Background: BMP signaling promotes mesendoderm differentiation and inhibits neural differentiation through downstream transcription factors.
Results: Ovol2 is up-regulated by BMP4, and its knockdown impairs BMP function in the neuroectodermal/mesendodermal cell fate decision.
Conclusion: Ovol2 acts as a novel BMP downstream target to inhibit neural differentiation and promote mesendodermal differentiation.
Significance: This study uncovers the mechanism of how BMP signaling regulates early cell fate decision.
Keywords: Bone Morphogenetic Protein (BMP), Embryo, Embryonic Stem Cell, Neurodevelopment, Neurodifferentiation, Ovol2, Mesendodermal Differentiation
Abstract
During early embryonic development, bone morphogenetic protein (BMP) signaling is essential for neural/non-neural cell fate decisions. BMP signaling inhibits precocious neural differentiation and allows for proper differentiation of mesoderm, endoderm, and epidermis. However, the mechanisms underlying the BMP pathway-mediated cell fate decision remain largely unknown. Here, we show that the expression of Ovol2, which encodes an evolutionarily conserved zinc finger transcription factor, is down-regulated during neural differentiation of mouse embryonic stem cells. Knockdown of Ovol2 in embryonic stem cells facilitates neural conversion and inhibits mesendodermal differentiation, whereas Ovol2 overexpression gives rise to the opposite phenotype. Moreover, Ovol2 knockdown partially rescues the neural inhibition and mesendodermal induction by BMP4. Mechanistic studies further show that BMP4 directly regulates Ovol2 expression through the binding of Smad1/5/8 to the second intron of the Ovol2 gene. In the chick embryo, cOvol2 expression is specifically excluded from neural territory and is up-regulated by BMP4. In addition, ectopic expression of cOvol2 in the prospective neural plate represses the expression of the definitive neural plate marker cSox2. Taken together, these results indicate that Ovol2 acts downstream of the BMP pathway in the cell fate decision between neuroectoderm and mesendoderm to ensure proper germ layer development.
Introduction
In early mouse embryogenesis, the three germ layers are generated through a process known as gastrulation, which commences at embryonic day 6.5 with the appearance of the primitive streak. During gastrulation, some epiblast cells ingress through the primitive streak to form the mesoderm and endoderm, whereas the remaining cells become ectoderm cells, which differentiate into either neuroectoderm or epidermis (1–3). It is generally believed that a bipotential mesendodermal population exists in the primitive streak as the common precursor of mesoderm and endoderm (4, 5). Because the germ layers contribute to all of the progenitor cells to form the whole embryo, the accurate control of cell fate decisions among different germ layers is crucial for proper embryonic development.
Many signaling pathways are involved in the early cell fate decision between neuroectoderm and mesendoderm (6), among which the bone morphogenic protein (BMP)2 pathway is one of the most important pathways. In mouse embryos, depletion of BMPRIA (BMP type IA receptor) leads to a premature loss of pluripotency, precocious neuroectoderm differentiation, and impaired mesoderm differentiation (7). In addition, the ablation of BMP4 prevents mesendodermal differentiation (8). Previous studies of embryonic stem cell (ESC) differentiation have also identified roles for BMP signaling in the inhibition of neural differentiation and the promotion of mesoderm and endoderm differentiation (9–13). In the chick embryo, ectopic expression of BMP4 represses the definitive neural marker cSox2 in the prospective neural plate, indicating that BMP4 inhibits neural induction in the chick (14). These results indicate that BMP signals are necessary to prevent precocious neuroectoderm differentiation and allow for proper development of mesoderm and endoderm. However, the mechanisms by which BMP signals control the cell fate decision remain largely unknown. Because BMPs exert their activity through the downstream Smad1/5/8-Smad4 transcriptional complex to activate or repress its target gene expression (15–17), we were interested in whether there are novel targets that mediate BMP regulation of the neuroectoderm/mesendoderm cell fate decision.
Despite the extensive study in signaling pathways, few transcription factors have been uncovered to play essential roles in regulating the decision between the neuroectoderm and mesendoderm/mesoderm cell fates. Tbx6 is essential for the regulation of Sox2 expression, which controls the cell fate decision between the caudal neural plate and the paraxial mesoderm in the mouse embryo (18). Moreover, SIP1 was found to inhibit mesendodermal differentiation and favor neural differentiation in human ESCs (19).
Ovol2 (Ovo-like 2) belongs to the Ovo gene family, which encodes an evolutionarily conserved group of C2H2 zinc finger DNA-binding proteins among various species (20, 21). The founding member of the Ovo gene family, the Drosophila ovo, plays vital roles in epidermal differentiation and female germ line development (22–24). Mouse Ovol1 is also essential for epidermis development and spermatogenesis, suggesting a functional conservation with its Drosophila ortholog (25). Ablation of the Ovol2 gene leads to embryonic lethality before embryonic day 10.5, indicating that Ovol2 is involved in early embryonic development (26, 27). In Ovol2-null mice, the neuroectoderm was expanded in the cranial region, which caused a failure of cranial neural tube closure (26). Furthermore, heart development and extraembryonic and embryonic vascularization were also severely affected (26, 27). However, the functions of Ovol2 in the early cell fate specification between neuroectoderm and mesendoderm have not been addressed. In human keratinocytes, OVOL1 was identified as a downstream target of the TGF-β/BMP7-Smad4 signaling pathway (28). It remains unknown whether Ovol2 is also regulated by BMP signals.
Here, we identify Ovol2 as a novel target gene downstream of BMP signaling to regulate the cell fate decision between neuroectoderm and mesendoderm. In mouse ESCs, Ovol2 is directly up-regulated by BMP4 and partially mediates BMP4 function to inhibit neural conversion and promote mesodermal and endodermal differentiation. In vivo, chick Ovol2 (cOvol2) expression is excluded from neural territory and is up-regulated by BMP4. Ectopic expression of cOvol2 in the prospective neural plate inhibits neural specification in the early chick embryo.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The mouse Ovol2A cDNA was inserted into pIRES2-EGFP and pcDNA3.1-myc. The Ovol2A-IRES-EGFP region was then subcloned into the lentiviral vector pFUGW-IRES-GFP for overexpression experiments. The mutant Ovol2 was generated by PCR using KOD-plus (Toyobo Biotechnology) and then subcloned into the lentiviral vector pFUGW-IRES-Dsred for rescue experiments. The pcDNA3.1-myc-Ovol2 construct was used to transiently express Ovol2 in HEK293T cells to detect the knockdown efficiency of Ovol2 shRNAs. Chick Ovol2 cDNA was amplified by PCR from an Hamburger and Hamilton stage 5 (HH5) chick cDNA library and cloned into pBluescript (for in situ probe preparation) and pCAGGS-IRES-GFP (for chick embryo electroporation). The 992-bp Ovol2 gene promoter flanking upstream of the translation start site (ATG) was amplified by PCR from mouse genomic DNA and was then inserted into the luciferase reporter vector pGL3-Basic to generate the pOvoP−992/−1 construct. The pOvoPEn+61/+1378 construct was generated by inserting a 1.3-kb region (+61/+1378) of the Ovol2 genomic DNA between the promoter and the luciferase gene of the pOvoP−992/−1 construct. Sequential deletion of the 1.3-kb enhancer region was performed either by enzymatic digestion or by PCR amplification. The generation of site-mutated luciferase constructs was performed by PCR using KOD-plus (Toyobo Biotechnology) following the manufacturer's instructions. All of the constructs were confirmed by sequencing.
Cell Culture and Treatment
The mouse ESC line R1 was maintained on a layer of mitomycin C-treated mouse embryonic fibroblast feeder cells in standard medium. Serum-free neural differentiation of ESCs was performed as described previously (13). For the unbiased differentiation of ESCs, single cells were aggregated in Petri dishes at a density of 1 × 105 cells/ml in differentiation medium containing serum. Differentiation day 0 indicates the day on which the ESCs were seeded to differentiate. In some experiments, recombinant human BMP4 protein (R&D Systems) was freshly added at a final concentration of 10 ng/ml or washed by fresh medium twice within the indicated length of time.
For the luciferase and ChIP assays, P19C6, a subclone of the mouse EC (embryonic carcinoma) cell line, was cultured as previously described (29).
Gene Knockdown and Overexpression in ESCs
A lentiviral vector pLenti-psilencer expressing shRNA was used for Ovol2 knockdown in ESCs. The control and Ovol2 shRNA sequences are listed in supplemental Table S1. For the overexpression experiments, Ovol2A full-length cDNA was cloned into the lentiviral expression vector pFUGW-IRES-EGFP (30). Lentiviral packaging and transfection of ESCs were performed as previously described (31). After lentiviral transfection, GFP-positive cells were sorted using a FACSAria I cell sorter (BD Biosciences) and propagated in ESC culture medium. The cells were then differentiated as embryoid bodies (EBs) and analyzed at the indicated time.
