Background: Salt-inducible kinase (SIK) 2 is an AMP-activated protein kinase family kinase that mediates hormonal and nutrient signaling but has no known link to cellular stress response.
Results: p300/CBP and HDAC6 reciprocally regulates Lys-53 acetylation of SIK2, consequently impacting its activity and function in autophagosome maturation.
Conclusion: SIK2 kinase activity, via a acetylation-based regulatory switch, contributes to autophagy progression.
Significance: SIK2 may be linked to neurodegenerative or protein aggregate disorders.
Keywords: AMP-activated Kinase (AMPK), Autophagy, CBP, Histone Deacetylase, Neurodegeneration, p300
Abstract
Salt-inducible kinase 2 (SIK2) is a serine/threonine protein kinase belonging to the AMP-activated protein kinase (AMPK) family. SIK2 has been shown to function in the insulin-signaling pathway during adipocyte differentiation and to modulate CREB-mediated gene expression in response to hormones and nutrients. However, molecular mechanisms underlying the regulation of SIK2 kinase activity remains largely elusive. Here we report a dynamic, post-translational regulation of its kinase activity that is coordinated by an acetylation-deaceytlation switch, p300/CBP-mediated Lys-53 acetylation inhibits SIK2 kinase activity, whereas HDAC6-mediated deacetylation restores the activity. Interestingly, overexpression of acetylation-mimetic mutant of SIK2 (SIK2-K53Q), but not the nonacetylatable K53R variant, resulted in accumulation of autophagosomes. Further consistent with a role in autophagy, knockdown of SIK2 abrogated autophagosome and lysosome fusion. Consequently, SIK2 and its kinase activity are indispensable for the removal of TDP-43Δ inclusion bodies. Our findings uncover SIK2 as a critical determinant in autophagy progression and further suggest a mechanism in which the interplay among kinase and deacetylase activities contributes to cellular protein pool homeostasis.
Introduction
Salt-inducible kinase 2 (SIK2)3 is a member of the AMP-activated protein kinase (AMPK) family, constituents of which are regarded as important mediators of energy and stress signaling. The kinase activity of SIK2 has been shown to regulate a number of signaling pathways and associated gene expression in response to hormones and nutrients. Particularly, its link to the insulin-signaling pathway was evidenced by phosphorylation of IRS-1 during adipocyte differentiation (1), and phosphorylation of coactivator transducer of regulated CREB activity (TORC2) during insulin-modulated gluconeogenesis (2, 3). Bricambert et al. (4) also showed that SIK2 inhibits ChREBP-mediated hepatic lipogenesis and steatosis in mice through inhibitory phosphorylation of p300 HAT on Ser-89. Furthermore, SIK2 is known to regulate the initiation of mitosis through phosphorylating the centrosome linker protein, C-Nap1 (5). An intimate link between SIK2 and the CREB coactivator TORC1/2 has also been established, particularly in the contexts of melanogenesis (6, 7), cerebral ischemia-associated neuronal survival (8), and corticotropin-releasing hormone transcription (9). However, as an AMPK family kinase, its possible functional link to cellular stress response has not been reported and requires further clarification.
In contrast to the activation of AMPK, in which the causal role of energy disturbance is well established, the physiological conditions that underlie the activation of SIK2 remain to be defined. Phosphorylation of SIK2 on Thr-175 is the hallmark of its kinase activation. Although LKB1 is known to activate 13 kinases of the AMPK family (10), a low level of SIK2-Thr-175 phosphorylation still persisted in the LKB1-null HeLa cells, thus implying additional regulatory mechanisms. Another potential key determinant of the post-translational regulation of SIK2 kinase lies in protein stability and protein-protein interaction. Interestingly, CaMK1-mediated phosphorylation on the Thr-484 residue was previously shown to negatively regulate the SIK2 protein level (8), whereas PKA modulates the interaction between SIK2 and 14-3-3 by Ser-358 phosphorylation (11). However, issues regarding other modes of modifications and regulation for this multifunctional kinase are currently unresolved.
