Background: Asymmetric dimethylarginine (ADMA) can induce endothelial nitric-oxide synthase (eNOS) redistribution from the plasma membrane to the mitochondria.
Results: AMDA induces nitration of Akt1 at Tyr350 within the client-binding domain, increasing its activation and enhancing eNOS phosphorylation.
Conclusion: Under physiologic conditions, Akt1-mediated redistribution of eNOS to the mitochondria enhances mitochondrial coupling.
Significance: Reducing Akt1 nitration may reduce the deleterious effects of Akt1 signaling in various pathologies.
Keywords: Akt, Bioenergetics, Endothelium, Mitochondria, Nitric-oxide Synthase
Abstract
We have recently demonstrated that asymmetric dimethylarginine (ADMA) induces the translocation of endothelial nitric-oxide synthase (eNOS) to the mitochondrion via a mechanism that requires protein nitration. Thus, the goal of this study was elucidate how eNOS redistributes to mitochondria and to identify the nitrated protein responsible for this event. Our data indicate that exposure of pulmonary arterial endothelial cells to ADMA enhanced eNOS phosphorylation at the Akt1-dependent phosphorylation sites Ser617 and Ser1179. Mutation of these serine residues to alanine (S617A and S1179A) inhibited nitration-mediated eNOS translocation to the mitochondria, whereas the phosphormimic mutations (S617D and S1179D) exhibited increased mitochondrial redistribution in the absence of ADMA. The overexpression of a dominant-negative Akt1 also attenuated ADMA-mediated eNOS mitochondrial translocation. Furthermore, ADMA enhanced Akt1 nitration and increased its activity. Mass spectrometry identified a single nitration site in Akt1 located at the tyrosine residue (Tyr350) located within the client-binding domain. Replacement of Tyr350 with phenylalanine abolished peroxynitrite-mediated eNOS translocation to mitochondria. We also found that in the absence of ADMA, eNOS translocation decreased mitochondrial oxygen consumption and superoxide production without altering cellular ATP level. This suggests that under physiologic conditions, eNOS translocation enhances mitochondria coupling. In conclusion, we have identified a new mechanism by which eNOS translocation to mitochondria is regulated by the phosphorylation of eNOS at Ser617 and Ser1179 by Akt1 and that this is enhanced when Akt1 becomes nitrated at Tyr350.
Introduction
Nitric oxide (NO) is a potent signaling molecule produced by NO synthase (NOS) and plays diverse roles in biological processes such as neurotransmission, inflammation, vasodilation, and vascular homeostasis (1–3). There are 3 isoforms of NOS: neuronal, endothelial, and inducible. Endothelial NOS is the principle isoform in the vasculature (4). NOS catalyzes the 5-electron oxidation of l-arginine to produce NO and l-citrulline (5). Asymmetric dimethylarginine (ADMA)2 is an endogenous inhibitor of eNOS that impairs NO generation by competing with its substrate, l-arginine. A number of studies demonstrate a relationship between elevated ADMA levels and cardiovascular diseases such as hypercholesterolemia, hyperhomocysteinemia, diabetes mellitus, pulmonary hypertension, chronic heart failure, and coronary artery disease (6–10). ADMA inhibits NO production causing vasoconstriction and the elevation of blood pressure leading to endothelial dysfunction (11, 12). We have recently shown that ADMA induces eNOS uncoupling and redistribution of eNOS from the plasma membrane to the mitochondrion (13). However, the molecular mechanisms by which this occurs remains unresolved. Previous studies have identified a region within eNOS contained within amino acids 628–632 as being important for docking of eNOS to the outer mitochondrial membrane in endothelial cells (14). However, the signaling molecules that are involved and the impact of ADMA on this process has not been elucidated. Mitochondria, which are the major sites of ATP generation in the cell, are also a site of production of reactive oxygen species (ROS) (15). Excessive production of ROS results in oxidative stress leading to mitochondrial damage and dysfunction, which contributes to endothelial dysfunction associated with cardiovascular disorders (16–20). The contribution of mitochondria to vascular pathophysiology has been gaining increasing attention.
In the current study, we have elucidated the molecular mechanisms by which ADMA induces mitochondrial translocation of eNOS. Herein we illustrate a novel pathway involving ADMA-mediated Akt nitration and its subsequent activation. We also identify the tyrosine residue of Akt that is nitrated and also investigate its significance in regulating eNOS translocation to mitochondria and its effect on mitochondrial function.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
Primary cultures of ovine pulmonary artery endothelial cells (PAEC) were isolated as described previously (21). Cells were maintained in DMEM containing phenol red supplemented with 10% fetal calf serum (Hyclone, Logan, UT), antibiotics, and antimycotics (MediaTech, Herndon, VA) at 37 °C in a humidified atmosphere with 5% CO2, 95% O2. Cells used in these experiments were between passage 8 and 12, seeded at ∼50% confluence, and utilized when fully confluent. HEK 293 cells were cultured in DMEM + 4.5 g of d-glucose containing 10% fetal calf serum. Cells were treated with ADMA or the peroxynitrite donor, 3-morpholinosydnonimine hydrochloride (SIN-1) for 1 h. SIN-1 was dissolved in PBS just before treatment.
Generation of Akt and eNOS Mutant Proteins and Cell Transfection
The mutation of the tyrosine residue located at position 350 in mouse Akt1 to phenylalanine and the mutations of lysine 612 of human eNOS to histidine (K612H eNOS), aspartic acid (K612D eNOS), and alanine (K612A eNOS) and Y350F in mouse Akt1 in pcDNA 3.1 plasmids were generated commercially (Retrogen). Bovine eNOS mutations at serine 617 to aspartic acid (S617D) or alanine (S617A) and serine 1179 to aspartic acid (S1179D) or alanine (S1179A) in pcDNA 3.1 plasmids were generated commercially by GENEWIZ. PAEC and HEK 293 cells were transfected with plasmids using Effectene reagent (Qiagen) for 48 h. Although both human and bovine eNOS mutants were utilized, for simplicity, the eNOS amino acids are identified using the bovine nomenclature and phosphorylation sites. The Akt1 dominant-negative mutant adenovirus (Akt1 K179A) was obtained from Biomyx. The eNOS-GFP construct, a gift from Dr. William Sessa (Yale University), was used as described previously (22).
Measurement of Nitrite Accumulation
To measure nitrite accumulation in the culture media we utilized a chemiluminescence method. 0.05 g of potassium iodide (KI) was dissolved in 7 ml of freshly prepared acetic acid. The KI/acetic acid reagent was added to a septum-sealed purge vessel and bubbled with nitrogen gas. The gas stream was connected via a trap containing 1 n NaOH to a Sievers 280i Nitric Oxide Analyzer (GE Healthcare). Samples were injected with a syringe through a silicone/Teflon septum. Results were analyzed by measuring the area under curve of the chemiluminescence signal using the Liquid software (GE Healthcare).
Live Cell Imaging
PAEC were grown until 50% confluence and transfected with the eNOS-GFP construct. Cells were allowed to grow for 24 h, collected by trypsinization, resuspended in media, seeded onto the Lab-Tek chamber slides (Termo Scientific), and allowed to attach overnight. PAEC were then incubated with 100 nm MitoTracker® Red FM (Invitrogen) for 30 min at 37 °C, the media was changed to low glucose DMEM without l-arginine (AthenaESTM) and cells were incubated for a further 24 h. Cells were then exposed to ADMA (0–10 μm) or SIN-1 (1 mm) and imaged over time using a computer-based confocal DeltaVision imaging system (Applied Precision Inc.) using appropriate excitation and emission wavelengths for red and green fluorescence. Cells were imaged every 10 min for the duration of the study. Software SoftWorx 3.6.2 was used to trace each cell and calculate the Pearson product-moment correlation coefficient. In additional sets of experiments, cells were pretreated (30 min) with NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (carboxy-PTIO, 100 μm), then exposed to either ADMA (10 μm) or SIN-1 (1 mm). The effect of the NO donor spermine NONOate (25 μm) was also studied using the same experimental design.
Immunoprecipitation and Western Blotting
Immunoprecipitation and immunoblotting were performed as described previously (23). Briefly, PAEC were serum-starved for 16 h in l-arginine-free DMEM, then exposed to ADMA (10 μm, 1 h). Lysis buffer containing 1% Triton X-100, 20 mm Tris, pH 7.4, 100 mm NaCl, 1 mm EDTA, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor mixture (Pierce) was then added and insoluble proteins were precipitated by centrifugation at 13,000 × g for 10 min at 4 °C. The supernatants were then incubated overnight with an anti-3-NT antibody (2 μg, 4 °C) followed by incubation in Protein G plus Protein A-agarose (Calbiochem) for 2 h at room temperature. The immune complexes were then precipitated by centrifugation, washed three times with lysis buffer, boiled in SDS sample buffer, and subjected to SDS-PAGE using 4–12% polyacrylamide gels and then transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween (TBST). The primary antibodies used for immunoblotting were anti-Akt (Cell Signaling), anti-phospho-GSK3β (Cell Signaling), anti-eNOS (BD Bioscience), anti-phospho-S1177 eNOS (Cell Signaling), and anti-phospho-S617 eNOS (Millipore). The membranes were then washed with TBST (3 times for 10 min), incubated with secondary antibodies coupled to horseradish peroxidase, washed again with TBST (3 times for 10 min), and the protein bands were visualized using the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) on a Kodak 440CF image station. Band intensity was quantified using Kodak one-dimensional image processing software. Protein loading was normalized by reprobing membranes with β-actin (Sigma). In the studies using purified mitochondrial fractions protein loading was normalized by reprobing membranes using an antibody specific for VDAC (Cell Signaling). Purity of the mitochondria fraction was assessed by reprobing membranes with NaK-ATPase (Cell Signaling) (cellular membrane marker) or lamin B1 (Cell Signaling) (nuclei membrane marker). Total cell lysates were used as a positive control for each antibody.
Analysis of Akt Activity
Recombinant Akt1 kinase activity was measured using the ELISA-based Akt/PKB kinase activity assay kit (Enzo Biochem). Briefly, the biotinylated peptide substrate for Akt was incubated with 1 μg of purified Akt1 for 30 min at 30 °C. After the kinase reaction was stopped, the samples were transferred to a streptavidin-coated 96-well plate and incubated with a phosphoserine detection antibody followed by incubation with a horseradish peroxidase (HRP)-conjugated antibody. Finally, the HRP substrate 3,3′,5,5′-tetramethylbenzidine was added to the wells and incubated for color development at room temperature for 1 h. Absorbance at 450 nm was then recorded and Akt activity was expressed as A450 nm/μg of Akt1 protein. In addition, Akt activity in PAEC was determined using an Akt kinase assay kit (Cell Signaling Technology) according to the manufacturer's protocol. Briefly, Akt1 was immunoprecipitated from cell extracts using an Akt1-specific antibody cross-linked to agarose beads. Then an in vitro kinase assay was performed using GSK-3 fusion protein as a substrate. Phosphorylation of GSK-3 was then measured by Western blotting using a phospho-GSK-3 (Ser21/9) antibody.
