Background: CALHM1 is an ion channel for which structural information is lacking.
Results: CALHM1 has poor ion selectivity and a wide (∼14 Å) pore and is a hexamer, with monomers having four transmembrane domains with cytoplasmic termini.
Conclusion: CALHM1 shares structural features with pannexins, connexins, and innexins.
Significance: CALHMs, connexins, and pannexins/innexins are three structurally related protein families with shared and distinct functional properties.
Keywords: Biophysics, Fluorescence Resonance Energy Transfer (FRET), Ion Channels, Neurobiology, Oocyte, Single Molecule Biophysics, Dye Transfer, Ion Permeation, Photobleaching, Topology
Abstract
CALHM1 (calcium homeostasis modulator 1) forms a plasma membrane ion channel that mediates neuronal excitability in response to changes in extracellular Ca2+ concentration. Six human CALHM homologs exist with no homology to other proteins, although CALHM1 is conserved across >20 species. Here we demonstrate that CALHM1 shares functional and quaternary and secondary structural similarities with connexins and evolutionarily distinct innexins and their vertebrate pannexin homologs. A CALHM1 channel is a hexamer, comprised of six monomers, each of which possesses four transmembrane domains, cytoplasmic amino and carboxyl termini, an amino-terminal helix, and conserved extracellular cysteines. The estimated pore diameter of the CALHM1 channel is ∼14 Å, enabling permeation of large charged molecules. Thus, CALHMs, connexins, and pannexins and innexins are structurally related protein families with shared and distinct functional properties.
Introduction
CALHM1 (calcium homeostasis modulator 1), a gene of unknown function, was identified as a possible modifier of the age of onset of Alzheimer disease (1, 2). CALHM1 encodes a glycosylated membrane protein expressed throughout the brain that lacks significant homology to other proteins. Six human CALHM homologs have been identified, with alternatively spliced variants and different expression patterns throughout the body, and CALHM1 is conserved across >20 species, including vertebrates as well as urochordates, hemichordates, and nematodes. Recently, CALHM1 was shown to form a novel Ca2+-permeable ion channel whose gating is allosterically regulated by both membrane voltage and extracellular Ca2+ concentration ([Ca2+]o) (3). CALHM1 ion channels were shown to mediate enhanced neuronal excitability in response to reduced [Ca2+]o that occurs in the brain in physiological and pathological situations (3). Cortical neurons from mice with Calhm1 genetically deleted have altered electrophysiological properties and fail to respond to reduced [Ca2+]o (3). Notably, CALHM1 is permeable to Ca2+ (1, 3) and may also participate in cytoplasmic Ca2+ homeostasis (1, 3–5).
The ion permeability properties of CALHM1 are unique: Ca2+ is only 10-fold selected for over Na+ (PCa/PNa ∼11), CALHM1 does not discriminate between Na+ and K+, and it allows significant monovalent anion permeation (PCl/PNa ∼0.5) (3). To understand the permeation properties of the CALHM1 ion channel in more detail, we examined permeation of different monovalent and divalent cations as well as different sized structurally similar ions using electrophysiological and optical imaging techniques. Both approaches indicate that the CALHM1 pore has a surprisingly wide diameter, comparable with that of gap junction-forming connexins. Furthermore, we show that, like connexons, a CALHM1 ion channel is a hexamer of CALHM1 monomers, each of which contains four transmembrane domains with cytoplasmic amino and carboxyl termini. Together, these functional and structural features suggest that CALHM1 belongs to a third protein family that is similar to connexins and pannexins/innexins.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Neuro-2a (N2a)2 mouse neuroblastoma cells were cultured in Eagle's minimum essential medium supplemented with 10% fetal bovine serum and 0.5× penicillin/streptomycin (Invitrogen) at 37 °C, 5% CO2. For all imaging experiments, cells were plated on glass coverslips 1 day prior to transfection. SH-SY5Y cells were maintained in a 1:1 mixture of ATCC-formulated Eagle's minimum essential medium and F-12 medium, supplemented with 10% FBS and 0.5× penicillin/streptomycin in 95% air, 5% CO2 at 37 °C. Human wild-type (WT) CALHM1 was subcloned into pIRES2-EGFP (Clontech), pEGFPN1 (Clontech), pcDNA3.1-Myc-His version B (Invitrogen), and pBF (provided by F. Ashcroft, Oxford, UK) vectors. CALHM1-GFP was subcloned from pEGFPN1 into pBF. N2a and SH-SY5Y cells were transfected using Lipofectamine 2000 (Invitrogen).
Electrophysiology
All electrophysiological recordings were performed at room temperature (20–23 °C). cRNA was in vitro transcribed from linearized plasmids with the mMessage mMachine kit (Ambion). Female Xenopus laevis were purchased from Xenopus One. Oocytes were defolliculated by treatment with collagenase (Worthington). At least 2 h after collagenase treatment, 1–5 ng of CALHM1 cRNA was injected into oocytes with 80 ng of Xenopus connexin 38 antisense oligonucleotide to inhibit endogenous connexin 38 (Cx38) currents (3, 6, 7). Oocytes were kept at 16 °C in a ND96 solution (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 2.5 mm sodium pyruvate, 5 mm HEPES, 1× penicillin/streptomycin, pH 7.6 (adjusted by NaOH)). Recordings were performed 3–5 days after injection. Oocytes used in two-electrode voltage clamp experiments were injected with a 50-nl mixture of 20 mm BAPTA and 10 mm Ca2+ at least 30 min prior to recording to clamp [Ca2+]i to ∼100 nm and minimize activation of endogenous Ca2+-activated Cl− currents (3). In divalent cation-free solutions, 0.5 mm EGTA and 0.5 mm EDTA were added to the bath solutions without adding divalent cations. In ion permeability experiments, sucrose was used as a substitute for NaCl or Ca2+ to maintain osmolarity.
Data were acquired with an OC-725C amplifier (Warner Instruments Corp.) at 1 kHz with 16-bit A/D converter (Instrutech ITC-16). Electrodes were made from thin walled TW100F-6 glass (World Precision Instruments, Inc.), filled with 3 m KCl, and connected by 3 m KCl agar bridges to the bath solution.
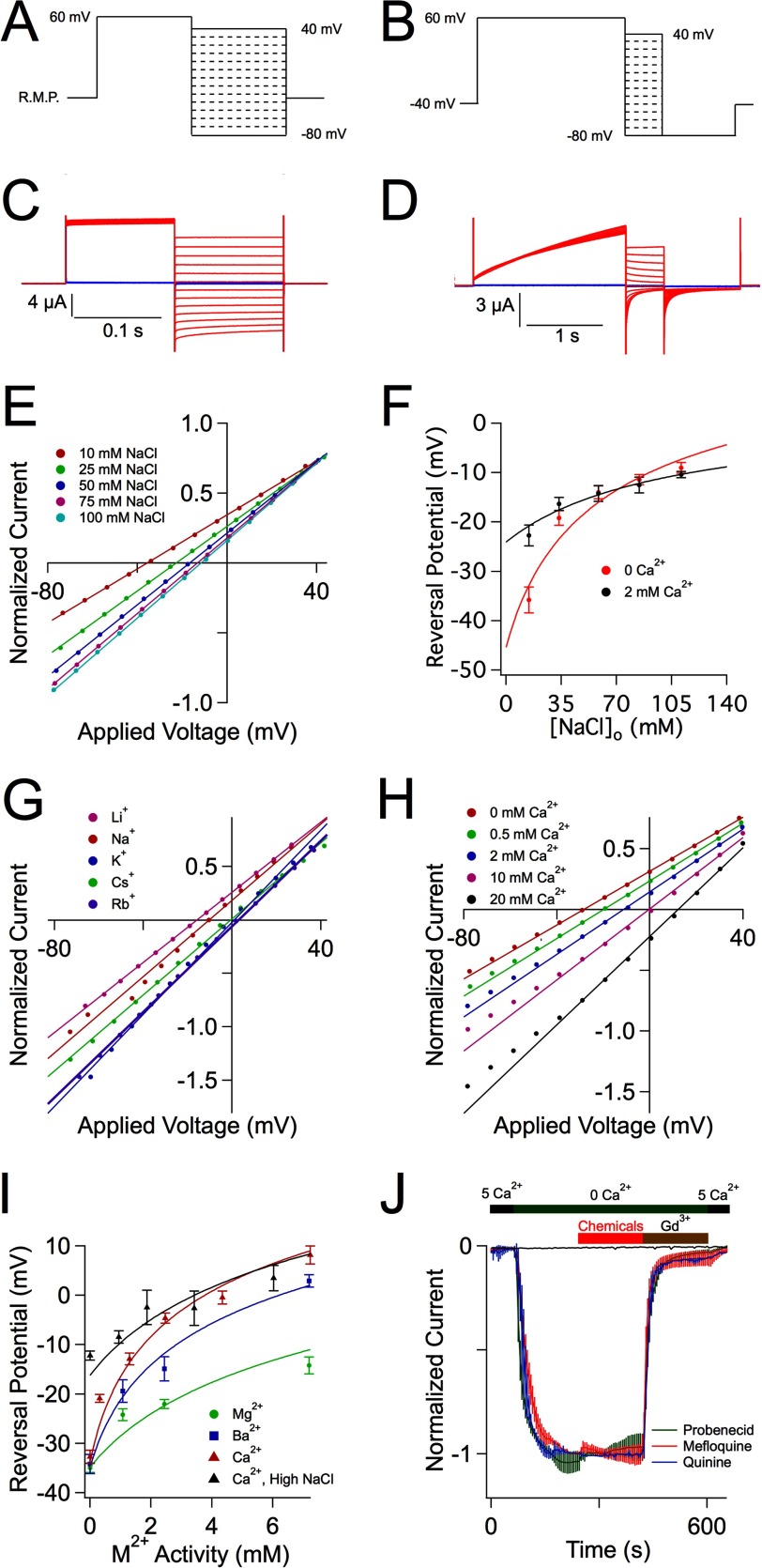
Two different voltage protocols were used during this study. In the absence of divalent cations, CALHM1 channels have fast activation gating and cannot be closed even at very negative voltages (3). A divalent-free voltage protocol (Fig. 1A) was designed to determine the instantaneous current-voltage relations. Oocytes were clamped at resting membrane potential, which was measured at the beginning of each experiment. A short (250-ms) depolarization to +60 mV from resting membrane potential activated CALHM1 channels. The voltage was then immediately stepped to test pulses ranging from −80 to +40 mV in 10-mV increments every 15 s to record the instantaneous currents. When oocytes are bathed in divalent cation-containing solutions, activation gating is slowed and CALHM1 is stabilized in closed states (3). A divalent cation voltage protocol (Fig. 1B) was used to determine the instantaneous current-voltage (I-V) relations. Oocytes were clamped at a holding potential of −40 mV to close CALHM1 channels. A 2000-ms depolarization to +60 mV from −40 mV activated CALHM1. The voltage was then immediately stepped to test potentials from −80 to +40 mV in 10-mV increments every 15 s to measure the instantaneous currents. One additional step to −80 mV for 1000 ms was applied to ensure a lack of nonspecific leak during this protocol.
FIGURE 1.