In this report, we found a hitherto unrecognized requirement of SIK2 activity for autophagosome maturation. Importantly, our work also revealed a novel post-translational regulation of its kinase activity, which is coordinated by p300/CBP and HDAC6. Collectively, these results extended the known cellular roles of SIK2 to critical functions in autophagy, and further highlight a mechanism by which the interplay among kinase and acetylase/deacetylase activities contribute to cellular protein pool homeostasis.
EXPERIMENTAL PROCEDURES
DNA Constructs and Mutagenesis
The pBluescript II vector encoding SIK2 sequence (KIAA0781) was from HUGE, Japan. The DNA fragment encoding SIK2 was excised using SalI and XhoI sites in the pBluescript-SIK2 vector and subcloned into pCMV-FLAG mammalian expression vector (Stratagene, La Jolla, CA). SIK2-K49M, SIK2-K53Q, and SIK2-K53R were generated with the site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The pCMV-FLAG-SIK2 plasmid was used as template. The following primers were synthesized for creating mutants: SIK2-KD (K49M), 5′-GGTGGCAATAATGATAATCGATAAG-3′; SIK2-K53R, 5′-GGCAATAAAAATAATCGATAGGTCTCAGCTGGATC-3′; and SIK2-K53Q, 5′-GGCAATAAAAATAATCGATCAGTCTCAGTCTCAGCTGGATGC-3′. The mutations were then confirmed by DNA sequencing. shRNAs against human SIK2 was generated by using the pSuper RNAi system (Oligoengine, Seattle, WA), with the target sequence for human SIK2 (NM_015191.1) 5′-GCAGTTGTTGTATGAACAA-3′. siRNAs targeting SIK1 and SIK2 were acquired from Dharmacon (Chicago, IL). All plasmids were verified by sequencing.
Antibodies and Recombinant Proteins
Anti-ubiquitin monoclonal antibody was from Millipore, and anti-GFP monoclonal antibody was from Clontech (Palo Alto, CA). Monoclonal antibodies to SIK2 (clone 15G10) and α-tubulin (clone 10D8), as well as rabbit-derived anti-FLAG tag, anti-β-actin, and anti-γ-tubulin antibodies were generated in the lab and affinity purified according to standard protocols. For SIK2, rabbit anti-Thr(P)-175 antibody were generated by keyhole limpet hemocyanin-conjugated phosphopeptide, ELLAT*WSGSPPYC. Recombinant HDAC1–8 proteins were isolated by immunoprecipitation of cell-free extracts from HEK293T cells transfected with FLAG-tagged recombinant expression plasmids.
Cell Culture and Transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (100 units/ml) at 37 °C in a 5% CO2 humidified atmosphere. Calcium phosphate-mediated transfection of HEK293T cells was performed according to a standard protocol.
Immunoprecipitation and Western Blot Analysis
Immunoprecipitation was performed as previously described (12). Cells expressing recombinant FLAG-SIK2 were harvested and rinsed with PBS. Lysis buffer containing 20 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 0.1% Triton X-100, 1 mm dithiothreitol (DTT), and protease inhibitors was used to prepare cell lysate. Cell lysates were immunoprecipitated with M2-agarose beads (Sigma-Aldrich) for 1 h at 4 °C. The agarose beads bound with FLAG-SIK2 were then washed 3 times with lysis buffer and subjected to PAGE. Western blot analysis was performed using the indicated antibodies and subsequently visualized using ECL chemiluminescence (PerkinElmer, Waltham, MA) and x-ray films (Eastman Kodak Co., Rochester, NY). Band signals were scanned before being quantified by the software Image Gauge (Fujifilm, Tokyo, Japan).