Determination of ATP Levels
ATP levels were estimated using the firefly luciferin-luciferase method utilizing a commercially available kit (Invitrogen). ATP is consumed and light is emitted when firefly luciferase catalyzes the oxidation of luciferin. The amount of light emitted during the reaction is proportional to the availability of ATP. Luminescence was measured using a Fluoroscan Ascent FL luminometer (Thermo Electron).
Determination of Mitochondrial Superoxide Levels
MitoSOXTM Red mitochondrial superoxide indicator (Molecular Probes), a fluorogenic dye for selective detection of superoxide in the mitochondria of live cells was used. Briefly, cells were washed with fresh media, and then incubated in media containing MitoSOX Red (2.5 μm) for 30 min at 37 °C in dark conditions, then subjected to fluorescence microscopy at an excitation of 510 nm and an emission at 580 nm. An Olympus IX51 microscope equipped with a CCD camera (Hamamatsu Photonics) was used for acquisition of fluorescent images. The average fluorescent intensities (to correct for differences in cell number) were quantified using ImagePro Plus version 5.0 imaging software (Media Cybernetics).
Mitochondrial Isolation
Mitochondria were isolated from cells using a Mitochondria Isolation kit (Pierce). Briefly, 2 × 107 cells were pelleted by centrifugation at 850 × g for 2 min, and Mitochondria Isolation Reagent A was added to the pellet. Cells were vortexed for 5 s and then incubated on ice for 2 min. Mitochondria Isolation Reagent B was then added followed by vortexing for 5 s. Samples were then were incubated on ice for 5 min with vortexing every minute. Mitochondria Isolation Reagent C was added, and the samples were inverted several times to mix. The supernatant obtained by centrifugation at 700 × g (10 min at 4 °C) was transferred to a new tube and centrifuged at 3,000 × g for 15 min. Mitochondria Isolation Reagent C was added to the pellet and centrifuged at 12,000 × g for 5 min. The pellet contains the mitochondrial fraction. The purity of the mitochondrial fraction and equal loading in the Western blot analyses was confirmed by reprobing the membranes with antibodies specific to NaK-ATPase (membrane fraction) and lamin B1 (nuclear fraction). To ensure NaK-ATPase and lamin B1 antibody immunoreactivity, we also ran total cell lysates.
Analysis of Mitochondrial Bioenergetics
The XF24 Analyzer (Seahorse Biosciences, North Billerica, MA) was used to estimate the effect of eNOS mitochondrial translocation on mitochondrial bioenergetics in HEK 293 cells. XF24 measures the oxygen consumption rate (OCR). Initial studies identified the optimum number of cells/well as 25,000/0.32 cm2. Cells were transfected with wild-type eNOS and eNOS mutants S617D and S1179D 48 h prior to the experiment and were seeded on XF24 culture microplates 4 h before measurements were begun. The culture medium was changed to Dulbecco's modified Eagle's medium (DMEM, pH 7.4) supplemented with 5 mm glucose and 2 mm sodium pyruvate. The XF24 culture microplates were then incubated in a CO2-free XF prep station at 37 °C for 40 min to allow temperature and pH calibration. We then determined basal mitochondrial respiration as picomoles/min of oxygen consumed.
In-gel Digestions of Akt1
HEK 293 cells overexpressing Akt1 were exposed or not to SIN-1 (0.5 mm) for 1 h, then lysed, Akt1 was then immunoprecipitated and subjected to SDS-gel electrophoresis. Protein bands were then visualized by Coomassie staining and the band corresponding to Akt1 was excised. Bands were destained and alkylated with iodoacetamide. The samples were then subjected to overnight in-gel digestion with trypsin (25 ng/μl) in 25 mm ammonium bicarbonate buffer (pH 7.8). Peptides were extracted with 5% formic acid, 50% acetonitrile and evaporated to near dryness. Mass spectrometry analysis of untreated and nitrated Akt was then performed.
MALDI-TOF-TOF Mass Spectrometry
Peptide calibration standards and matrix CHCA were from Applied Biosystems (Carlsbad, CA). All spectra were taken on an ABSciex 5800 MALDI-TOF-TOF mass spectrometer in positive reflector mode (10 kV) with a matrix of CHCA. At least 1000 laser shots were averaged to obtain each spectrum. Masses were calibrated to known peptide standards. Aliquots of the Akt1 tryptic digest (5 μl) were cleaned on a C18 ZipTip (Millipore, Bedford MA) as per the manufacturer's instructions. Bound peptides were desalted with two 5-μl washes of 0.1% TFA and then eluted with 2.5 μl of aqueous, acidic acetonitrile (75% CH3CN, 0.1% TFA). The eluate was mixed with 2.5 μl of freshly prepared CHCA stock solution (20 mg/ml of CHCA in aqueous acetonitrile as above), and 1.5-μl portions of this mixture were spotted onto a MALDI sample plate for air drying. Additionally, 1.5 μl of crude peptides were mixed with 1.5 μl of CHCA and spotted. MS/MS of the 1700.5 m/z peak was performed in positive reflector mode without collision-induced dissociation. MS and MS/MS spectra were analyzed in Protein Pilot 3.0, Mascot Distiller, and PEAKS software packages.
Molecular Modeling
To identify the interaction sites between Akt1 and eNOS we built a homology model of Akt1 with an attached consensus sequence of eNOS phosphorylation site. We used the available structure of Akt1 (PDB code 3CQU). This structure contains a gap between aa 446 and 463 and an inhibitory peptide bound to the active site. To model this structure we utilized Yasara software (24). The geometry of the reconstructed region of human Akt1 was automatically optimized using the steepest descent energy minimization algorithm in the solvent implicit model. In the obtained structure of Akt1, the peptide bound to the catalytic domain was mutated to the phosphorylation motif of eNOS at Ser617 (612KIRFNSIS617). To identify possible interference between phosphorylation of Ser617 and mitochondrial translocation sequence of eNOS, we reconstructed the human eNOS reductase domain. Utilizing the crystal structure of the nNOS reductase domain as a template (PDB code 1TTL) we built a homology model for the human eNOS sequence. The final geometry of the eNOS reductase domain was again minimized using the steepest descent energy minimization algorithm available in the automatic homology modeling of Yasara. Molecular dynamic (MD) simulations were performed using Yasara structure. The Amber 99 all-hydrogen force field was used in the runs (25). The simulations for 10 ns at 298 K were carried out using the structure with and without a nitro group at Tyr350 of Akt1. Simulations were carried out within a simulation cube filled with water molecules. All the atoms were free to move. Snapshots were saved every 2 ns. Final structures after MD were analyzed for contact energy between the eNOS peptide (612KIRFNSIS617) and Akt1 using the algorithm in Yasara and changes in the binding energy between nitrated and intact Tyr350 of Akt were identified. The eNOS autoinhibitory loop simulation were carried out in the same settings. MD simulations were utilized for the reductase domain of eNOS from homology modeling. All amino acids except the autoinhibitory loop (aa 600–643) were fixed. Two runs (2 ns) were performed for unmodified eNOS and for eNOS with phosphorylated Ser617 in the Yasara structure.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 4.01 (GraphPad Software, San Diego, CA). The mean ± S.D. or S.E. was calculated for all samples, and the significance was determined either by the unpaired t test (for 2 groups) or analysis of variance (for ≥3 groups). For the analysis of variance analyses, Newman-Kuels post hoc testing was employed. A value of p < 0.05 was considered significant.
RESULTS
ADMA Stimulates eNOS Mitochondrial Translocation in Pulmonary Arterial Endothelial Cells
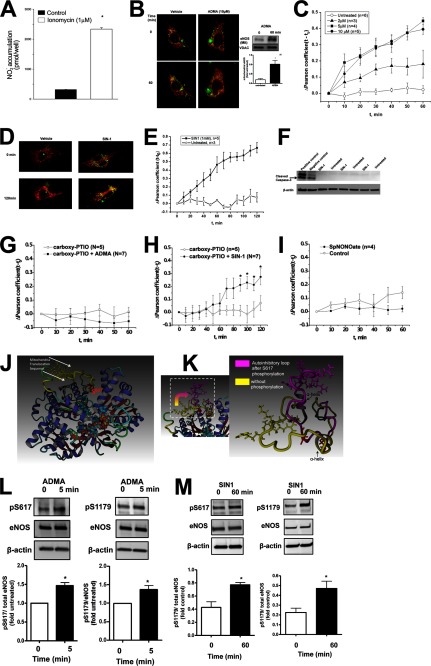
In initial experiments an eNOS-GFP fusion protein was transiently transfected into COS-7 cells and its ability to produce NO was confirmed when the cells were stimulated with ionomycin to increase intracellular Ca2+ levels (Fig. 1A). PAEC were then transiently transfected with a GFP-tagged eNOS construct and after 48 h the cells were pretreated with MitoTracker Red to visualize the mitochondria. Cells were then exposed to increasing concentrations of ADMA (0–10 μm, 0–60 min) for 1 h and monitored using live cell fluorescent microscopy. The Pearson product-moment correlation coefficient was then calculated for each ADMA concentration over time. Our results indicate that, compared with vehicle control, there is a significant increase in eNOS mitochondrial translocation with 5 and 10 μm ADMA as early as 10 min of exposure (Fig. 1, B and C). That endogenous eNOS behaves in a similar manner to that observed with live cell imaging of the eNOS-GFP protein was confirmed using Western blot analysis of isolated mitochondria from untransfected PAEC exposed to ADMA (Fig. 1B, inset). As we have previously shown that attenuating peroxynitrite generation attenuates ADMA-mediated eNOS mitochondrial translocation (13), we next determined if the peroxynitrite donor, SIN-1, could induce a similar mitochondrial redistribution of eNOS. Our results indicate that mitochondrial translocation of eNOS was also stimulated by SIN-1, but significant translocation only occurred after 50 min of exposure (Fig. 1, D and E), likely due to the time required for a threshold amount of peroxynitrite to be released as SIN-1 decomposes. There was no evidence of an induction of apoptotic events as indicated by a lack of cleaved caspase-3 in SIN-1-exposed PAEC (Fig. 1F). The requirement for both NO and superoxide in the redistribution of eNOS to the mitochondria was shown by the NO scavenger, carboxy-PTIO abolishing both ADMA (Fig. 1G) and SIN-1 (Fig. 1H)-induced eNOS translocation and the NO donor, spermine NONOate alone failing to induce eNOS redistribution to mitochondria (Fig. 1I).
FIGURE 1.