CALHM1 is a poorly selective ion channel. A, divalent-free voltage protocol. B, divalent voltage protocol. C, representative families of current traces from CALHM1-expressing (red) and control (blue) oocytes in response to the divalent-free voltage protocol shown in A and described under “Experimental Procedures.” Oocytes were bathed in a solution containing 100 mm NaCl, 10 mm HEPES, 0.5 mm EGTA, 0.5 mm EDTA, pH 7.3. D, representative families of current traces from CALHM1-expressing (red) and control (blue) oocytes in response to the divalent voltage protocol shown in B and described under “Experimental Procedures.” Oocytes were bathed in a solution containing 100 mm NaCl, 2 mm CaCl2, 10 mm HEPES, pH 7.3. E, normalized instantaneous I-V curves from representative CALHM1-expressing oocyte. Increasing [NaCl]o depolarized Erev. The solid lines are linear fits. F, permeabilities of K+, Cl−, and Ca2+ relative to Na+ were determined by plotting Erev from Erev versus [NaCl]o in the absence (red) or presence (black) of extracellular Ca2+, and the solid lines were calculated by fitting the data with either the Goldman-Hodgkin-Katz constant field equation (Equation 1) or the extended constant field equation (Equations 2–5), respectively. G, normalized instantaneous I-V curves from representative CALHM1-expressing oocytes bathed in 100 mm monovalent cation solutions. Shifts in Erev enabled calculation of relative permeabilities. Solid lines, linear fits. H, normalized instantaneous I-V curves from representative CALHM1-expressing oocytes bathed in various [Ca2+]o. Increasing [Ca2+]o depolarized Erev. Solid lines, linear fits. I, relative permeabilities of divalent cations (M2+) were determined by plotting Erev from H versus bath M2+ activity, and the solid lines were derived from fitting the data with the constant field equation (Equations 2–5). J, pharmacology of CALHM1 currents. Averaged, normalized currents (mean ± S.E.) are shown for 1 mm probenecid (green, n = 4), 30 μm mefloquine (red, n = 4), and 200 μm quinine (blue, n = 4). Black line, currents recorded in control oocyte injected only with Cx38 antisense oligonucleotide, normalized to the average CALHM1 currents (7.8 ± 1.1 μA, n = 12) from the same batch of oocytes.
Relative Permeability Estimation
Relative permeabilities of K+ and Cl− (PK and PCI, respectively) were estimated from reversal potentials (Erev) using the Goldman-Hodgkin-Katz constant field equation,
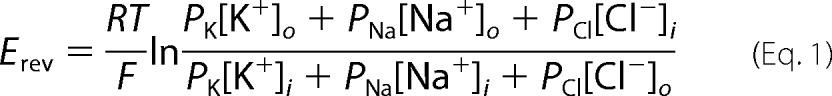 |
where subscripts i and o denote intracellular and extracellular, respectively; R, T, and F have their usual meanings, and PNa, PK, and PCl are the membrane permeabilities to Na+, K+, and Cl−, respectively. For estimations of the relative divalent cation (M2+) permeability, the Goldman-Hodgkin-Katz equation can be derived to an extended constant field equation,
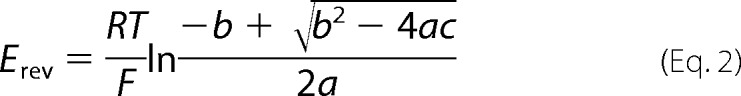 |
where
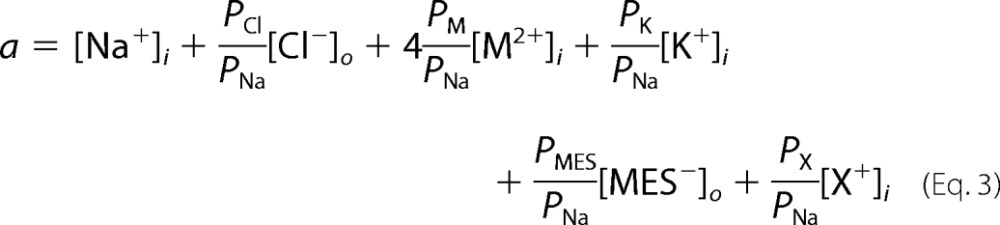 |
and
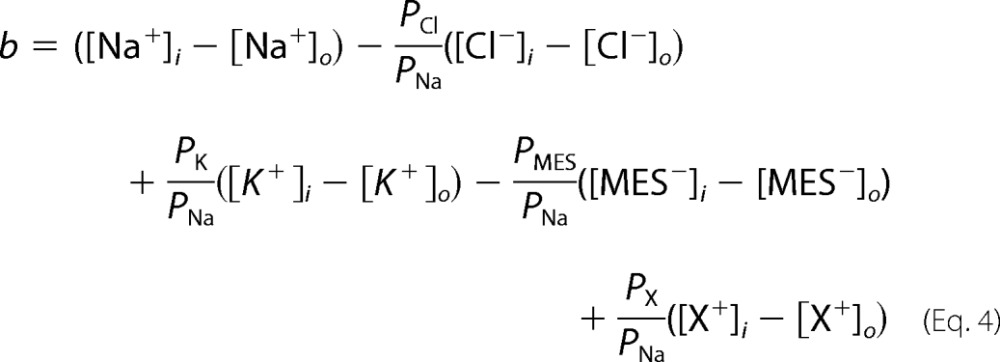 |
and
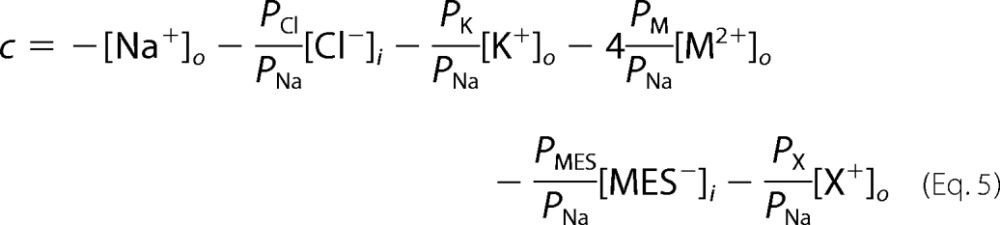 |
where X+ is any other monovalent cation except for Na+ and K+, and PNa, PK, PCl, PMES, PX, and PM are the membrane permeabilities to Na+, K+, Cl−, MES−, X+, and M2+, respectively. The cytoplasmic ionic concentrations of Na+, K+, Cl−, and Ca2+ were assumed to be 7 mm, 98 mm, 37 mm, and 3 × 10−4 mm, respectively (8). Both equations assume that the membrane is a homogeneous slab of material into which permeant particles partition instantaneously from the bulk solution, the ions cross the membrane independently, the electric field is constant (9), and the intracellular ionic concentrations do not change during the protocols.
Ionic Activity
When oocytes were bathed in solutions containing divalent cations, the ionic activities of the divalent cations were used instead of concentration. The ionic activity was calculated using Equation 6 (10),
where
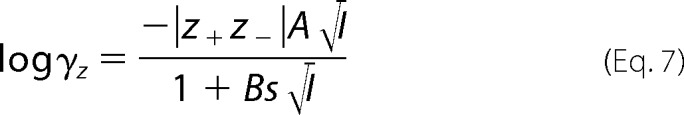 |
where γ is the activity coefficient, z is the valence of the ion, A and B are constants that equal 0.5108 and 0.3287 at 25 °C, respectively, s is ion radius and I is the ionic strength,
 |
where z is the ionic valence and c is the molar concentration.
Pore Size
Two estimates of pore diameter were made using the excluded volume model (11–14). Assuming a circular pore and spherical amines, the relationship between the relative permeability and ionic radius is as follows,
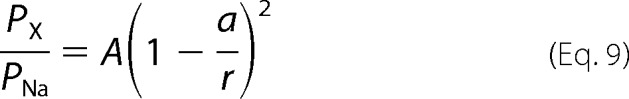 |
where PX/PNa is the relative permeability, a is the radius of the amine compound, r is the radius of the pore, and A is a scaling factor. The second model includes a term for the viscous drag of the amine as a function of the size of the amine (a). This model is defined as follows,
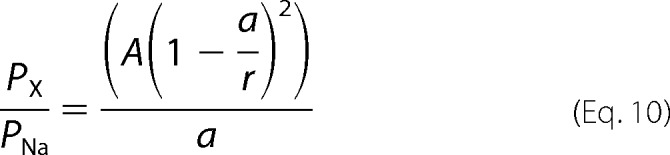 |
where all terms are the same as above.
CALHM1 Current Pharmacology
CALHM1-expressing oocytes bathed in 5 mm Ca2+ solution were clamped at a holding membrane potential of −15 mV. CALHM1 currents induced by exposure to a nominally Ca2+-free solution containing 1 mm Mg2+ were measured in response to test pulses to −80 mV for 25 ms every 5 s. Following 180 s of exposure to nominally Ca2+-free solution, test agents were perfused into the bath for 180 s. 100 μm Gd3+ was applied for 180 s after perfusion of the tested agents, and the currents were normalized to those immediately preceding application of the test agents for each experiment. Averaged normalized currents (mean ± S.E.) are shown.
Fluorescent Dye Uptake
N2a cells were transiently transfected with CALHM1 or empty vector. Experiments were performed 16–24 h post-transfection. Because extracellular Ca2+ modulates the gating of CALHM1, cells were incubated in dye-containing (0.1 mg/ml) Hanks' balanced saline solution (HBSS) for 8 min with Ca2+ (5 mm) to keep CALHM1 in closed states and thus inhibit dye uptake. The cells were then washed in 5 mm Ca2+ HBSS without dye to wash away excess extracellular dye. Alternatively, to activate CALHM1, cells were washed with HBSS containing 0.5 mm EGTA for 1 min. Then cells were incubated in nominally divalent cation-free HBSS containing dye (0.1 mg/ml) for 5 min. The cells were then incubated in dye-containing (0.1 mg/ml) HBSS with Ca2+ (5 mm) for 3 min to close CALHM1 and trap the dye inside the cells. Finally, the cells were washed in 5 mm Ca2+ HBSS without dye to remove excess extracellular dye. Differential interference contrast and fluorescence images were taken using the appropriate optical filters to visualize each dye.
Fluorescence images were analyzed using ImageJ. The plasma membranes of healthy cells were traced on the differential interference contrast images to establish the regions of interest of a particular field of cells. The same regions of interest were used in the corresponding fluorescence image to measure the average fluorescent intensity of each cell. The off-cell background fluorescence was subtracted from the fluorescence of each cell to correct for any residual dye that had not been sufficiently washed away. A box plot was generated for each cell category for each dye. Because the fluorescent intensities of the CALHM1-activated cells were not normally distributed, the Kruskal-Wallis test was used to determine statistical significance of median fluorescent intensities with an adjusted p value due to the multiple comparisons.
Western Blotting
16–24 h after transfection, cells were lysed in standard lysis buffer (250 mm NaCl, 50 mm Tris, pH 8.0, 1% Triton X-100, and a protease and phosphatase inhibitor mixture (PhosSTOP; Roche Applied Science)). Samples were bath-sonicated at 4 °C for 10 min and then centrifuged at 14,000 × g for 10 min. Supernatants were used to determine protein concentrations (Bio-Rad DC protein assay method). 4× sample buffer (8% SDS, 40% glycerol, 250 mm Tris, pH 6.8, and 0.002% bromphenol blue) was added to each sample before being boiled for 10 min. The percentage of β-mercaptoethanol (βME) was varied between 0.5 and 8%. Proteins were separated by SDS-PAGE on 10% Tris-HCl gels, transferred to nitrocellulose membranes, and blocked at room temperature for 1 h in Tris-buffered saline (50 mm Tris/HCl, 150 mm NaCl, pH 7.6) containing 0.1% Tween 20 (TBST) with 5% nonfat milk. Membranes were washed in TBST and incubated overnight at 4 °C with primary antibody in 5% milk/TBST. The CALHM1 antibody (gift from P. Marambaud; specifically recognizes the sequence 319EPPLMGNGWA328) concentration was 1:250, and the tubulin antibody (32-2600, Invitrogen) concentration was 1:2000. Membranes were incubated for 1 h at room temperature with secondary antibody (goat anti-mouse, 1:5000 dilution in 5% milk/TBST) conjugated to HRP, followed by chemiluminescence detection (ECL-Plus reagent, Pierce) and exposure to HyBlot CL (Denville Scientific). To ensure equal loading, membranes were submerged in stripping buffer (Restore Western blot stripping buffer, Pierce) at 37 °C for 20 min and then reprobed with a loading control antibody, β-tubulin. Films were scanned.
