Background: JMJD6 hydroxylates U2AF65, but its role in histone modification has been obscure.
Results: Our analysis of histones purified from JMJD6 knock-out mouse embryos reveals that JMJD6 hydroxylates histone lysyl residues.
Conclusion: JMJD6 mediates histone lysyl 5-hydroxylation, which is a novel histone modification.
Significance: Our study identifies a new function for Jumonji family proteins in epigenetic modification of histones.
Keywords: Enzyme Kinetics, Epigenetics, Gene Knock-out, Histone Modification, Hydroxylysine, 5-Hydroxylysine, JMJD6, Lysine 5-Hydroxylation
Abstract
JMJD6 is reported to hydroxylate lysyl residues of a splicing factor, U2AF65. In this study, we found that JMJD6 hydroxylates histone lysyl residues. In vitro experiments showed that JMJD6 has a binding affinity to histone proteins and hydroxylates multiple lysyl residues of histone H3 and H4 tails. Using JMJD6 knock-out mouse embryos, we revealed that JMJD6 hydroxylates lysyl residues of histones H2A/H2B and H3/H4 in vivo by amino acid composition analysis. 5-Hydroxylysine was detected at the highest level in histones purified from murine testis, which expressed JMJD6 at a significantly high level among various tissues examined, and JMJD6 overexpression increased the amount of 5-hydroxylysine in histones in human embryonic kidney 293 cells. These results indicate that histones are additional substrates of JMJD6 in vivo. Because 5-hydroxylation of lysyl residues inhibited N-acetylation and N-methylation by an acetyltransferase and a methyltransferase, respectively, in vitro, histone 5-hydroxylation may have important roles in epigenetic regulation of gene transcription or chromosomal rearrangement.
Introduction
Jumonji domain containing 6 (JMJD6),2 which possesses high binding affinity to single-stranded RNA, is reported to hydroxylate lysyl residues of an RNA splicing factor, U2AF65 (1, 2). JMJD6 contains a JmjC domain that catalyzes lysyl hydroxylation of proteins in the presence of 2-oxoglutarate, Fe(II), and ascorbate. Proteins belonging to the JmjC family are classified into 2-oxoglutarate oxygenases (3). Among the known 2-oxoglutarate oxygenases, PLOD3 (procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3) mediates hydroxylation of unmodified lysyl residues, yielding 5-hydroxylysine (4). Most JmjC family members catalyze hydroxylation of N-methyl groups at the ϵ-amino group of lysyl residues and generate hydroxymethyl groups, which are immediately processed to formaldehyde molecules, resulting in demethylation of methylated lysyl residues (5). However, JMJD6 does not add a hydroxyl group to the N-methyl group but adds it to one of the backbone carbons in a lysyl side chain and generates a stable 5-hydroxylysine (1). JMJD6 knock-out mice exhibited severe anemia, growth retardation, and a delay in terminal differentiation of the kidney, intestine, liver, and lung during embryogenesis, resulting in perinatal lethality (6, 7).
In this study, we first identified JMJD6 as a novel UHRF1 (ubiquitin-like with PHD and RING finger domains 1) interacting protein. UHRF1 has important roles in transferring DNA methylation status and recognizes histone modification status (8). Therefore, we thought that JMJD6 might hydroxylate histone molecules through interaction with UHRF1. Using JMJD6 knock-out mice, we revealed that JMJD6 hydroxylates histone lysyl residues and generates 5-hydroxylysine in vivo. 5-Hydroxylation is a novel histone lysyl modification. Because it interfered with N-acetylation and N-methylation by an acetyltransferase and a methyltransferase, respectively, the modification may regulate transcription through these interactions with other histone modifications.
EXPERIMENTAL PROCEDURES
JMJD6 Wild-type and Knock-out Mice
Details of the JMJD6 knock-out mice were described elsewhere (6). C57BL/6 mice were used as wild-type control mice. JMJD6 knock-out embryonic day 14.5 (E14.5) embryos were obtained by crossing heterozygous JMJD6 mutant mice.
Antibodies, Plasmids, and Cell Lines
The following antibodies were used: anti-JMJD6 rabbit polyclonal antibody (ab10526, Abcam), and anti-β-actin mouse monoclonal antibody (GTX26276, GeneTex). Human JMJD6 cDNA was cloned into pGEX-6p-1 (GE Healthcare) and pcDNA5/FRT/TO (Invitrogen). Doxycycline (Dox)-inducible JMJD6 stable cells were generated using the Flp-In T-REx system (Invitrogen) according to the manufacturer's instructions. JMJD6 expression was induced by Dox (final concentration, 0.5 μg/ml; TaKaRa, Tokyo, Japan). J1 mouse ES cells were obtained from ATCC (Manassas, VA) and maintained in DMEM with 15% fetal bovine serum (FBS), nonessential amino acids, 2-mercaptoethanol, and leukemia inhibitory factor. Flp-In T-Rex 293 cells were obtained from Invitrogen and the Dox-inducible JMJD6 stable 293 cells were maintained in DMEM with 10% FBS, 10% tetracycline-free FBS, hygromycin B (100 μg/ml), and blasticidin S (15 μg/ml).
