Background: Inteins are protein maturation machines that enable technologies for regulating enzymes and protein semisynthesis.
Results: Robust splicing was achieved with novel genetically selected exteins.
Conclusion: In contrast to commonly held perceptions, the natural extein was not the fastest splicing substrate.
Significance: Natural precursors balance extein- and intein-selective pressures. Specificity studies provide a predictive rubric for identifying new intein insertion sites.
Keywords: Enzyme Catalysis, Enzyme Kinetics, Protein Chemistry, Protein Engineering, Protein Processing, Npu DnaE Intein, Extein Specificity, Genetic Selection, Protein Splicing, Trans-splicing
Abstract
Inteins are naturally occurring intervening sequences that catalyze a protein splicing reaction resulting in intein excision and concatenation of the flanking polypeptides (exteins) with a native peptide bond. Inteins display a diversity of catalytic mechanisms within a highly conserved fold that is shared with hedgehog autoprocessing proteins. The unusual chemistry of inteins has afforded powerful biotechnology tools for controlling enzyme function upon splicing and allowing peptides of different origins to be coupled in a specific, time-defined manner. The extein sequences immediately flanking the intein affect splicing and can be defined as the intein substrate. Because of the enormous potential complexity of all possible flanking sequences, studying intein substrate specificity has been difficult. Therefore, we developed a genetic selection for splicing-dependent kanamycin resistance with no significant bias when six amino acids that immediately flanked the intein insertion site were randomized. We applied this selection to examine the sequence space of residues flanking the Nostoc punctiforme Npu DnaE intein and found that this intein efficiently splices a much wider range of sequences than previously thought, with little N-extein specificity and only two important C-extein positions. The novel selected extein sequences were sufficient to promote splicing in three unrelated proteins, confirming the generalizable nature of the specificity data and defining new potential insertion sites for any target. Kinetic analysis showed splicing rates with the selected exteins that were as fast or faster than the native extein, refuting past assumptions that the naturally selected flanking extein sequences are optimal for splicing.
Introduction
Inteins are a class of single turnover enzymes that catalyze a self-excision reaction out of a host protein-intein fusion (the precursor protein), resulting in a free intein and a mature host protein (the extein) where the splice junctions have been ligated with a standard peptide bond. Over 550 inteins have been identified in the InBase database, and they are ubiquitous in single cell organisms from all domains of life (1). Intein splicing mechanisms are diverse, with three distinct mechanisms identified to date (2). In each of these mechanisms, a series of three or four carefully coordinated displacement reactions was catalyzed in the absence of additional cofactors or energy sources by the intein plus the extein residue forming the C-terminal splice junction (2). Coordination of these multistep pathways is likely to require a precise architecture at the intein active site that can be affected by proximal extein residues.
Splicing in native host proteins is so rapid that the precursor protein is rarely, if ever, detected in nature, whereas splicing in heterologous host proteins often results in significant amounts of single splice junction cleavage byproducts or unreacted precursor. As a result, it has been proposed that the native flanking extein sequences are optimal for splicing of each intein (3, 4). However, inteins are generally located within highly conserved extein motifs and active sites of essential proteins, so presumably there is also selective pressure to maintain the residues at the intein insertion site for optimal extein function (5). Thus we hypothesize that the native precursor may not necessarily represent the most rapid splicing context for the intein but, rather, a balance of host protein sequence requirements, the specificity of the homing endonuclease (present in many inteins), and the need for a functional intein.
Because of their unique chemistry, inteins have proven useful in numerous applications such as protein tagging and purification, control of enzyme activity, assembling active proteins in vivo, protein semisynthesis in vitro, segmental isotope labeling for NMR, and many others (for review, see Refs. 6–9). The technologies enabled by inteins are growing, yet there remain some fundamental gaps in our understanding of how inteins work, especially in the specificity requirements for splicing (defined here as the flanking extein sequences that permit splicing). Previous studies characterizing the effect of varying a single amino acid flanking the N- or C-terminal splice junction demonstrated that the subset of extein residues that promote splicing or cleavage are specific to each intein (10–14). To our knowledge, no studies have examined the immense number of possible combinations when multiple extein residues are queried in a full splicing reaction. In lieu of such information, researchers design splicing experiments governed by the assumption that the natural host context will be the best and have, therefore, added three to five native flanking extein residues when placing an intein in a novel host protein. This limits the usefulness of these techniques by leaving behind a “scar” of flanking extein residues after splicing. Another approach to improving splicing in heterologous systems uses protein engineering strategies to change or relax intein specificity by intein mutation (4, 12, 15). Complementary tools that allow us to gauge the full scope of viable substrates of important inteins are highly desirable for understanding inteins as enzymes and expanding their usefulness in biotechnology by providing more options for intein insertion.
Here we present a genetic selection based on kanamycin (Kan)4 resistance that allows for comprehensive examination of all possible combinations of three proximal N-terminal extein residues and three proximal C-terminal extein residues. Unlike previous intein kanamycin selection systems, our system was optimized for maintenance of kanamycin resistance with an extremely broad range of post-splicing six-residue insertions to minimize bias due to the reporter protein. The system was tested with the Nostoc punctiforme Npu DnaE, the Mycobacterium tuberculosis Mtu-H37Rv RecA, and the MP-Be DnaB inteins. We further applied this system to study splicing of the Npu DnaE intein in detail. This intein is naturally split and works in trans but can be fused to splice in cis as well (16). The Npu DnaE intein is one of the fastest known inteins (17–20) and is important for biotechnology. Our results show that the Npu DnaE intein is able to efficiently splice multiple sequences of far different composition than its native flanking exteins, defining new potential insertion sites preceding Cys-Trp or Cys-Met. These data expand the usefulness of this intein and provide a new framework for predictable design of protein splicing experiments.
EXPERIMENTAL PROCEDURES
All enzymes and reagents were obtained from New England Biolabs (NEB, Ipswich MA) unless otherwise noted and used as described by the manufacturer. DNA sequencing was performed either by the NEB Core Sequencing Facility or by Beckman Coulter Genomics (Danvers, MA) for single pass automated sequencing in two directions using cultures provided in 96-well microtiter dishes.
PCR and Plasmid Construction
All PCR amplification steps used Phusion DNA polymerase (NEB) according to the manufacturer's instructions. The following two thermocycling protocols were used for most PCR amplifications: Protocol A (1 cycle at 98 °C for 30 s; 25 cycles at 98 °C for 10 s, 65 °C for 20 s, 72 °C for 120 s; 1 cycle at 72 °C for 5 min); Protocol B (1 cycle of 98 °C for 30 s; 25 cycles of 98 °C for 10 s, 65 °C for 20 s, 72 °C for 25 s; 1 cycle of 72 °C for 5 min). Restriction endonucleases and T4 DNA ligase were obtained from NEB. Chemically competent Escherichia coli NEB 10-beta cells were used for all plasmid recovery stages.