Blue Native PAGE
Blue Native PAGE (BN-PAGE) Western blotting was performed according to Invitrogen's recommended protocols. NativePAGETM reagents were acquired from Invitrogen. Transiently transfected SH-SY5Y cells were lysed in standard lysis solution (250 mm NaCl, 50 mm Tris, pH 8.0, and 1% Triton X-100). Appropriate volumes of NativePAGETM sample buffer (Invitrogen) were added, and an appropriate volume of NativePAGETM G-250 sample additive was introduced immediately before loading on a NativePAGETM Novex® 3–12% BisTris gel. NativeMarkTM unstained protein standard was added to the gel to estimate molecular mass. The Dark Blue Cathode Buffer was used until the dye front migrated through the first third of the gel. It was replaced with the Light Blue Cathode Buffer, and electrophoresis continued. The gel was run at 150 V for the first 60 min and then 250 V for the remainder of the time. Following electrophoresis, proteins were transferred to PVDF membranes using NuPAGE® transfer buffer at 180 mA for 60 min to transfer higher molecular mass proteins at 4 °C. The proteins were fixed on the membrane by shaking the membrane in 8% acetic acid for 15 min. Acetic acid was removed by rinsing the membrane in deionized water. Background dye was removed by shaking in methanol followed by washing in water. Standard Western blotting techniques, described above, were used to detect CALHM1.
Single Molecule Photobleaching
Single-molecule bleaching experiments were performed as described (15). Defolliculated Xenopus oocytes were injected with cRNAs for human CALHM1-GFP 12–24 h before imaging. Individual CALHM1-GFP puncta were visualized by total internal reflection fluorescence (TIRF) microscopy using a home-built system (16) based on an Olympus IX70 microscope equipped with a ×60, numerical aperture 1.45 TIRF objective. Devitellinated oocytes were allowed to settle on a coverglass forming the base of the recording chamber and were bathed in calcium-free Ringer's solution (containing 120 mm NaCl, 2 mm KCl, 5 mm MgCl2, 1 mm EGTA, 5 mm HEPES, pH 7.4). GFP-tagged molecules lying within the ∼100-nm evanescent field were excited by total internal reflection of a 488-nm laser beam incident through the microscope objective. Images (128 × 128 pixel; 1 pixel = 0.33 μm) were acquired at 10 frames s−1 by a Cascade 128+ electron multiplying CCD camera (Roper Scientific).
The resulting image stacks were processed in MetaMorph (Molecular Devices) by subtraction of a heavily smoothed (7 × 7-pixel) copy of each frame to correct for bleaching of autofluorescence and other background signals. Traces, such as those in Fig. 3D (black data points), were then obtained from selected spots, excluding those that showed obvious movement or where spots were too close to be unambiguously separated. The number of bleaching steps in each trace was determined using the StepFinder algorithm, devised originally to find steps of single molecular motors (17), which assumes only that the data is a train with steps of varying size and duration, hidden in Gaussian noise. Previous reports have shown that the amplitudes of successive bleaching steps recorded from multimeric tagged proteins vary (18–21). We also detected photobleaching steps by visual inspection (15, 18, 22, 23), with measurements restricted to those spots that showed complete bleaching and where fluorescence steps could be clearly resolved above the noise level. A bleaching step was defined as an abrupt (≤2 frames) change in fluorescence between dwell states during which the intensity fluctuated within the noise level around a stable value for ≥3 frames (18).
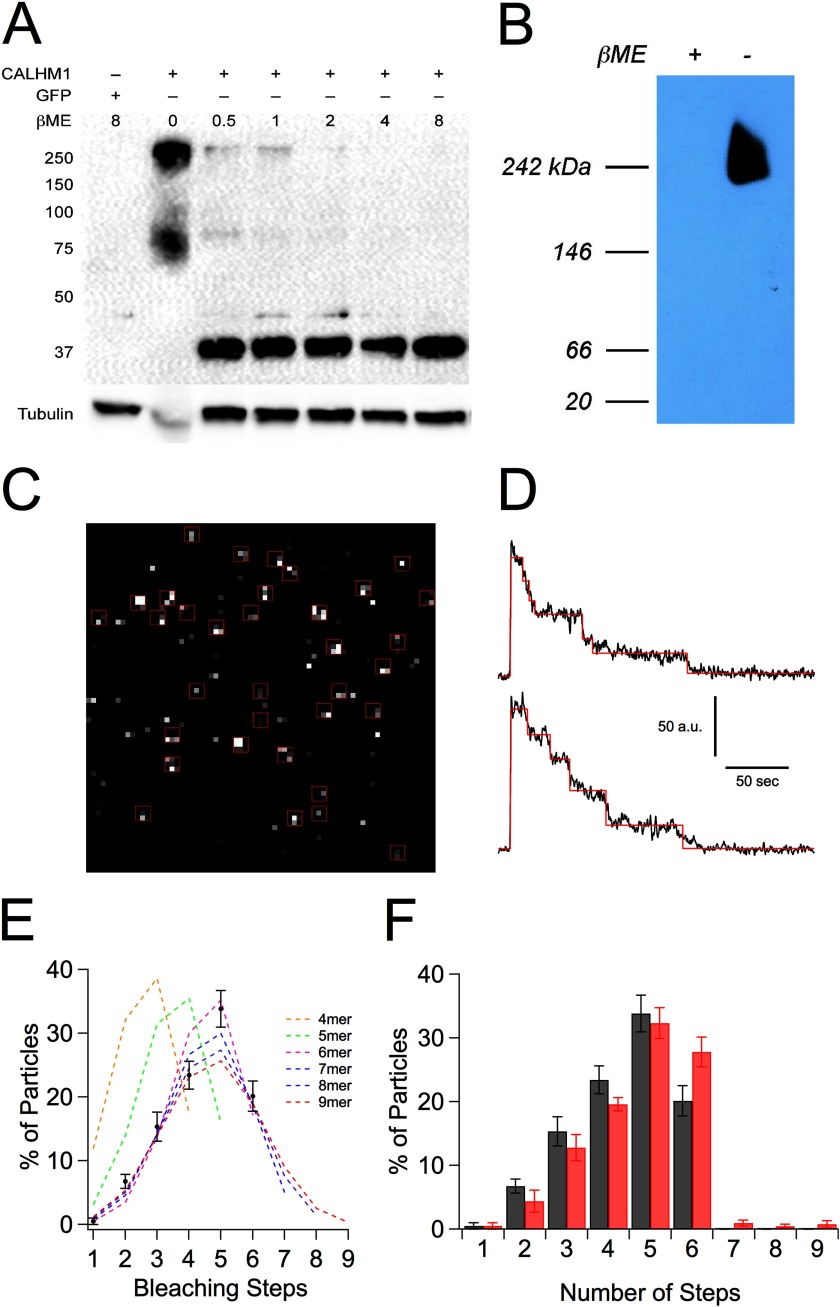
FIGURE 3.
CALHM1 channel is a hexamer. A, SDS-PAGE of N2a cells transiently transfected with GFP or CALHM1 with increasing concentrations of βME. B, BN-PAGE of SH-SY5Y cells transiently transfected with CALHM1 under reducing and non-reducing conditions. The absence of lower molecular mass bands in reducing conditions (+βME) is probably due to altered structure of CALHM1 that allows the reduced protein to migrate through the gel quickly. Because the experiment was optimized to detect higher order oligomers, monomeric CALHM1 probably runs through the gel. C, representative TIRF image of CALHM1-EGFP in oocyte plasma membrane acquired before photobleaching, showing fluorescent spots and regions of interest used to measure fluorescence intensity. D, representative fluorescence intensity traces from two fluorescent spots. Steps found using StepFinder (red line) are overlaid on the fluorescence intensity measurement (black line). After determining the number of steps in each bleaching trace, traces shown here were smoothed using a 5-point running average for display purposes. E, distribution of the number of bleaching steps observed from CALHM1-EGFP-expressing oocytes. Black data points, the average percentage of particles that bleached in each number of bleaching steps. Error bars, S.E. Data obtained from 271 particles, eight imaging records, and three oocytes were fitted with a binomial equation assuming the number of monomers in the complex and with the percentage of fluorescent GFP molecules being a free parameter (colored, dashed lines). F, distribution of the number of bleaching steps observed using different methods to identify bleaching steps. The same traces that were analyzed using StepFinder and summarized in E (black bars) were also analyzed by visual inspection (red bars) to identify bleaching steps. Error bars, S.E.
The observed distribution of the number of bleaching steps was fitted with a binomial distribution,
 |
where Total represents the total number of monomers in the multimer, Steps is the number of photobleaching steps, and Bright is the percentage of GFP molecules that are fluorescent.
Immunostaining
PC12 cells were transiently transfected with pIRES-CALHM1-EGFP2 or pIRES-EGFP2 alone. Two methods of immunostaining were used to distinguish between cytoplasmic or extracellular location of an epitope. For the permeabilized cell method, cells were washed twice with PBS at 4 °C and then fixed on ice with 4% paraformaldehyde for 20 min followed by permeabilization for 30 min with 0.2% saponin at room temperature. Cells were washed twice with PBS before incubating in primary antibody for 1 h at 4 °C and then washed three times in PBS and incubated in secondary antibody (Alexa-Fluor secondary antibody) for 1 h at room temperature. Finally, cells were washed three times in PBS at room temperature. Cells were counterstained with DAPI. Using the unpermeabilized cell immunostaining protocol, cells were washed twice in PBS + 1% BSA, incubated in primary antibody for 1 h at 4 °C, and washed twice in PBS before being fixed in 4% paraformaldehyde for 20 min at 4 °C. The cells were washed three times in PBS and then incubated in secondary antibody for 1 h at room temperature, washed three times in PBS at room temperature, and counterstained with DAPI. Control experiments, performed on cells expressing GFP (an intracellular epitope) or cells expressing 3xHA-OCA2 (an epitope that is known to be in extracellular regions of OCA2 (24)), validated the methods (data not shown).
Fluorescent Dye Transfer
Transiently transfected (GFP, pIRES-CALHM1-GFP, or GFP-tagged Cx30) N2a cells, plated on glass coverslips, were visualized on a fixed stage fluorescence microscope and identified by expression of GFP. The cells were bathed in physiological extracellular solution (150 mm Na+, 5.4 mm K+, 20 mm glucose, 1.5 mm Ca2+, 1 mm Mg2+, 150 mm Cl−, 10 mm HEPES, pH 7.4). Alexa350 (2 mm) was dissolved in a patch pipette solution (140 mm K+, 5 mm Na+, 10 mm Cl−, 2 mm MgCl2, 1 mm EGTA, 10 mm HEPES, 4 mm MgATP, 2 mm phosphocreatine, pH 7.3). The electrode tips were back-filled with dye solution and used to patch a single transfected cell within a cluster of transfected cells. After achieving the whole-cell configuration, dye diffused into the cell for 5 min while holding at −20 mV. The cells were imaged in epifluorescence microscopy 5–60 min after dye loading (340/10-nm excitation filter, 410-nm dichroic, 460/50-nm emission filter) to assess the extent of dye transfer. Dye was transferred if more than one cell in the cluster had Alexa350 fluorescence.