RNA Preparation and qRT-PCR Analysis
Total RNAs were extracted using TRIzol reagent (Pufei). Reverse transcription and qRT-PCR were performed as described previously (13).
The copy numbers of the Ovol2 splice forms Ovol2A, Ovol2B, and Ovol2C in ESC cDNA were quantified as previously described (32). Briefly, standard curves for Ovol2A, -B, and -C were generated using serial dilution of positive control templates and isoform-specific primers, respectively. The copy numbers of each of the three splice forms of Ovol2 expressed in ES cells were then derived from the standard curves. The primers used are listed in supplemental Table S2.
Immunofluorescence Assay
Immunocytochemistry was performed as described previously (29, 33). The following antibodies were used: mouse monoclonal antibodies β-III-tubulin (Tuj1) (1:500; Sigma) and Oct4 (1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA); mouse polyclonal antibody Flk1 (1:100; BD Pharmingen); rabbit polyclonal antibodies Nestin (1:200), group B1 Sox proteins (Sox1/(2)/3) (1:200) with a preference for Sox1 and Sox3 over Sox2 (34, 35), and GFP (1:1000; Molecular Probes); and goat polyclonal antibodies T (1:200; R&D Systems) and Gata6 (1:200; R&D Systems). Fluorescein isothiocyanate (FITC) (1:200)-conjugated, Cy3 (1:500)-conjugated, and Cy5 (1:200)-conjugated secondary antibodies were obtained from Jackson Immunoresearch Laboratories (West Grove, PA). Normal mouse and rabbit IgGs (Zymed Laboratories Inc.) were used as negative controls. The images were taken with Olympus BX50 fluorescence microscopy or Leica confocal microscopy.
Luciferase Assay
Luciferase assays in P19 cells were performed as described previously (36). Briefly, after transfection with Fugene HD (Roche Applied Science), cells were serum-starved in N2B27 (Invitrogen) medium for 18 h, followed by BMP4 (10 ng/ml) treatment for 6 h; the cells were then harvested for the measurement of luciferase activity. All assays were performed in triplicate, and all values were normalized by Renilla LUC (Promega).
Chromatin Immunoprecipitation
The ChIP assay was performed following the manufacturer's protocol (Upstate Biotechnology, Inc.), and detailed procedures were described previously (36). Immunoprecipitation was performed with 2 μg of rabbit polyclonal antibody against phosphorylated Smad1/5/8 (Cell Signaling). Normal rabbit IgG was used as a negative control. Quantitative PCR was used to amplify different regions of the mouse Ovol2 gene, and reaction parameters are available on request. The primers used for the ChIP assay (Ovo-BRE and Ovo-Ctrl) are listed in supplemental Table S3.
Chick Embryology
Fertilized hen's eggs (Shanghai Academy of Agricultural Sciences) were incubated at 38.5 °C to the desired stages (37). For electroporation, 1 μg/μl plasmid was delivered to HH stage 3+/4 chick embryos following the published protocol (38). Electroporation was performed with a NAPA GENE electroporator at the following settings: 4 V, 5 pulses (50 ms with a 500-ms interval). Whole mount in situ hybridization and immunocytochemistry were performed following published protocols (39, 40). Whole mount embryos were visualized using a Leica microscope.
Statistics
The results shown are representative of three independent experiments. The data are expressed as the mean ± S.E. The Student's t test was used to compare the effects of all treatments. Differences were considered statistically significant as follows: *, p < 0.05.
RESULTS
Ovol2 Is Down-regulated upon ESC Neural Differentiation
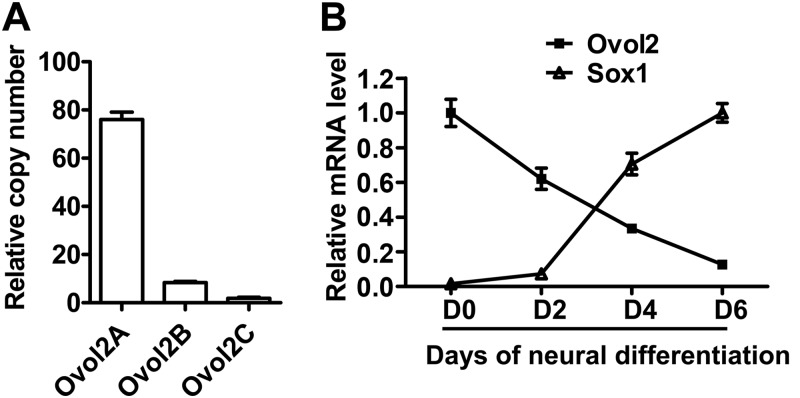
The mouse Ovol2 gene encodes three isoforms (Ovol2A, -B, and -C) with presumably different transcriptional properties; the different isoforms are generated by the usage of distinct transcriptional start sites and alternative splicing (41). Ovol2 has been shown to express in mouse ESCs (26, 42). qRT-PCR analysis showed that Ovol2A was the main isoform expressed in ESCs (Fig. 1A), and we used Ovol2 to represent Ovol2A in the following study. To explore the relationship between Ovol2 expression and ESC neural differentiation, we differentiated ESCs as EBs in serum-free KSR medium for 6 days. In this serum-free EB culture (SFEB) condition, ESCs efficiently converted to Sox+Oct4− neural progenitor cells (NPCs) (13). qRT-PCR analysis showed that the expression level of Ovol2 mRNA gradually declined with the increase of Sox1 mRNA (Fig. 1B), indicating that Ovol2 might be involved in the neural differentiation of ESCs.
FIGURE 1.
Ovol2 expression decreases with the neural differentiation of ESCs. A, qRT-PCR quantification of the copy numbers of three splicing isoforms of Ovol2 in ESCs. Values shown are the copy numbers for Ovol2A, Ovol2B, and Ovol2C in a 1-μl cDNA sample from ESC culture. B, qRT-PCR analysis of Ovol2 and Sox1 mRNA levels during ESC neural differentiation. ESCs were cultured in the SFEB condition for 6 days and subjected to analysis at differentiation day 0, 2, 4, and 6. For qRT-PCR analysis in all of the following experiments, the expression levels of Ovol2 and Sox1 were normalized to that of Gapdh, and the peak value for each gene was designated as 1, respectively. Error bars, S.E.