Examination of Potential Intein Insertion Sites in Aminoglycoside Phosphotransferase (Aph) for High Throughput Assay Development
Aminoglycoside phosphotransferases confer KanR to various bacteria and are good targets for genetic selection. A protein blast search using the Corynebacterium diphtheriae Aph sequence (accession number AY061891.1) as the query was done on the NCBI server at blast.ncbi.nlm.nih.gov. The 10 most similar unrelated bacterial Aph sequences identified by the search were aligned, and the consensus sequence was compared with the C. diphtheriae Aph gene. Five locations (A: Ala-62—Asn-63; B: Phe-87—Ile-88; C: Glu-118—Asn-119; D: Asp-144—Asg-145; E: Glu-181—Met-182) were chosen as possible intein insertions sites based on possessing low sequence conservation and being surface-exposed (21). Primers were designed to insert six variable residues at each site or to replace the six amino acids centered around each site. Mutations were made by inverted PCR using Protocol A with pACYC177 as the template. pACYC177 encodes both an ampicillin (Amp) and a kanamycin resistance marker. After PCR, the parental DNA was digested completely with 10 units of DpnI for 3 h. The resulting amplified DNA was ligated and transformed into NEB 10-beta cells. Equal quantities of cells were spread on LB agar plates containing either 100 μg/ml Amp or 100 μg/ml Kan. The ratio of cells that survived on Kan versus Amp plates was calculated after overnight incubation at 37 °C. The PCR control using primers with no mutations in KanR approximates the efficiency of the inverted PCR reaction and subsequent plating, and so was used to normalize the data.
Construction of KanR Plasmids for Intein Insertion
Silent mutations were made to encode BspEI and NheI restriction sites for cloning inteins into the KanR site C yielding plasmid p3A. This plasmid contained an additional NheI site at a distal position, so the entire Kan resistance gene and its endogenous promoter were subcloned from p3A to pJF119 (22) using KasI and KpnI restriction sites. The resulting p3AM1 plasmid was Kan- and Amp-resistant.
To enhance Western blot analysis of splicing variants, a His6 tag followed by a Gly-Gly-Ser-Gly linker was appended to the N terminus of the Aph gene by inverted PCR. The resulting plasmid was named pKanCH.
Functional and inactive variants of the Npu DnaE intein cis construct, the Mtu RecA intein, and the MP-Be DnaB intein were cloned into site C of pKanCH. Inactive inteins contained the following mutations at catalytic residues: Npu DnaE intein: C1A, N138D, C+1A; Mtu RecA intein: C1A, N440D, C+1A; MP-Be DnaB intein: C320S. All clones with these inactive inteins were kanamycin-sensitive.
Library Construction and Selection
Hand-mixed primers (forward, 5′-CT GAT TCC GGA GAA NNK NNK NNK TGC CTG AGC TAT GA; reverse, 5′-A CAC TGC TAG CGC ATC AAC AAT ATT MNN MNN GCA ATT AGA GGC AAT AA) were ordered from Integrated DNA Technologies (Coralville, IA) to introduce three variable codons flanking the intein N terminus and two variable codons flanking the intein C terminus after a fixed extein Cys residue (Fig. 1). In these primers N = A, T, G, or C, K = G or T, and M = C or A. Primers also included restriction enzyme sites for cloning into site C in the kanamycin resistance gene and sequences that matched the template, which was the cis construct of the Npu DnaE intein (16). PCR products using these primers were cloned into plasmids that contained mutations at the natural Aph ribosome binding site (RBS) changing GGGGTGTTATG to GAAGGTGTTCATG, which reduces the level of expression by ∼20-fold.5
FIGURE 1.
KanR selection construct used to study intein specificity for flanking extein sequences. KanR was used as a reporter for protein splicing with the Npu DnaE intein and three additional extein residues flanking both its N and C termini. The sequence of the native extein is shown inserted into KanR site C. Underlined amino acids were simultaneously randomized. In the numbering scheme used in this paper, residues in the KanR protein, the variable extein positions, and the intein were numbered independently as indicated. Positions in the KanR protein were numbered as in the original protein, ignoring insertions.
The resulting DNA was transformed into electrocompetent NEB 10-beta cells using a standard bacterial transformation protocol with up to 200 ng of DNA in a Gene Pulser Xcell (Bio-Rad). The cells were immediately resuspended in a total of 4 ml of LB media and allowed to recover for 1.5 h at 37 °C with shaking.
Freshly transformed cells were pelleted after recovery and resuspended in 1 ml of LB media before plating on Kan-selective plates. To estimate the size of the library, serial dilutions were plated on 100 μg/ml Amp plates, which yielded a library size of ∼5 × 107. The rest of the library was plated on 40 μg/ml Kan plates and incubated overnight at 37 °C. Colonies that grew on Kan-selective plates were arrayed into 96-well microtiter dishes containing LB media + 10% glycerol + 50 μg/ml Amp + 40 μg/ml Kan using the Genetix QPix II automated colony picker (Molecular Devices, Sunnyvale, CA). These microtiter plates were incubated overnight at 37 °C, a replica plate was made, and all plates were stored at −20 °C. Replica plates were sent to Beckman Coulter Genomics for automated sequencing.
Sequencing results were examined using an in-house automated program that compared the sequences of the RBS, the N- and C-terminal KanR fragments, and the intein. Only sequences that had unambiguous nucleotide assignments that did not contain any mutations and that maintained the KanR RBS knockdown sequence were included in our analysis of specificity. Only 483 of 768 clones sequenced yielded results that met these stringent criteria, mostly due to ambiguous nucleotide assignments.
Cloning into Luciferase and Western Blot Analysis of Expressed Proteins
To aid in cloning, XhoI and KpnI sites were introduced by silent mutagenesis to flank the firefly luciferase site Asn-230—Asp-234 using inverted PCR with the pEK4 (23) template.
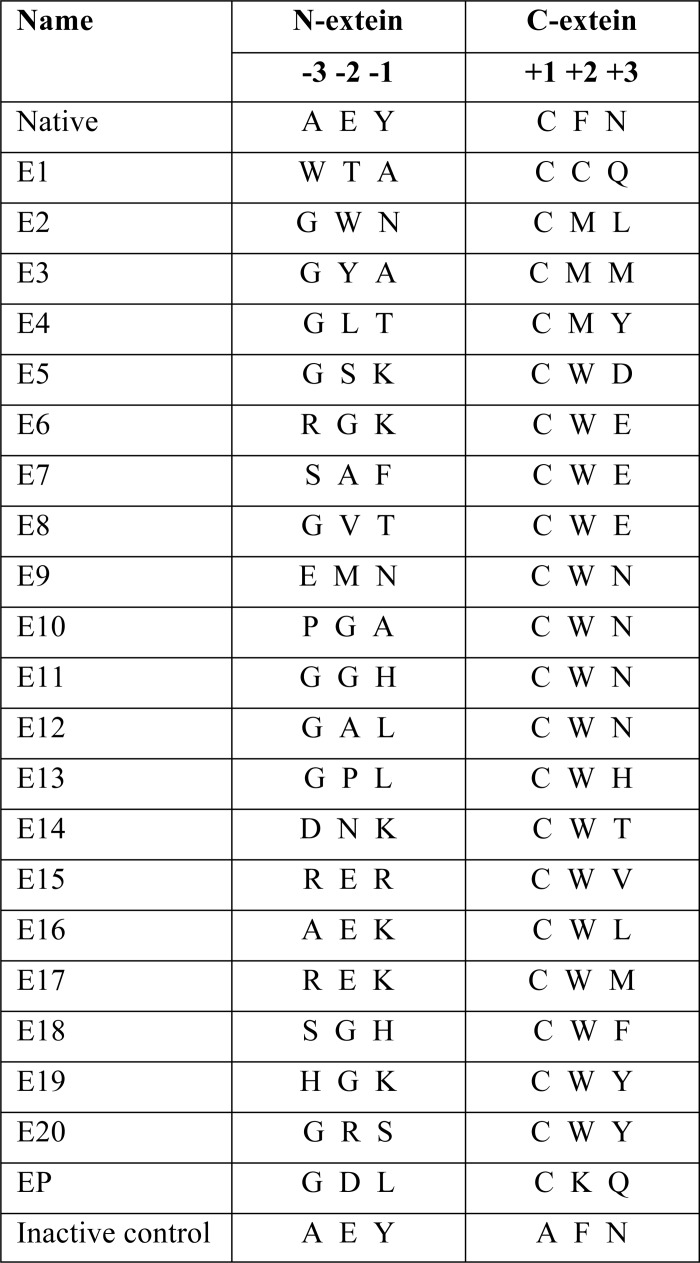
Insert DNA containing the Npu DnaE intein and the desired extein sequences were prepared using the pKanCH plasmid containing the Npu DnaE cis-intein (pKanCH_Npu) as template. The PCR primers used were forward (5′-GCAT GCT CGA GAT CCT ATT TTT GGC AAT XXX XXX XXX TGC CTG AGC TAT GAA ACC GAA AT) and reverse (5′-GAA TGG TAC CAC ACT TAA AAT CGC AGT XXX XXX GCA ATT AGA GGC AAT AAA GCC ATT TTT, where XXX represents the appropriate sequence to encode for selection hits E2, E4, E6, E8, E9, E12, E15 E16, E18, and E20 (see Table 2). The resultant plasmids were sequenced to verify the presence of the firefly luciferase and the intein with the desired flanking extein sequence.