ADMA induces phosphorylation of eNOS at Ser617 and Ser1179. The eNOS-GFP construct was transiently transfected into COS-7 cells. After, 48 h the cells were stimulated with ionomycin (1 μm, 45 min). The accumulation of nitrite in the medium was then measured by chemiluminescence. Background levels of nitrite were subtracted. Ionoycin significantly increases nitrite accumulation in the medium indicating the eNOS-GFP construct is active (A). For live cell imaging PAEC were transfected with eNOS-GFP (green). The mitochondria of these cells were then labeled with MitoTracker (red) and the cells were exposed to increasing concentrations of ADMA (0–10 μm, 1 h) or SIN-1 (1 mm, 2 h). Representative images are shown indicating that ADMA induces eNOS-GFP translocation to the mitochondria (yellow, B). Western blot analysis of mitochondrial fractions prepared from PAEC exposed to 10 μm ADMA (1 h) confirmed the mitochondrial redistribution of endogenous eNOS (B, inset). The Pearson coefficient was calculated for each dose of ADMA over the 1-h exposure time. Exposure of PAEC to 5 and 10 μm ADMA induces significant eNOS mitochondrial translocation (C). Similar changes were observed with SIN-1 (D and E), but significant mitochondrial translocation was not observed until 50 min of treatment (E). Whole cell extracts (10 μg) were also prepared from PAEC exposed or not to SIN-1 (1 mm, 2 h). Western blot analysis was performed using an antibody that recognizes only cleaved caspase-3. Loading was normalized by reprobing the membranes with an antibody specific to β-actin. Shown is a representative image containing three of six experiments (F). Also, included were total cell lysates from Jurkat cells that had been exposed or not to etoposide (25 μm, 5 h) as, respectively, positive and negative controls for cleaved caspase-3. No significant increases in the levels of cleaved caspase-3 were observed between any treatment groups. Pretreatment of PAEC with the NO scavenger, carboxy-PTIO (100 μm, 30 min), attenuated the redistribution of eNOS-GFP in response to both ADMA (G) and SIN-1 (H). The NO donor, spermine NONOate (SpNONOate, 25 μm, 1 h) failed to induce eNOS-GFP mitochondrial redistribution (I). Molecular dynamic simulation indicates that the mitochondrial translocation sequence is located within an unstructured autoinhibitory loop (J, yellow) and in close proximity to the Ser617 consensus Akt1 phosphorylation site (J, red). MD analysis also shows that Ser617 phosphorylation results in a conformational change in the autoinhibitory loop of eNOS exposing the mitochondrial translocation sequence (K, arrow). These changes may be driven by reorganization of the α helixes. Whole cell extracts exposed or not to ADMA (10 μm, 5 min) or SIN-1 (1 mm, 1 h) were analyzed to determine changes in eNOS phosphorylation at Ser617 and Ser1179. Representative images are shown (L and M). Both ADMA and SIN-1 increase the phosphorylation of eNOS at both Ser617 and Ser1179 (L and M). Data are mean ± S.E.; n = 3–7. *, p < 0.05 versus untreated.
ADMA Increases eNOS Phosphorylation at Serine 617 and 1179
A mitochondrial translocation sequence located in the reductase domain has been previously described for eNOS (14). To begin to identify how this sequence is regulated we developed a homology model of the human eNOS reductase domain using the crystallized nNOS reductase domain as a template. Analysis of this homology model identified an unstructured region located between aa 595 and 644. This sequence contains the autoinhibitory loop, the mitochondrial translocation sequence, and the Akt1 phosphorylation site at Ser617. Therefore, we further refined this structure using a 3-ns MD simulation in which we anchored the reductase domain with the exception of the residues within aa 595–644. The resulting structure indicated that the five amino acids that comprise the mitochondrial targeting sequence (RRKRK) are located within an unstructured autoinhibitory loop (Fig. 1J, loop shown in yellow) and that the mitochondrial targeting sequence lies very close to the Akt1 phosphorylation site located at Ser617 (Fig. 1J, shown in red). We hypothesized that the phosphorylation of Ser617 would change the conformation of the mitochondrial targeting sequence, which in turn could stimulate eNOS redistribution from the plasma membrane to the mitochondria. To test this we ran another 3-ns MD simulation in which the reductase domain was phosphorylated at Ser617 and then compared it to the unphosphorylated structure (Fig. 1J). Our simulation data indicate that phosphorylation at Ser617 induces a structural change in the autoinhibitory loop such that the mitochondrial translocation sequence becomes exposed (Fig. 1K) suggesting that phosphorylation of Ser617 is required for eNOS translocation to the mitochondria. Thus, we next determined if ADMA stimulates eNOS phosphorylation at serine 617 in PAEC. Our data indicate that ADMA treatment enhances eNOS phosphorylation at Ser617 as early as 5 min after exposure (Fig. 1L) and before we observed a significant increase in eNOS localization to the mitochondrion (Fig. 1C). We also found that the level of the other Akt1 phosphorylation sites on eNOS located at Ser1179 was also increased by ADMA exposure (Fig. 1L). The increase in both Ser617 and Ser1179 phosphorylation was maintained at 60 min of ADMA treatment (not shown). Exposure of PAEC to the peroxynitrite donor, SIN-1 also increased the phosphorylation of Ser617 and Ser1179 (Fig. 1M).
Peroxynitrite-mediated eNOS Mitochondrial Translocation Is Dependent on eNOS Phosphorylation at Ser617 and Ser1179
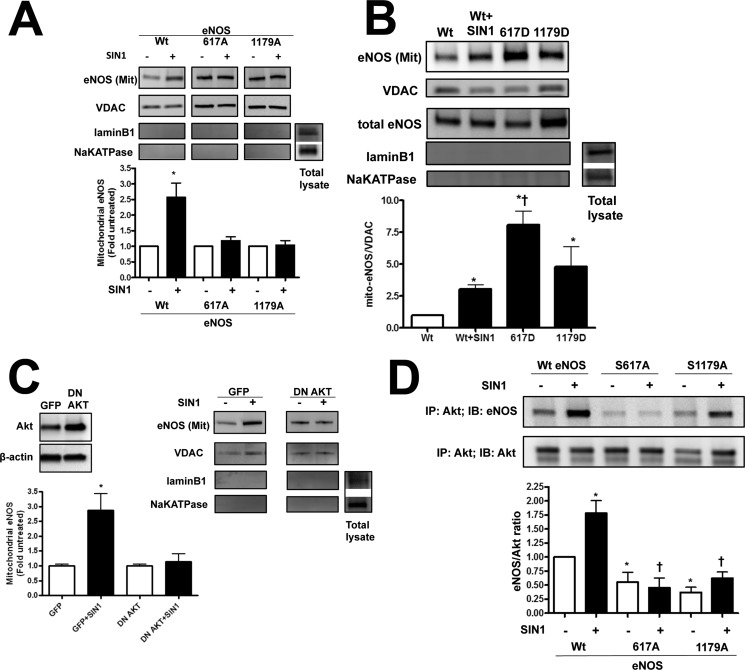
To examine the role of peroxynitrite-induced eNOS mitochondrial translocation of eNOS and the phosphorylation of Ser1179 and Ser617, HEK cells were transfected with either wild-type eNOS or phosphorylation deficient eNOS mutants (S617A and S1179A eNOS). Our data indicate that the peroxynitrite donor, SIN-1, enhances the mitochondrial translocation of wild-type eNOS, but not S617A or S1179A eNOS (Fig. 2A). This suggests that the efficient translocation requires phosphorylation at both Ser617 and Ser1179.
FIGURE 2.
Modulation of Ser617 and Ser1179 phosphorylation sites alters eNOS translocation. HEK 293 were transiently transfected with bovine eNOS expression plasmids containing wild-type eNOS (Wt eNOS) or mutations at serine 617 to alanine (S617A) or aspartic acid (S617D) and serine 1179 to alanine (S1179A) or aspartic acid (S1179D). After 48 h the cells were exposed or not to the peroxynitrite generator, SIN-1 (500 μm, 1 h). Mitochondrial fractions were then isolated and the level of eNOS protein accumulation in mitochondria was determined by Western blotting. Representative images are shown (A and B). SIN-1 induces the mitochondrial translocation of wt eNOS but this is blocked when either Ser617 or Ser1179 are mutated to alanine (A). Conversely, the mitochondrial localization of eNOS is significantly increased in cells expressing the S617D or S1179D eNOS mutants even in the absence of SIN-1 (B). Loading was normalized by reprobing with the mitochondrial protein, VDAC, and reprobed with antibodies raised against NaK-ATPase or lamin B1 to demonstrate no cross-contamination with the plasma membrane or nuclear fractions, respectively. PAEC were also transduced with a dominant-negative mutant of Akt1 (DN Akt, C, inset). The overexpression of DN Akt attenuates the SIN-1 mediated mitochondrial translocation of eNOS (C). Finally, we used immunoprecipitation (IP) to evaluate the interaction of Akt1 with WT eNOS and S617A and S1179A eNOS mutants in HEK 293 cells in the presence or absence of SIN-1 (500 μm, 1 h). There is an increase in the interaction of WT eNOS with Akt SIN-1 treatment (D). However, compared with WT eNOS, both phospho-null eNOS mutants (S617A and S1179A) exhibit a decrease in both basal and SIN-1 induced interactions with Akt1 (D). Data are mean ± S.E.; n = 4. *, p < 0.05 versus untreated, †, p < 0.05 versus Wt eNOS exposed to SIN-1 for (B) and p < 0.05 for Bonferroni comparison for untreated samples (D).
Effect of Phosphomimetic Mutations on eNOS Mitochondrial Translocation
To further determine the significance of phosphorylation of eNOS at Ser617 and Ser1179, we expressed wild-type and the phosphomimic eNOS mutants, S617D and S1179D, in HEK 293 cells. The wild-type eNOS expressing cells were then exposed to SIN-1 to stimulate eNOS mitochondrial translocation. The S617D and S1179D eNOS expressing cells were not exposed to SIN-1. Our data demonstrate that eNOS mitochondrial translocation was significantly higher in cells expressing S617D or S1179D eNOS in the absence of SIN-1 compared with SIN-1-stimulated cells expressing wild-type eNOS (Fig. 2B).
Endothelial NOS Mitochondrial Translocation Is Dependent on Akt1
Both Ser617 and Ser1179 are thought to be phosphorylated by Akt1. Thus, we next determined whether the mitochondrial translocation of eNOS is dependent on Akt1. To accomplish this we overexpressed a dominant-negative mutant of Akt1 (DN Akt1) in PAEC (Fig. 2C, inset). The level of eNOS in the mitochondria fraction was increased when cells were exposed to SIN-1 and this was attenuated by DN-Akt1 overexpression (Fig. 2C) indicating the involvement of Akt1 in regulating eNOS translocation to the mitochondria. Furthermore, when we evaluated the interaction of Akt1 with wild-type and the phospho-deficient eNOS mutants we found that SIN-1 stimulated Akt1/eNOS interactions only in wild-type eNOS (Fig. 2D). Surprisingly, in both eNOS phospho-null mutants (S617A and S1179A) we observed a decrease in their association with Akt1 compared with unstimulated wild-type eNOS expressing cells (Fig. 2D). This decreased interaction attenuates the nitration-mediated increase in Akt1 binding to eNOS suggesting that both serine residues may be required for efficient eNOS mitochondrial translocation.