Ovol2 Knockdown Promotes Neural Conversion and Inhibits Mesendodermal Differentiation of ESCs
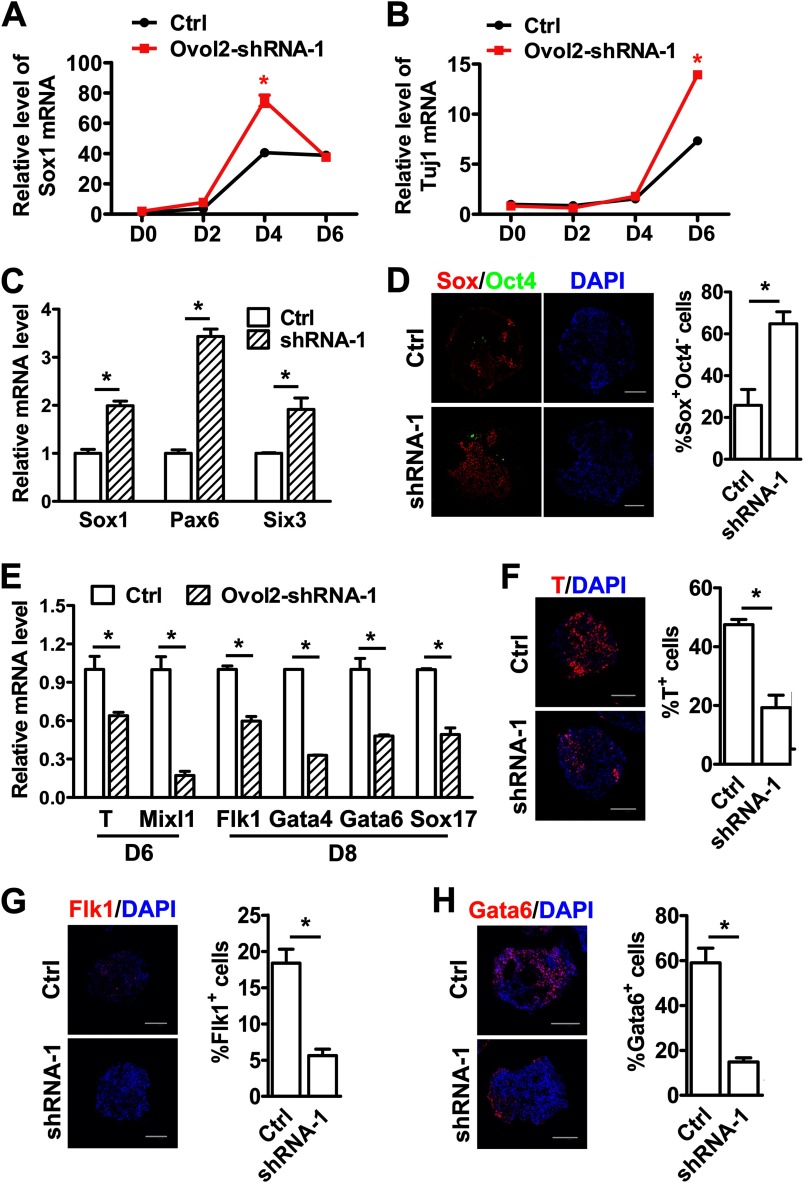
To study the function of Ovol2 in ESC differentiation, we established ESCs expressing shRNAs targeted against Ovol2 (Ovol2 shRNA ESCs) by lentiviral transduction and found that shRNA-1 could efficiently knock down Ovol2 expression (supplemental Fig. S1, A and B). qRT-PCR analysis showed that control shRNA ESCs and Ovol2 shRNA-1 ESCs expressed comparable levels of the pluripotency markers Oct4, Rex1, Nanog, and CRTR1 (supplemental Fig. S1C), suggesting that Ovol2 knockdown has no effect on the pluripotency of ESCs. The control and Ovol2 shRNA-1 ESCs were then differentiated in the SFEB condition for 6 days. We found that Ovol2 shRNA-1 ESCs expressed higher levels of the NPC marker Sox1 at day 4 (Fig. 2A) and the neuronal marker Tuj1 at day 6 (Fig. 2B) compared with the control ESCs, indicating that the knockdown of Ovol2 accelerates ESC neural differentiation.
FIGURE 2.
Knockdown of Ovol2 facilitates ESC neural conversion and impairs mesendodermal differentiation. A and B, control ESCs (Ctrl) or Ovol2 shRNA ESCs were induced in the SFEB condition for 6 days. The expression levels of Sox1 (A) and Tuj1 (B) at differentiation days 0, 2, 4, and 6 were analyzed by qRT-PCR and normalized to the expression of Gapdh. The value for each gene in day 0 control ESCs was designated as 1. C, EBs from control ESCs or Ovol2 shRNA ESCs were cultured in unbiased differentiation medium for 8 days. The expression levels of Sox1, Pax6, and Six3 were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for each marker in control EBs were designated as 1, respectively. D, double immunostaining for Sox (red) and Oct4 (green; artificial color) proteins in day 8 EBs cultured under the conditions described in C. For EB staining in all of the following experiments, sections from thousands of EB aggregates were stained, and statistical analysis were performed. Nuclei were stained with DAPI (blue). The diagram beside the images shows the percentage of Sox+Oct4− cells in the EBs. E, the expression levels of T and Mixl1 in day 6 EBs and of Flk1, Gata4, Gata6, and Sox17 in day 8 EBs cultured under the conditions described in C were analyzed by qRT-PCR. F–H, immunostaining for T in day 6 EBs (F), Flk1 (G), and Gata6 (H) in day 8 EBs. The percentages of T+, Flk1+, and Gata6+ cells were shown in the diagrams beside the images. Scale bar for all of the images, 100 μm. Error bars, S.E.
Because the SFEB system efficiently induces almost exclusively neural cell fates, we used an unbiased differentiation system to study the function of Ovol2 in early cell fate decisions (43). The ESCs were differentiated as EBs in serum-containing medium (10% FBS/DMEM) for 8 days. Compared with control ESCs, Ovol2 shRNA-1 ESCs expressed higher levels of the neuroectoderm markers Sox1, Pax6, and Six3 at day 8 (Fig. 2C). Immunostaining analysis further showed that Ovol2 shRNA-1 ESCs differentiated into a higher percentage of Sox+Oct4− NPCs compared with control ESCs (Fig. 2D), confirming that the knockdown of Ovol2 promoted the neural fate decision of ESCs. To test whether Ovol2 also functions in the differentiation of other germ layers, we examined germ layer markers by qRT-PCR and found that Ovol2 knockdown decreased the expression of the mesendoderm markers T and Mixl1 at day 6 and of the mesoderm marker Flk1 and the endoderm markers Gata4, Gata6, and Sox17 at day 8 (Fig. 2E). Consistently, immunostaining showed that the percentages of T+, Flk1+, and Gata6+ cells were reduced in Ovol2 shRNA-1 ESCs (Fig. 2, F–H), suggesting that Ovol2 knockdown inhibits mesendodermal differentiation of ESCs.
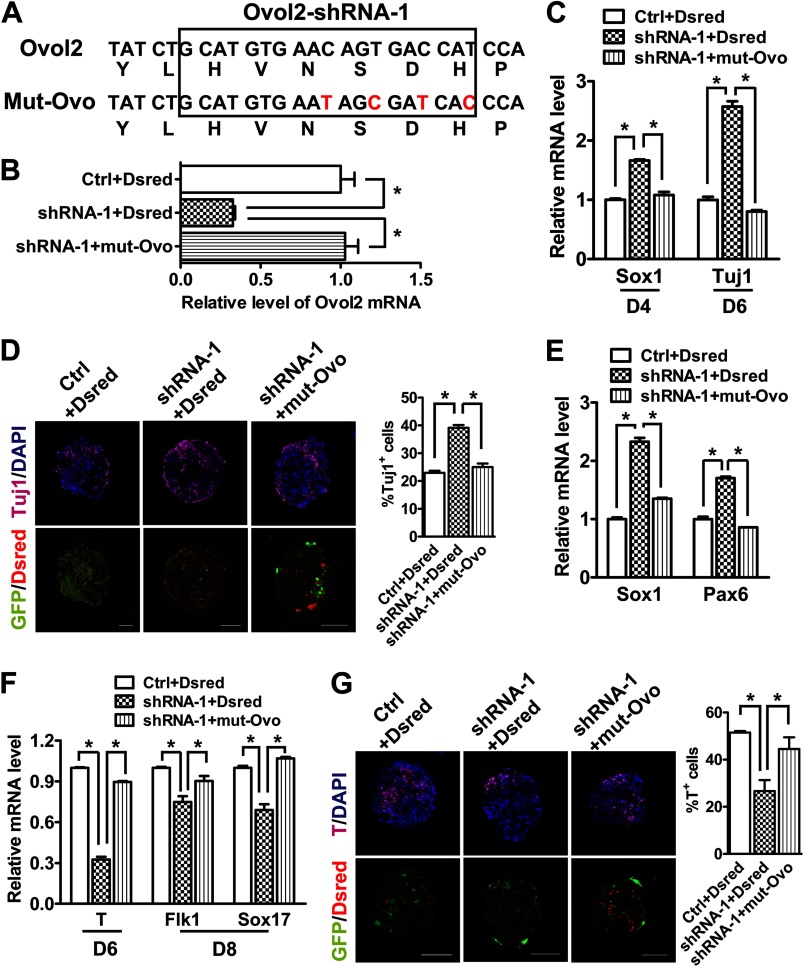
To confirm the specificity of Ovol2 knockdown, we carried out a rescue experiment using a mutant Ovol2 (mut-Ovo) with silent mutations in the target nucleotide sequence for the shRNA-1 (Fig. 3A). Ovol2 shRNA-1 ESCs were infected with lentivirus encoding mut-Ovo and red fluorescence protein (Dsred) gene for cell sorting. qRT-PCR analysis showed that the ESCs (shRNA-1 + mut-Ovo) expressed a similar level of total Ovol2 as the parental control ESCs (Ctrl + Dsred), suggesting that the mutant Ovol2 escaped the knockdown of shRNA-1 (Fig. 3B). During 6 days of neural differentiation in the SFEB system, expressing mut-Ovo in Ovol2 shRNA-1 ESCs rescued the knockdown phenotype of the up-regulation of the NPC marker Sox1 at day 4 and the neuronal marker Tuj1 at day 6 (Fig. 3C). Consistently, immunostaining showed that the percentage of Tuj1-positive neuronal cells was reduced to the control level by expression of mut-Ovo (Fig. 3D).
FIGURE 3.
