Abstract
Numerous molecular details of intracellular mRNA processing have been revealed in recent years. However, the export process of single native mRNA molecules, the actual translocation through the nuclear pore complex (NPC), could not yet be examined in vivo. The problem is observing mRNA molecules without interfering with their native behavior. We used a protein-based labeling approach to visualize single native mRNPs in live salivary gland cells of Chironomus tentans, an iconic system used for decades to study the mRNA life cycle. Recombinant hrp36, the C. tentans homolog of mammalian hnRNP A1, was fluorescence labeled and microinjected into living cells, where it was integrated into nascent mRNPs. Intranuclear trajectories of single mRNPs, including their NPC passage, were observed with high space and time resolution employing a custom-built light sheet fluorescence microscope. We analyzed the kinetics and dynamics of mRNP export and started to study its mechanism and regulation by measuring the turnover-kinetics of single Dbp5 at the NPC.
Keywords: Dbp5, cytoplasmic probing, light sheet microscope, mRNA labelling, mRNA-export, nuclear pore complex nuclear retention, nuclear probing, nucleocytoplasmic trafficking
Introduction
The spatial separation of transcription and translation in eukaryotes by the nuclear envelope necessitates an efficient transport route for mRNA from the nucleoplasmic transcription sites to the cytoplasmic ribosomes. Because of its importance this transport has been studied for decades. Already during transcription the nascent mRNA is covered by proteins forming an mRNA-protein particle, an mRNP. After release from the transcription site the mRNP diffuses through the nucleus, eventually attaches to a nuclear pore complex (NPC) and moves probably by thermal motion through its permeability barrier (for a review on the nuclear pore structure, see ref. 1). Similarly to the nucleocytoplasmic transport of proteins this passage requires transport receptors, but the receptor loading, Mex67/Mtr2 in yeast or NXF1/NXT1 in mammalian cells, is an integral part of the mRNP biogenesis. At the cytoplasmic face of the NPC the export factors are actively removed from the mRNP by the action of the DEAD box helicase Dbp5 together with Gle1 and IP6, supposably preventing the return of the mRNP into the nucleus.2-5
Recently, we studied the molecular details of mRNP export, namely the translocation of single native mRNPs through the nuclear pore complex (NPC), and combined this with a single molecule analysis of the turnover kinetics of Dbp5 at the NPC.6 Employing a custom-built light sheet fluorescence microscope (for review, see refs. 7–9) we visualized single endogenous mRNPs and Dbp5 molecules in live salivary gland cells of C. tentans, an iconic system used for decades to study aspects of the mRNA life cycle.10 The gland cell nuclei contain four polytene chromosomes, which are made up from thousands of perfectly aligned chromatids. Due to their polytene organization the nuclei exhibit large areas devoid of chromatin, where single mRNPs can be tracked without chromatin interference, which allows differentiation if an mRNP is bound to chromatin close to a NPC or to the NPC itself.
Labeling the Endogenous mRNPs and Dbp5
Messenger-RNPs may be fluorescently labeled in vivo by a number of different approaches (reviewed by Tyagi in ref. 11). We wanted to be as close as possible to the native situation, and used a protein, which is an intrinsic and stable compound of mRNPs, as indirect label. To this end we chose hrp36, the C.tentans homolog of hnRNPA1, which is tightly incorporated into the nascent mRNP during transcription and stays associated with it until translation.12 Since single molecule imaging is primarily dependent on the signal-to-noise ratio of the fluorescence signal it was desirable to render the respective mRNP as bright as possible. For covalent attachment often amino-reactive dyes are used, targeting the numerous lysine residues accessible on the protein surface. However, almost all lysines in the hrp36, and also the only existing cysteine, localize to the RNA-recognition motifs and their covalent modification might interfere with RNA binding. Therefore, we introduced an additional tetra-cysteine tag (tc, Cys-Cys-Pro-Gly-Cys-Cys13) to the hrp36 N-terminus, which allowed site-directed labeling with up to four maleimide-coupled AlexaFluor647 dye molecules. The tc-hrp36 was expressed in bacteria, purified and covalently labeled in vitro. After microinjection into the cytoplasm it was rapidly enriched in the nucleus by virtue of its endogenous M9 shuttling domain where it is incorporated into nascent mRNPs during transcription. In contrast to prior studies,14,15 which introduced artificial mRNAs genetically engineered to contain a tandem array of GFP tags, our approach resulted in an almost unmodified endogenous mRNP, both in terms of particle size and protein composition.