TABLE 2.
Selected extein sequences
The flanking extein sequences of clones representing the sequence diversity of selection hits are listed along with the flanking extein sequences in the native extein and in an inactive intein with mutations in essential residues C1A, the intein C-terminal Asn and C+1A. Cys+1 was held constant in the selection.
Western blots were done to assess the degree of splicing with each flanking extein sequence. Plasmids were transformed into NEB Express cells and incubated at 37 °C to an A600 nm of 0.6. Two ml of each culture was pelleted, resuspended in 250 μl of water, and then adjusted to an A600 nm of 5. Fifteen μl of these cells were mixed with 3× SDS sample buffer + DTT (NEB), boiled for 10 min, and electrophoresed on SDS-PAGE followed by transfer to nitrocellulose membranes using a Bio-Rad Trans-blot S.D. apparatus. After transfer to nitrocellulose, bands containing the firefly luciferase were detected with mouse IgG anti-firefly luciferase antibody (Abcam, Cambridge, MA) and IRDye 680 goat anti-mouse secondary antibody (LI-COR, Lincoln, NE).
Cloning Inteins into MBP-Intein-Paramyosin (MIP) Constructs for Trans-splicing Experiments
DNA encoding the Npu DnaE intein (cis construct) with three native extein residues flanking each side was amplified by PCR Protocol B using pKanCH_Npu as the template and primers containing XhoI and SpeI sites. Plasmid pMIP contains the E. coli maltose-binding protein (M) as the N-extein and the paramyosin-Δ Sal fragment (P) as the C-extein (2). The PCR insert and plasmid pMIP_MP-Be_His (2) were both digested with XhoI and SpeI, gel-purified, and ligated to yield plasmid pMIP_Npu, which was sequenced to verify that the intein was fused in between MBP and paramyosin-Δ Sal.
The cis-splicing precursor was then converted into a trans-splicing precursor. Plasmids containing M-IN and IC-PHis fragments were generated by inverted PCR with pMIP_Npu as the template and thermocycling Protocol A using M-IN primers (forward (5′-TGA CTG GGA AAA CCC TGG CGT TAC) and reverse (5′-ATT CGG CAG ATT ATC AAC ACG CAT CA)) and IC-PHis primers (forward (5′-ATG ATC AAA ATT GCC ACC CGT AAA TAT CTG) and reverse (5′-AAT CTA TGG TCC TTG TTG GTG AAG TGC TCG T)). After DpnI digestion, ligation, and transformation, plasmids pNpuDnaE_MBP_IN and pNpuDnaE_IC_para were obtained.
The desired N- and C-terminal flanking extein sequences were introduced using a final round of inverted PCR with primers for M-IN (forward (5′-GAG XXX XXX XXX TGC CTG AGC TAT GAA ACC GAA) and reverse (5′-GAG CGT ACC CCT TCC CTC GAT)) and for IC-Para (forward (5′-XXX XXX ACT AGT GGT GGC ATG CTT ACA GG) and reverse (5′-GCA ATT AGA GGC AAT AAA GCC ATT TTT CAG)), where XXX represents the appropriate codon for each flanking extein sequence listed in Table 3.
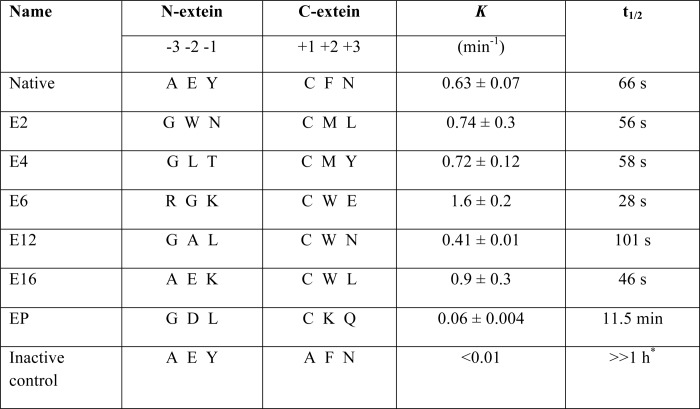
TABLE 3.
Kinetics of trans-splicing by the Npu DnaE intein with the listed flanking extein sequences in M-IN and IC-PHis
Kinetic parameters were measured as described under “Experimental Procedures” for selection hits (E2, E4, E6, E12, and E16), the native extein, the EP clone selected from a low stringency pilot experiment with the native RBS, and an inactive intein with mutations in essential residues C1A in M-IN and the intein C-terminal Asn plus C+1A in IC-PHis.
* No trans-splicing product was detected after 1 hour.
