes in nonreplicating cells, in replication foci and in DNA repair regions exhibit detectable differences (Fig. 1A–D). Fluorescence lifetimes depend on the immediate molecular environment and the motion of the molecule.32 Considering that PCNA is highly mobile in nonreplicating cells but immobile in replication and damaged DNA, the three corresponding fluorescence lifetimes can be expected to differ. A strong binding of PCNA to DNA during replication would be expected to result in a longer fluorescence lifetime. This agrees with our observation (Fig. 1).
Dynamics of the HP1-PCNA complex in nonreplicating nuclei
A comparison between the mobilities of the HP1-PCNA complex and GFP, PCNA and HP1β detected in nonreplicating cells demonstrates that the complex is less dynamic than PCNA and GFP and resembles the dynamic behavior of HP1β. Two explanations of this result (which are not mutually exclusive) are possible: (1) the complex is large; it contains not only HP1β and PCNA but also other proteins, therefore its mobility is restricted; (2) the complex binds transiently to some nuclear target.
In order to assess the possibility (1) the sizes of the known components of the complex were estimated. If only one PCNA is labeled, the whole protein trimer consists of approx. 1100 amino acids (aa) (if all three PCNA molecules are labeled, the trimer consists of more than 1500 amino acids, but this is a far less likely case). Similarly, GFP-HP1β dimer amounts to approx. 720 amino acids (and 1000 aa when both molecules of HP1 are tagged). The HP1-PCNA complex consists of at least 1500 amino acids. We assume, however, that the complex is likely to contain other proteins as well, hence it may embrace a lot more than 1500 amino acids. In a simplified approach, we can assume that PCNA trimer, HP1β dimer and the HP1-PCNA complex fold into a spherical shape with a known radius (r). In a first approximation a diffusion constant (D) depends only on the size of the protein complex (D = (1/f)kT, where f = 6πηr for spheres). In this case, one should expect at least 1.5 - 2 times lower diffusion constant of the HP1-PCNA complex in comparison with the HP1β dimer and a 30% slower diffusion of the complex in comparison with the PCNA trimer labeled with one molecule of GFP. The difference in diffusion constants between the complex and HP1β should be even greater if the complex embraced other proteins in addition to an HP1β dimer and a PCNA trimer. Since, despite the difference in size, the dynamics of the complex is similar to the dynamics of HP1β, we postulate that the size is not the main factor which determines the dynamics of the complex measured by FRAP. The mobility of the complex may thus reflect dynamic binding to some intranuclear target. The hypothesis (2), suggesting that the complex binds to chromatin via HP1β as an anchor, should be considered, even though its independent verification requires further experiments.
Regardless of the existence of binding sites for the HP1-PCNA complex in nonreplicating cells, an interesting question arises as to why subpoplulations of these proteins remain in a form of a complex at the time when no replication takes place. One of the possible explanations assumes that the complex is pre-assembled and always ready to assume its role in DNA damage response. This is plausible, since DNA repair is active even in the absence of external DNA damaging stimuli. For instance, it has been demonstrated that endogenous oxidants generate approximately 5,000 DNA single-strand lesions (SSLs) in one cell, during a single cell cycle of 24 h.33 These lesions are efficiently repaired. If the HP1-PCNA complex is always pre-assembled and involved in repair of these endogenous DNA lesions, an even more intriguing question concerns other putative partners of HP1 and PCNA in such a pre-existing protein assembly.
Dynamics of the HP1-PCNA complex at replication foci
Although we have shown that fluorescently tagged PCNA and the HP1-PCNA complex are bound in replication foci and exhibit no mobility, HP1β apparently shows a relatively high rate of exchange with the mobile pool. These observations might seem inconsistent with the notion that HP1 is a component of the chromatin-bound complex. There is no discrepancy here since the dynamics of HP1β recorded in replication foci is a convolution of the behaviors of the bound and the abundant, unbound forms, as described in Results.