RESULTS
CALHM1 Has Weak Ion Selectivity
To examine the permeability properties of CALHM1 channels, whole-cell instantaneous currents were recorded using either the divalent-free voltage (Fig. 1A) or divalent voltage (Fig. 1B) protocol from oocytes expressing CALHM1. Representative families of current traces elicited in the absence or presence of divalent cations are shown in Fig. 1, C and D, respectively. The instantaneous currents at the different test pulses were normalized to the current measured at +60 mV at the end of the protocol. The normalized instantaneous currents versus test pulse voltages were plotted for various [NaCl]o (Fig. 1E). The reversal potentials (Fig. 1F) were determined from linear fits to the instantaneous I-V relations (solid lines in Fig. 1E). The relative permeabilities estimated by Goldman-Hodgkin-Katz equations (solid lines in Fig. 1F) were PNa/PK/PCl = 1:1.14:0.52 and 1:1.46:0.88 in the absence and presence of 2 mm extracellular Ca2+, respectively, indicating that CALHM1 is poorly selective with a slight preference for monovalent cations over anions, in agreement with a previous study (3). To extend those studies, we examined the selectivity among monovalent cations. We observed that CALHM1 poorly selects among Group I monovalent cations, with PNa/PLi/PK/PRb/PCs = 1:0.77:1.54:1.57:1.53 (instantaneous I-V relations in Fig. 1G). Increasing [Ca2+]o depolarized Erev (Fig. 1H), demonstrating that Ca2+ permeates CALHM1. Whereas CALHM1 is selective for Ca2+ over Na+, its selectivity among divalent cations (M2+) is weak, with PNa/PMg/PCa/PBa = 1:3.1:13.8:8.6 (Fig. 1I). Taken together, these data indicate that CALHM1 is a poorly selective ion channel.
The CALHM1 Channel Pore Has a Wide Functional Diameter
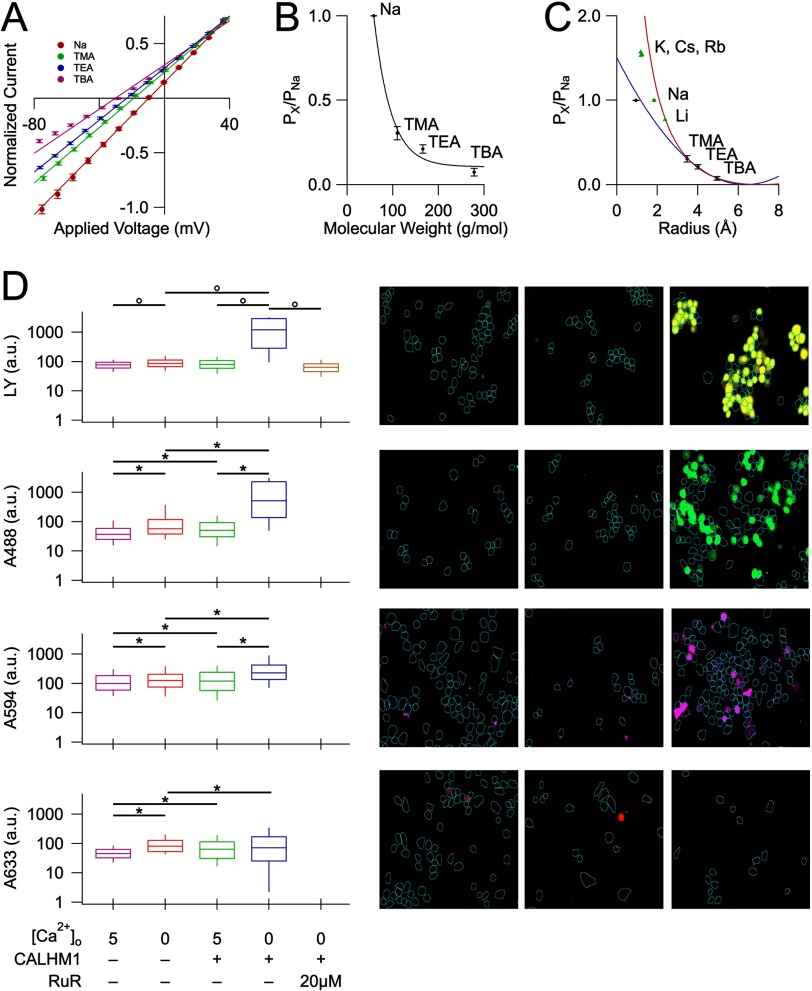
The relatively poor ion selectivity properties of CALHM1 suggested that its conducting pore might have a wide diameter. To probe the functional diameter of the CALHM1 pore, we examined permeation of various tetraalklylammonium monovalent cations with different sizes (tetramethylammonium (TMA+), tetraethylammonium (TEA+), and tetrabutylammonium (TBA+)) (25) (Table 1). Normalized instantaneous I-V relations were used to determine Erev of each tested ion (Fig. 2A), from which the relative permeability of each ion was estimated from an extended constant field equation (Equations 2–5) as PNa/PTMA/PTEA/PTBA = 1:0.31:0.21:0.07. Thus, CALHM1 has significant permeability to large cations. An exponential relationship between the relative permeability of each amine and its molecular mass (Fig. 2B) suggests that the size of the molecule, rather than binding within the pore, is the major determinant of its permeation (12–14, 26). Accordingly, the pore of CALHM1 behaves as a molecular sieve for monovalent cations (12–14, 26). By plotting the relative permeabilities against the Pauling radii of the amines, an excluded volume model can be used to estimate the functional diameter of the pore (12–14, 26) (Fig. 2C). This method estimates the functional diameter of the CALHM1 pore to be 12.8 Å (Equation 9). A better fit of the data, obtained by adding a term for viscous drag of each ion (Fig. 2C, red line), yielded an estimate of the CALHM1 functional pore diameter of 14.2 Å (Equation 10). Of interest, the relationship between the relative permeabilities of small monovalent cations (from Fig. 1G) and their Stokes radii was also well fitted by the volume exclusion model that incorporates viscous drag (Fig. 2C, green triangles). These results suggest that small monovalent cations are hydrated when they permeate through the CALHM1 pore. This can account for the lack of ion selectivity observed and further suggests that the pore is wide.
TABLE 1.
Ionic radii of monovalent cations
The Pauling and the Stokes radii (90) of each ion used to estimate the pore diameter of CALHM1 are shown.
| Ion | Pauling radius | Stokes radius |
|---|---|---|
| Li+ | 0.60 | 2.38 |
| Na+ | 0.95 | 1.84 |
| K+ | 1.33 | 1.25 |
| Rb+ | 1.48 | 1.18 |
| Cs+ | 1.69 | 1.19 |
| TMA+ | 3.47 | 2.05 |
| TEA+ | 4.00 | 2.82 |
| TBA+ | 4.94 | 4.72 |
FIGURE 2.
The ion-conducting pore of CALHM1 is wide. A, normalized instantaneous I-V curves of representative CALHM1-expressing oocytes bathed in various sized tetraalkylammonium cations (Table 2) using the divalent-free voltage protocol (Fig. 1A). Solid lines, linear fits; error bars, S.E. B, plot of relative permeability of each cation against its molecular mass is an exponential relationship. C, plot of relative permeability of each cation versus its respective ionic radius. Fitting data with the volume exclusion equation (blue curve; Equation 9) estimates a pore diameter of 12.8 Å. Fitting data with the volume exclusion equation including a term for the viscous drag of the ion (red curve; Equation 10) estimates a pore diameter of 14.2 Å. Relative permeabilities of monovalent cations from Fig. 1G are plotted versus their Stokes radii (green triangles). D, left, box plots of fluorescence intensities of intracellular Lucifer Yellow, Alexa488, Alexa594, and Alexa633 taken up by mock- and CALHM1-transfected cells in the presence (5 mm) or absence of extracellular Ca2+ to inhibit or activate CALHM1 opening, respectively (data plotted on log scale). Numbers of cells for each condition ranged from 316 to 1489. For each box plot, the middle line represents the median; upper and lower bounds of the box represent 75th and 25th percentiles, respectively; and upper and lower tails represent 90th and 10th percentiles, respectively. For each dye, median fluorescence intensities were compared between transfection conditions and extracellular calcium conditions only, using the Kruskal-Wallis test. Therefore, statistical significance was adjusted to correct for multiple comparisons; °, p < 0.01; *, p < 0.0125. Cells not expressing CALHM1 and cells incubated in solutions containing 5 mm Ca2+ showed background levels of fluorescence. Right, representative fluorescence images in dye uptake experiments. Regions of interest (blue traces) were drawn around morphologically normal cells in each field. Left, mock-transfected cells incubated in solutions containing 0 Ca2+. Middle, CALHM1-transfected cells incubated in solutions containing 5 mm Ca2+. Right, CALHM1-transfected cells incubated in solutions containing 0 Ca2+. RuR, Ruthenium Red. a.u., arbitrary units.
The estimated functional pore size of CALHM1 is large compared with traditional selective ion channels (3–6 Å) (25–28), whereas it is comparable with those of gap junction-forming connexins (29). The pore properties of connexins have been explored by imaging permeation of fluorescent dyes of different sizes (30, 31). As an independent approach to evaluate the CALHM1 functional pore diameter, we measured uptake into CALHM1-expressing neuroblastoma N2a cells of different sized fluorescent molecules with the same electrical valence (Alexa488 (A488), Alexa594 (A594), and Alexa633 (A633)) that are structurally related as well as Lucifer Yellow (LY) (Table 2). Significant uptake of LY, A488, and A594 was observed only in CALHM1-transfected cells and only under conditions in which CALHM1 channels were activated (Fig. 2D). Furthermore, dye uptake was inhibited by a CALHM1 blocker (3) (20 μm Ruthenium Red (RuR); Fig. 2D), consistent with CALHM1 mediating dye uptake. Uptake of LY and A488 in CALHM1-activated cells was robust, whereas uptake of A594 was less pronounced. A larger molecule, A633, did not permeate in any condition, indicating that its diameter exceeds that of the CALHM1 pore. These results suggest that the diameter of A594 (median axis 13.8 Å) approximates that of the CALHM1 pore. Importantly, this value is in good agreement with the functional pore diameter estimated by electrophysiology (14.2 Å). This independent method of estimating the size of the CALHM1 pore confirms a functional diameter of ∼14 Å.
TABLE 2.
Molecular dimensions of fluorescent dyes
The unhydrated dimensions of each dye are shown. Because the molecules are not spherical, the median axial diameter (shown in boldface type) was used to approximate the minimum diameter of the pore (30). This assumes that the longest axis aligns with the pore.
| Lucifer Yellow | Alexa350 | Alexa488 | Alexa594 | Alexa633 | |
|---|---|---|---|---|---|
| x | 12.2 | 13.0 | 11.3 | 16.6 | ? |
| y | 9.9 | 5.2 | 10.5 | 13.8 | ? |
| z | 2.6 | 3.2 | 9 | 9.3 | ? |
| Mass (kDa) | 443 | 350 | 570 | 760 | ∼1150 |
| Charge | −2 | −1 | −2 | −2 | ? |
The CALHM1 Channel Pharmacology Is Unique
A previous study demonstrated that CALHM1 currents were inhibited by Gd3+, Ruthenium Red, and Zn2+ and partially inhibited by 2-aminoethoxydiphenyl borate (3). Because the pore sizes of CALHM1 are similar to those of connexin and pannexin channels, we asked if the pharmacology of CALHM1 was similar to the pharmacology of those channels. We expanded the pharmacological characterization of CALHM1 channels to include probenecid (1 mm), mefloquine (30 μm), and quinine (200 μm), inhibitors of pannexins (32), and specific members of the connexin family (33–35), respectively. None of these compounds inhibited CALHM1 currents (Fig. 1J), indicating that CALHM1 has a distinct pharmacology compared with connexins and pannexins.