Trans-splicing Kinetic Assay in MIP
Plasmids containing M-IN and IC-PHis were transformed into NEB Express Iq cells and grown on LB + Amp plates. Ten-ml cultures were inoculated with single colonies and grown at 37 °C to an A600 nm of 0.6–0.8 at which point the temperature was reduced to 25 °C, and 0.5 mm isopropyl 1-thio-β-d-galactopyranoside was added. The cultures were then incubated for five more hours. One milliliter of each culture was harvested, and the cell pellet was stored frozen at −80 °C until kinetic assays were performed. Cell pellets were resuspended in 300 μl of reaction buffer K (100 mm Tris. pH 7.5, 5 mm DTT, 1 mm EDTA), lysed by sonication, and centrifuged for 1 min at maximum speed in a benchtop microcentrifuge. Because the M-IN fragment expresses many fold more efficiently than the IC-PHis fragment, the kinetic assays were done by mixing M-IN and IC-PHis-containing lysates in a 1:5 ratio at 30 °C over a 10-min time course. Under these conditions the amount of the M-IN fragment observed in the Western blot remains constant, whereas the IC-PHis fragment is consumed. Each 20-μl time point was stopped by the addition of 10 μl of 3 × SDS sample buffer + DTT. After boiling for 10 min, the samples were run on SDS-PAGE followed by transfer to a nitrocellulose membrane. The membrane was probed with Mouse IgG anti-His tag antibody (Novagen, EMD Chemicals, San Diego, CA) to detect IC-PHis or M-PHis and rabbit anti-MBP antisera (NEB) to detect M-IN or M-PHis. Probing with IRDye 680 goat anti-mouse and IRDye 800 Goat anti-Rabbit secondary antibodies (LI-COR) allowed for quantification of the spliced product on the LI-COR Odyssey. The percent conversion was calculated as 1–M-PHis formation divided by the initial substrate (IC-PHis) concentration. These data were fit to a first order decay reaction using KaleidaGraph (Synergy, Reading, PA) with the equation A = Aoe−kt, where A is the substrate concentration at t, Ao is the initial substrate concentration, k is the apparent first order rate constant, and t is time. The t½ was calculated as t½ = ln2/k. At least two independent replicates of each mutant were done, and the standard deviation of the fits was calculated.
RESULTS
Developing a Selection System to Probe Extein Specificity
We set out to develop an assay system that could rapidly examine variable extein sequences of up to 108 members. Because of the ease of use and high stringency of antibiotic-based selections, we focused our efforts on identifying sites within C. diphtheriae aminoglycoside phosphotransferase (Aph), an enzyme responsible for kanamycin resistance (KanR). Based on comparison of this protein to an alignment of other Aph proteins, five regions were chosen as candidates to develop a splicing selection system (A: Ala-62—Asn-63; B: Phe-87—Ile-88; C: Glu-118—Asn-119; D: Asp-144—Asg-145; E: Glu-181—Met-182). These locations were surface-exposed and possess low sequence conservation, suggesting that they would be able to tolerate multiple mutations and still retain KanR (data not shown). A plasmid was constructed that contained β-lactamase to confer Amp resistance and the C. diphtheriae Aph gene under the control of its endogenous promoter. Using Phusion polymerase-based inverted PCR, we created 10 libraries where either (a) six codons at a chosen site were replaced with six consecutive in-frame NNK codons or (b) six in-frame NNK codons were inserted into the middle of the chosen sequence, where N represents any nucleotide and K represents G or C. These libraries represent the scar that would be left in the KanR protein after splicing when three flanking amino acids are included at each end of the intein. An ideal site would have no sequence preference and no selection bias due to the scar sequence in KanR. The permissibility of each site for random sequence replacement or insertion was quantified based on the percent of colonies that recovered on Kan versus Amp plates. Two sites were tolerant for the randomized cassettes: site C with a 6-amino acid insertion between Glu-118 and Asn-119 had 91 ± 8% permissibility, and site D with a replacement of the codons between Asn-142 and Phe-147 had 47 ± 1% permissibility. Insertion site C had been previously identified as a useful site for intein insertion (24). Other sites in the Aph gene have also been used for intein insertion (4, 15) but were not tested for permissibility of random scar sequences.
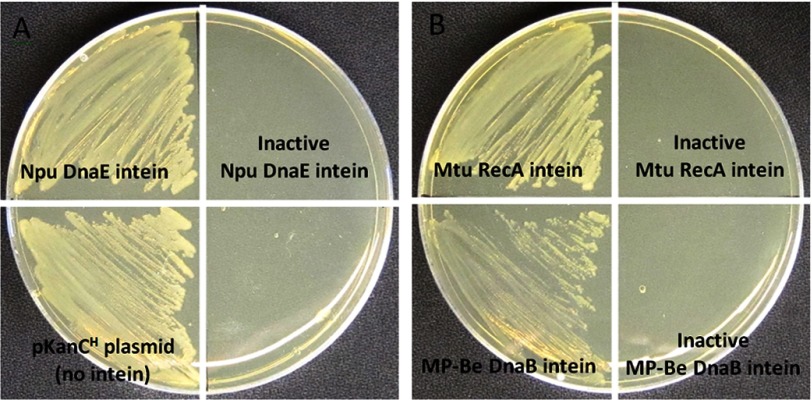
Inteins flanked by three native N- and C-terminal extein residues were cloned separately into site C or D and tested for the ability of splicing to restore KanR (Fig. 1). Catalytically inactivated inteins (see “Experimental Procedures”) were likewise inserted into the same sites to ensure that KanR required splicing. The inteins tested were the Mtu-H37Rv RecA intein flanked by N-terminal Lys-Asn-Lys and C-terminal Cys-Ser-Pro, the MP-Be DnaB intein flanked by N-terminal Gln-Asp-Gln and C-terminal Thr-Lys-Asn, and a cis construct of the naturally split Npu PCC73102 DnaE intein (16, 25) flanked by N-terminal Ala-Glu-Tyr and C-terminal Cys-Phe-Asn (Fig. 2 and data not shown). KanR was observed with the native inteins but not with the inactive inteins. These results validate the use of these two sites for assessing intein functionality. However, site C showed far greater tolerance for variable amino acid incorporation than site D and would thus make the best system for high throughput analysis of extein sequences.
FIGURE 2.
Kanamycin resistance is dependent on intein splicing in KanR site C. Three inteins were tested to ensure that resistance to kanamycin required a functional intein. E. coli were transformed with plasmids carrying the KanR gene with either an active or a catalytically inactive intein in site C. Growth after overnight incubation was then tested on plates with 40 μg/ml Kan. A, shown is growth at 37 °C with cells transformed with plasmids carrying the native Npu DnaE intein, an inactive Npu DnaE intein (C1A, N138D, C+1A), or a no intein control. B, shown is growth at 30 °C with cells transformed with plasmids carrying the native Mtu RecA intein, an inactive Mtu RecA intein (C1A, N440D, C+1A), the native MP-Be DnaB intein, or an inactive MP-Be DnaB intein (C320S).
Library Selection for Npu DnaE Intein Substrate Specificity
We constructed a library containing the intein flanked by three variable N-terminal extein residues labeled −3, −2, and −1 and two variable C-terminal extein residues labeled +2 and +3 (Fig. 1). NNK and MNN nucleotide sequences within the forward and reverse primers, respectively, were used to incorporate all possible amino acids at each variable codon. The +1 position cysteine (which immediately follows the intein C terminus) is a crucial nucleophile in the splicing mechanism, so this position was fixed. The maximal complexity of a library of 5 randomized amino acid is 3.2 × 106. A pilot study using the endogenous KanR promoter and the Npu DnaE intein resulted in ∼100,000 surviving colonies from a library of 1 × 107 clones when grown on 80 μg/ml Kan at 37 °C. Because we sought highly efficient splicing, we increased the stringency of the selection by altering the sequence of the ribosome binding site to reduce expression. By limiting production of the KanR precursor, we required efficient splicing to achieve the threshold needed to confer antibiotic resistance. Under these more stringent selection conditions, plating 5 × 107 transformants on 40 μg/ml Kan at 37 °C yielded ∼5000 colonies. To assess the diversity of this library, 124 plasmids were isolated from colonies that were not subjected to Kan selection. These non-selected sequences show all possible amino acids at each of the five variable positions including the naturally occurring extein residues, which indicates that the library was sufficiently diverse (data not shown).