Identification of Tyr350 as the Tyrosine Nitration Site on Akt1
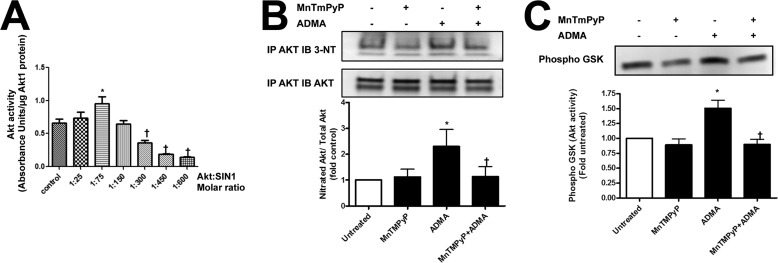
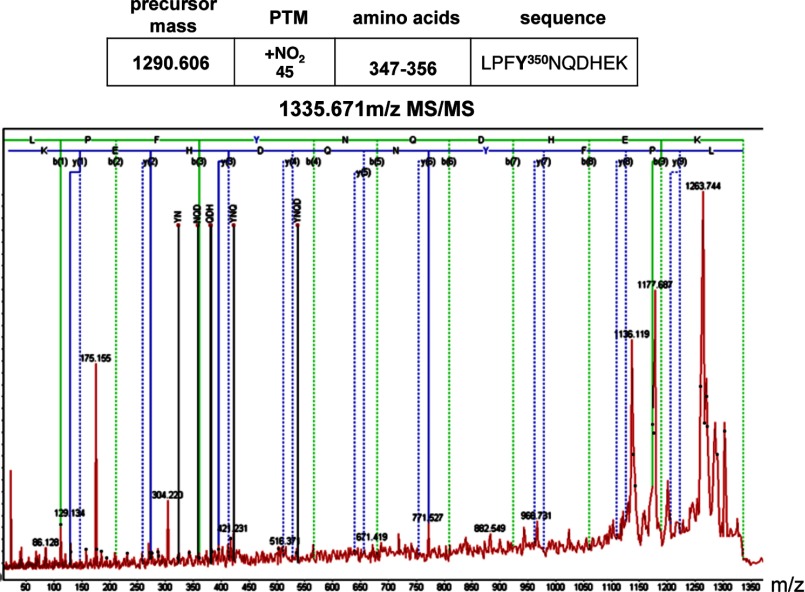
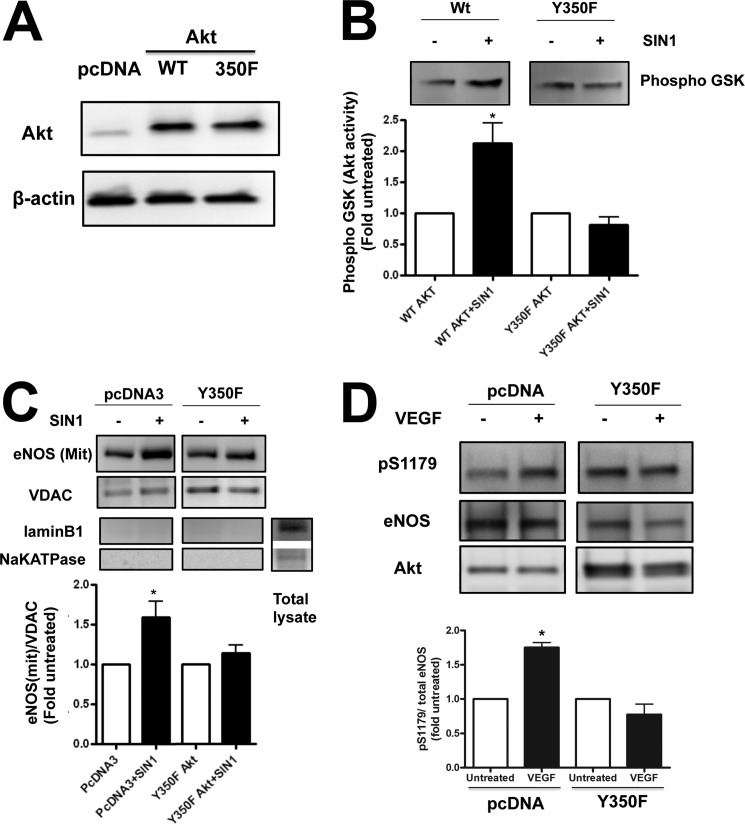
Utilizing recombinant Akt1 incubated with SIN-1 we identified a biphasic effect of peroxynitrite on Akt1 activity. At lower Akt1:SIN-1 molar ratios Akt1 activity is stimulated, whereas at a higher concentration of SIN-1 Akt1 activity is attenuated (Fig. 3A). Furthermore, we found that the exposure of PAEC to ADMA increased Akt1 nitration (Fig. 3B) and activity (Fig. 3C). Although these changes were blocked in the presence of the peroxynitrite scavenger MnTmPyP (Fig. 3, B and C). Interestingly, we observed two bands in our immunoprecipitation analyses. Using mass spectrometry analysis, both bands were identified as Akt1 (not shown). To identify the specific Akt1 nitration site Akt1 was overexpressed in HEK 293 cells and exposed to SIN-1 to induce protein nitration. Cell lysates from treated and untreated cells were then subjected to one-dimensional electrophoresis followed by in-gel tryptic digestion and mass spectroscopy. Searching for a signal of nitro-group addition +45 Da resulted in the peak with molecular mass of 1335.6 Da, which is 45 Da higher (nitro group) than the molecular mass of the unmodified peptide (1290.6 Da). This peptide was identified within Akt1 at 347LPFYNQDHEK356 (Fig. 4). An MS/MS spectrum from the 1335.6 m/z precursor peptide was obtained in positive reflector mode using air as the collision gas. The fragmentation pattern produced was fitted with the predicted sequence for nitration modified at Tyr350 Akt1 peptide LPFY(NO2)NQDHEK (Fig. 4). In particular, we found the following collision fragments: y1-H2O, y2, y3-H2O, y6, indicated as solid blue lines; b1, b3, and b9-H2O indicated as solid green lines; and immonium ions Y(NO2)N, NQD, QDH, Y(NO2)NQ and Y(NO2)NQD indicated as solid black lines. To confirm the importance of Tyr350 nitration, HEK 293 cells were transiently transfected with expression vectors containing either wild-type Akt1 or a mutant, in which the Tyr350 was replaced by phenylalanine (Y350F Akt1). Overexpression was confirmed by immunoblot analysis (Fig. 5A). We next determined the effect of SIN-1 on Akt activity. Our data demonstrate that SIN-1 increased Akt1 activity in wild-type, but not Y350F Akt1 overexpressing cells (Fig. 5B). Similarly when wild-type eNOS was co-expressed, SIN-1 enhanced eNOS translocation to the mitochondria in wild-type, but not in, Y350F Akt1 overexpressing cells (Fig. 5C). To test if the Y350F mutation exerts a dominant-negative affect on Akt1 signaling we examined the effect of its overexpression in PAEC exposed to VEGF. Our data demonstrate that overexpression of Y350F Akt1 inhibits the ability of VEGF to stimulate eNOS phosphorylation at Ser1179 indicating it has a dominant-negative phenotype (Fig. 5D).
FIGURE 3.
Nitration enhances the activity of Akt1. Purified recombinant Akt1 was incubated with different molar ratios of SIN-1 to induce enzyme nitration. The effect of SIN-1 was biphasic with a 1:75 molar ratio enhancing Akt1 activity and molar ratios of 1:300 or higher attenuating its activity (A). PAEC were also exposed to ADMA (10 μm, 1 h) in the presence or absence of the peroxynitrite scavenger, MnTMPyP (25 μm), and the level of Akt1 nitration was determined by immunoprecipitation (IP). Exposure of PAEC to ADMA increases Akt1 nitration and this is blocked by MnTMPyP (B). The ADMA-mediated increase in Akt1 nitration was also correlated with increased AKT activity and is again blocked by MnTMPyP (C). Data are mean ± S.E.; n = 3. *, p < 0.05 versus untreated; †, p < 0.05 versus 1:75 molar ratio of SIN-1 (A) or ADMA alone (B and C).
FIGURE 4.
Identification of tyrosine 350 as the nitration site on Akt1. Akt1 was immunoprecipitated from HEK 293 cells treated or not with SIN-1 (500 μm, 1 h). A tryptic digest of Akt1 was analyzed for tyrosine nitration sites not present in untreated cells. The peptide with m/z 1335.67 (parent peptide LPFYNQDHEK with m/z 1290.61 + 45 Da of nitro group) was further fragmented and MS/MS data analyzed. Solid lines represent predicted masses for the nitrated peptide found in MS/MS with error less than 0.3 ppm. MS/MS spectrum of the 1335.6 m/z ion was obtained in positive reflector mode fitted with peptide 347LPFY(NO2)NQDHEK356 from the Akt1 sequence.
FIGURE 5.
Expression of a Y350F Akt1 mutant protein attenuates eNOS mitochondrial translocation. HEK 293 were transiently transfected with either wild-type Akt1 (Wt Akt) or a mutant in which Tyr350 was replaced with phenylalanine (Y350F Akt). Initial Western blotting confirmed equal overexpression of the two constructs (A). The cells were also exposed or not to the peroxynitrite generator, SIN-1 (500 μm, 1 h), and the effect on Akt activity was determined. SIN-1 increased Akt1 activity only in the WT Akt overexpressing cells (B). HEK 293 cells were also co-transfected with WT eNOS and either the parental plasmid (pcDNA3) or Y350F Akt and the effect of SIN-1 on eNOS mitochondrial translocation was determined. The SIN-1-mediated increase in eNOS mitochondrial translocation was blocked by overexpression of Y350F Akt (C). Loading was normalized by reprobing with the mitochondrial protein, VDAC, and reprobed with antibodies raised against NaK-ATPase or lamin B1 to demonstrate no cross-contamination with the plasma membrane or nuclear fractions, respectively. PAEC were also transiently transfected with either the parental plasmid (pcDNA3) or the Y350F Akt mutant plasmid. After 48 h, the cells were exposed to the Akt activator VEGF (100 nm, 10 min) and then the effect on eNOS phosphorylation at Ser1179 was determined by Western blotting. Representative images are shown. VEGF increases eNOS Ser1179 phosphorylation, however, overexpression Y350F attenuates this affect (D). Blots were stripped and reprobed for eNOS and Akt1. Data are mean ± S.E.; n = 3. *, p < 0.05 versus untreated.