Ovol2 knockdown phenotype is rescued by expression of shRNA-resistant mutant Ovol2. A, silent mutations in mut-Ovo that prevented their knockdown by Ovol2 shRNA-1. B, qRT-PCR analysis of total Ovol2 mRNA levels from ESCs with control (Ctrl) or Ovol2 shRNA-1 or shRNA-1 ESCs expressing the mutant Ovol2. The vectors carry RFP that was used for cell sorting. C, the three groups of ESCs described in B were induced in the SFEB condition for 6 days. The expression levels of Sox1 at differentiation day 4 and that of Tuj1 at day 6 were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for each marker in control EBs were designated as 1, respectively. D, immunostaining and quantification of Tuj1 positive cells in day 6 EBs cultured under the conditions described in C. E, EBs from the three groups of ESCs were cultured in unbiased differentiation medium for 8 days. The expression levels of Sox1 and Pax6 were analyzed by qRT-PCR. F, qRT-PCR analysis of the mRNA levels of T (at day 6) and of Flk1 and Sox17 (at day 8) in the EBs cultured under the conditions described in E. G, immunostaining and quantification of T-positive cells in day 6 EBs described in E. Scale bar for all of the images, 100 μm. Error bars, S.E.
We also performed the rescue experiment in the unbiased differentiation system. Indeed, the levels of neural markers Sox1 and Pax6 were reduced to the control levels (Fig. 3E), and the levels of mesendodermal marker T, the mesoderm marker Flk1, and the endoderm marker Sox17 were increased to the control levels (Fig. 3F). Immunostaining for T confirmed that mut-Ovo significantly rescued the Ovol2 knockdown-induced defect in mesendodermal differentiation (Fig. 3G). Taken together, these data suggest that Ovol2 knockdown promotes neural conversion and impairs mesendodermal differentiation of ESCs, and this phenotype can be rescued by overexpressing knockdown-resistant Ovol2.
Ovol2 Overexpression Inhibits Neural Conversion and Promotes Mesendodermal Differentiation of ESCs
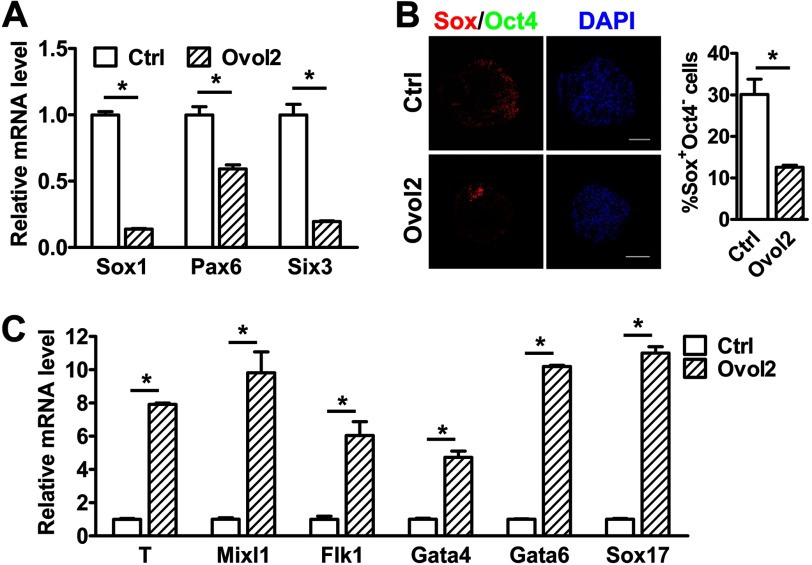
To further confirm the function of Ovol2 in cell fate decisions, we overexpressed Ovol2 in ESCs through lentiviral transduction (supplemental Fig. S2A) and found no significant change in the expression of pluripotency markers in Ovol2-overexpressing ESCs (Ovol2-ESCs) (supplemental Fig. S2B). After 8 days of unbiased differentiation in serum-containing medium, Ovol2-ESCs displayed reduced expression of the neural markers Sox1, Pax6, and Six3 compared with control GFP-ESCs (Fig. 4A). Immunostaining analysis further showed that Ovol2-overexpressing ESCs differentiated into fewer Sox+Oct4− NPCs than control ESCs (Fig. 4B). On the other hand, Ovol2-overexpressing ESCs expressed much higher levels of the mesoderm markers T, Mixl1, and Flk1 as well as the endoderm markers Gata4, Gata6, and Sox17 than control ESCs after 4 days of differentiation (Fig. 4C). Together, these results suggest that Ovol2 inhibits neural conversion and promotes mesendodermal differentiation of ESCs.
FIGURE 4.
Overexpression of Ovol2 represses neural conversion and promotes mesendodermal differentiation of ESCs. A, EBs from control GFP-ESCs or Ovol2-overexpressed ESCs were cultured in unbiased differentiation medium for 8 days. The expression levels of Sox1, Pax6, and Six3 were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for each marker in the control EBs (Ctrl) were designated as 1, respectively. B, double immunostaining for Sox (red) and Oct4 (green; artificial color) proteins and the percentage of Sox+Oct4− cells in day 8 EBs described in A. Scale bar, 100 μm. C, the expression levels of T, Mixl1, Flk1, Gata4, Gata6, and Sox17 in day 4 EBs cultured under the conditions described in A were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for each marker in control EBs were designated as 1, respectively. Error bars, S.E.
Ovol2 Partially Mediates BMP Function during ESC Differentiation
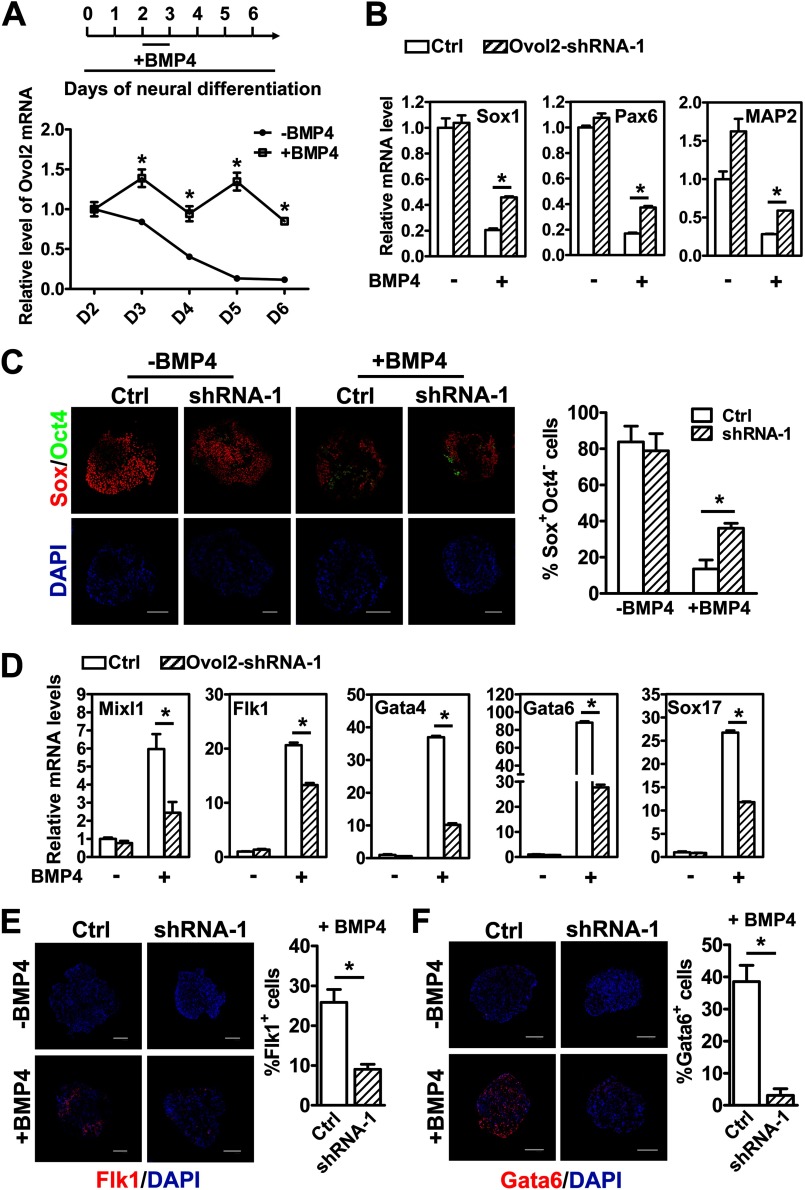
Previously, we showed that BMP signaling inhibits neural differentiation and promotes non-neural lineages during day 2–3 of ESC neural differentiation (13). Because Ovol2 has similar functions as the BMP pathway in the neural/mesendodermal cell fate decision, and OVOL1 is up-regulated by BMP7/Smad4 signaling in human keratinocytes (28), we hypothesized that Ovol2 might also be involved in the BMP pathway to regulate the neural/mesendodermal cell fate decision of ESCs. To this end, we first examined whether Ovol2 is regulated by BMP signals. ESCs were differentiated in the SFEB system, and BMP4 (10 ng/ml) was added at day 2–3. qRT-PCR analysis showed that the addition of BMP4 could prevent the decrease in Ovol2 expression during SFEB-induced ESC neural differentiation (Fig. 5A), suggesting that Ovol2 is up-regulated by BMP4.