The CALHM1 Ion Channel Is a Hexamer
Because the pore sizes of CALHM1 and connexins are similar, we asked if CALHM1 has structural similarities with connexins. Connexin monomers oligomerize to form a hexamer called a connexon (29, 36). Whereas CALHM1 was previously suggested to oligomerize to form tetramers (1, 2), we reinvestigated the oligomeric status of CALHM1 in transiently transfected mouse N2a cells, first using SDS-PAGE. Under non-reducing conditions, two bands were present: one at ∼80 kDa and another at ∼250 kDa, corresponding to ∼2 and 6 times the mass of a CALHM1 monomer. Both bands disappeared upon the addition of the reducing agent βME, with a single band at 40 kDa (Fig. 3A), the expected mass of monomeric CALHM1, becoming dominant. To obtain another estimation of the oligomeric mass of CALHM1, BN-PAGE was employed (37, 38). One CALHM1-immunoreactive band was observed at ∼240 kDa under non-reducing conditions specifically in cells transfected with CALHM1 (Fig. 3B), similar to the molecular mass of the band observed in non-reducing SDS-PAGE. Because a CALHM1 monomer has a molecular mass of ∼40 kDa, these results suggest that CALHM1 might be larger than a tetramer, perhaps a hexamer.
A caveat of BN-PAGE or non-reducing SDS-PAGE for determination of molecular stoichiometry is that the protein might be associated with other proteins that contribute to the observed mass. We therefore used single-molecule subunit counting (21) to determine the number of monomers that form a CALHM1 channel. CALHM1-EGFP was expressed in Xenopus oocytes and imaged at the plasma membrane using TIRF microscopy (Fig. 3C). Expression of carboxyl-terminal EGFP-tagged CALHM1 generated ionic currents similar to untagged CALHM1 (not shown). The fluorescence intensities of immobile fluorescent particles were measured over time. Only particles that bleached completely and displayed at least one discrete bleaching step were analyzed (as in Fig. 3D). Using the StepFinder algorithm (17) to objectively determine the number of bleaching steps, most of the immobile fluorescent spots bleached in five steps, with a significant fraction bleaching in six steps (Fig. 3E). None of the particles bleached in more than six steps. Because not every EGFP molecule is fluorescent (18, 21–23, 39), the distribution of bleaching steps follows a binomial distribution with a probability (p) of EGFP being fluorescent. The data were best fitted by a binomial distribution assuming a 6-mer, with p = 74.6% (Fig. 3E and Equation 11), similar to previous studies that determined that ∼80% of EGFP molecules are fluorescent (21, 23, 39). Visual inspection analysis (15, 18, 22, 23) revealed a similar distribution, with the data well fitted by a binomial distribution assuming a 6-mer, with p = 80.2% (Fig. 3F and Equation 11). These results indicate that a CALHM1 channel is a hexamer of CALHM1 monomers, consistent with the conclusion reached from the SDS- and BN-PAGE determinations.
CALHM1 Has Four Transmembrane Helices with Cytoplasmic Termini
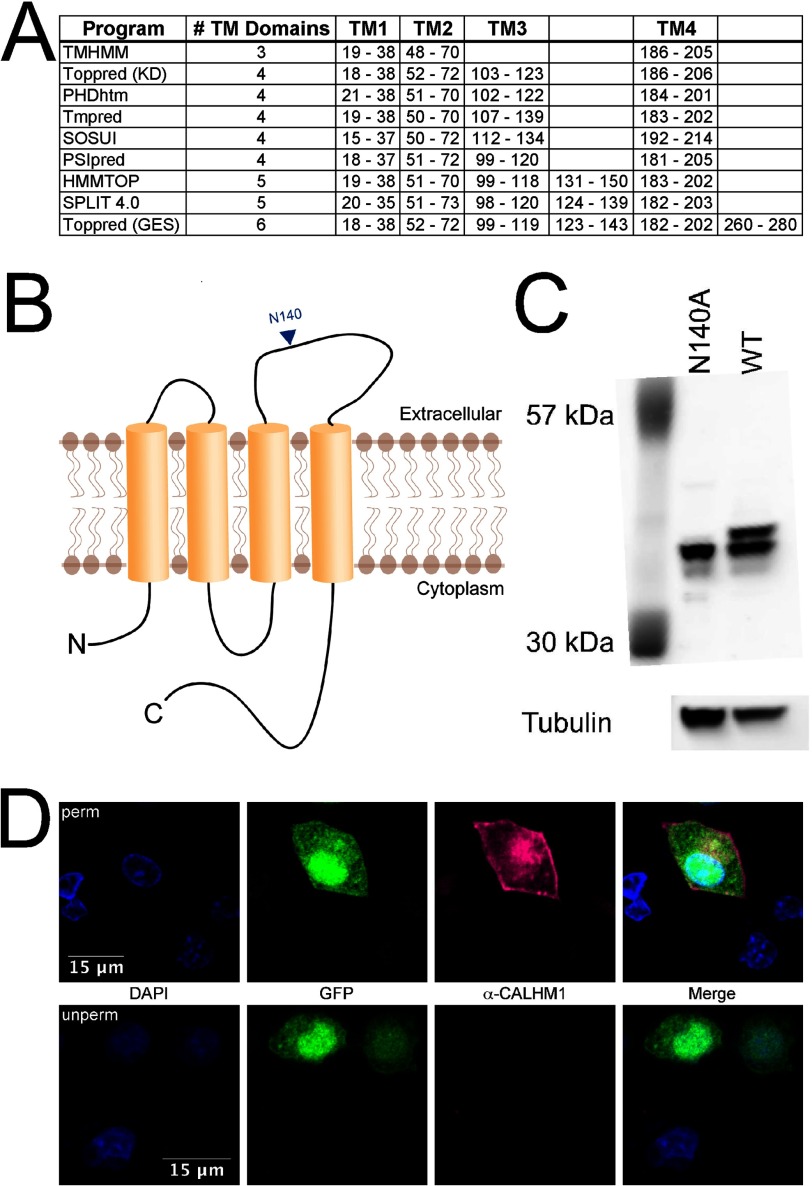
These results indicate that a CALHM1 ion channel is a hexamer with a large pore diameter. These structural features are similar to those of gap junction-forming connexons. We asked whether structural similarities between CALHM1 and connexons extended to the secondary structures of their monomers. Connexins have four membrane-spanning helices (TM) (29, 36). Although CALHM1 was originally proposed to have three TM and one re-entrant hydrophobic loop (1), TM prediction algorithms (40–45) suggest that human CALHM1 and other CALHMs have 3–6 hydrophobic domains (Fig. 4A), with the majority predicting four TM with cytoplasmic amino and carboxyl termini (Fig. 4B). Three prediction programs propose an additional TM domain between residues 123 and 150. This stretch of residues contains a putative N-glycosylation site, Asn-140. N-Glycosylation of this residue would indicate that this region is not in a TM domain but is instead located in an extracellular loop in the protein. We therefore confirmed that Asn-140 is N-glycosylated (1) (Fig. 4C), indicating that this region is not a TM but is part of an extracellular loop between TM3 and TM4. We determined the localization of the CALHM1 carboxyl terminus by exposing permeabilized or non-permeabilized transiently transfected PC12 cells to an antibody directed against the CALHM1 carboxyl terminus. CALHM1 was detected only in cells exposed to the permeabilized immunostaining protocol (Fig. 4D), suggesting that the carboxyl terminus is located in the cytoplasm. These results are consistent with a CALHM1 topology of four TM with cytoplasmic amino and carboxyl termini (Fig. 4B), similar to connexins.
FIGURE 4.
Membrane topology of CALHM1. A, predicted membrane-spanning regions from nine membrane topology prediction programs. B, model membrane topology of four TM domains; three cytoplasmic domains, including the amino and carboxyl termini; and two extracellular domains. C, CALHM1 is N-glycosylated at Asn-140. Western blot of wild type CALHM1 reveals two bands, whereas CALHM1 with Asn-140 mutated to alanine (N140A) reveals only the lower band, indicating that CALHM1 is glycosylated at Asn-140, indicating extracellular localization. D, the carboxyl terminus is cytoplasmic. Top, permeabilized PC12 cells expressing CALHM1 were exposed to CALHM1 carboxyl-terminal antibody, revealing a positive immunostaining signal (right center panel). Bottom, unpermeabilized PC12 cells expressing CALHM1 exposed to the same antibody had no immunostaining signal (right center panel). Transfected cells were identified by EGFP expressed by the same vector. A similar strategy to localize the amino terminus was unsuccessful because insertion of an epitope tag prevented CALHM1 expression.
CALHM1 Does Not Form Gap Junctions
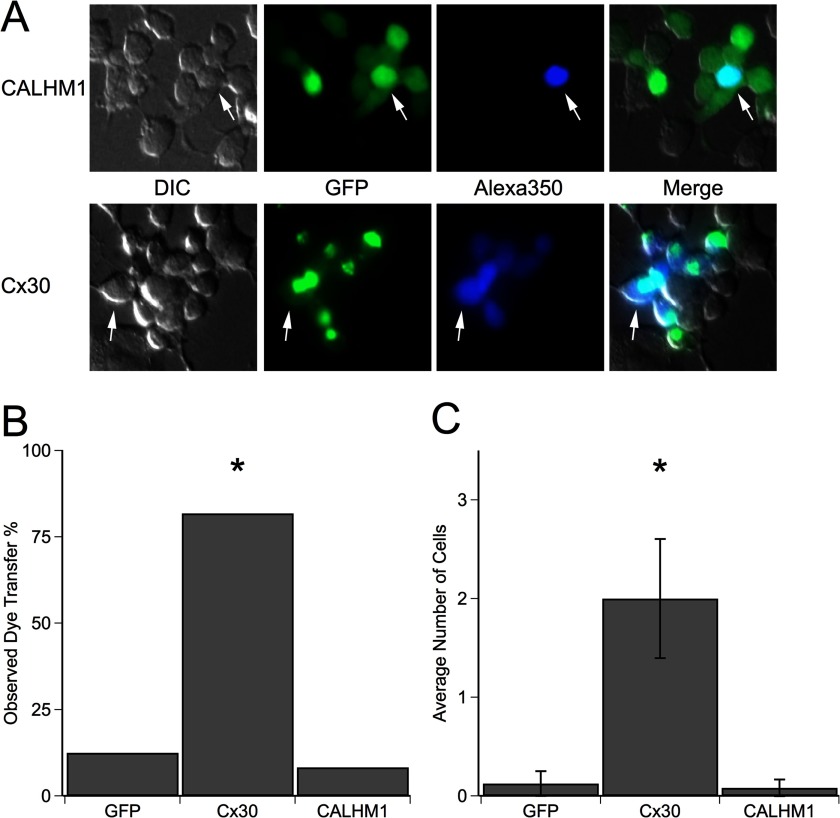
The structural similarities between CALHM1 and connexins suggest that the two proteins may have a similar functions. We asked if CALHM1 can form gap junctions in a manner similar to connexons. The ability of CALHM1 or Cx30 to form gap junctions was tested by using a microelectrode back-filled with Alexa350 dye (Table 2) to fill one transfected cell in a cluster of transfected cells with dye and measuring the extent of dye transfer to other cells in the cluster (Fig. 5A). 9 of 11 (81.8%) Cx30-transfected dye-loaded cells transferred dye to at least one other cell. Furthermore, Cx30 dye-loaded cells transferred dye to 2.4 ± 0.6 other cells. These results confirm that Cx30 forms gap junctions (46). In contrast, only 1 of 12 (8.3%) dye-loaded CALHM1-transfected cells transferred dye to another cell, similar to dye-loaded GFP-transfected cells (Fig. 5B). The lone CALHM1-transfected cell that did transfer dye transferred it to only one other cell (Fig. 5C). We considered that CALHM1 might form gap junctions but that the presence of extracellular Ca2+ in these experiments kept CALHM1 closed, preventing dye transfer. To test this, experiments were also performed in the absence of extracellular Ca2+. CALHM1 activation by low [Ca2+]o (3) resulted in dye leakage through CALHM1 to the extracellular space, but no dye transfer to adjacent CALHM1-expressing cells was observed (data not shown). These data, together with the observation that Asp-140 is N-glycosylated, strongly suggest that CALHM1 does not form gap junctions but instead functions as an unapposed plasma membrane ion channel (3).