Elucidation of the Molecular Mechanism of Nitration-mediated Akt1 Activation
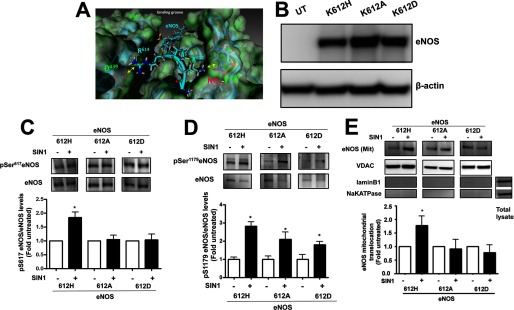
To better understand the mechanism by which the nitration of Tyr350 enhances Akt1 activation, we developed a homology model of Akt1 with the eNOS peptide containing the Akt phosphorylation site at Ser617 present in the Akt1 substrate-binding groove. Analysis of this model revealed that Tyr350 is located in close proximity to lysine 612 in the phosphorylation motif of eNOS. Furthermore, the introduction of a nitro group adds a negative charge at Tyr350 that is predicted to form a salt bridge to the positively charged lysine residue located at aa 612 (Lys612) in the eNOS-derived peptide (Fig. 6A). We predicted that this would enhance the binding of nitrated Akt with eNOS and result in increased phosphorylation of eNOS Ser617. To test this hypothesis, HEK cells were transfected with eNOS mutants where the Lys612 residue was mutated to histidine (K612H, positively charged), alanine (K612A, neutral), or aspartic acid (K612D, negatively charged) to alter the charge on the residue that interacts with Tyr350. Immunoblotting was performed to confirm equal expression of each eNOS mutant (Fig. 6B). The effect of SIN-1 treatment on eNOS phosphorylation at Ser617 and eNOS translocation to mitochondria was then analyzed. We found that SIN-1 induced eNOS phosphorylation at Ser617 in K612H-transfected cells, but not in the alanine or aspartic acid mutants (Fig. 6C). However, under the same conditions, phosphorylation of Ser1179 was increased in all the eNOS mutant proteins (Fig. 6D), suggesting that the mutation of Lys612 does not alter the ability of Akt1 to bind to eNOS. The mitochondrial translocation in the K612H eNOS mutant was also stimulated by SIN-1 treatment but this did not occur in cells expressing the K612A and K612D eNOS mutants (Fig. 6E). Together these data indicate that replacement of Lys612 with an uncharged or negatively charged amino acid significantly attenuates the ability of eNOS to translocate to the mitochondrion and also provides further supporting evidence for the importance of phosphorylation of Ser617 in regulating eNOS mitochondrial translocation.
FIGURE 6.
Role of Lys612 in the interaction of eNOS with nitrated Akt1. Analysis of the interactions between the Akt1 client protein binding groove and eNOS at Ser617 indicates the pre-positioning of a positively charged lysine residue located at 612 (K612) in eNOS with Tyr350 of Akt1. As tyrosine nitration results in the addition of a negative charge this suggests that there might be an enhanced interaction with eNOS via the positively charged Lys612 (A). HEK 293 cells were transfected with eNOS mutants in which different charged residues were introduced at position 612: K612H, slightly positive charge; K612A, neutral; and K612D, negative charge. Initial Western blotting confirmed the equal overexpression of the three constructs (B). Cells were also exposed or not to the peroxynitrite generator, SIN-1 (500 μm, 1 h), and the level of phosphorylation at Ser617 was determined by Western blotting. Representative images are shown. SIN-1 increases Ser617 phosphorylation only in the K612H eNOS mutant (C). Blots were stripped and reprobed for eNOS to normalize loading. However, under the same conditions, phosphorylation of Ser1179 was increased in all the eNOS mutant proteins (D). Mitochondrial fractions were also isolated and the level of eNOS protein accumulation in mitochondria was determined by Western blotting. Representative images are shown. Only the K612H eNOS mutant is redistributed to the mitochondria in response to SIN-1 (E). Data are mean ± S.E.; n = 3. *, p < 0.05 versus untreated. UT, untransfected.
Physiologic Role of eNOS Translocation on Mitochondrial Function
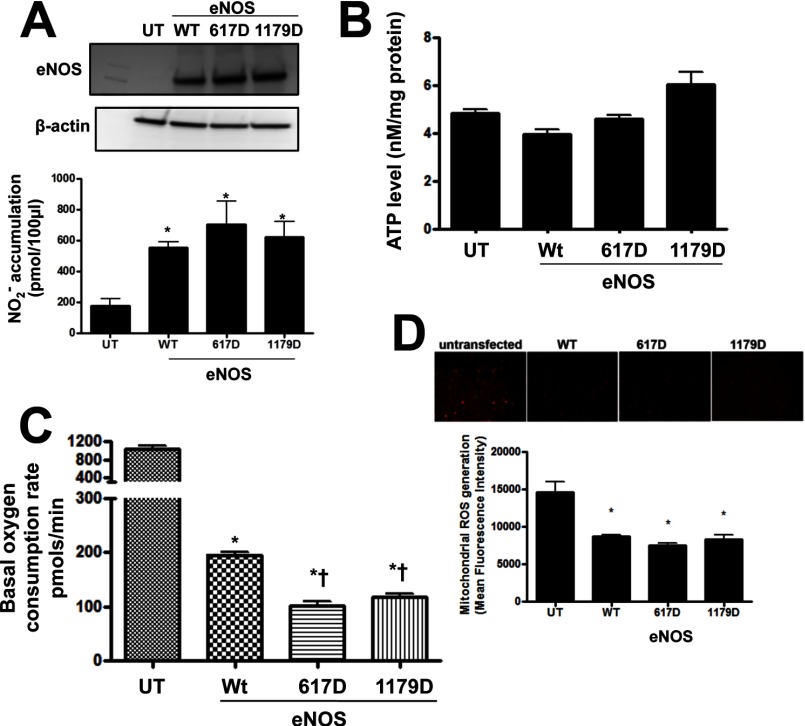
Next we addressed the question regarding the actual physiologic role of eNOS on mitochondrial function. To accomplish this we examined the effect of wild-type and S617D and S1179D eNOS overexpression on mitochondrial bioenergetics, superoxide production, and ATP production in HEK 293 cells. Initial studies confirmed that each construct exhibited similar levels of expression (Fig. 7A, upper panel) and generated similar levels of NO (Fig. 7A, lower panel). Interestingly, we did not observe the expected increase in NO production in the S1179D mutant perhaps due to the fact that our studies were carried out under nonstimulated conditions. Furthermore, ATP levels were not altered by wild-type or phosphomimic eNOS overexpression (Fig. 7B). However, the basal mitochondrial OCR was dramatically reduced in wild-type eNOS-transfected cells and reduced even further by overexpression of S617D or S1179D eNOS (Fig. 7C). Finally, to evaluate the effect of the overexpression of mitochondria-targeted eNOS on mitochondrial oxidative stress we utilized live cell imaging in cells exposed to the mitochondrial superoxide production indicator: MitoSOX. Our data demonstrate that all three eNOS proteins attenuated mitochondrial superoxide levels (Fig. 7D). Together these data suggest that the eNOS mitochondrial translocation leads to a decrease in basal respiration rate with decreased superoxide production, but without affecting cellular ATP levels.
FIGURE 7.
The effect of eNOS translocation on mitochondrial function. HEK 293 were transiently transfected with bovine eNOS expression plasmids containing wild-type eNOS (Wt eNOS) or mutations at serine 617 to aspartic acid (S617D) or serine 1179 to aspartic acid (S1179D). Initial Western blotting confirmed equal overexpression of the three constructs (A, upper panel). In addition, the expression of all three constructs results in a significant increase in NO production (A, lower panel). The expression of these constructs does not change the total cellular ATP levels (B). However, the basal OCR dropped significantly in WT eNOS-transfected cells and this is further decreased in cells overexpressing the S617D and S1179D eNOS mutants (C). The effect on mitochondrial ROS was also determined, using MitoSOX red fluorescence. Representative images are shown. The expression of all three constructs causes a significant decrease in MitoSOX fluorescence (D). Data are mean ± S.E.; n = 5. *, p < 0.05 versus untransfected cells; †, p < 0.05 versus WT eNOS. UT, untransfected.
DISCUSSION
ADMA is an endogenous competitor of l-arginine for the NOS active site. It has been well documented that increased levels of ADMA accompanies many pathological conditions (26–30) causing both oxidative and nitrosative stress (13, 31–33). One of the proposed mechanisms for these events is eNOS uncoupling (13, 34). Furthermore, due to the rapid reaction between the superoxide and NO produced by uncoupled eNOS, the local concentration of peroxynitrite is likely to be markedly increased making it probable that the proteins that interact with eNOS will be preferentially targeted for nitration. Indeed this has been shown for Hsp90 (35). Our previously published data indicate that ADMA-mediated increases in peroxynitrite generation leads to the translocation of eNOS to the mitochondrion resulting in mitochondrial dysfunction (13). Here we identify Akt1 as a novel target of ADMA-induced protein nitration that is involved in eNOS trafficking. Our data indicate that nitration of Akt1 leads to its activation in agreement with a recent study in which Akt1 activation has been shown to occur as an acute response to peroxynitrite exposure in the PDGF signaling cascade (36).
Akt1 belongs to the serine/threonine protein kinase family and is known to phosphorylate eNOS at Ser617 (37) and Ser1179 (38). Our data indicate that ADMA stimulates the activity of Akt1 resulting in increased eNOS phosphorylation at Ser617 and Ser1179. Phosphorylation at both sites is thought to enhance the activity of the enzyme likely through the reversal of the enzyme suppression associated with the autoinhibitory loop and reducing the concentration of calcium required for activation (39–41). Interestingly, introducing an aspartic acid to mimic the phosphorylation at Ser617 and Ser1179 enhanced eNOS mitochondrial translocation. Aspartic acid mutants expressed in HEK cells without additional stimulation produced the same amount of NO as wild-type eNOS, thus, it is unlikely that these mutations are producing significantly different levels of peroxynitrite. Conversely, the S1179A and S617A eNOS mutants were resistant to nitration-mediated mitochondrial translocation. Serine 617 is located adjacent to the region of eNOS (627–631) previously shown to be required for eNOS to redistribute to the mitochondrion (14). An examination of our homology model of the eNOS reductase domain suggests that the positively charged RRKRK region of the mitochondrial translocation sequence lies within the autoinhibitory loop of eNOS. From our molecular model of eNOS it also appears that when Ser617 is not phosphorylated this sequence is located deep within the eNOS protein and so it is not likely to be accessible for phosphorylation. However, our MD simulations predict that the introduction of a phosphate group at Ser617 increases solvent exposure of the loop such that the mitochondrial translocation sequence flexes outward, which increases its exposure. Our data also demonstrate the importance of Ser1179 phosphorylation in eNOS mitochondrial translocation. Ser1179 is located far from the mitochondrial translocation sequence and it is unclear how Ser1179 is involved in the translocation process. We speculate, based on our homology model and previously published data, that Ser1179 is more accessible to Akt1 than Ser617 and that its phosphorylation induces a conformational shift in eNOS that makes Ser617 more accessible to Akt1. After phosphorylation of Ser617, the loop undergoes a conformational shift exposing the mitochondrial translocation sequence, allowing eNOS to redistribute. If true, this hypothesis would predict that although Ser1179 phosphorylation is important for the mitochondrial translocation of eNOS it is the phosphorylation of Ser617 that actually drives the process. Our data do support this hypothesis as we found that Akt1/eNOS interactions are decreased in the S617A and S1179A mutants, whereas the mitochondrial translocation of S617D is significantly higher than that of S1179D.