Recombinant C. tentans Dbp5 was prepared and fluorescence labeled in a conventional manner. It was expressed as GST fusion protein in E. coli, and after removal of the GST moiety labeled with the NHS-ester derivative of AlexaFluor647.
Imaging Single mRNPs and Dbp5 Molecules in C. tentans Salivary Glands
To examine the export of single mRNPs we co-injected tc-tagged NTF2 fluorescently labeled with a second color to colocalize the nuclear envelope.16,17 NTF2, a transport receptor unrelated to mRNA export, was enriched at the NPCs and due to the high NPC density of C. tentans salivary gland cells the nuclear envelope then appeared as a continuous line. The salivary glands were kept in hemolymph to ensure almost physiological conditions. This considerably increased background due to the significant hemolymph autofluorescence. This in turn, together with the complex and light scattering morphology of the gland tissue, prevented the use of standard epifluorescent microscopy and prompted us to use a custom-built light sheet fluorescence microscope (LSFM) to overcome the given problems.
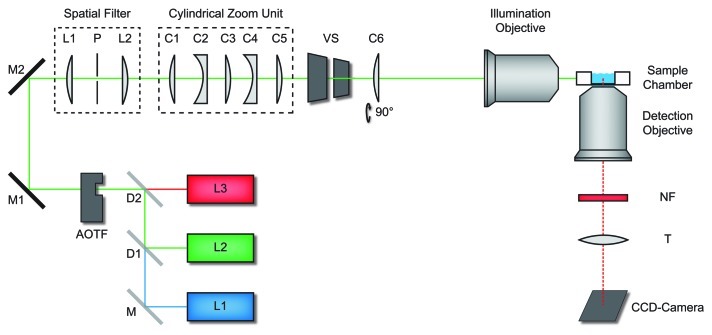
The key feature of a LSFM is the perpendicular configuration of illumination and detection pathway (Fig. 1). In this configuration the focal plane of the detection objective is selectively illuminated. This provides an optical sectioning effect that largely eliminates background fluorescence, because fluorophors outside the observation plane are not excited. Beyond that, photobleaching is minimized by the small illuminated sample volume. In a LSFM the whole field of view is illuminated and imaged simultaneously. Therefore, this elegant technique allows to record image data in a parallel manner, and not sequentially like in classical confocal laser scanning systems. This enables sensitive imaging at high imaging rates. All three LFSM features are especially beneficial for single molecule imaging in spatially extended systems.18-20
Figure 1. Light sheet fluorescence microscope. Light from various laser sources (L1 - L3) was directed into the illumination beam path via dichroic beam splitters (D1 and D2), and mirrors (M, M1, M2). Laser source and intensity was controlled by an acousto-optical-tunable filter (AOTF). Two achromatic lenses L1 and L2 (f = 30 mm) and the pinhole P formed a spatial filter. A cylindrical zoom unit comprised five lenses (C1 - C5). The cylindrical lens (C6) oriented perpendicular to the zoom lens unit focused the beam into the back focal plane of the illumination objective, which generated the light sheet in the sample chamber. A vertical slit (VS) controlled the width of the light sheet. The sample can be moved by a motorized sample scanner. Fluorescence detection was performed by the detection objective, a set of notch filters (NF), the tube lens (T) and an electron multiplying CCD-camera. Figure according to Ritter et al.18
The key to obtain high contrast single molecule signals is the use of low, i.e., picomolar, concentrations of labeled molecules, bright and photostable dyes and reduction of the fluorescence background.21 Using LSFM we could record movies showing the pathways of single fluorescent mRNPs and Dbp5 molecules toward and across the nuclear envelope in realtime. We achieved a localization precision of up to 10 nm at a temporal resolution of 20 ms in these experiments. For measurements of the Dbp5 kinetics we acquired 1,300 movies from 12 different glands and for mRNP export we recorded 7,180 movies from 70 different glands with an imaging rate of 50 Hz and a duration of 20 sec. The latter corresponded to 40 h non-stop observation time. The enormous amount of data and the relatively rare occurrence of events (since only a minute fraction of mRNPs were fluorescent and only a small nuclear envelope section could be monitored in the microscope) demanded a semi-automated image data evaluation process. Custom-written ImageJ plugins enabled us to extract the nuclear envelope position in every movie and to automatically create kymographs, which then were used for fast screening of the raw data and extraction of the respective NPC dwell times. The molecular trajectories at the nuclear envelope were manually evaluated using established data analysis techniques based on custom-developed Matlab routines.6,21,22 The respective programs may be downloaded from http://www.chemie.uni-bonn.de/pctc/kubitscheck/downloads.