FIGURE 5.
CALHM1 does not form gap junctions. A, representative images of dye transfer in N2a cells transiently transfected with either GFP-tagged CALHM1 (top row) or GFP-tagged Cx30 (bottom row). In a group of transfected cells, a single cell was loaded with Alexa350 by a patch clamp whole-cell dialysis for 5 min (arrowhead). Fluorescence images were taken 5–60 min after loading to determine the extent of Alexa350 transfer to other cells. B, percentage of injected cells that transferred dye to neighboring transfected cells. Only one of 12 dye-loaded CALHM1-transfected cells examined transferred dye, and it was transferred to only one other cell, probably a daughter cell that had not yet completed cytokinesis (89), similar to what was observed in the GFP-transfected cells. Conversely, 9 of 11 of the Cx30-transfected cells transferred dye to neighboring transfected cells. Furthermore, the dye was transferred to multiple cells in six of eight cases. Eight GFP-expressing or untransfected, 11 Cx30-expressing, and 12 CALHM1-expressing cells were injected with dye. Statistical significance was determined using Fisher's exact test, p < 0.001. C, average number of cells into which dye transferred. The one injected control cell and one injected CALHM1-transfected cell that transferred dye transferred it to only one neighboring cell. Alternatively, when Cx30-transfected cells transferred dye, there were on average 2.44 recipient cells (Student's unpaired t test with unequal variance, p < 0.01). DIC, differential interference contrast.
DISCUSSION
The present study is the first structural investigation of the CALHM1 ion channel. Based on our results, we conclude that CALHM1 has four transmembrane domains with a cytoplasmic amino and carboxyl termini and that it forms a hexameric, nonselective ion channel with a wide permeation pore. The structural characteristics of CALHM1 are similar to those of connexins, pannexins, and innexins. However, unlike connexins and innexins, CALHM1 does not form gap junctions.
The Permeation Pathway of CALHM1
Our electrophysiological analyses of mono- and divalent cation permeation indicate that CALHM1 forms a nonselective ion channel with only weak charge selectivity, with Pdivalent cation > Pmonovalent cation > Pmonovalent anion. Of note, the selectivity sequence of CALHM1 is remarkably similar to that of connexins rCx40 and rCx43 (47) and rCx43 (48).
Two independent methods demonstrated that the pore of CALHM1 has a ∼7-Å functional radius at its narrowest region (Fig. 2). In the first method, the relative permeabilities of various tetraalkylammonium ions were determined and then used to estimate the functional pore radius. These ions have similar shapes and valence, differing only in their radii. Fitting the data with a volume exclusion model estimated a functional pore radius of ∼7 Å. Although the parabolic dependence of the volume exclusion model predicts that molecules larger than 7.1 Å would permeate CALHM1, this behavior was not observed. Notably, this model is also able to predict the relative permeabilities of the Group I monovalent cations examined (Fig. 1G) when plotted versus their respective Stokes' radii. The Stokes' radius accounts for the hydration but not the shape of the ion, which could lead to an underestimation of the true radius of the ions because most ions are not perfectly spherical.
These results suggest that small ions remain at least partially hydrated as they permeate the pore. The hydration shell can shield the ion from the amino acids that line the pore, minimizing interactions that could promote selectivity. Larger ions that approach the size of the pore may be able to interact with the pore lining residues, resulting in increased selectivity among big ions.
In the second method, we employed optical imaging of fluorescent dye uptake using dyes of the same valence and similar structure with different sizes. The smallest dyes, LY and A488 (Table 2), easily permeated through CALHM1. The influx of A594 was significantly less robust, suggesting that the dimensions of A594 (median axis 13.8 Å) approach the diameter of the pore. The largest dye, A633, did not permeate through CALHM1. Structural information about A633 is unavailable because it is considered proprietary by its manufacturer. Conversations with representatives of the manufacturer revealed that its structure is similar to those of the other Alexa dyes with a mass of ∼1150 g/mol. Assuming that the larger mass is correlated with an increase in its molecular dimensions, it is reasonable to conclude that the diameter of A633 is >13.8 Å. These results also suggest that the CALHM1 functional pore diameter is ∼14 Å.
The good agreement between two independent experimental approaches suggests that the CALHM1 functional pore diameter is ∼14.2 Å. The CALHM1 pore is therefore quite large compared with most ion channels (e.g. nicotinic receptors (9.2 Å (12, 49)), voltage-gated Ca2+ channels (6 Å (26)), NMDA receptors (7.2 Å (50)), Orai1 (3.8 Å (51)), Orai3 (3.8Å (52)), and voltage-gated K+ channels (4.7 Å (53)), voltage-gated Na+ channels (6.1 Å (25)), and cyclic nucleotide-gated channels (9.2 Å (54)). The pore diameter of CALHM1 is comparable with those of connexons (Cx26 (14 Å (29)), Cx43 (12.6 Å (48)), Cx40 (13.2 Å (47)), and Cx32 (between 12 and 14 Å (55)), whereas it is smaller than those of Panx1 and Panx2 (17–21 Å and 30 Å, respectively (56)).
Although the pore radius of CALHM1 is large, its unitary monovalent cation conductance is only 24 picosiemens (3). The unitary conductances of connexon channels are not highly correlated with their pore diameters (reviewed in Ref. 57). Lack of correlation between the pore diameter and unitary conductance can occur if the narrowest region of a wide pore extends for a relatively long distance. An ion traversing the pore would be able to interact with multiple sites in the pore, retarding its diffusion and reducing its conductance.
The CALHM1 Ion Channel Is a Hexamer
CALHM1 was previously shown to homo-oligomerize (1). In the previous study, only monomeric CALHM1 (∼40 kDa) was observed under reducing conditions, whereas ∼160 and 80 kDa immunoreactive bands were observed under non-reducing conditions (1), consistent with the proposal that CALHM1 can form tetramers and dimers, respectively. We noted that the gel employed previously spanned a limited range of protein mass, 70 to ∼150 kDa, which could preclude detection of possible oligomers larger than a tetramer. To determine if CALHM1 can form complexes larger than 160 kDa, we repeated SDS-PAGE experiments using gels that span a wider range of molecular mass (Fig. 3A). In the absence of βME, we observed prominent bands at ∼250 and ∼80 kDa, suggesting that CALHM1 can form oligomers larger than a tetramer. In the presence of βME, the intensity of the higher molecular weight band diminished, and a single prominent band at ∼40 kDa appeared. Accordingly, SDS and boiling alone are insufficient to denature the CALHM1 complex. Instead, reducing agents must be used to denature the CALHM1 complex. In the absence of a reducing agent, the molecular mass markers may slightly overestimate the molecular mass of the CALHM1 complex because the markers are denatured proteins that probably migrate faster than non-denatured, non-reduced CALHM1 complex. Thus, the observed ∼250 kDa band is consistent with the expected 240-kDa mass of a CALHM1 hexamer.
Another estimate of the mass of the CALHM1 complex was obtained using BN-PAGE (Fig. 3B). BN-PAGE uses Coomassie® G-250 as a charge shift molecule instead of SDS, enabling proteins to remain in their native conformations as they migrate through the gel. In the absence of βME, a single CALHM1 band was present at ∼240 kDa. This is a more accurate estimation because the molecular mass markers are also in their native conformations. Despite repeated attempts with a range of concentrations, we were unable to detect lower molecular weight CALHM1 species when the sample was reduced by βME. The inability to detect even monomeric CALHM1 is likely because denatured CALHM1 migrates rapidly through the BN-polyacrylamide gel. Nevertheless, the detection of a ∼250-kDa species by both non-reducing SDS-PAGE and non-denaturing BN-PAGE suggests that native CALHM1 is probably composed of more than four, possibly six, CALHM1 monomers.
The major caveat of non-reducing SDS-PAGE as well as BN-PAGE is that other proteins can contribute mass to the CALHM1 complex. Thus, a complex with mass of ∼240 kDa does not necessarily contain six CALHM1 monomers. Furthermore, proteins with native conformations with acidic isoelectric points and compact structures may migrate faster than other proteins in non-reducing PAGE, introducing errors of up to 15% in the mass estimation (37).
Because many factors, including size, charge, and shape of the protein, determine how quickly it migrates through a gel, we employed an alternate approach to determine the subunit stoichiometry of a CALHM1 channel. Single-molecule photobleaching experiments were used to determine the number of CALHM1 monomers in a functional CALHM1 ion channel complex. This method takes advantage of the discrete nature of GFP photobleaching to directly determine the number of subunits in membrane proteins by counting the bleaching steps of single fluorescent proteins (21). Because CALHM1 is genetically fused to GFP, the number of bleaching steps represents the number of GFP-tagged subunits in the complex. This technique has been used previously to determine the stoichiometry of membrane proteins that have up to seven monomers in the complex (58). The low expression level of the protein reduced the probability of aggregation of multiple complexes in the same pixel region.
Consistent with the photobleaching traces shown in Fig. 3D, previous reports have shown that the amplitude of successive bleaching steps recorded from multimeric tagged proteins can vary (18–21). There are four main reasons for this behavior (19). First, the orientation of different fluorophores, even in the same complex, is different. Second, fluorophores with the same orientation can interact with various surface impurities or charges, which can lead to variability in fluorescence emission intensity. Third, the location of the oligomer relative to the focus of the excitation laser beam can produce different fluorescence intensities. Finally, because we employed a TIRF microscope, the fluorophore's distance into the cell can alter its fluorescence emission intensity. Therefore, the number of bleaching steps cannot be determined by dividing the total fluorescence intensity by the intensity of a single bleaching step. As a result, we are not able to detect multiple bleaching steps if they photobleach simultaneously.
Using both StepFinder and visual inspection to identify photobleaching steps, the highest percentage of particles bleached in five steps, with a significant percentage bleaching in six steps. The distribution of CALHM1-EGFP bleaching steps should approximate a binomial distribution with two parameters: total number of molecules in the complex and the percentage of EGFP molecules that initially fluoresce. Fitting the data with a binomial equation that assumed the total number of molecules in the complex but allowed the percentage of fluorescing EGFP molecules to be free, the StepFinder distribution was fitted best assuming a 6-mer with 74.6% of the GFP molecules initially fluorescent. By visual inspection, the distribution of bleaching steps was fitted well by a 6-mer with 80.2% of the GFP molecules fluorescent. Previous similar experiments (21, 23, 39) found that ∼20% of the GFPs are in a non-fluorescing state, possibly due to misfolding or incomplete maturation of the GFP. Our determinations are in good agreement with those results, which is perhaps not unexpected because we used the same fluorophore from the same source as those studies (EGFP from Clontech pEGFP-N1). Accordingly, the distribution of bleaching steps can be fitted best assuming that CALHM1 is a hexamer.
The two methods used to identify bleaching steps had similar results. The visual inspection method identified some very fast bleaching steps that StepFinder did not identify, because the magnitude of the bleaching steps was less than the noise of the measurement, causing a right shift of the distribution of bleaching steps. The visual inspection method also identified a small percentage of particles that bleached in more than six steps, probably due to more than one CALHM1 complex being in the same pixel region.