Another question that arises from this study is what proteins or protein complexes are involved in eNOS trafficking? Although the identity of this protein is as yet unknown it is possible that this will prove to be one of the molecular chaperones shown to play key roles in the translocation of nuclear-encoded proteins into the mitochondrion (42, 43). For example, it is well established that the chaperone, Hsp70 maintains mitochondrial targeted proteins in a translocation-competent state (42) and its co-chaperone, C-terminal Hsc70-interacting protein (CHIP) is known to associate with eNOS and to induce changes in its subcellular distribution (44). CHIP has been shown to facilitate the assembly of translocation complexes (45). It is interesting to note that CHIP contains a region that is highly negatively charged located between aa 185 and 190 (EGDEDD) (46). It is possible that this sequence could interact with the positively charged eNOS mitochondrial translocation loop located at aa 627–631 (RRKRK) (14). Furthermore, we have recently shown in an animal model of pulmonary hypertension associated with increased pulmonary blood flow that increases in ADMA correlates with increased activities of CHIP and Hsp70 (47) and increased translocation of eNOS to the mitochondrion (48). However, further studies will be required to determine whether Hsp70 and/or CHIP play a role in regulating eNOS translocation to the mitochondrion.
Akt1 activation occurs when PtdInsP3 (a product of PI 3-kinase) binds to the pleckstrin homology domain of Akt1 inducing a translocation to the plasma membrane. In addition, phosphorylation events are required for full Akt1 activation (49). However, the phosphorylation-dependent activation of Akt1 is complex and not fully understood. The main activation events appear to be the phosphorylation of Thr308 in the activation loop of the kinase and Ser473 at the COOH terminus (50). However, other studies suggest that tyrosine phosphorylation of Akt1 regulates enzyme activity (51–53). Our mass spectroscopy analysis identified a single nitration site in Akt1 located at Tyr350. Molecular modeling of Akt1 revealed that Tyr350 is located in the binding groove within the kinase domain of Akt1, suggesting that Tyr350 nitration may modulate client protein binding. Studies by Chen et al. (51) have also identified, by sequence alignment of subdomains VII and VIII of Akt-1, -2, and -3, conserved tyrosine residues located at Tyr315, Tyr326, and Tyr340. Mutations of each of these implicated Tyr315 and Tyr326 as being important for full Akt1 activity (51). So far no studies have suggested that Tyr350 is susceptible to phosphorylation. However, as several of the amino acids preceding Tyr326 are predominantly negatively charged amino acids, and that the preferred substrates for Akt1 contain the positively charged, RXRXXS region, Tyr326 may be involved in stabilizing the interaction of Akt1 with its client proteins (51). This is similar to what we suggest occurs when Tyr350 is nitrated, as nitration also induces a negative charge on the tyrosine residue. However, unlike Tyr326 phosphorylation and Tyr350 nitration, phosphorylation of Tyr315 is predicted to affect the activation loop in the kinase domain of Akt1, similar to that observed when Thr308 is phosphorylated (51, 52). Another study failed to identify phosphorylation of Tyr315 and Tyr326 in Akt1 but identified Tyr474 as being phosphorylated (53). In this study it was postulated that Tyr474 phosphorylation enhances the affinity of the hydrophobic domain with the NH2-terminal lobe within Akt1 enabling the phosphorylation of Thr308 (53), stimulating enzyme activity. Whether nitration of Tyr350 can modulate or interact with these other tyrosine residues and alter Akt1 activity will require further study.
All Akt1 client proteins so far identified contain a consensus phosphorylation motif, RXRXX(S/T) (54), with the eNOS consensus adjacent to the mitochondrial targeting sequence being 612KXRXXS617 (37). Thus, the eNOS Akt phosphorylation sequence contains two conserved positively charged amino acids, Lys612 and Arg614 (bovine sequence). Our computational analysis indicates that nitration of the nitro group on Tyr350 introduces a negative charge to this residue. This makes it likely that a salt bridge with the Lys612 would be formed and binding energy calculations using the sequence KXRXXS predicts enhanced binding when Tyr350 is nitrated. This in turn was predicted to lead to increased phosphorylation of Ser617. Using site-directed mutagenesis of Lys612, we introduced differently charged amino acids within the Akt1 phosphorylation motif of eNOS. We introduced amino acids with a slight positive charge (K612H), a neutral charge (K612A), and a negative charge (K612D). The phosphorylation of Ser617 and mitochondrial translocation were regulated in accordance with the charge at amino acid 612 such that K612H > K612A = K612D. Furthermore, we found that Ser617 phosphorylation and mitochondrial translocation was enhanced in the K612H mutants only when cells were stimulated by SIN-1. This suggests that the formation of the salt bridge is dependent on the introduction of the negative charge at Tyr350 when Akt1 is nitrated. Interestingly, phosphorylation of Ser1179 was increased in all mutants upon SIN-1 stimulation. This suggests that in our eNOS mutants only the interaction between Akt1 and Ser617 is modified and provides further evidence for the importance of Ser617 phosphorylation in eNOS trafficking to the mitochondria. Alternatively as the Akt substrate motif in eNOS 612KIRFNS* the disruption of the substrate motif with K612A or K612D may decrease Ser617 phosphorylation independently of binding and also independent of phosphorylation of Ser1179. Furthermore, we found that the Y350F Akt1 mutation abrogates peroxynitrite-mediated eNOS phosphorylation at Ser617 (and Ser1179) and translocation to mitochondria. Thus, our data provide clear evidence that the positive charge at Lys612 is critical for a salt bridge formation between eNOS and nitrated Akt1. Interestingly, Tyr350 is a conserved residue in Akt1 isolated from different species (human, rat, bovine, canine, mouse, chicken, frog, and fly) but also between the three isoforms of Akt: 1, 2, and 3. Thus, we speculate that a similar mechanism of nitration-mediated activation may be shared between the different Akt isoforms. However, further studies will be required to test this possibility.
Our observations indicate that eNOS uncoupling or a peroxynitrite donor stimulate eNOS translocation to mitochondria via the nitration of Akt1. As both active Akt1 and eNOS localize predominantly to the plasma membrane it is likely that, at least under conditions of increased ADMA, that the peroxynitrite required for Akt1 activation is derived from uncoupled eNOS. Thus, we speculate that redistribution of eNOS is independent of mitochondria-derived superoxide. However, when Akt1 is activated by nitration it will likely phosphorylate eNOS at Ser1179 and Ser617 independent of whether it is coupled or not. Thus, the mitochondria will be enriched with eNOS in accordance with the current cellular state and both coupled and uncoupled eNOS will be directed to the mitochondria. As the mitochondria are a significant source of superoxide in the cell it is possible that even when coupled eNOS is redistributed the generation of NO in the mitochondria will react with the superoxide present in the mitochondria to increase peroxynitrite generation. Indeed, we have previously shown that when eNOS is redistributed to the mitochondria in response to ADMA the levels of nitrated proteins in the mitochondria increase (13).
The physiological role of eNOS mitochondrial translocation is still uncertain. NO is known to modulate mitochondrial function by reversibly inhibiting cytochrome c oxidase, which decreases mitochondrial energy production (55, 56). However, NO may play an adaptive role in some situations such as at the fetal to newborn transition through its ability to modulate oxidative phosphorylation (57). This may protect the cell by decreasing mitochondrial derived ROS. In turn, this would attenuate mitochondrial oxygen utilization through the electron transport chain leading to better coupling between oxygen utilization and ATP generation (58). Our data strongly support these findings as the overexpression of wild-type eNOS and the phosphomimic mutants S617D and S1179D, which exhibit enhanced mitochondrial translocation, all decreased basal OCR and mitochondrial ROS production. However, this marked decrease in respiration rate did not alter ATP production. This may be explained by enhanced coupling of the electron transport chain that enables the same degree of ATP generation with decreased oxygen demand. Although, it is not clear which are the key proteins through which NO regulates mitochondrial respiration, several have been identified in previous studies. These include Complexes I and III of the electron transport chain (60–62), manganese-superoxide dismutase (MnSOD) (63, 64), cytochrome c (64, 65), mitochondrial creatine kinase (66, 67), and aconitase (68, 69). In addition, we have shown that carnitine acetyltransferase is also nitrated resulting in its inactivation (70). As carnitine acetyltransferase plays an important role in cellular energy metabolism through the regulation of fatty acid biosynthesis, alterations in its activity can have profound effects on mitochondrial function (71). Alterations in carnitine acetyltransferase activity and increased levels of ADMA are also associated with the development of pulmonary hypertension (70). Alternatively, it is possible that eNOS translocation stimulates glycolysis in the cell. This so-called “Warburg effect” was first observed in cancer cells (72) and has been recently shown to occur in both SMC (73) and EC (74) isolated from pulmonary hypertensive patients. However, further studies will be required to determine the exact mechanism by which the mitochondrial targeting of eNOS can reduce mitochondrial OCR but maintain cellular ATP levels.
In conclusion, our data indicate that nitration of Akt1 at Tyr350 leads to its activation, resulting in enhanced eNOS phosphorylation and subsequent mitochondrial translocation. Increased activation of Akt1 has been implicated in cancer cell progression through its ability to inhibit apoptotic signaling and promote cell survival (75, 76). Although, how this activation of Akt1 occurs has not been fully elucidated it is tempting to speculate that it may be due to its enhanced nitration especially as increased levels of nitration have been identified in a number of cancers (59, 77, 78). Thus, preventing Akt1 nitration may serve as a potential strategy to alleviate deleterious effects of Akt1 activation in various pathologies.
This work was supported, in whole or in part, by National Institutes of Health Grants HL60190 (to S. M. B.), HL67841 (to S. M. B.), and HL101902 (to S. M. B. and D. F.) and a Transatlantic Network Development Grant from the LeDucq Foundation (to S. M. B.).
- ADMA
- asymmetric dimethylarginine
- ROS
- reactive oxygen species
- PAEC
- pulmonary artery endothelial cell
- SIN-1
- 3-morpholinosydnonimine hydrochloride
- carboxy-PTIO
- 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- OCR
- oxygen consumption rate
- MD
- molecular dynamic
- CHIP
- C-terminal Hsc70-interacting protein.