Quantitative RT-PCR
For qRT-PCR reactions, specific primers and probes for mouse JMJD6 (forward, 5′-GACCCGGCACAACTACTACG-3′; reverse, 5′-CTCTTGTGCATTGAGCAGAAC-3′) and mouse GAPDH (forward, 5′-CCATGTTTGTGATGGGTGTG-3′ and reverse, 5′-ACTGTGGTCATGAGCCCTTC-3′) were used. PCR reactions were performed using the TaKaRa Thermal Cycler Dice® Real Time System Single following the manufacturer's instructions. Amplification conditions were 30 s at 95 °C and then 40 cycles each consisting of 5 s at 95 °C and 30 s at 60 °C.
Purification of GST-JMJD6 and in Vitro Binding Assay
Recombinant GST-JMJD6 was expressed in BL21-CodonPlus DE3-RIL cells. The transformed bacteria were incubated in l-Broth media with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C overnight. Following this, the bacteria were lysed in sonication buffer (150 mm NaCl, 20 mm Tris-HCl (pH 7.5), 2 mm EDTA, 10% glycerol, 1% Triton X, and 0.8 mg/ml lysozyme) by sonication. GST-JMJD6 was purified using glutathione-Sepharose 4FF (GE Healthcare) and eluted by glutathione. The purified proteins were incubated with biotin-labeled histone H31–21 peptides (12–405, Millipore, Billerica, MA) or recombinant full-length histone H4 (14–697, Millipore) in 0.1% Nonidet P-40 lysis buffer (150 mm NaCl, 0.1% Nonidet P-40, and 50 mm Tris-HCl (pH 8.0)) for 1 h at 4 °C. The biotin-labeled histone H31–21 peptides were pulled down with interacting proteins by streptavidin Sepharose (S951, Invitrogen). Full-length histone H4 was immunoprecipitated with anti-JMJD6 rabbit polyclonal antibody (ab10526, Abcam), which was also used for Western blotting.
In Vitro Hydroxylation Assay
To perform the enzyme assay, GST-JMJD6 was prepared as described above. Extracted GST-JMJD6 was concentrated using a 50 K column (Millipore), and its buffer was replaced with 50 mm Tris-HCl (pH 7.5) by dialysis using EasySep (TOMY, Tokyo, JAPAN). Purity of GST-JMJD6 was assessed by Coomassie Brilliant Blue staining. The enzyme assay was performed in 50 mm Tris-HCl (pH 7.5) buffer containing 500 μm α-ketoglutarate, 100 μm L-ascorbate, 100 μm Fe(NH4)2SO4, 10 μm GST-JMJD6, and 20 μm histone peptides. Protein purification and the enzyme assay were performed on the same day to avoid reduction of enzymatic activity of JMJD6.
MS Analysis
Peptides treated with JMJD6 were acidified with trifluoroacetic acid (TFA; final concentration, 0.5%) and absorbed with ZipTipC18. The captured peptides were washed with 0.1% TFA with 2% acetonitrile once and eluted with 0.5 μl of the matrix solution (4 mg/ml cyano-4-hydroxycinnamic acid, 0.1% TFA, 70% acetonitrile) onto the MALDI target plate (AB Sciex, Foster City, CA). The spotted samples were analyzed with the reflectron mode of 4800 plus MALDI-TOF-TOF mass spectrometer (AB Sciex).
Purification of Histones and Detection of 5-Hydroxylysine by Amino Acid Composition Analysis
Histones H2A/H2B and H3/H4 were separately purified from tissues or culturing cells using a histone purification kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. The extracted histones were separated by SDS-PAGE, transferred to a membrane (Immobilon-PSQ, Millipore), and stained by Coomassie Brilliant Blue. The transferred histones were used for amino acid composition analysis to detect 5-hydroxylysine.
The JMJD6-treated peptides or the purified histones were hydrolyzed in 6 n HCl vapor at 110 °C for 20 h. The acid hydrolysates of the peptides were derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and 6-aminoquinolylcarbamyl amino acid thus obtained was quantified by ion-pair chromatography using tetramethylammonium bromide on a C18-reversed phase column (L-column 2, 3.0 mm, inner diameter × 250 mm, 3 μm, CERI, Tokyo, Japan) (9). Each amino acid was separated by HPLC. The acid hydrolysates of the histones were purified on a graphitic carbon column (Hypercarb, 2.1 mm, inner diameter × 100 mm, 3 μm, Thermo Fisher Scientific, Inc., Waltham, USA), and a fraction including 5-hydroxylysine was derivatized with 6-aminoquinolylcarbamyl. The 6-aminoquinolylcarbamyl amino acids were separated on a C18-reversed phase column (Inertsustain C18HP, 3.0 mm, inner diameter × 250 mm, 3 μm, GL Sciences, Tokyo, Japan) and quantified. Synthetic racemic mixture of dl-5-hydroxylysine (catalog no. H0377, Sigma-Aldrich), and 2S,5R-hydroxylysine (catalog no. 55501, Sigma-Aldrich) were used as standards.