The Structure of CALHM1 Is Similar to Those of Connexins and Pannexins/Innexins
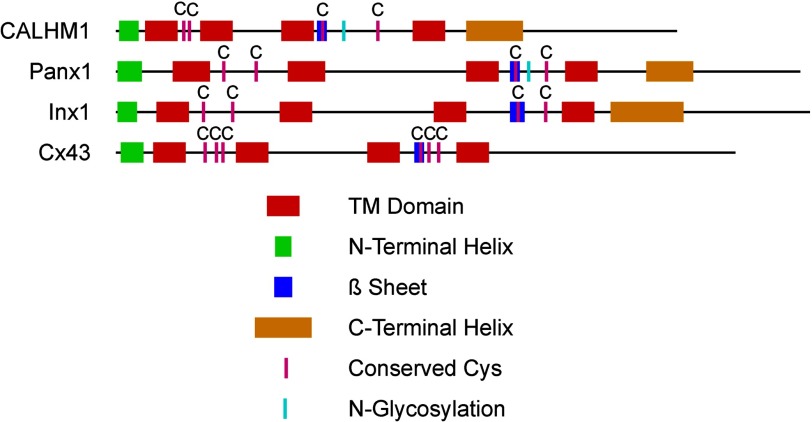
Connexins comprise a large family of gap junction proteins in vertebrates (59, 60). Functional studies have identified innexins as the proteins responsible for forming functional hemichannels and gap junctions in invertebrates (61–64). However, innexins and connexins have no evolutionary relationship (59, 61, 62, 65, 66). In contrast, pannexins are vertebrate homologs of innexins (59, 67, 68), but they do not form gap junctions, functioning instead as unapposed plasma membrane ion channels (69). Connexons (29, 36) and pannexins (Panx1 (70)) are hexameric assemblies of subunits. The amino acid sequences of Panx1 and Cx43, which have no evolutionary relationship, are 18.7% similar. By the same analysis, the CALHM1 amino acid sequence is 18.3 and 17.9% similar to Panx1 and Cx43, respectively, suggesting that CALHM1 shares no evolutionary relationship with these or other protein families (71, 72). Nevertheless, like connexons (29, 36) and Panx1 (70), the CALHM1 ion channel is comprised of a hexamer of subunits that surround a wide central pore. Furthermore, we note that CALHMs, connexins, and innexins/pannexins share secondary structural features (73) (Fig. 6). In addition to the presence of four TM domains, all three families possess an amino-terminal α-helix, a β-sheet in the second extracellular loop, and, except for connexins, α-helical regions in the carboxyl termini that align well (Fig. 4E). Conserved cysteines in the two extracellular loops are present in all three families. Connexins have three conserved Cys residues in each extracellular loop, pannexins and innexins have two Cys residues in each loop (74, 75), and CALHM proteins also have two conserved Cys residues in the first extracellular loop, and although there are multiple Cys residues in the second extracellular loop, only two are conserved among all members of the CALHM family. Pannexins and CALHMs are distinguished from the gap junction-forming connexins and innexins by being N-glycosylated (76), a post-translational modification that when introduced into connexins prevents gap junction formation (Cx32 (77, 78)) and when removed from Panx1 increases junctional conductance in paired Panx1-expressing oocytes (77). CALHM1 does not appear to form gap junctions (Fig. 5), but instead it functions as an unapposed plasma membrane ion channel (3).
FIGURE 6.
Schematic depiction of the alignment of the secondary structures of CALHM1, Panx1, Cx43, and Inx1. Conserved cysteines and glycosylation sites are also shown.
CALHM1, connexins, pannexins, and innexins have similar structural features that confer both shared and distinct functional properties. Like connexons (79–81) and pannexin channels (74, 82–85), CALHM1 is activated by membrane depolarization (3), although their kinetic responses are distinct. Reduced [Ca2+]o activates connexons (79–81) and CALHM1 (3) but not pannexins (83). Pannexins and connexins share pharmacological features that are absent in CALHM1 (3, 83, 86) (Fig. 1J). The predominant function of connexins is to form gap junctions that allow the passage of molecules of up to 1 kDa between cells. Plasma membrane hemichannels exist, although their physiological roles are not clear. Pannexins and connexons have been proposed to mediate cellular release of large molecules, including adenine nucleotides and prostaglandins, that could act as autocrine and paracrine signaling factors (57, 82, 87, 88). The similar structural features of CALHM1 suggest that in addition to acting as a sensor of extracellular Ca2+ in the brain (3), it may also participate in similar signaling functions. Although sequence analysis suggests that CALHMs, connexins, and pannexins share no evolutionary relationship, the structural and functional similarities of CALHM1 with the connexin, pannexin, and innexin protein families defines CALHM as a new family within a superfamily of proteins related by structural homoplasy that have shared and distinct functional properties and roles in cellular physiology.
Acknowledgments
We thank Drs. M. S. Marks and A. Sitaram for the 3xHA-OCA2 plasmid; Dr. P. Marambaud for the CALHM1 antibody; Dr. S. W. Yum for the Cx30 plasmid; Drs. Y. E. Goldman and Y. Sun for help with preliminary photobleaching studies; Drs. N. A. Cohen and R. J. Lee for use of the microscope; and Dr. C. Deutsch, Dr. T. Hoshi, Dr. B. Salzberg, Dr. D. Ren, Dr. S. Baylor, Dr. H. Vais, Dr. D. D.-O. Mak, and M. Muller for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants MH059937 and GM056328 (to J. K. F.) and GM048071 (to I. P.).
- N2a
- Neuro-2a
- HBSS
- Hanks' balanced saline solution
- βME
- β-mercaptoethanol
- BN-PAGE
- blue native PAGE
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TIRF
- total internal reflection fluorescence
- EGFP
- enhanced green fluorescent protein
- Cx
- connexin
- TMA
- tetramethylammonium
- TEA
- tetraethylammonium
- TBA
- tetrabutylammonium
- TM
- transmembrane
- LY
- Lucifer Yellow
- A488
- A594, and A633, Alexa488, -594, and -633, respectively.
REFERENCES
- 1. Dreses-Werringloer U., Lambert J. C., Vingtdeux V., Zhao H., Vais H., Siebert A., Jain A., Koppel J., Rovelet-Lecrux A., Hannequin D., Pasquier F., Galimberti D., Scarpini E., Mann D., Lendon C., Campion D., Amouyel P., Davies P., Foskett J. K., Campagne F., Marambaud P. (2008) A polymorphism in CALHM1 influences Ca2+ homeostasis, Aβ levels, and Alzheimer's disease risk. Cell 133, 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert J. C., Amouyel P. (2007) Genetic heterogeneity of Alzheimer's disease. Complexity and advances. Psychoneuroendocrinology 32, S62–S70 [DOI] [PubMed] [Google Scholar]
- 3. Ma Z., Siebert A. P., Cheung K. H., Lee R. J., Johnson B., Cohen A. S., Vingtdeux V., Marambaud P., Foskett J. K. (2012) Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc. Natl. Acad. Sci. U.S.A. 109, E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gallego-Sandín S., Alonso M. T., García-Sancho J. (2011) Calcium homoeostasis modulator 1 (CALHM1) reduces the calcium content of the endoplasmic reticulum (ER) and triggers ER stress. Biochem. J. 437, 469–475 [DOI] [PubMed] [Google Scholar]
- 5. Moreno-Ortega A. J., Ruíz-Nuño A., García A. G., Cano-Abad M. F. (2010) Mitochondria sense with different kinetics the calcium entering into HeLa cells through calcium channels CALHM1 and mutated P86L-CALHM1. Biochem. Biophys. Res. Commun. 391, 722–726 [DOI] [PubMed] [Google Scholar]
- 6. Bahima L., Aleu J., Elias M., Martín-Satué M., Muhaisen A., Blasi J., Marsal J., Solsona C. (2006) Endogenous hemichannels play a role in the release of ATP from Xenopus oocytes. J. Cell Physiol. 206, 95–102 [DOI] [PubMed] [Google Scholar]
- 7. Ebihara L. (1996) Xenopus connexin38 forms hemi-gap-junctional channels in the nonjunctional plasma membrane of Xenopus oocytes. Biophys. J. 71, 742–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weber W. (1999) Ion currents of Xenopus laevis oocytes. State of the art. Biochim. Biophys. Acta 1421, 213–233 [DOI] [PubMed] [Google Scholar]
- 9. Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., Sinauer, Sunderland, MA [Google Scholar]
- 10. Hamer W. J., Wu Y.-C. (1972) Osmotic coefficients and mean activity coefficients of univalent electrolytes in water at 25 °C. J. Phys. Chem. Ref. Data 1, 1047–1099 [Google Scholar]
- 11. Adams D. J., Dwyer T. M., Hille B. (1980) The permeability of endplate channels to monovalent and divalent metal cations. J. Gen. Physiol. 75, 493–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dwyer T. M., Adams D. J., Hille B. (1980) The permeability of the endplate channel to organic cations in frog muscle. J. Gen. Physiol. 75, 469–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farris H. E., LeBlanc C. L., Goswami J., Ricci A. J. (2004) Probing the pore of the auditory hair cell mechanotransducer channel in turtle. J. Physiol. 558, 769–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y. M., Favre I., Schild L., Moczydlowski E. (1997) On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. Effect of alanine mutations at the DEKA locus on selectivity, inhibition by Ca2+ and H+, and molecular sieving. J. Gen. Physiol. 110, 693–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Penna A., Demuro A., Yeromin A. V., Zhang S. L., Safrina O., Parker I., Cahalan M. D. (2008) The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 456, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demuro A., Parker I. (2005) Optical single-channel recording. Imaging Ca2+ flux through individual ion channels with high temporal and spatial resolution. J. Biomed. Opt. 10, 11002. [DOI] [PubMed] [Google Scholar]
- 17. Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 18. Demuro A., Penna A., Safrina O., Yeromin A. V., Amcheslavsky A., Cahalan M. D., Parker I. (2011) Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc. Natl. Acad. Sci. U.S.A. 108, 17832–17837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding H., Wong P. T., Lee E. L., Gafni A., Steel D. G. (2009) Determination of the oligomer size of amyloidogenic protein β-amyloid(1–40) by single-molecule spectroscopy. Biophys. J. 97, 912–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ji W., Xu P., Li Z., Lu J., Liu L., Zhan Y., Chen Y., Hille B., Xu T., Chen L. (2008) Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. U.S.A. 105, 13668–13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ulbrich M. H., Isacoff E. Y. (2007) Subunit counting in membrane-bound proteins. Nat. Methods 4, 319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakajo K., Ulbrich M. H., Kubo Y., Isacoff E. Y. (2010) Stoichiometry of the KCNQ1-KCNE1 ion channel complex. Proc. Natl. Acad. Sci. U.S.A. 107, 18862–18867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu Y., Ulbrich M. H., Li M. H., Buraei Z., Chen X. Z., Ong A. C., Tong L., Isacoff E. Y., Yang J. (2009) Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sitaram A., Piccirillo R., Palmisano I., Harper D. C., Dell'Angelica E. C., Schiaffino M. V., Marks M. S. (2009) Localization to mature melanosomes by virtue of cytoplasmic dileucine motifs is required for human OCA2 function. Mol. Biol. Cell 20, 1464–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hille B. (1971) The permeability of the sodium channel to organic cations in myelinated nerve. J. Gen. Physiol. 58, 599–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCleskey E. W., Almers W. (1985) The Ca channel in skeletal muscle is a large pore. Proc. Natl. Acad. Sci. U.S.A. 82, 7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meuser D., Splitt H., Wagner R., Schrempf H. (1999) Exploring the open pore of the potassium channel from Streptomyces lividans. FEBS Lett. 462, 447–452 [DOI] [PubMed] [Google Scholar]
- 28. Nishida M., MacKinnon R. (2002) Structural basis of inward rectification. Cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 Å resolution. Cell 111, 957–965 [DOI] [PubMed] [Google Scholar]
- 29. Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T. (2009) Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602 [DOI] [PubMed] [Google Scholar]