REFERENCES
- 1. Moncada S., Palmer R. M., Higgs E. A. (1991) Nitric oxide. Physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 43, 109–142 [PubMed] [Google Scholar]
- 2. Kubes P., Suzuki M., Granger D. N. (1991) Nitric oxide. An endogenous modulator of leukocyte adhesion. Proc. Natl. Acad. Sci. U.S.A. 88, 4651–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ignarro L. J. (2002) Nitric oxide as a unique signaling molecule in the vascular system. A historical overview. J. Physiol. Pharmacol. 53, 503–514 [PubMed] [Google Scholar]
- 4. Shaul P. W., North A. J., Brannon T. S., Ujiie K., Wells L. B., Nisen P. A., Lowenstein C. J., Snyder S. H., Star R. A. (1995) Prolonged in vivo hypoxia enhances nitric-oxide synthase type I and type III gene expression in adult rat lung. Am. J. Respir. Cell Mol. Biol. 13, 167–174 [DOI] [PubMed] [Google Scholar]
- 5. Abu-Soud H. M., Feldman P. L., Clark P., Stuehr D. J. (1994) Electron transfer in the nitric-oxide synthases. Characterization of l-arginine analogs that block heme iron reduction. J. Biol. Chem. 269, 32318–32326 [PubMed] [Google Scholar]
- 6. Hedner T., Himmelmann A., Hansson L. (2002) Homocysteine and ADMA. Emerging risk factors for cardiovascular disease? Blood Press. 11, 197–200 [DOI] [PubMed] [Google Scholar]
- 7. Nash D. T. (2002) Insulin resistance, ADMA levels, and cardiovascular disease. J. Am. Med. Assoc. 287, 1451–1452 [DOI] [PubMed] [Google Scholar]
- 8. Lentz S. R., Rodionov R. N., Dayal S. (2003) Hyperhomocysteinemia, endothelial dysfunction, and cardiovascular risk. The potential role of ADMA. Atheroscler. Suppl. 4, 61–65 [DOI] [PubMed] [Google Scholar]
- 9. Cavusoglu E., Ruwende C., Chopra V., Yanamadala S., Eng C., Pinsky D. J., Marmur J. D. (2009) Relationship of baseline plasma ADMA levels to cardiovascular outcomes at 2 years in men with acute coronary syndrome referred for coronary angiography. Coron. Artery Dis. 20, 112–117 [DOI] [PubMed] [Google Scholar]
- 10. Gorenflo M., Zheng C., Werle E., Fiehn W., Ulmer H. E. (2001) Plasma levels of asymmetrical dimethyl-l-arginine in patients with congenital heart disease and pulmonary hypertension. J. Cardiovasc. Pharmacol. 37, 489–492 [DOI] [PubMed] [Google Scholar]
- 11. Cooke J. P. (2004) Asymmetrical dimethylarginine. The Uber marker? Circulation 109, 1813–1818 [DOI] [PubMed] [Google Scholar]
- 12. Vallance P., Leiper J. (2004) Cardiovascular biology of the asymmetric dimethylarginine:dimethylarginine dimethylaminohydrolase pathway. Arterioscler. Thromb. Vasc. Biol. 24, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 13. Sud N., Wells S. M., Sharma S., Wiseman D. A., Wilham J., Black S. M. (2008) Asymmetric dimethylarginine inhibits HSP90 activity in pulmonary arterial endothelial cells. Role of mitochondrial dysfunction. Am. J. Physiol. Cell Physiol. 294, C1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao S., Chen J., Brodsky S. V., Huang H., Adler S., Lee J. H., Dhadwal N., Cohen-Gould L., Gross S. S., Goligorsky M. S. (2004) Docking of endothelial nitric-oxide synthase (eNOS) to the mitochondrial outer membrane. A pentabasic amino acid sequence in the autoinhibitory domain of eNOS targets a proteinase K-cleavable peptide on the cytoplasmic face of mitochondria. J. Biol. Chem. 279, 15968–15974 [DOI] [PubMed] [Google Scholar]
- 15. St-Pierre J., Buckingham J. A., Roebuck S. J., Brand M. D. (2002) Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 277, 44784–44790 [DOI] [PubMed] [Google Scholar]
- 16. Penckofer S., Schwertz D., Florczak K. (2002) Oxidative stress and cardiovascular disease in type 2 diabetes. The role of antioxidants and pro-oxidants. J. Cardiovasc. Nurs. 16, 68–85 [DOI] [PubMed] [Google Scholar]
- 17. Puddu P., Puddu G. M., Galletti L., Cravero E., Muscari A. (2005) Mitochondrial dysfunction as an initiating event in atherogenesis. A plausible hypothesis. Cardiology 103, 137–141 [DOI] [PubMed] [Google Scholar]
- 18. Ballinger S. W. (2005) Mitochondrial dysfunction in cardiovascular disease. Free Radic. Biol. Med. 38, 1278–1295 [DOI] [PubMed] [Google Scholar]
- 19. Cai L., Kang Y. J. (2001) Oxidative stress and diabetic cardiomyopathy. A brief review. Cardiovasc. Toxicol. 1, 181–193 [DOI] [PubMed] [Google Scholar]
- 20. Li H., Samouilov A., Liu X., Zweier J. L. (2003) Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrate reduction. Evaluation of its role in nitrite and nitric oxide generation in anoxic tissues. Biochemistry 42, 1150–1159 [DOI] [PubMed] [Google Scholar]
- 21. Wedgwood S., Mitchell C. J., Fineman J. R., Black S. M. (2003) Developmental differences in the shear stress-induced expression of endothelial NO synthase. Changing role of AP-1. Am. J. Physiol. Lung Cell. Physiol. 284, L650–662 [DOI] [PubMed] [Google Scholar]
- 22. Tian J., Hou Y., Lu Q., Wiseman D. A., Vasconcelos Fonsesca F., Elms S., Fulton D. J., Black S. M. (2010) A novel role for caveolin-1 in regulating endothelial nitric-oxide synthase activation in response to H2O2 and shear stress. Free Radic. Biol. Med. 49, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sud N., Sharma S., Wiseman D. A., Harmon C., Kumar S., Venema R. C., Fineman J. R., Black S. M. (2007) Nitric oxide and superoxide generation from endothelial NOS. Modulation by HSP90. Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L1444–1453 [DOI] [PubMed] [Google Scholar]
- 24. Krieger E., Darden T., Nabuurs S. B., Finkelstein A., Vriend G. (2004) Making optimal use of empirical energy functions. Force-field parameterization in crystal space. Proteins 57, 678–683 [DOI] [PubMed] [Google Scholar]
- 25. Zhou R. (2003) Free energy landscape of protein folding in water. Explicit versus implicit solvent. Proteins 53, 148–161 [DOI] [PubMed] [Google Scholar]
- 26. Nakamura T., Sato E., Fujiwara N., Kawagoe Y., Ueda Y., Suzuki T., Ueda S., Adachi H., Okuda S., Yamagishi S. (2009) Ezetimibe decreases serum levels of asymmetric dimethylarginine (ADMA) and ameliorates renal injury in non-diabetic chronic kidney disease patients in a cholesterol-independent manner. Pharmacol. Res. 60, 525–528 [DOI] [PubMed] [Google Scholar]
- 27. Zinellu A., Pinna A., Sotgia S., Zinellu E., Usai M. F., Carta F., Gaspa L., Deiana L., Carru C. (2008) Increased plasma asymmetric dimethylarginine (ADMA) levels in retinal venous occlusive disease. Clin. Chem. Lab. Med. 46, 387–392 [DOI] [PubMed] [Google Scholar]
- 28. Schulze F., Lenzen H., Hanefeld C., Bartling A., Osterziel K. J., Goudeva L., Schmidt-Lucke C., Kusus M., Maas R., Schwedhelm E., Strodter D., Simon B. C., Mugge A., Daniel W. G., Tillmanns H., Maisch B., Streichert T., Boger R. H. (2006) Asymmetric dimethylarginine is an independent risk factor for coronary heart disease. Results from the multicenter Coronary Artery Risk Determination investigating the Influence of ADMA Concentration (CARDIAC) study. Am. Heart J. 152, 493.e1–8 [DOI] [PubMed] [Google Scholar]
- 29. Schnog J. B., Teerlink T., van der Dijs F. P., Duits A. J., Muskiet F. A. (2005) Plasma levels of asymmetric dimethylarginine (ADMA), an endogenous nitric-oxide synthase inhibitor, are elevated in sickle cell disease. Ann. Hematol. 84, 282–286 [DOI] [PubMed] [Google Scholar]
- 30. Böger R. H. (2005) Asymmetric dimethylarginine (ADMA) and cardiovascular disease. Insights from prospective clinical trials. Vasc. Med. 10, S19–25 [DOI] [PubMed] [Google Scholar]
- 31. Sydow K., Münzel T. (2003) ADMA and oxidative stress. Atherosclerosis 4, 41–51 [DOI] [PubMed] [Google Scholar]
- 32. Pitocco D., Zaccardi F., Di Stasio E., Romitelli F., Martini F., Scaglione G. L., Speranza D., Santini S., Zuppi C., Ghirlanda G. (2009) Role of asymmetric-dimethyl-l-arginine (ADMA) and nitrite/nitrate (NOx) in the pathogenesis of oxidative stress in female subjects with uncomplicated type 1 diabetes mellitus. Diabetes Res. Clin. Pract. 86, 173–176 [DOI] [PubMed] [Google Scholar]
- 33. Sharma S., Smith A., Kumar S., Aggarwal S., Rehmani I., Snead C., Harmon C., Fineman J., Fulton D., Catravas J. D., Black S. M. (2010) Mechanisms of nitric-oxide synthase uncoupling in endotoxin-induced acute lung injury. Role of asymmetric dimethylarginine. Vascul. Pharmacol. 52, 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Antoniades C., Shirodaria C., Leeson P., Antonopoulos A., Warrick N., Van-Assche T., Cunnington C., Tousoulis D., Pillai R., Ratnatunga C., Stefanadis C., Channon K. M. (2009) Association of plasma asymmetrical dimethylarginine (ADMA) with elevated vascular superoxide production and endothelial nitric-oxide synthase uncoupling. Implications for endothelial function in human atherosclerosis. Eur. Heart J. 30, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 35. Kawasaki H., Ikeda K., Shigenaga A., Baba T., Takamori K., Ogawa H., Yamakura F. (2011) Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells. Free Radic. Biol. Med. 50, 419–427 [DOI] [PubMed] [Google Scholar]
- 36. Klotz L. O., Schieke S. M., Sies H., Holbrook N. J. (2000) Peroxynitrite activates the phosphoinositide 3-kinase/Akt pathway in human skin primary fibroblasts. Biochem. J. 352, 219–225 [PMC free article] [PubMed] [Google Scholar]
- 37. Michell B. J., Harris M. B., Chen Z. P., Ju H., Venema V. J., Blackstone M. A., Huang W., Venema R. C., Kemp B. E. (2002) Identification of regulatory sites of phosphorylation of the bovine endothelial nitric-oxide synthase at serine 617 and serine 635. J. Biol. Chem. 277, 42344–42351 [DOI] [PubMed] [Google Scholar]