FIGURE 5.
Ovol2 partially mediates BMP function to inhibit neural cell fates and promote mesendodermal cell fates during ESC differentiation. A, ESCs were induced in SFEB condition, with or without BMP4 (10 ng/ml) treatment at day 2–3. The expression levels of Ovol2 in EBs during the differentiation process were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for day 2 EBs without BMP4 treatment were designated as 1. B, control shRNA ESCs (Ctrl) or Ovol2 shRNA-1 ESCs were differentiated as described in A. The expression levels of neural markers (Sox1, Pax6, and MAP2) in day 6 EBs were analyzed by qRT-PCR and normalized to the expression of Gapdh. The values for control EBs without BMP4 treatment for each gene were designated as 1. C, double immunostaining for Sox (red) and Oct4 (green; artificial color) and the percentages of Sox+Oct4− cells in day 6 EBs described in B. D, qRT-PCR analysis of the expression levels of Mixl1, Flk1, Gata4, Gata6, and Sox17 in day 6 EBs described in B. E and F, immunostaining for Flk1 (E) and Gata6 (F) and the percentages of Flk1+ and Gata6+ cells in day 6 EBs described in B. Scale bar for all of the images, 100 μm. Error bars, S.E.
To determine whether BMP4 regulates neural/mesendoderm differentiation through Ovol2, we differentiated control shRNA ESCs or Ovol2 shRNA-1 ESCs in the SFEB system, with or without the addition of BMP4 at day 2–3 of the differentiation, and harvested cells at day 6 for qRT-PCR and immunostaining analysis. We found that in control ESCs, BMP4 significantly repressed the neural markers Sox1, Pax6, and MAP2 (Fig. 5B) and greatly reduced the percentage of Sox+Oct4− NPCs (Fig. 5C). However, knockdown of Ovol2 partially rescued the down-regulation of Sox1, Pax6, and MAP2 (Fig. 5B) and the decrease of NPCs (Fig. 5C), indicating that BMP signals inhibited neural differentiation partially through Ovol2. Moreover, compared with control ESCs, knockdown of Ovol2 impaired the ability of BMP4 to induce the expression of mesoderm and endoderm markers (Mixl1, Flk1, Gata4, Gata6, and Sox17) (Fig. 5D). Immunostaining of Flk1 and Gata6 also confirmed this observation (Fig. 5, E and F). Taken together, these results suggest that Ovol2 partially mediates the function of BMP4 to inhibit neural cell fates and promote mesendodermal cell fates during ESC differentiation.
Ovol2 Is a Direct Downstream Target of BMP Signaling Pathway
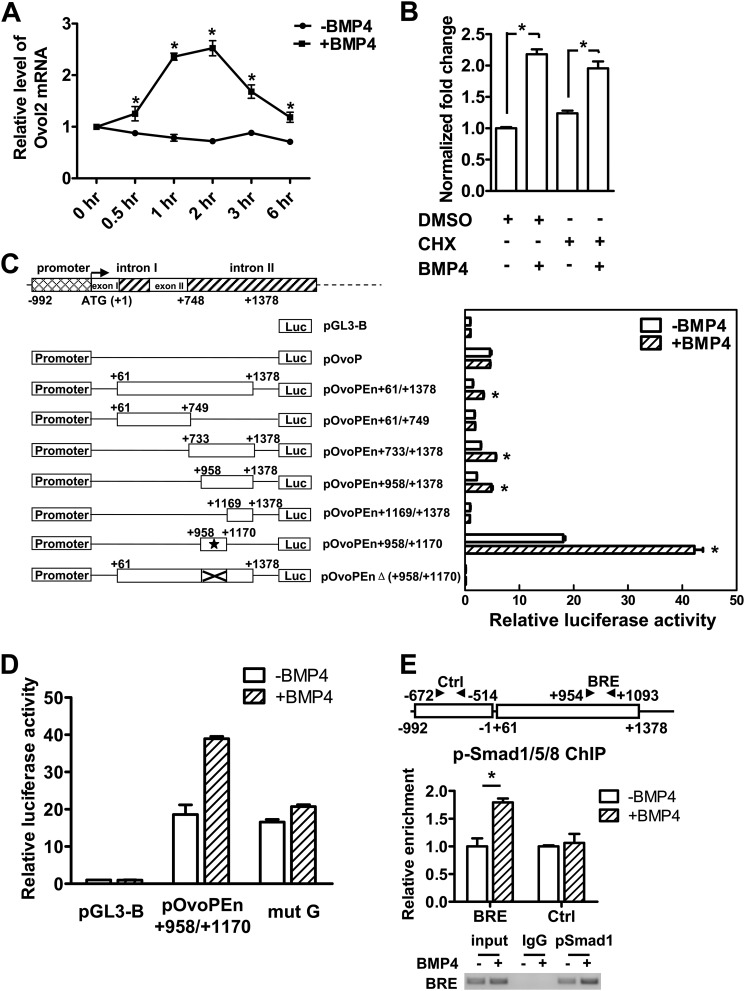
To test whether Ovol2 is directly regulated by BMP signals, we stimulated serum-starved ESCs with BMP4 for 0.5–6 h and measured Ovol2 transcription level. We found that Ovol2 transcription was elevated within 1 h of treatment (Fig. 6A), suggesting that Ovol2 is an early response BMP target. The up-regulation of Ovol2 mRNA by BMP4 was maintained even in the presence of the de novo protein synthesis inhibitor cyclohexamide (Fig. 6B). The direct BMP target genes Id1 and Msx2 (44) as well as the indirect target gene Wnt3 (45) were used as the positive and negative controls, respectively (supplemental Fig. S3). These data suggest that Ovol2 is a direct target gene of the BMP pathway.
FIGURE 6.
Ovol2 is directly regulated by BMP4. A, serum-starved ESCs were stimulated with BMP4 (10 ng/ml) for the indicated lengths of time, after which the cells were harvested for qRT-PCR analysis of Ovol2 expression. The value in the untreated ESCs was designated as 1. B, serum-starved ESCs were pretreated with cyclohexamide (CHX) for 4 h and then stimulated with BMP4 in the presence of cyclohexamide for 2 h, after which cells were harvested for qRT-PCR analysis for Ovol2. The value in the mock-treated ESCs was designated as 1. C, the localization of the minimal enhancer region in the Ovol2 gene. P19 cells were transiently transfected with constructs as indicated in the left panel for 24 h and serum-starved for 12 h, followed by treatment with or without BMP4 (10 ng/ml) for 6 h, and then the luciferase activity was determined. The activity of each construct was shown relative to that of the pGL3-Basic vector. The minimal enhancer region is indicated by an asterisk. D, P19 cells were transfected with pGL3-Basic, pOvoPEn+958/+1170, and site-mutated construct (mut G), respectively, and treated as described in C, and the luciferase activity was determined. E, ChIP assay of the p-Smad1/5/8 binding on Ovol2 enhancer. P19 cells were transiently transfected with pOvoPEn+61/+1378 for 24 h, serum-starved for 12 h, and then stimulated with BMP4 (10 ng/ml) (+BMP4) or mock (−BMP4) for 2 h before the cells were harvested for the ChIP assay. An antibody against p-Smad1/5/8 (Cell Signaling) was used, and the immunoprecipitates were analyzed by quantitative PCR for enrichment of Ovo-BRE and the control region (Ctrl). The data were normalized to the inputs and are represented as one of three independent experiments. The electrophoresis image of the ChIP product is also shown. Error bars, S.E.