mRNP-Export Kinetics
Since we used fluorescent hrp36 as an indirect marker any endogenous mRNPs were potentially labeled, and therefore we expected a broad distribution of measured export times. Indeed, the observed export durations ranged from 20 ms, which was the lower limit of our time resolution, up to about 6 sec. We took only those export events into account, where we could explicitly observe the approach to the NE, an interaction with NPCs, and the dissociation into the cytoplasm (total number 121). The distribution of the observed mRNP export durations could well be described by a biexponential decay function. Thus we determined two time constants, τ1 ≈65 ms and τ2 ≈350 ms. Notably, the majority (87%) of observed export events was fast (65 ms). Furthermore, we observed 3 export events that lasted between 1 and 6 sec. We assumed that these long events corresponded to export of especially large transcripts, namely the Balbiani Ring (BR) mRNAs.
Previously, a study of the mRNP export kinetics in mammalian cells was published by Gruenwald and Singer.14 These authors introduced an array of 24 MS2 stem loops into the β-actin mRNA. Coexpression of the respective MS2 coat protein fused to GFP (MCP-GFP) then resulted in fluorescent mRNPs. This mRNA:GFP probe was preferentially seen at both the nuclear and cytoplasmic faces of the NPC and only rarely localized to the central pore region. Using an elaborate statistical analysis they managed to extract a dwell time estimate for the central channel of 5–20 ms and an overall translocation time of 180 ms for their mRNA construct. The size of the β-actin mRNA containing the stem loops (3.3 kb) was larger than that of an average transcript (2.2 kb):23 Hence, the transport duration of 180 ms appears somewhat long compared with our results, since we observed that the majority of translocation events was clearly shorter (65 ms).
The transcription of a typical eukaryotic gene with 30 kb requires about 6–50 min and the synthesis of an average protein with 450 amino acids takes about 90 sec.24-27 Hence, compared with the duration of these elaborate synthesis processes the mRNP export itself is rapid and not rate-limiting to gene expression.
At the Gate
First explicit observations of mRNA molecules during export were obtained by electron microscopy (EM) in C. tentans salivary gland cells. Following the large BR mRNPs with a diameter of close to 50 nm allowed visualization of single native mRNPs during NPC transition,28-30 and provided a detailed view of this process (reviewed in refs. 10 and 31). From these data no kinetic information could be derived, but they suggested an initial docking step of the mRNP in the basket area of the NPC prior to translocation. Our data clearly supported this conception.
Several long export events (> 300 ms) provided us with detailed, multiple-step export trajectories and allowed us to scrutinize the actual NPC translocation. Since our mRNP:NPC colocalization precision did not allow direct determinations of exact mRNP positions inside the pore we adopted the approach of Lowe et al.,32 and calculated the best superposition of the trajectories with the known structure of the C. tentans NPC.30,32 We found that most mRNP trajectory positions sampled a confined area of 50x30 nm2, and this region aligned very well with the basket and nuclear entry of the NPC (Fig. 2A). This conspicuous accumulation suggested an early rate-limiting step in export. Such a transient delay could very well be related to the rearrangement of large mRNPs to allow 5′ entry of the mRNA in the central channel.