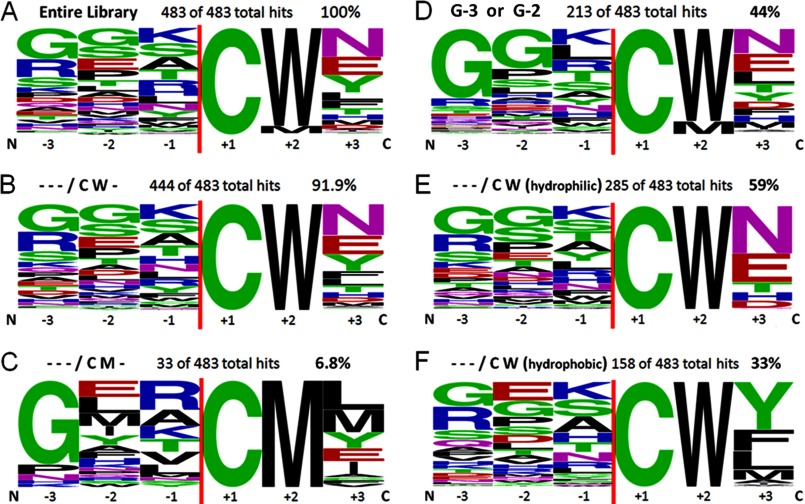
Because it is not practical to individually sequence all of the selected clones, the entire Aph and Npu DnaE intein precursor gene was sequenced from a representative, randomly chosen set of clones. Using an automated program we verified that 483 of the selected plasmids contained unambiguous sequences with no mutations in the Aph and the Npu DnaE intein genes. Examination of these extein sequences showed evidence of selection at some positions but not others. The largest specificity preferences were observed at the +2 and +3 positions of the C- extein (Table 1 and Fig. 3A). The natural splice site (Phe+2 and Asn+3) was not observed in the 483 sequenced clones. Instead, 92% of the selected sequences possessed Trp+2, and 7% contained Met+2 (Table 1 and Fig. 3). Phe, Val, Cys, and Lys were observed once, and Tyr was observed twice at the +2 position. The non-selected library confirmed that all residues were present at the +2 position in the expected quantities (data not shown), but Trp was selected over the naturally occurring Phe+2. The preference at the +3 position was for Tyr and residues similar to the natural Asn+3, with Asn+3, Asp+3, Gln+3, and Glu+3 representing 49% of the total hits (Table 1). The range of N-terminal extein sequences that allow splicing was very broad; however, it was evidently important to maintain flexibility at the N terminus because the selected hits have an abundance of Gly at the −3 and −2 positions (Table 1 and Fig. 3D).
TABLE 1.
Aggregate data for the plasmid population that survived KanR selection
Data for 483 randomly chosen clones selected for kanamycin resistance after the Npu DnaE intein with three flanking N- and C-terminal extcin amino acids at KanR site C was expressed using the RBS knockdown sequence. The theoretical occurrence value represents the number of times an amino acid should be observed in an unbiased library of 483 clones based on codon redundancy using NNK primers. The native flanking extein residues for the Npu DnaE intein residue are labeled with asterisks.
FIGURE 3.
Logo diagrams of flanking extein sequences for 483 randomly chosen positive hits selected for splicing of the Npu DnaE intein in KanR site C were made using the Weblogo program (28). The vertical red line indicates the position of the intein. The +1 position was fixed as Cys. A, shown is the entire library of 483 clones. B, shown are all clones with Trp+2. C, shown are all clones with Met+2. D, shown are all clones with Gly at −3 or −2. E, shown are all clones with Trp+2 and a hydrophilic residue at +3. F, shown is all clones with Trp+2 and a hydrophobic residue at +3.
Direct Observation of Splicing of the KanR-Npu DnaE Intein Fusion
Twenty positive hits representative of the range of sequence diversity found at the N- and C-terminal exteins were chosen for further analysis (Table 2). Cells containing these plasmids were grown in LB media plus Amp overnight, and Western blotting was used to directly confirm whether splicing occurred. Antibody probing based on the presence of an N-terminal His tag was used to detect precursor and the mature KanR product. All hits tested displayed robust in vivo splicing (∼95%) with no detectable amount of cleavage products (Fig. 4 and data not shown).
FIGURE 4.
Splicing of the Npu DnaE intein with the extein sequences listed in Table 2 fused to either KanR or luciferase. A, shown are KanR-Npu DnaE intein fusions E1–E8, the native extein (NE), or KanR with an N-terminal His tag but no intein (Kan). B, shown are KanR-Npu DnaE intein fusions E10–E20. Lane L contains the NEB prestained broad range (10–230 kDa) Ladder. Western blots A and B were probed with Mouse IgG anti-His tag antibody followed by detection with LI-COR IRDye 680 goat anti-mouse secondary antibody. C, shown are luciferase-Npu DnaE intein fusions with selected or native (NE) flanking extein sequences. Lane D, a mutated, catalytically inactive intein fusion is shown. The asterisk band is the result of proteolysis of the precursor in the inactive intein sample. Western blot C was probed with Mouse IgG anti-firefly luciferase antibody followed by detection with LI-COR IRDye 680 goat anti-mouse secondary antibody.
Transferability of Selected Sequences to Two Unrelated Precursors
To address the question of whether the KanR selection results provide details about the actual specificity of the intein or reflect the unique selective pressures found within KanR, we cloned a subset of the selection hits into firefly luciferase. These included the Npu DnaE intein plus the three natural N- and C-terminal extein residues, an inactive intein mutant, and nine selection hits (Fig. 4C). Western blot analysis with anti-luciferase antibody was used to ascertain the degree of splicing when precursors were expressed in NEB Express cells at 37 °C. All KanR selection hits tested showed a high degree of splicing in this unrelated enzyme.
Five selection hits that represent the most commonly selected motifs in our library were cloned into a third context, a split MIP system (described below). Here again, the selected extein sequences promoted efficient protein splicing (Table 3), indicating that these hits represent general specificity criteria for the Npu DnaE intein rather than a solution specific to KanR site C.
Kinetics of Trans-splicing of the Npu DnaE Intein with Selected Exteins
To further validate the selected extein sequences, we examined the kinetics of trans-splicing of the naturally split Npu DnaE intein using either the native flanking extein residues or sequences that represent commonly selected motifs (Table 3). As controls, an inactive Npu DnaE intein mutant and a variant identified in the low stringency pilot selection (EP) were also tested (Fig. 5). M-IN and IC-PHis fragments (Fig. 6A) were expressed separately in NEB Express cells by induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C for 4 h. Cells were harvested, and the kinetics of trans-splicing were assayed at 30 °C over a 10-min time course using the soluble fraction of the lysate. Samples taken at various time points were electrophoresed on SDS-PAGE followed by Western blot analysis to quantify spliced products (Fig. 6B). In the homologous Ssp DnaE intein, association of the IN and IC fragments was shown to approach diffusion-controlled limits (26, 27). Assuming that the Npu DnaE intein IN and IC fragments associate in a similarly rapid manor, the kinetics of trans-splicing can be fit to a first order decay reaction when the concentration of M-IN is maintained at excess over the concentration of IC-PHis.