Dynamics of the HP1-PCNA complex at DNA repair sites
The t1/2 values obtained for PCNA and the HP1-PCNA complex at DNA repair sites resemble those calculated in nonreplicating nuclei. This may seem incompatible with the postulated binding of the HP1-PCNA complex in the regions of damaged DNA. We note, however, that in both cases a small bleached out spot was observed (Fig. 2C, inset, arrowhead) in the first seconds of the FRAP experiment. This was not the case for GFP, which indicates that both PCNA and the HP1-PCNA complex were immobilized within the time scale of the experiment. Subsequent fluorescence recovery, which we observed, should thus be assigned to a large mobile fraction.
Because of the size of experimental errors and a complexity of the system under study, it is not straightforward to estimate the mobile vs. immobile fraction of HP1β, PCNA and the HP1-PCNA complex in the regions of DNA repair. FRAP curves recorded for PCNA and the complex at replication sites can be considered fairly accurate. Based on these curves, 84% and 89% of the PCNA and the complex pool, respectively, is bound to DNA and forms a pronounced immobile fraction with no signs of fluorescence recovery. When discussing the proportions of mobile vs. immobile fractions one can notice (Fig. 2E), that the values obtained at DNA repair sites for PCNA and the HP1-PCNA complex fall into the range between the mobile proteins (GFP and HP1β at repair sites) and the completely immobilized ones (PCNA and the HP1-PCNA complex at replication foci). This once again suggests the existence of two differently behaving populations of the complex and is consistent with a notion that only a subpopulation of the complex detected in the volume subjected to FRAP is bound and presumably involved in DNA repair processes.
The role of HP1 in DNA replication and repair
Over two decades of studies on HP1 protein led to characterization of the protein main functions. It is now known that HP1 is not only involved in heterochromatinization and gene silencing, but also in regulation of gene transcription. The latter role is complex and most likely involve various, largely unknown mechanisms. Nevertheless, the recently published list of HP1 interacting partners34 suggests that HP1 contributes to all key nuclear processes. In many cases, the role of HP1 results from posttranslational modifications of the target molecule or the HP1.35
The results presented in this report support the concept of multifunctional nature of HP1. We demonstrate new evidence showing involvement of HP1 in DNA replication and repair. It is known that HP1 can bind directly to p150 subunit of chromatin assembly factor 1 (CAF-1) complex,36 which is essential for coupling chromatin assembly to replication and thus for cell proliferation.37 The same was reported regarding p150 direct binding to PCNA.38 Thus, it seems plausible that the HP1-PCNA complex may serve as a loading and supporting platform recruiting various factors involved in DNA replication.
Materials and Methods
Cell cultures and transfection
HeLa 21–4 cells were cultured in Dulbecco’s modified Eagle medium (Sigma-Aldrich, D5523) supplemented with 10% fetal bovine serum (Sigma-Aldrich, F7524) and antibiotics. Cells were grown on 18 mm diameter, 0.17 mm thick coverslips (Menzel-Glasser) placed in 12-well cell culture multiwell plates (TPP). Transient transfections were performed 24 h after seeding, according to the protocol and with the use of FuGene HD reagent (Promega Corp., E2311). Experiments were performed 24–72 h after transfection. Plasmids encoding GFP-HP1β, GFP-PCNA, mCherry-HP1β, YN-PCNA and YC-HP1β fusion proteins were described previously.14 Free mCherry protein was encoded by pmCherry-N1 (Clontech, 632523) and free GFP by pEGFP-N1 vector (Clontech, 6085–1), monomerized as described previously.14
We use the term “reassociated YFP” when referring to YFP composed of two nonfluorescent polypeptides, to which the functional FP was cut as described in Hu et al.39 This manipulation, known as a BiFC (Bimolecular Fluorescence Complementation) technique, was used to visualize HP1-PCNA complex (Fig. 2) using YN-PCNA and YC-HP1β plasmids.
Confocal microscopy and analysis
Images were acquired using a Leica TCS SP5 II SMD confocal system (Leica Microsystems GmbH) integrated with FCS/FLIM TCSPC module (PicoQuant GmbH). In all the experiments, a 63x oil immersion NA 1.4 objective lens was used. During imaging, cells growing on a coverslip were maintained at 37°C in DMEM-F12 (Sigma-Aldrich, D2906) culture medium supplemented with 2% FBS, without Phenol Red. The confocal pinhole size was set at 1 Airy unit for FRET-FLIM and at 2 Airy units for FRAP experiments. Image analysis was performed using LAS AF (Leica Microsystems GmbH) and SymPhoTime II (PicoQuant GmbH) software.