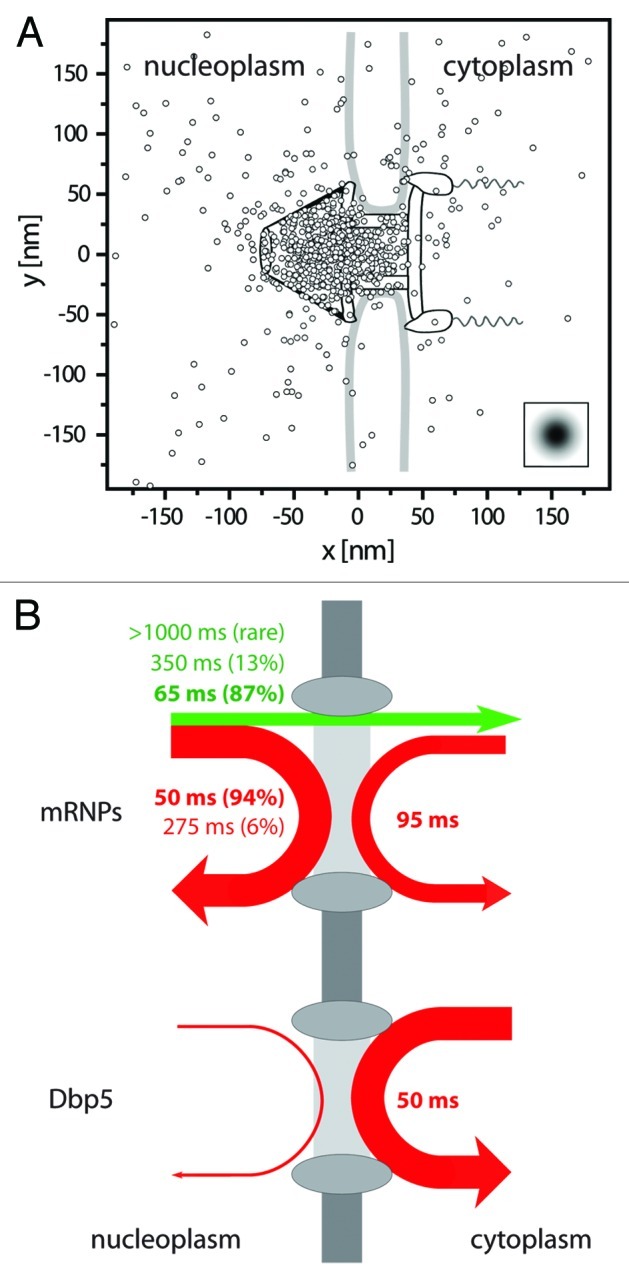
Figure 2. Binding sites and interaction durations with the nuclear pore complexes. (A) Distribution of binding sites of mRNP trajectories during export across the NPC. The figure shows a superposition of export trajectory positions with durations > 300 ms. The trajectories were aligned for an optimal overlay with the known NPC structure.30 The graphics shows the single positions superimposed onto the NPC scheme drawn to scale. The insert indicates the experimental single molecule localization precision. Figure according to Siebrasse et al.6 (B) Illustration of the interactions of mRNPs (labeled by hrp36) and the RNA helicase Dbp5 with the nuclear envelope. The line width indicates the relative occurrence of the respective process, and the color a successful (green) or unsuccessful (red) translocation. Upper graph: about 25% of the approaching mRNPs were successfully translocated across the nuclear envelope (straight green arrow), whereas 75% returned into the nucleus (red arrow). The numbers give the average interaction duration with the nuclear envelope, and the size of the relative fractions is reported in brackets. More than twice as many mRNPs encountered the nuclear envelope coming from nucleoplasm than from cytoplasm. Still, the latter events suggested that a fraction of mRNPs was still mobile in the cytoplasm, and was not immediately engaged in translation. Lower graph: most encounters of Dbp5 (91%) with the nuclear envelope began and ended in the cytoplasm, the interaction time was just a bit shorter than that of the majority of export events. Nuclear import of Dbp5 could not be observed at the used time resolution (20 ms), since protein import occurs fast, namely in 4 to 10 ms.17
Notably, Grünwald and Singer observed an additional 80 ms arrest of their β-actin:GFP mRNPs at the cytoplasmic face of the NPC, which was not seen in our experiments using native mRNPs.14 Indeed, Tyagi11 already noted that the use of the MS2 coat protein fused to GFP and a nuclear localization signal for RNA labeling potentially perturbates the mRNP export process and kinetics. The observation of mRNP arrest at the cytoplasmic face of the NPC could be caused by this effect.