FIGURE 5.
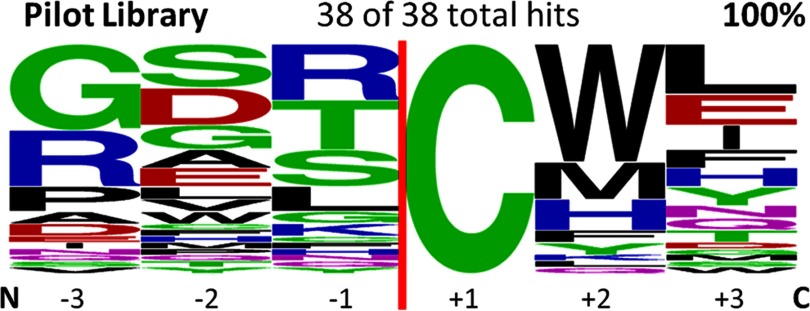
Logo diagram of flanking extein sequences identified in the low stringency pilot study with the native KanR RBS reveals expanded specificity at the +2 and +3 positions. This figure was made using the Weblogo program (28) and shows 38 randomly chosen positive hits selected for splicing of the Npu DnaE intein in KanR site C. This construct results in expression of more KanR protein than needed for resistance and, therefore, allows selection of precursors that splice slowly or with a low percentage of spliced product (data not shown). The vertical red line indicates the position of the intein. The +1 position was fixed as Cys.
FIGURE 6.
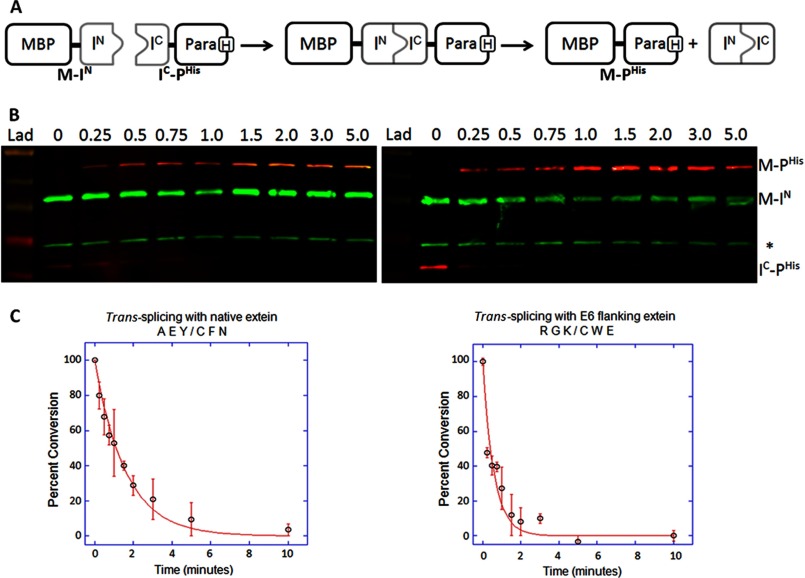
Kinetic analysis of trans-splicing inteins with selected flanking extein sequences. A, shown is a schematic representation of the trans-splicing reaction. The N-terminal precursor fragment (M-IN) consists of the maltose binding protein (M or MBP) followed by a short linker, three native or selected extein residues, and the N-terminal half of the Npu DnaE intein (IN: residues 1–102). The C-terminal precursor fragment (IC-PHis) consists of the C-terminal half of the Npu DnaE intein (IC; residues 103–138), three native or selected extein residues, a short linker, the ΔSal fragment of paramyosin (P) and a C-terminal His tag. M-IN and IC-PHis spontaneously assemble when mixed resulting in intein activation and protein splicing. Any unreacted precursor complex and the IN-IC product complex dissociate during SDS-PAGE sample preparation. B, Western blots show the time course of trans-splicing with native (Ala-Glu-Tyr/Cys-Phe-Asn, left panel) or E6 (Arg-Gly-Lys/Cys-Trp-Glu, right panel) flanking extein residues. Time points from 0 to 5 min are listed across the top of each blot. The nitrocellulose membranes were simultaneously probed with mouse IgG anti-His tag antibody and rabbit anti-MBP antiserum followed by detection with LI-COR IRDye 680 goat anti-mouse (red) and IRDye 800 goat anti-rabbit secondary antibodies (green). The band labeled M-PHis was quantified to measure the rate of spliced product formation. The asterisk marks endogenous E. coli MBP. Lane Lad contains the NEB prestained broad range (10–230 kDa) ladder. C, shown are plots of the percent conversion of precursor to product during the trans-splicing time courses with native (left panel) or E6 (right panel) flanking extein residues. The red line represents the fit of the data to a first order decay reaction, and the S.D. is indicated by the error bars.
In our system the Npu DnaE intein with native flanking extein residues has fast kinetics for trans-splicing, with an apparent first order rate constant of 0.63 ± 0.07 min−1 and a t½ of 66 s at 30 °C (Table 3). This value is in good agreement with the previously published rate constant of 0.66 ± 0.12 min−1 and a t½ of 63 s at 37 °C (18, 20). The inactive intein showed no observable M-PHis spliced product or disappearance of starting materials M-IN or IC-PHis over a 1-h time course. The EP sample from the low stringency pilot study yielded incomplete splicing with a k of 0.06 ± 0.004 min−1 and a t½ of 11.5 min (Table 3). The majority of the assayed hits (E2, E4, E6, and E16) displayed comparable splicing rates to the Npu DnaE intein with its native extein sequence (Table 3). The apparent first order rate constant for E6 (extein sequence Arg-Gly-Lys/Cys-Trp-Glu) is 1.6 ± 0.2 min−1 with a t½ of 28 s at 30 °C, which is 2.5 times faster than that of the native extein.
DISCUSSION
One of the greatest challenges in the study of inteins is separating the contribution to splicing rates of the extein versus the intein. To address this problem, we designed a high throughput system where a random six-amino acid substitution did not detectably inhibit host protein function, hypothesizing that this would allow us to study intein specificity requirements without significant bias from the extein. However, finding such sites is complicated, as most locations that meet these criteria fail for other reasons: (a) splicing does not occur, (b) activity of the host protein is not disrupted when the intein is present, or (c) splicing does not rescue host protein activity. In a study of 12 sites in firefly luciferase (9 replacement and 11 insertion libraries) none of the tested sites fully satisfied these criteria (data not shown). However one site in KanR met all criteria for a specificity assay, which was critical to the success of this study.
The Npu DnaE intein specificity results in KanR were validated by (a) the absence of bias as evidenced by 91% of KanR C site random cassettes maintaining kanamycin resistance, (b) transfer of selected extein sequences to two unrelated host proteins promoted similar levels of splicing, and (c) splicing rates equivalent to or greater than the naturally occurring exteins in trans-splicing experiments in a different extein protein.
Our specificity data provide a predictive model to scan available target protein sequence space to find or engineer intein insertion sites where we have confidence that splicing should occur. It appears that efficient splicing requires the native C-extein Cys-Phe-Asn or the genetically selected C-extein Cys-Trp and Cys-Met plus one of several acceptable +3 residues. Splicing experiments of the Npu DnaE intein within luciferase in principle validates the use of this predictive approach, albeit the targeted extein sequence was artificially inserted into the enzyme. However, the specificity data alone do not solve all problems when inserting inteins in heterologous proteins. Intein insertion is a complicated balance between expression, stability, and folding in addition to the requirements for splicing. These criteria must still be experimentally probed, but the specificity data provides more opportunities to find sites that permit splicing.