Because de novo protein synthesis was not required for the BMP-induced transcription of Ovol2, we proposed that BMP4 up-regulates Ovol2 through phosphorylated Smad1/5/8 binding to the promoter/enhancer region of the Ovol2 gene. Based on this hypothesis, we sought to identify the regulatory sequence of the Ovol2 gene responsive to BMP signals using a luciferase reporter assay in P19 cells. Although the 5′ upstream promoter (pOvoP−992/−1, pOvoP) displayed basic promoter activity, it failed to show a response to BMP4 (Fig. 6C). Through serial deletion, we mapped a 213-bp region in the second intron (pOvoPEn+958/+1170) of the mouse Ovol2 gene that is essential for the responsiveness to BMP signals (Fig. 6C). This 213-bp region was GC-rich (77% GC content) and highly conserved between mice and humans (supplemental Fig. S4). A reporter construct carrying a mutation of GCCC to TTTT in this region showed significantly impaired enhancer activity (Fig. 6D), suggesting that p-Smad1/5/8 might bind to this GC-rich region to regulate Ovol2 expression. To confirm this, we transfected P19 cells with the pOvoPEn+61/+1378 plasmid and treated them with or without BMP4 for 2 h. The ChIP assay using an antibody against p-Smad1/5/8 showed that in the presence of BMP4, p-Smad1/5/8 was recruited to the 213-bp region compared with the control region, which lies in the upstream promoter (Fig. 6E). Together, these data suggest that Ovol2 is directly regulated by BMP signaling through the binding of p-Smad1/5/8 to its enhancer region.
Chick Ovol2 Is Exclusively Expressed in Non-neural Domains in Early Chick Embryos
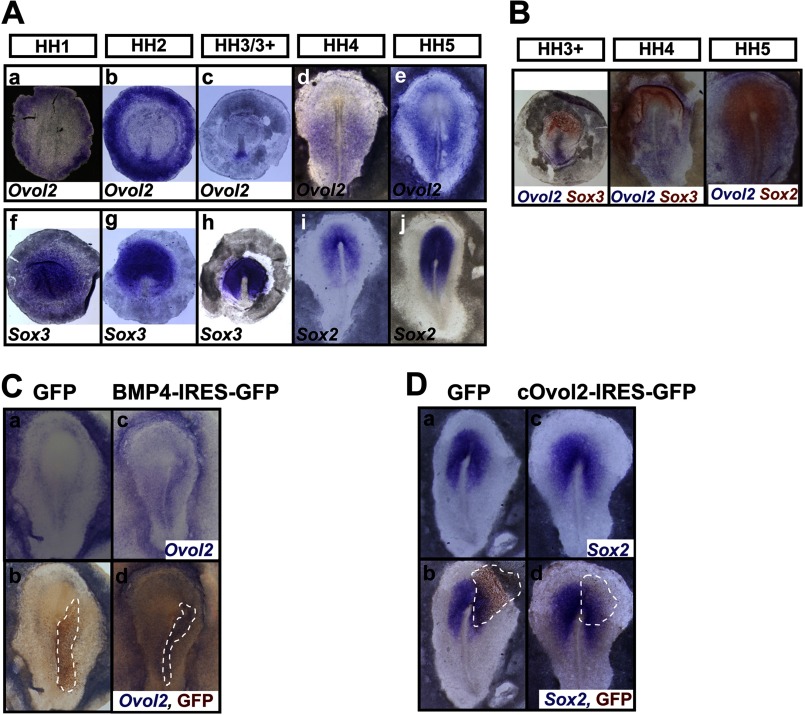
To further investigate the in vivo function of Ovol2 in the neural fate decision, we used early chick embryos. Because neural specification occurs before HH stage 5 of chick development, we used whole-mount in situ hybridization (ISH) in early chick embryos from HH stage 1 through 5 to compare the expression of cOvol2 with that of cSox2 and cSox3. Consistent with a previous study (46), cSox3 marked the epiblast in the area pellucida at HH stage 1 and the future neural plate before HH stage 3/3+ (Fig. 7A, f–h), and cSox2 marked the neural plate at HH stages 4 and 5 (Fig. 7A, i and j). At HH stage 1, cOvol2 mRNA was initially expressed in the epiblast of the area opaca (Fig. 7A, a), and this expression was sustained throughout the neural specification stages (Fig. 7A, b–e). At HH stages 2 and 3/3+, strong hybridization signal was also detected in the caudal primitive streak (Fig. 7A, b and c) and later expanded to the lateral caudal region along the primitive streak at HH stage 4 (Fig. 7A, d). At HH stage 5, cOvol2 transcripts were observed in the wing region surrounding the forming neural plate marked by cSox2 (Fig. 7A, e). Double ISH further confirmed that cOvol2 exhibited an expression pattern complementary to that of the preneural marker cSox3 and the neural plate marker cSox2 (Fig. 7B). These results suggest that Ovol2 might play a negative role in neural specification in chick embryos.
FIGURE 7.
Ovol2 is up-regulated by BMP4 and inhibits neural specification in the chick embryo. A, ISH analysis of cOvol2 (a–e), cSox3 (f–h), and cSox2 (i and j) in HH stage 1–5 chick embryos. B, double ISH for cOvol2 (blue) and cSox2/3 (magenta) in HH stage 3+ to 5 chick embryo. C, the expression of cOvol2 (blue) in the chick embryos electroporated with control GFP or BMP4-IRES-GFP plasmid was examined by ISH (a and c), and subsequent immunostaining for GFP (brown) marked the electroporated cells in the same embryos (b and d). D, the expression of cSox2 (blue) in the chick embryos electroporated with control GFP or cOvol2-IRES-GFP plasmid was examined as described in C. The electroporated field is highlighted by broken lines.
cOvol2 Is Up-regulated by BMP4 and Inhibits Neural Specification in Chick Embryos
Given that the expression pattern of cOvol2 is very similar to that of cBMP4 and cBMP7 (47, 48), which primarily resides in the caudal primitive streak and epidermal ectoderm (supplemental Fig. S5), we asked whether cOvol2 could be up-regulated by BMP4 in chick embryos. We electroporated BMP4 expression plasmids in the region lateral to the primitive streak of chick embryos at HH stage 3/3+ and harvested the electroporated embryos at HH stage 5 for ISH and immunostaining analysis (supplemental Fig. S6A). Indeed, BMP4 up-regulated the expression of cOvol2 in chick embryos (Fig. 7C, BMP4 (13 of 17) and control (0 of 17)). Furthermore, the ectopic electroporation of cOvol2-IRES-GFP expression plasmids into the prospective neural plate (supplemental Fig. S6B) repressed the expression of cSox2 in the electroporated domain (Fig. 7D, cOvol2 (12 of 15) and control (1 of 13)). Together, these results indicate that BMP4 up-regulates the expression of cOvol2 and that ectopic expression of cOvol2 inhibits neural specification in chick embryos.
DISCUSSION
In this study, we showed that Ovol2 mRNA is down-regulated during mouse ESC neural differentiation (Fig. 1B) and is up-regulated by BMP signals (Fig. 5A). Gain and loss of function assays showed that Ovol2 promotes mesendodermal differentiation and inhibits neural conversion (Figs. 2–4). Moreover, knockdown of Ovol2 impaired the ability of BMP to repress neural conversion and induce mesodermal and endodermal differentiation (Fig. 5). An in depth study showed that BMP recruits p-Smad1/5/8 to bind to the enhancer region of Ovol2 to directly regulate Ovol2 expression (Fig. 6). In early chick embryos, cOvol2 is exclusively expressed in non-neural regions and overlaps with the expression pattern of cBMP4/7 (Fig. 7, A and B and supplemental Fig. S5). cOvol2 is up-regulated by BMP4, and ectopic expression of cOvol2 represses the expression of the neural plate marker cSox2 in chick embryos (Fig. 7, C and D). Therefore, these results support the notion that Ovol2 acts as a novel BMP downstream target in the cell fate decision between neuroectoderm and mesendoderm.
It has been previously reported that ablation of Ovol2 leads to an expansion of neuroectoderm in the cranial region, abnormal development of the heart, and defect vascularization (26, 27). By clarifying the function of Ovol2 in early cell fate decisions, we speculate that the multiple defects observed in Ovol2-null mice are caused by the loss of balance between the differentiation of the neuroectoderm and mesendoderm. However, Ovol2 might not function as a switch to initiate the mesendodermal fate. We attempted to overexpress Ovol2 in the primitive streak of early chick embryos but failed to induce the expression of the early mesendoderm marker gene T (data not shown). This result indicates that although Ovol2 is required for mesoderm and endoderm differentiation in mouse ESCs, it might not be sufficient for the induction and initiation of mesendoderm in chick embryos.