Unsuccessful Translocation Events
Only the minority of mRNPs that hit the nuclear envelope was actually exported. We observed three times more events, where mRNPs encountered the NE, but then returned into nucleoplasm than completed export events. We called the latter process “nuclear probing.” The kinetics of nuclear probing exhibited two time constants, τ1 = 50 ms and τ2 = 275 ms. Most of the nuclear probing events were short (50 ms, see Figure 2B). We assumed that these observations corresponded to short-term interactions of mRNPs with nucleoplasmic structures of the NPC.
But what might be the nature of the long interactions? Are these failed or interrupted transports? Or does the longer time constant correspond to mRNPs, which are transiently caught at the basket due to some necessary processing? Incorrectly spliced mRNAs are retained in the nucleus. For yeast it was shown that the nuclear basket is essential for this retention, and inhibiting this last quality control checkpoint lead to a high number of unspliced transcripts in the cytoplasm.33 Therefore we suspect that the long nuclear probing corresponded to the interaction of not fully processed or incorrectly transcribed mRNPs with an mRNA quality checkpoint at the nuclear basket of the NPC, and final dismiss of translocation.34,35 Also, τ2 might be related to unsuccessful export attempts of mRNPs that entered the nuclear basket, but failed to find the correct orientation for entry, or encountered an occupied NPC channel.
Last not least we observed a frequent cytoplasmic probing of mRNPs taking almost 100 ms on the average. This duration was clearly longer than the short nuclear probing and the majority of the export events. We concluded that mRNPs after their release from the NPC still exhibited a certain affinity to structures of the NPC. Still, a transit back into the nucleus was usually not observed, although it occurred very rarely (5 events).
The ATP-dependent Helicase Dbp5
DEAD-box proteins (DBP) are ATP-dependent RNA helicases, and one or two of these enzymes are involved in almost every major step of mRNP biogenesis from transcription to translation.36,37 Dbp5 (DDX19 in human) is believed to determine the directionality of the export by detaching the transport receptors from the mRNP at the cytoplasmic face of the NPC.
Based on genetic, biochemical and X-ray cristallographic studies several models for the ATPase cycle of Dbp5 during mRNA export have been developed(reviewed in refs. 3, 5 and 37).38-41 The postulated models differ in several important aspects like the exact location of the rate-limiting step and display distinctions in their proposed Dbp5/Gle1/Nup214 or mRNA/nucleotide intermediates.38-40 The models agree with regard to the location of Dbp5 action: Dbp5 and Gle1 are directly located at the cytoplasmic face of the NPC via NUP214 (NUP159 in yeast) and hCG1 (NUP42), respectively.
Until recently, no in vivo kinetic data were available to align the ATPase cyle scheme to the real cellular context. Noble et al.40 provided fluorescence recovery after photobleaching (FRAP) data of GFP-tagged Dbp5 in yeast, but FRAP is an ensemble measurement, which allows rather rough estimates only, in particular in yeast. Therefore, we focused on the kinetics of Dbp5 in our recent study.