- 38. Fulton D., Gratton J. P., McCabe T. J., Fontana J., Fujio Y., Walsh K., Franke T. F., Papapetropoulos A., Sessa W. C. (1999) Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature 399, 597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tran Q. K., Leonard J., Black D. J., Persechini A. (2008) Phosphorylation within an autoinhibitory domain in endothelial nitric-oxide synthase reduces the Ca2+ concentrations required for calmodulin to bind and activate the enzyme. Biochemistry 47, 7557–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tran Q. K., Leonard J., Black D. J., Nadeau O. W., Boulatnikov I. G., Persechini A. (2009) Effects of combined phosphorylation at Ser-617 and Ser-1179 in endothelial nitric-oxide synthase on EC50(Ca2+) values for calmodulin binding and enzyme activation. J. Biol. Chem. 284, 11892–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ritchie S. A., Kohlhaas C. F., Boyd A. R., Yalla K. C., Walsh K., Connell J. M., Salt I. P. (2010) Insulin-stimulated phosphorylation of endothelial nitric-oxide synthase at serine-615 contributes to nitric oxide synthesis. Biochem. J. 426, 85–90 [DOI] [PubMed] [Google Scholar]
- 42. Kang P. J., Ostermann J., Shilling J., Neupert W., Craig E. A., Pfanner N. (1990) Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348, 137–143 [DOI] [PubMed] [Google Scholar]
- 43. Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem. 66, 863–917 [DOI] [PubMed] [Google Scholar]
- 44. Jiang J., Cyr D., Babbitt R. W., Sessa W. C., Patterson C. (2003) Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J. Biol. Chem. 278, 49332–49341 [DOI] [PubMed] [Google Scholar]
- 45. Li H. M., Niki T., Taira T., Iguchi-Ariga S. M., Ariga H. (2005) Association of DJ-1 with chaperones and enhanced association and colocalization with mitochondrial Hsp70 by oxidative stress. Free Radic. Res. 39, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 46. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun X., Fratz S., Sharma S., Hou Y., Rafikov R., Kumar S., Rehmani I., Tian J., Smith A., Schreiber C., Reiser J., Naumann S., Haag S., Hess J., Catravas J. D., Patterson C., Fineman J. R., Black S. M. (2011) C terminus of heat shock protein 70-interacting protein-dependent GTP cyclohydrolase I degradation in lambs with increased pulmonary blood flow. Am. J. Respir. Cell Mol. Biol. 45, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun X., Sharma S., Fratz S., Kumar S., Rafikov R., Aggarwal S., Rafikova O., Lu Q., Burns T., Dasarathy S., Wright J., Schreiber C., Fineman J. R., Black S. M. (2012) Disruption of endothelial cell mitochondrial bioenergetics in lamb with increased pulmonary blood flow. Antioxid. Redox Signal., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cameron A. J., De Rycker M., Calleja V., Alcor D., Kjaer S., Kostelecky B., Saurin A., Faisal A., Laguerre M., Hemmings B. A., McDonald N., Larijani B., Parker P. J. (2007) Protein kinases, from B to C. Biochem. Soc. Trans. 35, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 50. Dummler B., Hemmings B. A. (2007) Physiological roles of PKB/Akt isoforms in development and disease. Biochem. Soc. Trans. 35, 231–235 [DOI] [PubMed] [Google Scholar]
- 51. Chen R., Kim O., Yang J., Sato K., Eisenmann K. M., McCarthy J., Chen H., Qiu Y. (2001) Regulation of Akt/PKB activation by tyrosine phosphorylation. J. Biol. Chem. 276, 31858–31862 [DOI] [PubMed] [Google Scholar]
- 52. Scheid M. P., Marignani P. A., Woodgett J. R. (2002) Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol. Cell Biol. 22, 6247–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Conus N. M., Hannan K. M., Cristiano B. E., Hemmings B. A., Pearson R. B. (2002) Direct identification of tyrosine 474 as a regulatory phosphorylation site for the Akt protein kinase. J. Biol. Chem. 277, 38021–38028 [DOI] [PubMed] [Google Scholar]
- 54. Obata T., Yaffe M. B., Leparc G. G., Piro E. T., Maegawa H., Kashiwagi A., Kikkawa R., Cantley L. C. (2000) Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275, 36108–36115 [DOI] [PubMed] [Google Scholar]
- 55. Brown G. C. (1999) Nitric oxide and mitochondrial respiration. Biochim. Biophys. Acta 1411, 351–369 [DOI] [PubMed] [Google Scholar]
- 56. Brown G. C. (1997) Nitric oxide inhibition of cytochrome oxidase and mitochondrial respiration. Implications for inflammatory, neurodegenerative and ischaemic pathologies. Mol. Cell Biochem. 174, 189–192 [PubMed] [Google Scholar]
- 57. Singer M. (2007) Mitochondrial function in sepsis. Acute phase versus multiple organ failure. Crit. Care Med. 35, S441–448 [DOI] [PubMed] [Google Scholar]
- 58. Chandrasekar I., Eis A., Konduri G. G. (2008) Betamethasone attenuates oxidant stress in endothelial cells from fetal lambs with persistent pulmonary hypertension. Pediatr. Res. 63, 67–72 [DOI] [PubMed] [Google Scholar]
- 59. Riobó N. A., Clementi E., Melani M., Boveris A., Cadenas E., Moncada S., Poderoso J. J. (2001) Nitric oxide inhibits mitochondrial NADH:ubiquinone reductase activity through peroxynitrite formation. Biochem. J. 359, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhan X., Desiderio D. M. (2009) Mass spectrometric identification of in vivo nitrotyrosine sites in the human pituitary tumor proteome. Methods Mol. Biol. 566, 137–163 [DOI] [PubMed] [Google Scholar]
- 61. Brooks K. J., Hargreaves I. P., Bates T. E. (2000) Nitric-oxide-induced inhibition of mitochondrial complexes following aglycaemic hypoxia in neonatal cortical rat brain slices. Dev. Neurosci. 22, 359–365 [DOI] [PubMed] [Google Scholar]
- 62. Shiva S., Brookes P. S., Patel R. P., Anderson P. G., Darley-Usmar V. M. (2001) Nitric oxide partitioning into mitochondrial membranes and the control of respiration at cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 98, 7212–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. MacMillan-Crow L. A., Crow J. P., Kerby J. D., Beckman J. S., Thompson J. A. (1996) Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc. Natl. Acad. Sci. U.S.A. 93, 11853–11858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hortelano S., Alvarez A. M., Boscá L. (1999) Nitric oxide induces tyrosine nitration and release of cytochrome c preceding an increase of mitochondrial transmembrane potential in macrophages. FASEB J. 13, 2311–2317 [DOI] [PubMed] [Google Scholar]
- 65. Cassina A. M., Hodara R., Souza J. M., Thomson L., Castro L., Ischiropoulos H., Freeman B. A., Radi R. (2000) Cytochrome c nitration by peroxynitrite. J. Biol. Chem. 275, 21409–21415 [DOI] [PubMed] [Google Scholar]
- 66. Kanski J., Schöneich C. (2005) Protein nitration in biological aging. Proteomic and tandem mass spectrometric characterization of nitrated sites. Methods Enzymol. 396, 160–171 [DOI] [PubMed] [Google Scholar]
- 67. Wendt S., Schlattner U., Wallimann T. (2003) Differential effects of peroxynitrite on human mitochondrial creatine kinase isoenzymes. Inactivation, octamer destabilization, and identification of involved residues. J. Biol. Chem. 278, 1125–1130 [DOI] [PubMed] [Google Scholar]
- 68. Hausladen A., Fridovich I. (1996) Measuring nitric oxide and superoxide. Rate constants for aconitase reactivity. Methods Enzymol. 269, 37–41 [DOI] [PubMed] [Google Scholar]
- 69. Castro L. A., Robalinho R. L., Cayota A., Meneghini R., Radi R. (1998) Nitric oxide and peroxynitrite-dependent aconitase inactivation and iron-regulatory protein-1 activation in mammalian fibroblasts. Arch. Biochem. Biophys. 359, 215–224 [DOI] [PubMed] [Google Scholar]
- 70. Sharma S., Sud N., Wiseman D. A., Carter A. L., Kumar S., Hou Y., Rau T., Wilham J., Harmon C., Oishi P., Fineman J. R., Black S. M. (2008) Altered carnitine homeostasis is associated with decreased mitochondrial function and altered nitric oxide signaling in lambs with pulmonary hypertension. Am. J. Physiol. Lung Cell. Physiol. 294, L46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jogl G., Hsiao Y. S., Tong L. (2004) Structure and function of carnitine acyltransferases. Ann. N.Y. Acad. Sci. 1033, 17–29 [DOI] [PubMed] [Google Scholar]
- 72. Warburg O. (1956) On respiratory impairment in cancer cells. Science 124, 269–270 [PubMed] [Google Scholar]
- 73. Marsboom G., Toth P. T., Ryan J. J., Hong Z., Wu X., Fang Y. H., Thenappan T., Piao L., Zhang H. J., Pogoriler J., Chen Y., Morrow E., Weir E. K., Rehman J., Archer S. L. (2012) Dynamin-related protein 1-mediated mitochondrial mitotic fission permits hyperproliferation of vascular smooth muscle cells and offers a novel therapeutic target in pulmonary hypertension. Circ. Res. 110, 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu W., Koeck T., Lara A. R., Neumann D., DiFilippo F. P., Koo M., Janocha A. J., Masri F. A., Arroliga A. C., Jennings C., Dweik R. A., Tuder R. M., Stuehr D. J., Erzurum S. C. (2007) Alterations of cellular bioenergetics in pulmonary artery endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 104, 1342–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kauffmann-Zeh A., Rodriguez-Viciana P., Ulrich E., Gilbert C., Coffer P., Downward J., Evan G. (1997) Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature 385, 544–548 [DOI] [PubMed] [Google Scholar]
- 76. Lane D., Robert V., Grondin R., Rancourt C., Piché A. (2007) Malignant ascites protect against TRAIL-induced apoptosis by activating the PI3K/Akt pathway in human ovarian carcinoma cells. Int. J. Cancer 121, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 77. MacMillan-Crow L. A., Greendorfer J. S., Vickers S. M., Thompson J. A. (2000) Tyrosine nitration of c-SRC tyrosine kinase in human pancreatic ductal adenocarcinoma. Arch. Biochem. Biophys. 377, 350–356 [DOI] [PubMed] [Google Scholar]
- 78. Vickers S. M., MacMillan-Crow L. A., Green M., Ellis C., Thompson J. A. (1999) Association of increased immunostaining for inducible nitric-oxide synthase and nitrotyrosine with fibroblast growth factor transformation in pancreatic cancer. Arch. Surg. 134, 245–251 [DOI] [PubMed] [Google Scholar]