In Vitro Histone Acetyltransferase (HAT) Assay
The in vitro p300 colorimetric HAT assay was performed according to a protocol from BIOMOL (Plymouth Meeting, PA). In brief, the catalytic domain of human p300 (catalog no. SE-451, BIOMOL) and the indicated amount of control histone H41–23 peptides or 5-hydroxylysine containing histone H41–23 peptides, in which all lysines were substituted to 5-hydroxylysine (Sigma-Genosys, Hokkaido, Japan), were incubated in 50 μl of assay buffer (50 mm HEPES/NaOH (pH 7.9), 0.1 mm EDTA, 50 μg/ml BSA) in the presence of acetyl-coenzyme A (CoA, Sigma-Aldrich) at 37 °C. The reaction was stopped by adding 100 μl of quenching buffer (3.2 m guanidinium HCl, 100 mm Na2HPO4/NaH2PO4 (pH6.8)) at the indicated times. Following this, 50 μl of 2 mm 5,5′-dithiobis-2-nitrobenzoic acid (Sigma-Aldrich) in 100 mm Na2HPO4/NaH2PO4 (pH 6.8) was added, and absorbance at 405 nm was read by a spectrophotometer (ARVO MX/Light 1420 Multilabel/Luminescence Counter, PerkinElmer Life Sciences). The transfer of an acetyl group from an acetyl-CoA to the ϵ-amino group of lysine residues was quantified by measurement of the thiol group of CoA. A standard curve was generated using β2-mercaptoethanol.
In Vitro Histone Methyltransferase Assay
The in vitro histone methyltransferase assay was performed as described previously (10), except for slight modifications. In brief, a fixed amount of purified baculovirus-produced recombinant SMYD3 (1 μm) was incubated with indicated histone peptides, which were also used for the in vitro HAT assay, and 1 μCi of S-adenosyl-l-methionine (AdoMet; GE Healthcare) as the methyl donor in a mixture of 60 μl of methylase activity buffer (50 mm Tris-HCl (pH 8.5), 100 mm NaCl, 10 mm dithiothreitol) at 30 °C. The incorporated 3H-labeled methyl groups in the substrates were measured by a scintillation counter after filter binding (units, cpm). The concentration (nm) of the methylated substrate was calculated based on the basis of radioactivity.
RESULTS
JMJD6 Effectively Hydroxylates Histone Lysyl Residues in Vitro
During screening of UHRF1-interacting proteins, we identified JMJD6 as a novel binding partner of UHRF1 (data not shown). Because UHRF1 recognizes hemimethylated DNA and histone modifications, we assumed that JMJD6 might be recruited by UHRF1 to nucleosomes and modify histone lysyl residues. In vitro experiments showed that recombinant GST-JMJD6 possessed the ability to bind to histone H31–20 tail and histone H4 (Fig. 1, A and B) and hydroxylate multiple lysyl residues in the N-terminal tails of histone H31–20 and H41–30, which was detected as of 16, 32, or 48 Da shifts by MS analysis (Fig. 1, C and D); subsequent MS/MS analysis revealed that JMJD6 mediates monohydroxylation of lysyl residues. As indicated by Webby et al. (1), JMJD6 preferentially hydroxylated lysyl residues in the basic peptides, and no apparent sequence preference was observed in vitro (data not shown).
FIGURE 1.
JMJD6 interacts with and hydroxylates histone H3 and H4 in vitro. A and B, in vitro pulldown assay. Biotin-labeled histone H31–21 peptides (A) or recombinant histone H4 (B) were incubated with or without GST-JMJD6, pulled down by streptavidin-Sepharose, and detected by dot blot using streptavidin-HRP (A) or Coomassie Brilliant Blue (CBB) staining (B). Pulled down GST-JMJD6 was detected by Western blotting using anti-JMJD6 antibody (A) or Coomassie Brilliant Blue staining (B). C and D, enzymatic activity of GST-JMJD6 was measured by MS analysis. Histone H31–20 (C) and H41–30 (D) peptides were served as substrates. BSA was used as a negative control. IB, immunoblot.