The Preferred Substrate Is Not the Native Extein
All orthologs of the DnaE intein have the same flanking extein sequence (1, 20). Previous studies suggested that DnaE inteins have minimal specificity requirements for the N-terminal extein, but the Cys-Phe-Asn C-terminal extein sequence is required for efficient catalysis (4). The Npu DnaE intein can also support splicing when the +2 position was mutated to Gly, Glu, or Arg, although with significantly lower efficiencies (20). When Iwai et al. (17) examined the cross-reactivity of Npu DnaE IN with Ssp DnaE IC where the +2 position was mutated to all 20 amino acids, trans-splicing was observed with most uncharged residues after a 6-h induction. Our study is consistent with these findings while also defining alternative C-extein sequences that permit very efficient splicing.
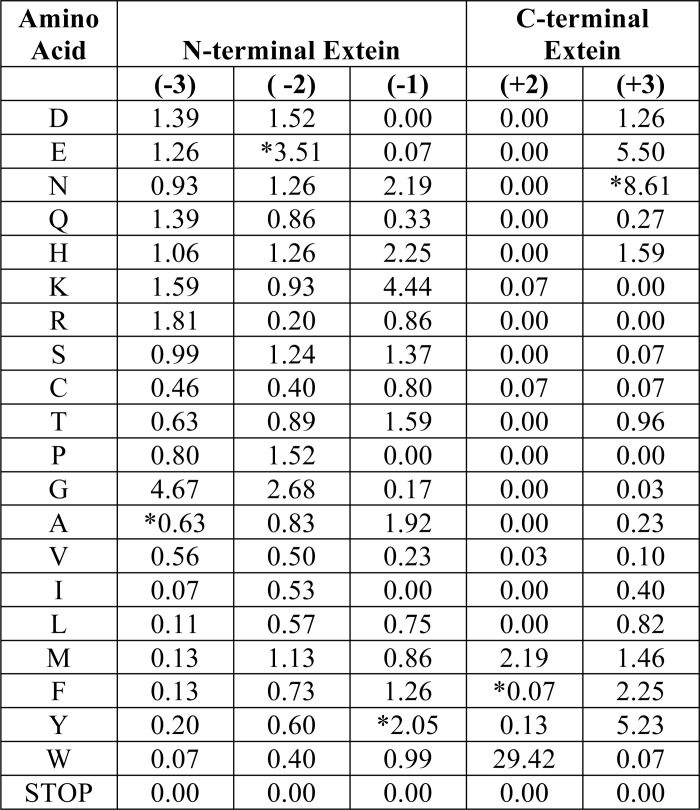
To aid in comparison of selective pressure at each position, we calculated a selectivity ratio (Table 4) that equals the observed frequency of an amino acid divided by the theoretical frequency based on codon degeneracy. A selectivity ratio of ∼1.0 indicates an unbiased representation, whereas higher values indicate greater selective pressure for the given amino acid, and lower values indicate selective pressure against that residue or overwhelming selective pressure for another amino acid. The most pronounced substrate specificity requirements for the Npu DnaE intein were found at the +2 and +3 positions. To our surprise, the specificity was for Trp+2 rather than the naturally occurring Phe+2, although all residues were present in the non-selected sample (data not shown). The selectivity ratio for Trp+2 was 29.4, whereas Phe+2 was 0.07 (Table 4). A lower stringency pilot selection yielded a relaxed specificity at +2 and +3 (Fig. 5) that is consistent with the findings of Iwai et al. (17).
TABLE 4.
Selectivity ratio for the plasmid population that survived kanamycin selection
Data for the 483 randomly chosen clones in Table 1 are shown. The selectivity ratio is defined as the observed frequency divided by the theoretical frequency. Selectivity ratio values near or equal to 1 indicate no selection. Higher selectivity ratios indicate stronger selective pressure for that amino acid, whereas values under 1 indicate negative selective pressure for that amino acid or high selective pressure for another amino acid. The native flanking Npu DnaE extein residues are labeled with asterisks. The +1 position was fixed to the native Cys residue and was thus not included in this table. The intein is present between the −1 and +1 position.
The +3 position shows significant selective pressure for Asn, Glu, and Tyr reflected in selectivity ratios of 8.61, 5.5, and 5.23 respectively (Table 4). Glu is similar to the native Asn+3 residue, whereas Tyr may reflect a distinct specificity. The charge or polarity of the +3 amino acid does not have a significant effect on the identity of any position in the N-terminal extein (Fig. 3, E and F), suggesting that there is no direct amino acid side chain cross-talk between the two halves of the extein or that such cross-talk is highly context-dependent. The only pattern that emerges is the abundance of Gly in the N-terminal extein (Fig. 3D and Table 4), with 44% of the selected population containing Gly at either −3 or −2. These two positions have an unbiased frequency for Gly in the unselected control library. Presumably, the intein active site needs some flexibility in the adjacent N-extein to facilitate splicing.
A second specificity group representing only ∼7% of the selected clones has Met at +2. The Met+2 specificity group has a different requirement than the Trp+2 group, preferring a large hydrophobic residue at +3 and a very strong preference for Gly, but only at −3 (Fig. 3C).
The relatively broad specificity observed in the Npu DnaE intein is not unprecedented (20). Selective substrate sequestration is critical for enhancing catalytic efficiency and preventing undesirable side reactions in multiple turnover enzymes. In contrast, such sophisticated molecular recognition strategies are not necessary in single turnover enzymes where the substrate is covalently linked to the enzyme, allowing inteins more potential variability in substrate specificity. Furthermore, selective pressure on mobile elements such as inteins may require a higher degree of substrate promiscuity to enable invasion of new sites.
A Paradigm Shift; the Native Extein Is Not Always the Best
In most enzyme systems the natural substrate is the most efficient substrate for the enzyme. However, selection with the Npu DnaE intein did not pull out the native extein sequence. Even more surprising is that we identified extein sequences that surpass the rate of splicing measured with the naturally occurring N- and C- terminal extein. Clearly the paradigm that the optimal splicing site for an intein is its natural context does not hold true in this case. Because natural intein insertion sites are an evolutionary balance between the specificity requirements of the intein, the need to retain a functional host protein, and the specificity requirement of the homing endonuclease, the natural splice site is narrowly defined. However, this narrow specificity does not necessarily reflect the optimal splicing condition for the intein. Our study decouples the specificity requirements of the intein from effects caused by the host protein or homing endonuclease and so provides a more accurate picture of this intein preferred substrates. These data expand the range of known splicable sequences for the Npu DnaE intein and should prove valuable for the reproducible design of heterologous splicing experiments.
Although our studies focused on only one intein, the conclusion that the natural intein insertion sequence only reflects a subset of efficiently splicable extein sequences can likely be generalized to other inteins. As such, it is important to reexamine the substrate specificity of commonly used inteins to understand their true catalytic potential.