Local DNA damage
Local DNA damage was induced as described previously,14 using 405nm pulse laser light; no exogenous photosensitizers were used (Fig. 1D and 2C).31
FRAP protocols and analysis
In FRAP experiments (bleaching and recording) a 100 mW Ar ion laser was used. For each experiment 3–5 pre-bleach images were collected and pixel size was set at 96 nm × 96 nm. The experimental conditions of FRAP experiments are listed in Table 1.
Table 1. Conditions of FRAP protocols used in experiments in nonreplicating nuclei, replication foci and DNA repair sites.
| Nonreplicating nuclei | Replication foci | DNA repair sites | Parameter | |
|---|---|---|---|---|
| GFP-labeled proteins (free GFP, HP1β, PCNA) |
2.36 Hz |
4.83 Hz |
4.88 Hz |
Pre and post bleach scanning frequency |
| 244 μW of 488 nm |
Imaging laser power |
|||
| Square 3 μm × 3 μm |
1 μm diameter circle |
ROI |
||
| For 1.2 sec 4.6 mW of 488 nm + 1.6 mW of 476 nm |
For 10 ms 1.8 mW of 488 nm + 600 μW of 476 nm |
For 10 ms 120 μW of 488 nm |
Bleaching insult |
|
| YFP-labeled HP1-PCNA complex |
2.36 Hz |
0.483 Hz |
4.88 Hz (recovery) 0.49 Hz (plateau) |
Pre and post bleach scanning frequency |
| 680 μW of 514 nm |
Imaging laser power |
|||
| Square 3 μm × 3 μm |
1 μm diameter circle |
ROI |
||
| For 1.2 sec 0.95 mW of 496 nm + 4.65 mW of 514 nm |
For 10 ms 1.8 mW of 514 nm |
For 10 ms 210 μW of 514 nm |
Bleaching insult | |
Analysis of the fluorescence recovery curves was performed as described previously.40 Briefly: the background signals were subtracted from each point in the recorded fluorescence recovery curves. A correction was applied for photobleaching, which occurred during recording of fluorescence recovery, as described previously.40 The background and photobleaching-corrected FRAP curves were normalized to the average value of fluorescence of the 3–5 pre-bleach images. The levels of post-bleach fluorescence were normalized to zero value. Fitting of the fluorescence recovery curves was done using the OriginLab software. The half-recovery times (t1/2) were calculated as the value on the abscissa, at the level where the ordinate values reached a half of the value of the last fitted point of the FRAP curve (i.e., in the plateau region). The t1/2 values were averaged, and the error value was calculated.
The choice of a particular FRAP protocol was determined by several factors, including preliminarily estimates of protein mobility, photobleaching rates and subcellular distribution of a protein under the selected conditions. The common denominator of the three groups of FRAP experiments was the free GFP. The differences in FRAP protocols did not influence the estimates of GFP mobility; this enabled comparisons between t1/2 values measured in different the groups.
FRET-FLIM protocols
FRET was detected by measuring a decrease of fluorescence lifetime of the donor in cells transiently expressing GFP-PCNA (donor-only) or GFP-PCNA and mCherry-HP1β (donor-acceptor). Monomerized EGFP (MEGFP) protein41 was used to overcome problems arising from dimerization of EGFP. When cotransfected, only cells with similar levels of fluorescence intensities were selected for analysis. The concentration of the HP1β fusion protein was estimated to be approximately three times higher than the tagged PCNA, however this difference did not influence the value of the detected resonant transfer of energy.42 In FLIM measurements a 470 nm pulse laser (40 MHz) and a 500–550 band-pass filter were used. Fluorescence lifetimes, collected from pixels indicated by arrowheads on Figure 1, were summed up. The total number of pixels taken for further analysis was adjusted for each experiment. The baseline level of the decay greater or equal to 10 photon counts was considered a prerequisite allowing proper curve fitting. Decays were tail-fitted with a two-exponential decay model using SymPhoTime II software (PicoQuant GmbH). Energy transfer was calculated on the basis of the mean fluorescence lifetimes weighted by amplitudes,43 according to the formula E = 1-τDA/τD. As a reference, fluorescence decay of fluorescein solution in 70% EtOH was measured. In all the experiments a monoexponential tail-fit provided fluorescein lifetime of 3.94 ± 0.05 ns.