- 30. Weber P. A., Chang H. C., Spaeth K. E., Nitsche J. M., Nicholson B. J. (2004) The permeability of gap junction channels to probes of different size is dependent on connexin composition and permeant-pore affinities. Biophys. J. 87, 958–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elfgang C., Eckert R., Lichtenberg-Fraté H., Butterweck A., Traub O., Klein R. A., Hülser D. F., Willecke K. (1995) Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J. Cell Biol. 129, 805–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silverman W., Locovei S., Dahl G. (2008) Probenecid, a gout remedy, inhibits pannexin 1 channels. Am. J. Physiol. Cell Physiol. 295, C761–C767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cruikshank S. J., Hopperstad M., Younger M., Connors B. W., Spray D. C., Srinivas M. (2004) Potent block of Cx36 and Cx50 gap junction channels by mefloquine. Proc. Natl. Acad. Sci. U.S.A. 101, 12364–12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Wittinghan F. J., Srinivas M., Sellitto C., White T. W., Mathias R. T. (2006) Mefloquine effects on the lens suggest cooperative gating of gap junction channels. J. Membr. Biol. 211, 163–171 [DOI] [PubMed] [Google Scholar]
- 35. Srinivas M., Hopperstad M. G., Spray D. C. (2001) Quinine blocks specific gap junction channel subtypes. Proc. Natl. Acad. Sci. U.S.A. 98, 10942–10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maeda S., Tsukihara T. (2011) Structure of the gap junction channel and its implications for its biological functions. Cell Mol. Life Sci. 68, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schägger H., Cramer W. A., von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 38. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 39. Tombola F., Ulbrich M. H., Isacoff E. Y. (2008) The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron 58, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Claros M. G., von Heijne G. (1994) TopPred II. An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686 [DOI] [PubMed] [Google Scholar]
- 41. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 42. Rost B., Fariselli P., Casadio R. (1996) Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5, 1704–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonnhammer E. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 44. Tusnády G. E., Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 45. von Heijne G. (1992) Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494 [DOI] [PubMed] [Google Scholar]
- 46. Common J. E., Becker D., Di W. L., Leigh I. M., O'Toole E. A., Kelsell D. P. (2002) Functional studies of human skin disease- and deafness-associated connexin 30 mutations. Biochem. Biophys. Res. Commun. 298, 651–656 [DOI] [PubMed] [Google Scholar]
- 47. Beblo D. A., Veenstra R. D. (1997) Monovalent cation permeation through the connexin40 gap junction channel. Cs, Rb, K, Na, Li, TEA, TMA, TBA, and effects of anions Br, Cl, F, acetate, aspartate, glutamate, and NO3. J. Gen. Physiol. 109, 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H. Z., Veenstra R. D. (1997) Monovalent ion selectivity sequences of the rat connexin43 gap junction channel. J. Gen. Physiol. 109, 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dani J. A. (1989) Open channel structure and ion binding sites of the nicotinic acetylcholine receptor channel. J. Neurosci. 9, 884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zarei M. M., Dani J. A. (1995) Structural basis for explaining open-channel blockade of the NMDA receptor. J. Neurosci. 15, 1446–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamashita M., Navarro-Borelly L., McNally B. A., Prakriya M. (2007) Orai1 mutations alter ion permeation and Ca2+-dependent fast inactivation of CRAC channels. Evidence for coupling of permeation and gating. J. Gen. Physiol. 130, 525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schindl R., Bergsmann J., Frischauf I., Derler I., Fahrner M., Muik M., Fritsch R., Groschner K., Romanin C. (2008) 2-Aminoethoxydiphenyl borate alters selectivity of Orai3 channels by increasing their pore size. J. Biol. Chem. 283, 20261–20267 [DOI] [PubMed] [Google Scholar]
- 53. Bezanilla F., Armstrong C. M. (1972) Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J. Gen. Physiol. 60, 588–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balasubramanian S., Lynch J. W., Barry P. H. (1995) The permeation of organic cations through cAMP-gated channels in mammalian olfactory receptor neurons. J. Membr. Biol. 146, 177–191 [DOI] [PubMed] [Google Scholar]
- 55. Oh S., Ri Y., Bennett M. V., Trexler E. B., Verselis V. K., Bargiello T. A. (1997) Changes in permeability caused by connexin 32 mutations underlie X-linked Charcot-Marie-Tooth disease. Neuron 19, 927–938 [DOI] [PubMed] [Google Scholar]
- 56. Ambrosi C., Gassmann O., Pranskevich J. N., Boassa D., Smock A., Wang J., Dahl G., Steinem C., Sosinsky G. E. (2010) Pannexin1 and Pannexin2 channels show quaternary similarities to connexons and different oligomerization numbers from each other. J. Biol. Chem. 285, 24420–24431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harris A. L. (2007) Connexin channel permeability to cytoplasmic molecules. Prog. Biophys. Mol. Biol. 94, 120–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Das S. K., Darshi M., Cheley S., Wallace M. I., Bayley H. (2007) Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. Chembiochem 8, 994–999 [DOI] [PubMed] [Google Scholar]
- 59. Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. (2000) A ubiquitous family of putative gap junction molecules. Curr. Biol. 10, R473–R474 [DOI] [PubMed] [Google Scholar]
- 60. Fushiki D., Hamada Y., Yoshimura R., Endo Y. (2010) Phylogenetic and bioinformatic analysis of gap junction-related proteins, innexins, pannexins and connexins. Biomed. Res. 31, 133–142 [DOI] [PubMed] [Google Scholar]
- 61. Phelan P., Bacon J. P., Davies J. A., Stebbings L. A., Todman M. G., Avery L., Baines R. A., Barnes T. M., Ford C., Hekimi S., Lee R., Shaw J. E., Starich T. A., Curtin K. D., Sun Y. A., Wyman R. J. (1998) Innexins. A family of invertebrate gap-junction proteins. Trends Genet. 14, 348–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barnes T. M. (1994) OPUS. A growing family of gap junction proteins? Trends Genet. 10, 303–305 [DOI] [PubMed] [Google Scholar]
- 63. Starich T. A., Lee R. Y., Panzarella C., Avery L., Shaw J. E. (1996) eat-5 and unc-7 represent a multigene family in Caenorhabditis elegans involved in cell-cell coupling. J. Cell Biol. 134, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Landesman Y., White T. W., Starich T. A., Shaw J. E., Goodenough D. A., Paul D. L. (1999) J. Cell Sci. 112, 2391–2396 [DOI] [PubMed] [Google Scholar]
- 65. Bruzzone R., White T. W., Paul D. L. (1996) Connections with connexins. The molecular basis of direct intercellular signaling. Eur. J. Biochem. 238, 1–27 [DOI] [PubMed] [Google Scholar]
- 66. White T. W., Deans M. R., O'Brien J., Al-Ubaidi M. R., Goodenough D. A., Ripps H., Bruzzone R. (1999) Functional characteristics of skate connexin35, a member of the γ subfamily of connexins expressed in the vertebrate retina. Eur. J. Neurosci. 11, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 67. Baranova A., Ivanov D., Petrash N., Pestova A., Skoblov M., Kelmanson I., Shagin D., Nazarenko S., Geraymovych E., Litvin O., Tiunova A., Born T. L., Usman N., Staroverov D., Lukyanov S., Panchin Y. (2004) The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 83, 706–716 [DOI] [PubMed] [Google Scholar]
- 68. Yen M. R., Saier M. H., Jr. (2007) Gap junctional proteins of animals. The innexin/pannexin superfamily. Prog. Biophys. Mol. Biol. 94, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sosinsky G. E., Boassa D., Dermietzel R., Duffy H. S., Laird D. W., MacVicar B., Naus C. C., Penuela S., Scemes E., Spray D. C., Thompson R. J., Zhao H. B., Dahl G. (2011) Pannexin channels are not gap junction hemichannels. Channels 5, 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boassa D., Ambrosi C., Qiu F., Dahl G., Gaietta G., Sosinsky G. (2007) Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J. Biol. Chem. 282, 31733–31743 [DOI] [PubMed] [Google Scholar]
- 71. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 72. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pei J., Grishin N. V. (2007) PROMALS. Towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23, 802–808 [DOI] [PubMed] [Google Scholar]
- 74. Barbe M. T., Monyer H., Bruzzone R. (2006) Cell-cell communication beyond connexins. The pannexin channels. Physiology 21, 103–114 [DOI] [PubMed] [Google Scholar]
- 75. Hua V. B., Chang A. B., Tchieu J. H., Kumar N. M., Nielsen P. A., Saier M. H., Jr. (2003) Sequence and phylogenetic analyses of 4 TMS junctional proteins of animals. Connexins, innexins, claudins and occludins. J. Membr. Biol. 194, 59–76 [DOI] [PubMed] [Google Scholar]
- 76. Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. (2007) Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J. Cell Sci. 120, 3772–3783 [DOI] [PubMed] [Google Scholar]
- 77. Boassa D., Qiu F., Dahl G., Sosinsky G. (2008) Trafficking dynamics of glycosylated pannexin 1 proteins. Cell Commun. Adhes. 15, 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dahl G., Nonner W., Werner R. (1994) Attempts to define functional domains of gap junction proteins with synthetic peptides. Biophys. J. 67, 1816–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Srinivas M., Calderon D. P., Kronengold J., Verselis V. K. (2006) Regulation of connexin hemichannels by monovalent cations. J. Gen. Physiol. 127, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Verselis V. K., Srinivas M. (2008) Divalent cations regulate connexin hemichannels by modulating intrinsic voltage-dependent gating. J. Gen. Physiol. 132, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ebihara L. (2003) New roles for connexons. News Physiol. Sci. 18, 100–103 [DOI] [PubMed] [Google Scholar]
- 82. Bao L., Locovei S., Dahl G. (2004) Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 572, 65–68 [DOI] [PubMed] [Google Scholar]
- 83. Bruzzone R., Barbe M. T., Jakob N. J., Monyer H. (2005) Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J. Neurochem. 92, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 84. Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H. (2003) Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. U.S.A. 100, 13644–13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Locovei S., Wang J., Dahl G. (2006) Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 580, 239–244 [DOI] [PubMed] [Google Scholar]
- 86. Eskandari S., Zampighi G. A., Leung D. W., Wright E. M., Loo D. D. (2002) Inhibition of gap junction hemichannels by chloride channel blockers. J. Membr. Biol. 185, 93–102 [DOI] [PubMed] [Google Scholar]
- 87. Qiu F., Dahl G. (2009) A permeant regulating its permeation pore. Inhibition of pannexin 1 channels by ATP. Am. J. Physiol. Cell Physiol. 296, C250–C255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shestopalov V. I., Panchin Y. (2008) Pannexins and gap junction protein diversity. Cell Mol. Life Sci. 65, 376–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Veenstra R. D., Wang H. Z., Beyer E. C., Ramanan S. V., Brink P. R. (1994) Connexin37 forms high conductance gap junction channels with subconductance state activity and selective dye and ionic permeabilities. Biophys. J. 66, 1915–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nightingale E. R., Jr. (1959) Phenomenological theory of ion solvation. Effective radii of hydrated ions. J. Phys. Chem. 63, 1381–1387 [Google Scholar]