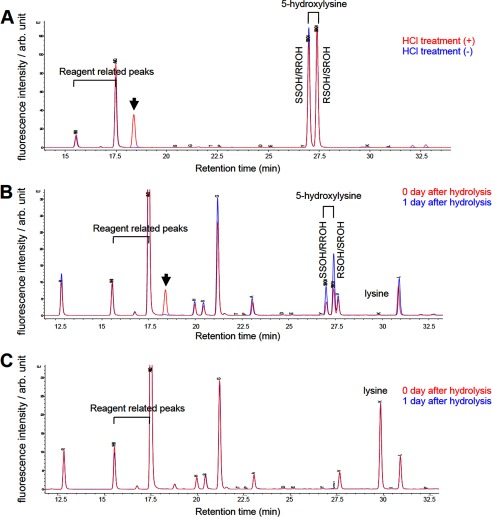
Next, we established a sensitive hydroxylysine detection method based on amino acid composition analysis as an alternative to the MS-based method. For amino acid composition analysis, we briefly hydrolyzed peptides or proteins with HCl and separated each amino acid residue by reversed phase HPLC to detect 5-hydroxylysine. To evaluate this method, we first performed reversed phase HPLC using simplicial synthetic 2S,5R-hydroxylysine and synthetic racemic mixture of 5-hydroxylysine composed of 2S,5S (SS)-, 2R,5R (RR)-, 2R,5S (RS)-, and 2S,5R (SR)-stereoisomers (Fig. 2A). We detected two peaks corresponding to SS/RR- and RS/SR-hydroxylysine by analyzing these synthetic 5-hydroxylysines without HCl treatment (Fig. 2A). After HCl treatment of these synthetic 5-hydroxylysines, another peak was appeared (Fig. 2A, arrow). This peak possibly corresponds to a lactone derivative, 3-amino-6-(aminomethyl)oxan-2-one, generated by dehydration condensation between C5 hydroxyl group and carboxyl group, which is described in a previous report (11). Next, we evaluated the method using unmodified H41–23 peptides and 5-hydroxylysine containing H41–23 peptides in which all the lysines at positions 5, 8, 12, and 20 were substituted with 5-hydroxylysines. After hydrolysis of these peptides, we detected two peaks corresponding to SS/RR- and RS/SR-hydroxylysine only in the 5-hydroxylysine containing peptides but not in the unmodified peptides (Fig. 2, B and C). We also detected the peak of the possible lactone derivative in the 5-hydroxylysine containing peptides by reversed phase HPLC performed in the same day of hydrolysis, but the peak disappeared in the next day of hydrolysis, indicating that the derivative is unstable. Because quantification of the derivative is technically difficult, we only quantified SS/RR- and RS/SR-hydroxylysine.
FIGURE 2.
Establishment of amino acid composition analysis for detecting 5-hydroxylysine. A, analysis of simplicial synthetic SR-hydroxylysine and synthetic racemic mixture (SS/RR/RS/SR) of 5-hydroxylysine either treated with (red) or without (blue) HCl. B and C, analysis of H41–20 peptides including synthetic 5-hydroxylysine (B) and unmodified H41–20 peptides (C). The peptides were analyzed in the same day of hydrolysis (red) or next day of hydrolysis (blue). The arrow indicates a 5-hydroxylysine derived peak, which possibly corresponds to a lactone derivative, 3-amino-6-(aminomethyl)oxan-2-one. SSOH/RROH, 2S,5S-/2R,5R-hydroxylysine; RSOH/SROH, 2R,5S-/2S5R-hydroxylysine; arb. unit, arbitrary units.
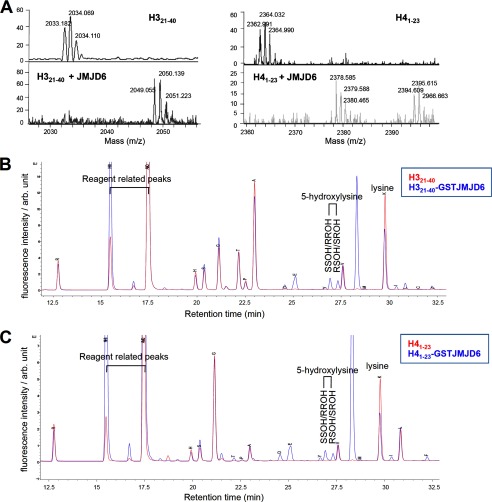
Using this method, we analyzed H321–40 and H41–23 peptides treated with or without recombinant GST-JMJD6. First, we confirmed hydroxylation of the peptides by GST-JMJD6 by MS analysis (Fig. 3A). Then, the peptides were separated from the enzyme reaction mixture, by reversed phase HPLC. The separated peptides were treated with HCl, and each amino acid residue was separated by reversed phase HPLC (Fig. 3, B and C). Comparison of the chromatograph between amino acids derived from the JMJD6-treated and -untreated peptides identified two additional peaks in the peptides treated with JMJD6, which are matched with the standard synthetic 5-hydroxylysine (Fig. 3, B and C).
FIGURE 3.
JMJD6 hydroxylates histone H3 and H4 peptides detected by amino acid composition analysis. A, hydroxylation of H321–40 and H41–23 peptides by GST-JMJD6 was confirmed by MS analysis. B and C, results of amino acid composition analysis of H321–40 (B) and H41–23 (C) peptides treated with (blue) or without (red) GST-JMJD6. SSOH/RROH, 2S,5S/2R,5R-hydroxylysine; RSOH/SROH, 2R,5S-/2S,5R-hydroxylysine; arb. unit, arbitrary units.