In-gel Tryptic Digestion and Mass Spectrometry
In-gel tryptic digestion was performed with the previously described procedure (13). The peptides resuspended in 0.1% formic acid were analyzed using a quadrupole ion trap mass spectrometer (Esquire 3000 plus, Bruker-Daltonics, Leipzig, Germany) interfaced with a high-pressure liquid chromatograph system (UltiMate, LC Packings, San Francisco, CA). A 150 × 0.5-mm C18 column (ZorbaxSB, Agilent Technologies, Palo Alto, CA), and mobile phases consisting of 0.1% formic acid in water and 0.1% formic acid in acetonitrile were used. An acetonitrile gradient from 15 to 60% in 60 min was performed to elute peptides from the column. The acquired mass spectra were then analyzed. Phosphorylation identification was also performed with LTQ-FT (linear quadrupole ion trap-Fourier transform ion cyclotron resonance) mass spectrometer (ThermoFisher Scientific, Inc., Waltham, MA), an Agilent 1100 Series binary HPLC pump (Agilent Technologies), and a Famos autosampler (LC Packings).
In Vitro Kinase Assay
The FLAG-SIK2 immune complexes were washed with lysis buffer three times and once with kinase buffer (20 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 50 mm NaCl, 1 mm DTT, and 0.5 mm NaF). Kinase reaction was performed by adding kinase buffer plus 50 μm ATP and 5 μCi of [γ-32P]ATP into the precipitates, followed by incubation at 30 °C for 15 min. The reaction was stopped by adding sample buffer, followed by SDS-PAGE and autoradiography.
In Vitro Deacetylation Assay
For the in vitro deacetylation assay, HDAC6 immunoprecipitated with M2 beads were washed with lysis buffer three times and twice with deacetylation buffer (10 mm Tris-HCl, pH 8.0, and 150 mm NaCl). The immune complexes were subsequently incubated with purified SIK2 in 10 μl of deacetylation buffer for 2 h at 37 °C. The reaction was terminated with SDS sample buffer, and subjected to SDS-PAGE and immunoblotting.
[γ-35S]ATP Binding Assay
Binding of [γ-35S]ATP by wild-type and mutant proteins was performed in 10 μl of binding buffer containing 25 mm HEPES, pH 8.1, 100 mm NaCl, 5 mm MgCl2, 1 mm DTT, 11% glycerol, and 9.6 μm [γ-35S]ATP (65 Ci/mmol, 12.5 mCi/ml) at 37 °C for 20 min. [γ-35S]ATP was cross-linked to proteins by a 5-min UV irradiation on ice in a UV cross-linker. The reactions were then resolved by SDS-PAGE and visualized by autoradiography.
Immunofluorescence Staining and Confocal Microscopy
Cells grown on coverslips were washed with PBS and fixed with 4% formaldehyde in PBS. The cells were permeabilized with 0.5% Triton X-100 in PBS and blocked by 1% BSA prior to being probed with the indicated antibodies. Alexa 488-conjugated goat anti-rabbit IgG, Alexa 594-conjugated goat anti-rabbit IgG, and Alexa 594-conjugated goat anti-mouse IgG (Invitrogen) were used as secondary antibodies. The cells were counterstained with Hoechst to visualize DNA. For the cells expressing GFP-SIK2 and red fluorescent ER marker, they were stained with DAPI immediately after the fixation step. Images were acquired using an inverted confocal microscopy (LSM-510, Zeiss, Thornwood, NY) installed with a ×63/NA 1.4 oil immersion objective lens. For quantification, five to 10 fields were randomly selected from each sample for analysis.
Electron Microscopy
The sample preparation procedure followed a common standard protocol with modification. Briefly, after fixation, the cells were neutralized with 0.1 m ammonium chloride and embedded with LR-Gold reagent (London Resin Co., Reading, Berkshire, UK). The ultrathin sections of the embedded samples were stained with uranyl acetate and lead citrate. Samples were examined using either a Jeol JEM 2100F (JEOL Ltd., Tokyo, Japan) or an FEI Tecnai T12 electron microscope (FEI Co., Eindhoven, Netherlands).