BMP signaling is essential for the inhibition of precocious neural differentiation, thereby ensuring normal development of mesoderm and endoderm (7). However, it remains challenging to clarify how BMP signaling regulates the early cell fate decisions through downstream transcription factors, although several studies have contributed to our understanding of this process. In ESCs, BMP signals induce the expression of Id genes to sustain pluripotency and inhibit neural differentiation (9). In Xenopus, BMP signals inhibit neural induction through the downstream genes Msx1 and Dlx3 (49–51), whereas Msx1 and Dlx5 participate in the establishment and maintenance of the neural/epidermal boundary in chick embryos (52–54). During mouse gastrulation, Tlx2 is required for mesoderm formation downstream of BMP signaling (55). We have shown that Id1 and Id2 partially mediate the function of BMPs to promote ESC differentiation into non-neural cell fates (13). Recently, we also showed that another BMP downstream target gene, AP2γ, specifically functions in epidermal versus neural differentiation in both ESCs and chick embryos (56). In this study, we showed that Ovol2 partially mediates BMP function in the neuroectoderm/mesendoderm cell fate decision (Fig. 5). In chick embryos, the inhibitory effect of cOvol2 on neural specification is less severe than that of BMP4 (Fig. 7D) (data not shown) (14), suggesting again that Ovol2 only partially mediates BMP-induced neural inhibition. A similar phenomenon is observed in Msx1/Dlx5 double knock-out mice, which display no significant abnormality in nervous system development (57). This suggests the co-existence of specificity and redundancy in BMP downstream targets to guarantee the normal progress of development. Because Smads bind to DNA with low specificity and affinity (58), it is generally believed that the BMP signaling pathway exerts its specific effect by choosing cell type-specific DNA-binding cofactors to regulate its downstream target genes (15). Although we observed the direct interaction of Smad1 with the enhancer of Ovol2 gene (Fig. 6E), we speculate that some specific DNA-binding Smad partners may participate in regulation of Ovol2 transcription. It is a challenge to clarify the mechanism of this specific regulation, and we will try to address this issue in our future research.
Extensive studies have shown that the expression of the ovo family gene is regulated by the Wnt signaling pathway. In Drosophila, ovo/svb integrates the Wg (Drosophila Wnt) and DER (Drosophila EGF receptor) pathways to control denticle formation in the epidermis (22). Ovol1, another mouse homolog of Ovol2, is directly regulated by β-catenin/LEF1 downstream of the Wnt pathway and plays a crucial role in hair formation and spermatogenesis (59). We stimulated ESCs with the GSK3β inhibitor CHIR99021 for 2 h to activate the Wnt/β-catenin signaling pathway and observed a significant increase in Ovol2 expression (data not shown). Moreover, we scanned the 5′ region of the mouse Ovol2 gene and found several putative LEF/TCF binding sites, suggesting that Wnt-induced regulation of Ovo gene expression might be evolutionarily conserved.
BMP signals and Wnt signals often play similar roles in early embryonic development. Overexpression of Wnt1 in ESCs inhibits neural differentiation, whereas Wnt antagonist Sfrp2 promotes ESC differentiation into neural progenitor cells (60). Moreover, the specification of the neural plate border in chick embryo is dependent on the cooperation of the Wnt and BMP pathways (61). These studies suggest that Wnt signals play similar or synergic roles with the BMP pathway in neural induction and patterning. The function of BMPs and Wnts in mesendoderm development is also similar. Wnt3 knock-out mice could form the egg cylinder but failed to form a primitive streak and mesoderm (62). The addition of the Wnt inhibitor Dkk1 significantly suppresses the differentiation of ESCs into the primitive streak, mesoderm, and endoderm fates (63). Therefore, the Wnt/β-catenin and BMP signaling pathways have similar functions in the regulation of neural and mesendodermal differentiation. Given the fact that Ovol2 is directly regulated by BMP signals (Fig. 6), it will be interesting to test whether Ovol2 acts as a dual effector downstream of both BMP and Wnt signals to regulate the cell fate decision between neuroectoderm and mesendoderm.
Supplementary Material
Acknowledgments
We thank Dr. Xing Dai (University of California, Irvine, CA) for mouse Ovol2 cDNA, Dr. Claudio D. Stern (University College London) for the Xenopus BMP4 expression plasmid and the kind guidance on chick electroporation, Dr. Paul J. Scotting (University of Nottingham) for in situ probes cSox2 and cSox3, and Dr. Yuqiang Ding (Tongji University School of Medicine, China) for pCAGGS-IRES-GFP vector. We also thank the cell biology core facilities for the confocal and FACS study (Shanghai Institutes for Biological Sciences, China) and members of the Jing laboratory for critical reading of the manuscript.
This work was supported in part by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDA01010201; National Key Basic Research and Development Program of China Grant 2009CB941100; National Natural Science Foundation of China Grants 30830034, 90919046, and 91219303; and Council of Shanghai Municipal Government for Science and Technology Grant 11ZR1443200.

This article contains supplemental Tables S1–S3 and Figs. S1–S6.
- BMP
- bone morphogenetic protein
- ESC
- embryonic stem cell
- EB
- embryoid body
- qRT-PCR
- quantitative RT-PCR
- SFEB
- serum-free EB culture
- NPC
- neural progenitor cell
- p-Smad1/5/8
- phosphorylated Smad1/5/8
- HH
- Hamburger and Hamilton
- ISH
- in situ hybridization
- mut-Ovo
- mutant Ovol2.
REFERENCES
- 1. Gadue P., Huber T. L., Nostro M. C., Kattman S., Keller G. M. (2005) Germ layer induction from embryonic stem cells. Exp. Hematol. 33, 955–964 [DOI] [PubMed] [Google Scholar]
- 2. Lu C. C., Brennan J., Robertson E. J. (2001) From fertilization to gastrulation. Axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 11, 384–392 [DOI] [PubMed] [Google Scholar]
- 3. Tam P. P., Loebel D. A. (2007) Gene function in mouse embryogenesis. Get set for gastrulation. Nat. Rev. Genet. 8, 368–381 [DOI] [PubMed] [Google Scholar]
- 4. Kimelman D., Griffin K. J. (2000) Vertebrate mesendoderm induction and patterning. Curr. Opin. Genet. Dev. 10, 350–356 [DOI] [PubMed] [Google Scholar]
- 5. Rodaway A., Patient R. (2001) Mesendoderm. An ancient germ layer? Cell 105, 169–172 [DOI] [PubMed] [Google Scholar]
- 6. Watabe T., Miyazono K. (2009) Roles of TGF-β family signaling in stem cell renewal and differentiation. Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 7. Di-Gregorio A., Sancho M., Stuckey D. W., Crompton L. A., Godwin J., Mishina Y., Rodriguez T. A. (2007) BMP signalling inhibits premature neural differentiation in the mouse embryo. Development 134, 3359–3369 [DOI] [PubMed] [Google Scholar]
- 8. Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995) Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105–2116 [DOI] [PubMed] [Google Scholar]
- 9. Ying Q. L., Nichols J., Chambers I., Smith A. (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 10. Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 [DOI] [PubMed] [Google Scholar]
- 11. Park C., Afrikanova I., Chung Y. S., Zhang W. J., Arentson E., Fong G. G., Rosendahl A., Choi K. (2004) A hierarchical order of factors in the generation of FLK1- and SCL-expressing hematopoietic and endothelial progenitors from embryonic stem cells. Development 131, 2749–2762 [DOI] [PubMed] [Google Scholar]
- 12. Pearson S., Sroczynska P., Lacaud G., Kouskoff V. (2008) The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development 135, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 13. Zhang K., Li L., Huang C., Shen C., Tan F., Xia C., Liu P., Rossant J., Jing N. (2010) Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development 137, 2095–2105 [DOI] [PubMed] [Google Scholar]
- 14. Linker C., Stern C. D. (2004) Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development 131, 5671–5681 [DOI] [PubMed] [Google Scholar]
- 15. Massagué J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 16. Feng X. H., Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 17. Shi Y., Massagué J. (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 18. Takemoto T., Uchikawa M., Yoshida M., Bell D. M., Lovell-Badge R., Papaioannou V. E., Kondoh H. (2011) Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chng Z., Teo A., Pedersen R. A., Vallier L. (2010) SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell 6, 59–70 [DOI] [PubMed] [Google Scholar]