JMJD6 Hydroxylates Histone Lysyl Residues in Vivo
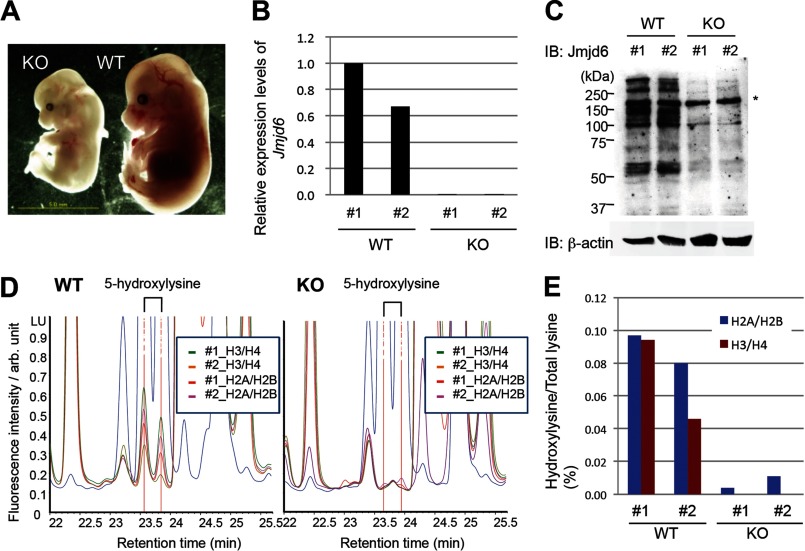
To investigate histone lysyl hydroxylation in vivo, we performed the amino acid composition analysis for analyzing a mixture of histone H2A/H2B and a mixture of histone H3/H4 proteins isolated from two JMJD6 wild-type and two JMJD6 knock-out whole embryos at E14.5 (Fig. 4A). JMJD6 knock-out was confirmed by qRT-PCR and Western blotting (Fig. 4, B and C). The results showed that 0.097 and 0.080% of total lysyl residues in histone H2A/H2B and 0.094 and 0.046% of those in histone H3/H4 were 5-hydroxylated in each of the two JMJD6 wild-type mice (Fig. 4, D and E), whereas 0.004 and 0.011% of total lysyl residues in histone H2A/H2B and 0.000 and 0.000% of those in histone H3/H4 were 5-hydroxylated in each of the two JMJD6 knock-out mice (Fig. 4, D and E), indicating that JMJD6 hydroxylates histone lysyl residues in vivo.
FIGURE 4.
JMJD6 hydroxylates histones H2A/H2B and H3/H4 in mouse embryos. A, a representative image of JMJD6 knock-out and wild-type E14.5 embryos. B, JMJD6 knock-out was confirmed by qRT-PCR. GAPDH was used as an internal control. C, JMJD6 knock-out was confirmed by Western blotting. The asterisk indicates a nonspecific band. β-Actin was used as a loading control. D, result of amino acid composition analysis of histones derived from two Jmjd6 wild-type (left) and knock-out (right) E14.5 embryos. E, % of 5-hydroxylysine in total lysine of histones H2A/H2B (blue) and H3/H4 (red) was calculated from the HPLC data (D). IB, immunoblot; arb. unit, arbitrary units.
We also generated Dox-inducible JMJD6 stable HEK293 cells. JMJD6 induction by Dox was confirmed by qRT-PCR and Western blotting (Fig. 5, A and B) and increased 5-hydroxylation levels of histone lysyl residues (Fig. 5C). In addition, we purified histones from a 6-month-old JMJD6 wild-type mouse testis, which expressed JMJD6 at the highest level among various tissues and cells (Fig. 5, D and E). In the testis, 0.238 and 0.054% of total lysyl residues in histone H2A/H2B and H3/H4, respectively, were 5-hydroxylated (Fig. 5F). In the mouse J1 ES cells, 0.053 and 0.020% of total lysyl residues in histone H2A/H2B and H3/H4, respectively, were 5-hydroxylated (Fig. 5F).
FIGURE 5.
Amount of 5-hydroxylysine in JMJD6 overexpressed HEK 293 cells, mouse testis, and ES cells. A, expression levels of JMJD6 in Dox-inducible JMJD6 stable cells were examined by qRT-PCR before and after 48-h Dox induction. B, induction of JMJD6 by Dox in the cells was confirmed by Western blotting. C, amino acid composition analysis of histones derived from Dox-inducible JMJD6 stable cell lines. The blue and red bars indicate % of 5-hydroxylysine in the total lysine of histone H2A/H2B and in the H3/H4, respectively. D, relative expression levels of Jmjd6 in various mouse tissues and cells were examined by qRT-PCR. E, expression of Jmjd6 in a 6-month-old mouse testis was examined by immunohistochemistry. F, amino acid composition analysis of histones derived from 6-month-old mouse testis and J1 ES cells. The blue and red bars indicate % of 5-hydroxylysine in the total lysine of histone H2A/H2B and in the H3/H4, respectively.