Acknowledgments
We gratefully acknowledge financial assistance of National Committee for Science (UMO-2011/01/B/NZ3/00609). The confocal microscope was purchased through an EU structural funds grant BMZ no. POIG.02.01.00–12–064/08. We thank Dr. Peter Hemmerich (FLI, Jena, Germany) for sharing the GFP-HP1β encoding plasmid and Mrs. Ewa Błasiak (Jagiellonian University) for help with preparation of plasmids.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23683
References
- 1.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–72. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–10. doi: 10.1016/S0959-437X(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 3.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–5. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 5.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–97. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–99. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolderson E, Savage KI, Mahen R, Pisupati V, Graham ME, Richard DJ, et al. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1β (HP1-β) mobilization and DNA repair in heterochromatin. J Biol Chem. 2012;287:28122–31. doi: 10.1074/jbc.M112.368381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomberk G, Wallrath L, Urrutia R. The heterochromatin protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 2006;5:2842–51. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- 10.LeRoy G, Weston JT, Zee BM, Young NL. Plazas- Mayorca MD, Garcia BA. Heterochromatin protein 1 is extensively decorated with histone code-like posttranslational modifications. Mol Cell Proteomics. 2009;8:2432–42. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarębski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A. 2009;75:619–25. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- 12.Mortusewicz O, Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin DS, Bai W, Yao N, O’Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci U S A. 1997;94:12863–8. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trembecka-Lucas DO, Dobrucki JW. A heterochromatin protein 1 (HP1) dimer and a proliferating cell nuclear antigen (PCNA) protein interact in vivo and are parts of a multiprotein complex involved in DNA replication and DNA repair. Cell Cycle. 2012;11:2170–5. doi: 10.4161/cc.20673. [DOI] [PubMed] [Google Scholar]
- 15.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–86. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swaminathan R, Hoang CP, Verkman AS. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys J. 1997;72:1900–7. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verkman AS. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem Sci. 2002;27:27–33. doi: 10.1016/S0968-0004(01)02003-5. [DOI] [PubMed] [Google Scholar]
- 18.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–9. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 19.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, et al. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol. 2005;25:9350–9. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, et al. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–21. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 21.Krishna TSR, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–43. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 22.Kelman Z, O’Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995;23:3613–20. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, et al. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–13. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 24.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-δ. Nature. 1987;326:515–7. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 25.Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–54. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–33. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 29.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 30.Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2945–50. doi: 10.4161/cc.8.18.9486. [DOI] [PubMed] [Google Scholar]
- 31.Solarczyk KJ, Zarębski M, Dobrucki JW. Inducing local DNA damage by visible light to study chromatin repair. DNA Repair (Amst) 2012;11:996–1002. doi: 10.1016/j.dnarep.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Lakowicz JR. Principles of Fluorescence Spectroscopy, Springer, 2006, p. 954. [Google Scholar]
- 33.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci U S A. 2003;100:12871–6. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon SH, Workman JL. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays. 2011;33:280–9. doi: 10.1002/bies.201000138. [DOI] [PubMed] [Google Scholar]
- 35.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–40. doi: 10.1016/S1097-2765(00)80204-X. [DOI] [PubMed] [Google Scholar]
- 37.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–8. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–85. doi: 10.1016/S0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 39.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–98. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 40.Trembecka DO, Kuzak M, Dobrucki JW. Conditions for using FRAP as a quantitative technique--influence of the bleaching protocol. Cytometry A. 2010;77:366–70. doi: 10.1002/cyto.a.20866. [DOI] [PubMed] [Google Scholar]
- 41.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–6. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 42.Periasamy A. FLIM Microscopy in Biology and Medicine. Taylot & Francis-CRC Press, 2010, p. 407. [Google Scholar]
- 43.Sillen A, Engelborghs Y. The Correct Use of “Average” Fluorescence Parameters. Photochem Photobiol. 1998;67:475–86. [Google Scholar]