We measured the interaction of Dbp5 and NPCs with temporal resolutions of 5 and 20 ms yielding a single dwell time of single wild-type Dbp5 molecule at NPCs of about 50 ms. However, a simultaneous observation of Dbp5 and mRNPs at the NPC could not be achieved. Thus, we cannot directly link the measured time constant to a certain export process. Nevertheless, the majority of the observed mRNP exports (87%) showed a short translocation time (65 ± 5 ms), and therefore it is reasonable to assume that each Dbp5 interacts with one mRNP molecule. Notably, this does not exclude that a single mRNP requires several Dbp5 acting simultaneously in parallel to complete the remodelling. The situation could be similar for the remaining export events (13%) with a longer time constant (350 ms, up to several seconds). Since we observed a transport delay in the basket, but not at the cytoplasmic face of the NPC, it is not probable that export of larger mRNPs requires multiple Dbp5 molecules acting in sequence. Rather, Dbp5 might just be required for the decisive final exit step leading to mRNP release from the pore, and the major part of the translocation time might be needed for mRNP entry into the pore or for accommodation to its topology (or vice versa). However, these considerations are speculative, and an answer to these questions requires certainly deeper study. It is unlikely, however, that a specific Dbp5 acts on several mRNPs sequentially, because this would generally imply a longer binding duration.
Noteworthy, Zhao et al.42 observed in C.tentans cells that Dbp5 can also be recruited to the nascent BR mRNP. A related observation was made by Estruch and Cole,43 who described a direct interaction of Dbp5 with transcription factors in yeast. Therefore, it was speculated that Dbp5 is cotranscriptionally loaded on mRNPs, is associated with them during export, and even during translation. However, we most often observed single Dbp5 approaching the NPC from cytoplasm and not from the nuclear side. We never observed an exit of Dbp5 across the NPC on the time frame of mRNP export. Furthermore, we could show that the NPC localization of Dbp5 does not require a previous entry into the nucleus, and requires only the NUP214 interaction to concentrate on the NPCs.6 These results argue against the idea that Dbp5 binding to mRNPs in the nucleus is essential for its export, at least for the majority of mRNPs. It is known that Dbp5 has functions in transcription, export and translation (reviewed in ref. 5). All these functions could well be performed by different Dbp5 pools.
The location of Dbp5 and the components of its enzymatic cycle3 together with the results of Dbp5 inhibition experiments38 suggest that Dbp5 effects the asymmetric exit of mRNPs from the NPC at its cytoplasmic face.
Future Directions
The export of native mRNPs can be analyzed in vivo with a minimal perturbation of the process. Obviously, mRNA export is not a rate-limiting step in gene expression. Examination of long export events suggested that mRNPs are delayed in the basket of the NPC, probably for quality control purposes. Presumably, a single Dbp5 is involved in a single mRNP export process only, but in contrast, a single mRNP may require several Dbp5 to achieve exit from the NPC.
What are the necessary next steps to advance our understanding of mRNA export? The molecular organization of the FG-repeats inside the pore is still debated, and several different models are used to explain the observed properties of the permeability barrier. Analysis of mRNP movement through NPCs at higher spatial resolution and colocalization precision will help to understand the structure and function of the permeability barrier in greater detail.44 An improved spatial resolution in combination with application of splicing inhibitors will allow to verify the role of the observed delay in the nuclear basket before translocation as quality control process. Further insights into the regulation of nucleocytoplasmic transport will be obtained by examining the effect of mRNA export inhibition on passive transport and active protein import.
To fully understand the directionality of mRNA export, the details of the enzymatic cycle of Dbp5 and its reaction partners, and especially the sequence and kinetics of the single steps therein must be clarified. For this, a kinetic analysis of selected Dbp5 mutants at the NPC or NPC components will be required. And finally, it is important to understand the mechanistic effect of Dbp5 on the mRNP in detail, what may be achieved by state-of-the-art ultrastructural approaches.