5-Hydroxylation Prevents N-Acetylation and N-Methylation of Histone Lysyl Residues in Vitro
Because lysyl residues in histone tails are often subjected to N-acetylation and N-methylation, we examined whether 5-hydroxylation of lysyl residues affects modifications at the ϵ-amino groups. First, we examined the effect of lysyl 5-hydroxylation on histone H4 N-acetylation by p300, which catalyzes N-acetylation of the ϵ-amino group of lysyl residues, including histone H4K5 and H4K8, through its HAT domain (12). Kinetic analysis using the unmodified and the 5-hydroxylysine containing H41–23 peptides in which all the lysines were substituted with 5-hydroxylysines as substrates revealed that 5-hydroxylation largely interfered with the HAT activity of p300 in vitro (Fig. 6, A–C). Lineweaver-Burk plot analysis was performed to calculate the maximum velocity (Vmax) and Michaelis constant (Km) values (Table 1; Fig. 6C, R2 = 0.9981 and 0.9902). Vmax of the reactions in which p300 acetylated the 5-hydroxylysine-containing peptides (H41–23OH) was 5-fold less than that of the control peptides (6.95 ± 0.45 and 35.34 ± 1.65 μm/min, respectively), whereas Km of the two reactions was quite similar (204.51 ± 10.12 and 200.37 ± 14.32 μm, respectively), indicating that 5-hydroxylation does not inhibit binding of lysyl residues to p300 but reduces the catalytic efficiency.
FIGURE 6.
5-Hydroxylation of lysyl residue impairs N-acetylation and N-methylation in vitro. A–C, the in vitro colorimetric HAT assay was performed using a fixed amount of p300 (0.44 μm) and control H41–23 peptides (●) or 5-hydroxylysine-containing peptides (H41–23 OH, ■). BSA was used as a negative control (♦, ▴). After the reactions, absorbance (405 nm) of the coproduct (CoA) was measured. A, reactions were terminated at the indicated time points, and the concentration of CoA was calculated on the basis of a standard curve that was generated from β2-mercaptoethanol. B and E, substrate concentration-velocity (s-v) plot. C and F, Lineweaver-Burk plot. The vertical axis is 1/velocity [v], and the horizontal axis is 1/substrate concentration [S]. D–F, the in vitro histone methyltransferase assay was performed using a fixed amount of SMYD3 (1 μm) and control H41–23 peptides (●) or 5-hydroxylysine-containing peptides (H41–23OH, ■). AdoMet was used as a methyl donor. After the reactions, radioactivity (cpm) of the 3H-methylated substrates was measured. The concentration of incorporated 3H-methyl groups (nm) was calculated based on the basis of radioactivity (1 cpm was 0.02563 nm in the reaction). D, reactions were performed with fixed amounts of the peptides (141 μm) and terminated at the indicated time points.
TABLE 1.
Effect of 5-hydroxylation on N-acetylation of ϵ-amino group of lysyl residues
Lineweaver-Burk plots were used for estimation of the kinetic constants, Vmax, and Km. R2 is the determination coefficient (see Fig. 6C).
| Vmax | Km | |
|---|---|---|
| μm/min | μm | |
| H41–23 + p300 | 35.34 ± 1.65 (R2 = 0.9981) | 204.51 ± 10.12 |
| H41–23 OH + p300 | 6.95 ± 0.45 (R2 = 0.9902) | 200.37 ± 14.32 |
We also examined the effect of lysyl 5-hydroxylation on the histone methyltransferase activity of SMYD3, which catalyzes lysyl N-methylation of histone H3 (13) and also H4 (data not shown) through its SET (su(var) 3–9 enhancer-of-zeste trithorax) domain by the histone methyltransferase assay. The results showed that 5-hydroxylation at lysyl residues almost completely inhibited N-methylation catalyzed by SMYD3 (Table 2 and Fig. 6, D–F); Vmax values of the reactions with the control peptides (H41–23) and the 5-hydroxylysine-containing peptides (H41–23OH) as substrates were 10.90 ± 0.92 and 0.48 ± 0.26 nm/min, respectively. Similar to the HAT assay, Km values of the two reactions were ∼80.63 ± 16.26 and 75.26 ± 9.60 μm, respectively.
TABLE 2.
Effect of 5-hydroxylation on N-methylation of ϵ-amino group of lysyl residues
Lineweaver-Burk plots were used for estimation of the kinetic constants, Vmax, and Km. R2 is the determination coefficient (see Fig. 6F).
| Vmax | Km | |
|---|---|---|
| nm/min | ||
| H41–23 + SMYD3 | 10.90 ± 0.92 (R2 = 0.9918) | 80.63 ± 16.26 μm |
| H41–23 OH + SMYD3 | 0.48 ± 0.26 (R2 = 0.9366) | 75.26 ± 9.60 μm |
Subsequently, we performed reciprocal experiments using H41–23 or H49–23 peptides, in which all the lysines are either unmodified, N-acetylated, or N-monomethylated. JMJD6 effectively hydroxylated the control peptides (Fig. 7, A and B, left panels); however, N-acetylation and N-monomethylation at the ϵ-amino group of the lysines completely blocked 5-hydroxylation by JMJD6 (Fig. 7, A and B, right panels).