- 20. Li B., Dai Q., Li L., Nair M., Mackay D. R., Dai X. (2002) Ovol2, a mammalian homolog of Drosophila ovo. Gene structure, chromosomal mapping, and aberrant expression in blind-sterile mice. Genomics 80, 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson A. D., Fitzsimmons D., Hagman J., Chamberlin H. M. (2001) EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 128, 2857–2865 [DOI] [PubMed] [Google Scholar]
- 22. Payre F., Vincent A., Carreno S. (1999) ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature 400, 271–275 [DOI] [PubMed] [Google Scholar]
- 23. Oliver B., Perrimon N., Mahowald A. P. (1987) The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev. 1, 913–923 [DOI] [PubMed] [Google Scholar]
- 24. Mével-Ninio M., Terracol R., Kafatos F. C. (1991) The ovo gene of Drosophila encodes a zinc finger protein required for female germ line development. EMBO J. 10, 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dai X., Schonbaum C., Degenstein L., Bai W., Mahowald A., Fuchs E. (1998) The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 12, 3452–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mackay D. R., Hu M., Li B., Rhéaume C., Dai X. (2006) The mouse Ovol2 gene is required for cranial neural tube development. Dev. Biol. 291, 38–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Unezaki S., Horai R., Sudo K., Iwakura Y., Ito S. (2007) Ovol2/Movo, a homologue of Drosophila ovo, is required for angiogenesis, heart formation, and placental development in mice. Genes Cells 12, 773–785 [DOI] [PubMed] [Google Scholar]
- 28. Kowanetz M., Valcourt U., Bergström R., Heldin C. H., Moustakas A. (2004) Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor β and bone morphogenetic protein. Mol. Cell Biol. 24, 4241–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C., Xia C., Bian W., Liu L., Lin W., Chen Y. G., Ang S. L., Jing N. (2006) Cell aggregation-induced FGF8 elevation is essential for P19 cell neural differentiation. Mol. Biol. Cell 17, 3075–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naldini L., Blömer U., Gage F. H., Trono D., Verma I. M. (1996) Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. U.S.A. 93, 11382–11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tiscornia G., Singer O., Verma I. M. (2006) Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 [DOI] [PubMed] [Google Scholar]
- 32. Ginzinger D. G. (2002) Gene quantification using real-time quantitative PCR. An emerging technology hits the mainstream. Exp. Hematol. 30, 503–512 [DOI] [PubMed] [Google Scholar]
- 33. Gao X., Bian W., Yang J., Tang K., Kitani H., Atsumi T., Jing N. (2001) A role of N-cadherin in neuronal differentiation of embryonic carcinoma P19 cells. Biochem. Biophys. Res. Commun. 284, 1098–1103 [DOI] [PubMed] [Google Scholar]
- 34. Okada Y., Shimazaki T., Sobue G., Okano H. (2004) Retinoic acid concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 275, 124–142 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka S., Kamachi Y., Tanouchi A., Hamada H., Jing N., Kondoh H. (2004) Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol. Cell Biol. 24, 8834–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin Z., Liu L., Bian W., Chen Y., Xu G., Cheng L., Jing N. (2009) Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J. Biol. Chem. 284, 8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hamburger V., Hamilton H. L. (1992) A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 195, 231–272 [DOI] [PubMed] [Google Scholar]
- 38. Voiculescu O., Papanayotou C., Stern C. D. (2008) Spatially and temporally controlled electroporation of early chick embryos. Nat. Protoc. 3, 419–426 [DOI] [PubMed] [Google Scholar]
- 39. Streit A., Stern C. D. (2001) Combined whole-mount in situ hybridization and immunohistochemistry in avian embryos. Methods 23, 339–344 [DOI] [PubMed] [Google Scholar]
- 40. Stern C. D. (1998) Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr. Top. Dev. Biol. 36, 223–243 [DOI] [PubMed] [Google Scholar]
- 41. Unezaki S., Nishizawa M., Okuda-Ashitaka E., Masu Y., Mukai M., Kobayashi S., Sawamoto K., Okano H., Ito S. (2004) Characterization of the isoforms of MOVO zinc finger protein, a mouse homologue of Drosophila Ovo, as transcription factors. Gene 336, 47–58 [DOI] [PubMed] [Google Scholar]
- 42. Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D., Robson P. (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 [DOI] [PubMed] [Google Scholar]
- 43. Schroeder I. S., Rolletschek A., Blyszczuk P., Kania G., Wobus A. M. (2006) Differentiation of mouse embryonic stem cells to insulin-producing cells. Nat. Protoc. 1, 495–507 [DOI] [PubMed] [Google Scholar]
- 44. Hollnagel A., Oehlmann V., Heymer J., Rüther U., Nordheim A. (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 274, 19838–19845 [DOI] [PubMed] [Google Scholar]
- 45. Nostro M. C., Cheng X., Keller G. M., Gadue P. (2008) Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rex M., Orme A., Uwanogho D., Tointon K., Wigmore P. M., Sharpe P. T., Scotting P. J. (1997) Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev. Dyn. 209, 323–332 [DOI] [PubMed] [Google Scholar]
- 47. Streit A., Lee K. J., Woo I., Roberts C., Jessell T. M., Stern C. D. (1998) Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development 125, 507–519 [DOI] [PubMed] [Google Scholar]
- 48. Chapman S. C., Schubert F. R., Schoenwolf G. C., Lumsden A. (2002) Analysis of spatial and temporal gene expression patterns in blastula and gastrula stage chick embryos. Dev. Biol. 245, 187–199 [DOI] [PubMed] [Google Scholar]
- 49. Feledy J. A., Beanan M. J., Sandoval J. J., Goodrich J. S., Lim J. H., Matsuo-Takasaki M., Sato S. M., Sargent T. D. (1999) Inhibitory patterning of the anterior neural plate in Xenopus by homeodomain factors Dlx3 and Msx1. Dev. Biol. 212, 455–464 [DOI] [PubMed] [Google Scholar]
- 50. Ishimura A., Maeda R., Takeda M., Kikkawa M., Daar I. O., Maéno M. (2000) Involvement of BMP-4/msx-1 and FGF pathways in neural induction in the Xenopus embryo. Dev. Growth Differ. 42, 307–316 [DOI] [PubMed] [Google Scholar]
- 51. Suzuki A., Ueno N., Hemmati-Brivanlou A. (1997) Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development 124, 3037–3044 [DOI] [PubMed] [Google Scholar]
- 52. Yang L., Zhang H., Hu G., Wang H., Abate-Shen C., Shen M. M. (1998) An early phase of embryonic Dlx5 expression defines the rostral boundary of the neural plate. J. Neurosci. 18, 8322–8330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pera E., Stein S., Kessel M. (1999) Ectodermal patterning in the avian embryo. Epidermis versus neural plate. Development 126, 63–73 [DOI] [PubMed] [Google Scholar]
- 54. Streit A., Stern C. D. (1999) Establishment and maintenance of the border of the neural plate in the chick. Involvement of FGF and BMP activity. Mech. Dev. 82, 51–66 [DOI] [PubMed] [Google Scholar]
- 55. Tang S. J., Hoodless P. A., Lu Z., Breitman M. L., McInnes R. R., Wrana J. L., Buchwald M. (1998) The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development 125, 1877–1887 [DOI] [PubMed] [Google Scholar]
- 56. Qiao Y., Zhu Y., Sheng N., Chen J., Tao R., Zhu Q., Zhang T., Qian C., Jing N. (2012) AP2γ regulates neural and epidermal development downstream of the BMP pathway at early stages of ectodermal patterning. Cell Res. 22, 1546–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Levi G., Mantero S., Barbieri O., Cantatore D., Paleari L., Beverdam A., Genova F., Robert B., Merlo G. R. (2006) Msx1 and Dlx5 act independently in development of craniofacial skeleton, but converge on the regulation of Bmp signaling in palate formation. Mech. Dev. 123, 3–16 [DOI] [PubMed] [Google Scholar]
- 58. Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., Pavletich N. P. (1998) Crystal structure of a Smad MH1 domain bound to DNA. Insights on DNA binding in TGF-β signaling. Cell 94, 585–594 [DOI] [PubMed] [Google Scholar]
- 59. Li B., Mackay D. R., Dai Q., Li T. W., Nair M., Fallahi M., Schonbaum C. P., Fantes J., Mahowald A. P., Waterman M. L., Fuchs E., Dai X. (2002) The LEF1/β-catenin complex activates movo1, a mouse homolog of Drosophila ovo required for epidermal appendage differentiation. Proc. Natl. Acad. Sci. U.S.A. 99, 6064–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aubert J., Dunstan H., Chambers I., Smith A. (2002) Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 20, 1240–1245 [DOI] [PubMed] [Google Scholar]
- 61. Patthey C., Edlund T., Gunhaga L. (2009) Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development 136, 73–83 [DOI] [PubMed] [Google Scholar]
- 62. Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R., Bradley A. (1999) Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365 [DOI] [PubMed] [Google Scholar]
- 63. Lindsley R. C., Gill J. G., Kyba M., Murphy T. L., Murphy K. M. (2006) Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133, 3787–3796 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.