Acknowledgments
UK acknowledges financial support by the DFG Grant Ku 2474/7–1, and T.K. gratefully acknowledges financial support by the German National Academic Foundation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23386
References
- 1.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 2.Björk P, Wieslander L. Nucleocytoplasmic mRNP export is an integral part of mRNP biogenesis. Chromosoma. 2011;120:23–38. doi: 10.1007/s00412-010-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–8. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grünwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–41. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tieg B, Krebber H. Dbp5 - From nuclear export to translation. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.10.010. In press. [DOI] [PubMed] [Google Scholar]
- 6.Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Natl Acad Sci U S A. 2012;109:9426–31. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller PJ, Stelzer EH. Quantitative in vivo imaging of entire embryos with Digital Scanned Laser Light Sheet Fluorescence Microscopy. Curr Opin Neurobiol. 2008;18:624–32. doi: 10.1016/j.conb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Reynaud EG, Krzic U, Greger K, Stelzer EH. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2008;2:266–75. doi: 10.2976/1.2974980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santi PA. Light sheet fluorescence microscopy: a review. J Histochem Cytochem. 2011;59:129–38. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–8. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 12.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–72. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 14.Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–7. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–52. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 16.Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci U S A. 1994;91:10212–6. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubitscheck U, Grünwald D, Hoekstra A, Rohleder D, Kues T, Siebrasse JP, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–43. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritter JG, Spille J-H, Kaminski T, Kubitscheck U. A cylindrical zoom lens unit for adjustable optical sectioning in light sheet microscopy. Biomed Opt Express. 2010;2:185–93. doi: 10.1364/BOE.2.000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritter JG, Veith R, Siebrasse J-P, Kubitscheck U. High-contrast single-particle tracking by selective focal plane illumination microscopy. Opt Express. 2008;16:7142–52. doi: 10.1364/OE.16.007142. [DOI] [PubMed] [Google Scholar]
- 20.Spille JH, Kaminski T, Königshoven HP, Kubitscheck U. Dynamic three-dimensional tracking of single fluorescent nanoparticles deep inside living tissue. Opt Express. 2012;20:19697–707. doi: 10.1364/OE.20.019697. [DOI] [PubMed] [Google Scholar]
- 21.Kaminski T, Spille JH, Nietzel C, Siebrasse J-P, Kubitscheck U. Nuclear trafficking and export of single, native mRNPs in Chironomus tentans salivary gland cells. Methods Mol Biol. 2013 doi: 10.1007/978-1-62703-526-2_6. In press. [DOI] [PubMed] [Google Scholar]
- 22.Siebrasse JP, Veith R, Dobay A, Leonhardt H, Daneholt B, Kubitscheck U. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Natl Acad Sci U S A. 2008;105:20291–6. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.AJF G. WM G, Miller J, Lewontin R. Modern Genetic Analysis. New York: W.H. Freeman, 1999. [Google Scholar]
- 24.Ben-Tabou de-Leon S, Davidson EH. Modeling the dynamics of transcriptional gene regulatory networks for animal development. Dev Biol. 2009;325:317–28. doi: 10.1016/j.ydbio.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olofsson SO, Bjursell G, Boström K, Carlsson P, Elovson J, Protter AA, et al. Apolipoprotein B: structure, biosynthesis and role in the lipoprotein assembly process. Atherosclerosis. 1987;68:1–17. doi: 10.1016/0021-9150(87)90088-8. [DOI] [PubMed] [Google Scholar]
- 27.Xu L, Chen H, Hu X, Zhang R, Zhang Z, Luo ZW. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol Biol Evol. 2006;23:1107–8. doi: 10.1093/molbev/msk019. [DOI] [PubMed] [Google Scholar]
- 28.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–13. doi: 10.1016/0092-8674(92)90224-Z. [DOI] [PubMed] [Google Scholar]
- 29.Kylberg K, Björk P, Fomproix N, Ivarsson B, Wieslander L, Daneholt B. Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp Cell Res. 2010;316:1028–38. doi: 10.1016/j.yexcr.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111:223–36. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 31.Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–8. doi: 10.1016/S0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 32.Lowe AR, Siegel JJ, Kalab P, Siu M, Weis K, Liphardt JT. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–3. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/S0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- 34.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 35.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–29. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 36.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Ledoux S, Guthrie C. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes Dev. 2011;25:1109–14. doi: 10.1101/gad.2062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–64. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–42. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25:1065–77. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Jin SB, Björkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–87. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Estruch F, Cole CN. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell. 2003;14:1664–76. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goryaynov A, Ma J, Yang W. Single-molecule studies of nucleocytoplasmic transport: from one dimension to three dimensions. Integr Biol (Camb) 2012;4:10–21. doi: 10.1039/c1ib00041a. [DOI] [PMC free article] [PubMed] [Google Scholar]