FIGURE 7.
N-Acetylation and N-methylation of lysyl residues impairs 5-hydroxylation by JMJD6 in vitro. A, the in vitro hydroxylation assay was performed using GST-JMJD6 (10 μm) and 85 μm of control H41–23 peptides or N-acetyl-lysine-containing peptides. B, the in vitro hydroxylation assay was performed using GST-JMJD6 (10 μm) and 85 μm control H41–23 peptides or N-monomethyl-lysine-containing peptides. BSA was used as a negative control. 5-Hydroxylation by JMJD6 was detected by MS analysis.
DISCUSSION
We found a novel histone modification, 5-hydroxylation, by JMJD6. JMJD6 reportedly hydroxylates a splicing factor, U2AF65 (1). That study and another report (1, 14) stated that evidence of histone lysyl hydroxylation was not found by MS-based analysis in vivo. In the present study, we developed an alternative method, amino acid composition analysis, to detect 5-hydroxylation of histone lysyl residues. As reported previously, we have not detected clear evidence of 5-hydroxylation of histone lysyl residues by MS-based analysis. We think that there are several causes for this. 1) The amount of 5-hydroxylysine is too small to detect by MS-based analysis. 2) Artificial methionine oxidation during preparation of samples for MS analysis makes detection of 5-hydroxylysine difficult. 3) 5-Hydroxylysine could be an intermediate form as it is in collagen, and a further unknown modification(s) such as glycosylation could be added; the final product of collagen is glucosylgalactosyl hydroxylysine (4). The collagen hydroxylase, PLOD3, possesses galactosyltransferase and glucosyltransferase activities. Unlike PLOD3, JMJD6 does not appear to possess any other enzymatic activities by domain search; therefore, it is difficult to assume possible further modification(s) by its protein structure. We may have been able to detect 5-hydroxylation in histone lysyl residues by amino acid composition analysis but not by the MS-based analysis because many modifications such as glycosylation or galactosylation could be removed during the hydrolysis process of amino acid composition analysis. By this analysis, we detected both SS/RR- and SR/RS-hydroxylysine in JMJD6-treated histone peptides and also in JMJD6 wild-type E14.5 embryos, ES cells, and the Dox-inducible JMJD6 stable HEK293 cells. Because the relationship between RR and RS and also between SS and RS is a diastereomer, we were able to distinguish them. However, because a relationship between SS and RR, and also between RS and SR is an enantiomer, we were not able to separate them by this method. Despite this, these two peaks are most likely SS- and RS-hydroxylysine because JMJD6 is reported to generate SS-hydroxylysine (11). The RS-hydroxylysine could be generated from SS-hydroxylysine through the lactone derivative, 3-amino-6-(aminomethyl)oxan-2-one, which is unstable and difficult to be quantified. Because of this difficulty, we only quantified SS/RR- and RS/SR-hydroxylysine in this report. Therefore, actual quantity of 5-hydroxylysine in the samples examined here could be a little higher.
Because we detected 5-hydroxylysines in the UHRF1 KO ES cells (data not shown), UHRF1 is not required for 5-hydroxylation of histone lysyl residues by JMJD6. Therefore, biological significance of the interaction between UHRF1 and JMJD6 remains unclear. Further analysis is also required to determine the biological significance of 5-hydroxylation of histone lysyl residues. In vitro experiments suggest that 5-hydroxylation can inhibit N-acetylation and N-methylation by p300 and SMYD3. The active site structure of the p300 and general control of amino acid synthesis 5 (GCN5) HAT domains showed that the side chain of the 5-hydroxylysine can invade the catalytic pocket; however, the 5-hydroxyl group may disturb active form formation of the substrate (Fig. 8, A and B). The catalytic site of SET domains of Neurospora crassa Dim-5 and human SETD7, which are structurally similar to SMYD3 (15), suggested that the side chain of 5-hydroxylysine can invade the catalytic pocket; however, the 5-hydroxyl group may disturb an active form formation of the substrate (16, 17) similar to that in HAT domains (Fig. 8, C and D). Therefore, 5-hydroxylation could be important in the context of the histone code. It is known that histones H2A and H2B move more dynamically between the nucleosome and nucleoplasm. 5-Hydroxylation of these histones may have some effects for the movement because the modification was detected more in histones H2A/H2B than in histones H3/H4. The expression pattern of JMJD6 is also interesting. JMJD6 may play important role(s) in the testis, such as a role in histone-protamine exchange. We believe that our present finding provides a novel insight into epigenetic regulations of gene transcription and/or chromosomal rearrangement.
FIGURE 8.