Conclusion
We examined the substrate specificity of the Npu DnaE intein under highly stringent conditions and demonstrated that the fastest splicing rates were obtained with extein sequences that are significantly different from the native extein. Whereas previous studies suggested that a C-terminal Cys-Phe-Asn sequence was necessary to achieve highly efficient and rapid splicing (4, 20), our data show that sequences containing Trp and Met at +2 can splice as efficiently as the native extein. These data indicate that the selective pressure to maintain the natural extein sequence overrides the pressure to optimize catalytic efficiency of this intein. These data expand the range of sequences known to allow splicing of this important intein and provide a rubric for evaluating potential intein insertion sites for protein splicing technologies without having to mutate the intein to splice at a desired insertion site sequence.
Acknowledgments
We thank Prof. Philip Farabaugh (University of Maryland, Baltimore County) for generously donating the pEK4 plasmid. We thank Richard Roberts, Gregory Lohman, Paul Balbo, and Andy Gardner (all from NEB) for reading the manuscript and helpful discussion. We thank Don Comb (NEB) for support.
M. Cheriyan and F. Perler, unpublished data.
- Kan
- kanamycin
- KanR
- kanamycin resistance protein or kanamycin resistance
- Aph
- aminoglycoside phosphotransferase
- Amp
- ampicillin
- EP
- a selected clone from the low stringency pilot selection experiment
- IC
- the C-terminal fragment of the split Npu DnaE intein
- IC-PHis
- IC fused to the Δ Sal fragment of paramyosin with a C-terminal His tag
- IN
- the N-terminal fragment of the split Npu DnaE intein
- M or MBP
- E. coli maltose-binding protein
- M-IN
- maltose-binding protein fused to IN
- M-PHis
- spliced product where the maltose-binding protein is fused to the ΔSal fragment of paramyosin
- MIP
- maltose-binding protein fused to an intein fused to the ΔSal fragment of paramyosin
- MP-Be
- Mycobacteriophage Bethlehem
- Mtu
- Mycobacterium tuberculosis
- NEB
- New England Biolabs
- Npu
- Nostoc punctiforme
- RBS
- ribosome binding site.
REFERENCES
- 1. Perler F. B. (2002) InBase. The intein database. Nucleic Acids Res. 30, 383–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tori K., Dassa B., Johnson M. A., Southworth M. W., Brace L. E., Ishino Y., Pietrokovski S., Perler F. B. (2010) Splicing of the mycobacteriophage Bethlehem DnaB intein. Identification of a new mechanistic class of inteins that contain an obligate block F nucleophile. J. Biol. Chem. 285, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gangopadhyay J. P., Jiang S. Q., van Berkel P., Paulus H. (2003) In vitro splicing of erythropoietin by the Mycobacterium tuberculosis RecA intein without substituting amino acids at the splice junctions. Biochim. Biophys. Acta 1619, 193–200 [DOI] [PubMed] [Google Scholar]
- 4. Lockless S. W., Muir T. W. (2009) Traceless protein splicing utilizing evolved split inteins. Proc. Natl. Acad. Sci. U.S.A. 106, 10999–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dalgaard J. Z., Moser M. J., Hughey R., Mian I. S. (1997) Statistical modeling, phylogenetic analysis, and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J. Comput. Biol. 4, 193–214 [DOI] [PubMed] [Google Scholar]
- 6. Flavell R. R., Muir T. W. (2009) Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc. Chem. Res. 42, 107–116 [DOI] [PubMed] [Google Scholar]
- 7. Fong B. A., Wu W. Y., Wood D. W. (2010) The potential role of self-cleaving purification tags in commercial-scale processes. Trends Biotechnol. 28, 272–279 [DOI] [PubMed] [Google Scholar]
- 8. Volkmann G., Iwaï H. (2010) Protein trans-splicing and its use in structural biology. Opportunities and limitations. Mol. Biosyst. 6, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 9. Mootz H. D. (2009) Split inteins as versatile tools for protein semisynthesis. Chembiochem 10, 2579–2589 [DOI] [PubMed] [Google Scholar]
- 10. Amitai G., Callahan B. P., Stanger M. J., Belfort G., Belfort M. (2009) Modulation of intein activity by its neighboring extein substrates. Proc. Natl. Acad. Sci. U.S.A. 106, 11005–11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lew B. M., Paulus H. (2002) An in vivo screening system against protein splicing useful for the isolation of non-splicing mutants or inhibitors of the RecA intein of Mycobacterium tuberculosis. Gene 282, 169–177 [DOI] [PubMed] [Google Scholar]
- 12. Oeemig J. S., Zhou D., Kajander T., Wlodawer A., Iwaï H. (2012) NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J. Mol. Biol. 421, 85–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shemella P. T., Topilina N. I., Soga I., Pereira B., Belfort G., Belfort M., Nayak S. K. (2011) Electronic structure of neighboring extein residue modulates intein C-terminal cleavage activity. Biophys. J. 100, 2217–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chong S., Montello G. E., Zhang A., Cantor E. J., Liao W., Xu M. Q., Benner J. (1998) Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acids Res. 26, 5109–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Appleby-Tagoe J. H., Thiel I. V., Wang Y., Wang Y., Mootz H. D., Liu X. Q. (2011) Highly efficient and more general cis- and trans-splicing inteins through sequential directed evolution. J. Biol. Chem. 286, 34440–34447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oeemig J. S., Aranko A. S., Djupsjöbacka J., Heinämäki K., Iwaï H. (2009) Solution structure of DnaE intein from Nostoc punctiforme. Structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 583, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 17. Iwai H., Züger S., Jin J., Tam P. H. (2006) Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 18. Zettler J., Schütz V., Mootz H. D. (2009) The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 [DOI] [PubMed] [Google Scholar]
- 19. Carvajal-Vallejos P., Pallissé R., Mootz H. D., Schmidt S. R. (2012) Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J. Biol. Chem. 287, 28686–28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shah N. H., Dann G. P., Vila-Perelló M., Liu Z., Muir T. W. (2012) Ultrafast protein splicing is common among cyanobacterial split inteins. Implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nurizzo D., Shewry S. C., Perlin M. H., Brown S. A., Dholakia J. N., Fuchs R. L., Deva T., Baker E. N., Smith C. A. (2003) The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327, 491–506 [DOI] [PubMed] [Google Scholar]
- 22. Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. (1986) Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131 [DOI] [PubMed] [Google Scholar]
- 23. Kramer E. B., Farabaugh P. J. (2007) The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daugelat S., Jacobs W. R., Jr. (1999) The Mycobacterium tuberculosis recA intein can be used in an ORFTRAP to select for open reading frames. Protein Sci. 8, 644–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tori K., Cheriyan M., Pedamallu C. S., Contreras M. A., Perler F. B. (2012) The Thermococcus kodakaraensis Tko CDC21–1 intein activates its N-terminal splice junction in the absence of a conserved histidine by a compensatory mechanism. Biochemistry 51, 2496–2505 [DOI] [PubMed] [Google Scholar]
- 26. Shi J., Muir T. W. (2005) Development of a tandem protein trans-splicing system based on native and engineered split inteins. J. Am. Chem. Soc. 127, 6198–6206 [DOI] [PubMed] [Google Scholar]
- 27. Martin D. D., Xu M. Q., Evans T. C., Jr. (2001) Characterization of a naturally occurring trans-splicing intein from Synechocystis sp. PCC6803. Biochemistry 40, 1393–1402 [DOI] [PubMed] [Google Scholar]
- 28. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]