Structure around the active site of HAT domains and SET domains, a lysine side chain, and an S-adenosyl methionine (AdoMet). A, spatial localization among the HAT domain of GCN5 (gray), a lysine side chain (magenta), and CoA (cyan) (Protein Data Bank code 1QSN). The 5-hydroxyl group may locate close to acetyl-CoA, indicating that this may work as a steric barrier and prevent effective N-acetylation by the catalytic domain. B, structure of the HAT domain of p300 (gray) in complex with inhibitor, Lys-CoA (magenta) (Protein Data Bank code 3BIY). The 5-hydroxyl group may restrict the conformation of lysine side chain in the catalytic pocket of p300. The figures were prepared by using program PyMOL. C, spatial localization among the SET domain of N. crassa Dim-5 (green), a histone H3 peptide (magenta), and AdoMet (cyan) (Protein Data Bank code 1PEG). D, spatial association among the SET domain of human SETD7/9 (green), a histone H3 peptide containing monomethylated Lys-4 (magenta), and AdoMet (cyan) (Protein Data Bank code 1O9S). A side chain of a lysine residue locates in tightly hydrophobic pocket of the SET domains (C and D). Hydroxylation at position C5 of the chain may prevent a lysine side chain to locate in the pocket, causing inhibition of N-methylation by SET domains.
Acknowledgments
We thank Dr. Haruhiko Koseki and Dr. Jafar Sharif for providing us UHRF1 KO ES cells; Professor Shoji Tajima, Dr. Isao Suetake, Dr. Yoichi Shinkai, Dr. Kenji Ichiyanagi, Dr. Fumiyuki Sanematsu, and Dr. Hyun-Soo Cho for useful advice; and Yuichi Mishima and Dr. Atsuhiko Toyama for technical assistance.
This work was supported by JSPS KAKENHI Grant 22700867 and Kyushu University interdisciplinary programs in education and projects in research development.
- JMJD6
- Jumonji domain containing 6
- qRT-PCR
- quantitative RT-PCR
- HAT
- histone acetyltransferase
- E14.5
- embryonic day 14.5
- AdoMet
- S-adenosyl-l-methionine.
REFERENCES
- 1. Webby C. J., Wolf A., Gromak N., Dreger M., Kramer H., Kessler B., Nielsen M. L., Schmitz C., Butler D. S., Yates J. R., 3rd, Delahunty C. M., Hahn P., Lengeling A., Mann M., Proudfoot N. J., Schofield C. J., Böttger A. (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 [DOI] [PubMed] [Google Scholar]
- 2. Hong X., Zang J., White J., Wang C., Pan C. H., Zhao R., Murphy R. C., Dai S., Henson P., Kappler J. W., Hagman J., Zhang G. (2010) Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 107, 14568–14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loenarz C., Schofield C. J. (2008) Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4, 152–156 [DOI] [PubMed] [Google Scholar]
- 4. Myllylä R., Wang C., Heikkinen J., Juffer A., Lampela O., Risteli M., Ruotsalainen H., Salo A., Sipilä L. (2007) Expanding the lysyl hydroxylase toolbox: new insights into the localization and activities of lysyl hydroxylase 3 (LH3). J. Cell. Physiol. 212, 323–329 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y., Whetstine J. R. (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 6. Kunisaki Y., Masuko S., Noda M., Inayoshi A., Sanui T., Harada M., Sasazuki T., Fukui Y. (2004) Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood 103, 3362–3364 [DOI] [PubMed] [Google Scholar]
- 7. Böse J., Gruber A. D., Helming L., Schiebe S., Wegener I., Hafner M., Beales M., Köntgen F., Lengeling A. (2004) The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Unoki M., Brunet J., Mousli M. (2009) Drug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosis. Biochem. Pharmacol. 78, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 9. Masuda A., Dohmae N. (2010) Automated Protein Hydrolysis Delivering Sample to a Solid Acid Catalyst for Amino Acid Analysis. Anal. Chem. 82, 8939–8945 [DOI] [PubMed] [Google Scholar]
- 10. Strahl B. D., Ohba R., Cook R. G., Allis C. D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 96, 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mantri M., Loik N. D., Hamed R. B., Claridge T. D., McCullagh J. S., Schofield C. J. (2011) The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. Chembiochem. 12, 531–534 [DOI] [PubMed] [Google Scholar]
- 12. Schiltz R. L., Mizzen C. A., Vassilev A., Cook R. G., Allis C. D., Nakatani Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 13. Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F. P., Li M., Yagyu R., Nakamura Y. (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 6, 731–740 [DOI] [PubMed] [Google Scholar]
- 14. Han G., Li J., Wang Y., Li X., Mao H., Liu Y., Chen C. D. (2012) The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 113, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 15. Dillon S. C., Zhang X., Trievel R. C., Cheng X. (2005) The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., Tamaru H., Khan S. I., Horton J. R., Keefe L. J., Selker E. U., Cheng X. (2002) Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subramanian K., Jia D., Kapoor-Vazirani P., Powell D. R., Collins R. E., Sharma D., Peng J., Cheng X., Vertino P. M. (2008) Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]