Preparation of Soluble/Insoluble Fractions and Slot-blot Analysis
Cells were harvested and extracted with whole cell extraction buffer (0.15 m NaCl, 20 mm Tris-HCl, pH 7.6, 0.1% Triton X-100) containing protease inhibitors and phosphatase inhibitors (1 mm PMSF, 1 μg/ml of leupeptin, 1 μg/ml of pepstain A, 1 mm DTT, 5 μm NaF, 1 mm Na2VO3) and 1 mm sodium butyrate. Lysates were passed through 25-guage needle 10 times using a syringe. Soluble and insoluble fractions were separated by centrifugations at 12,000 × g for 20 min. The insoluble fraction was resuspended in 6 m urea and sonicated for 15 s. Protein concentrations were determined by a Bio-Rad Protein Assay (Bio-Rad). Ten μg of soluble and insoluble fractions were mixed with SDS sample buffer and separated by SDS-PAGE for analysis. For slot-blot analysis, the supernatant of the urea-dissolved insoluble fraction was collected upon centrifugation and filtered through 0.22-μm nitrocellulose membrane with Minifold II Slot-Blot System (Schleicher and Schuell, Keene, NH).
Statistical Analysis
The data were analyzed with Student's t test. Data were mean ± S.D. p values <0.05 were considered significant.
RESULTS
Lys-53 Is a Major Acetylation Site of SIK2 That Is Catalyzed by p300 and CBP
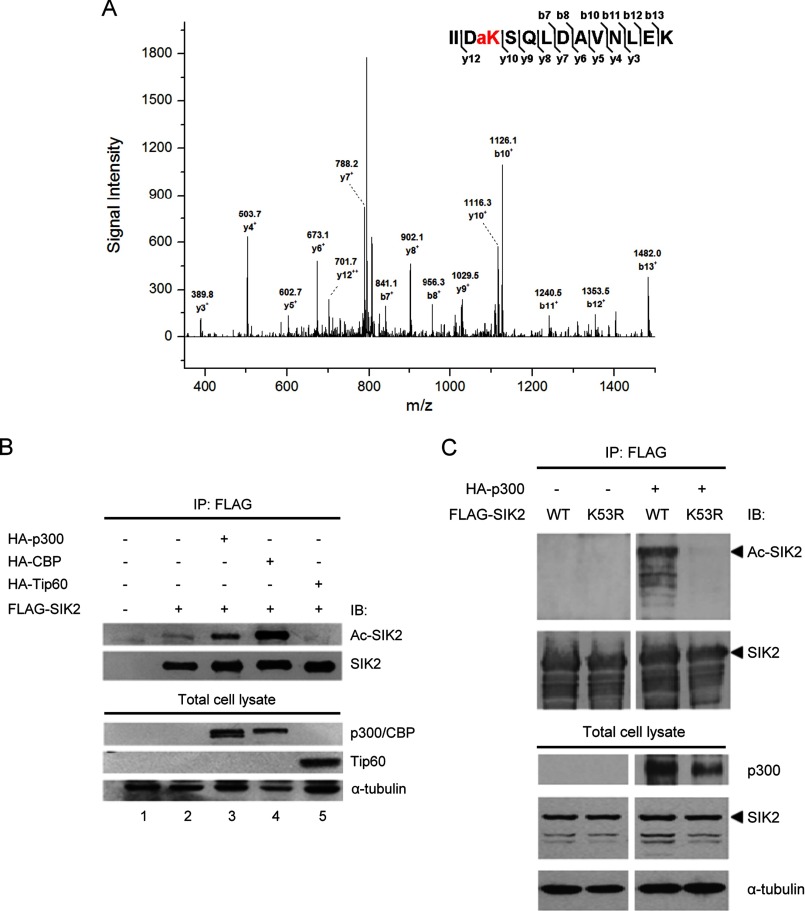
To deduce the regulatory mechanisms of SIK2 kinase activity, we first aimed to characterize post-translational modifications on SIK2 through mass spectrometric analysis of recombinant SIK2-WT protein recovered from HEK293T cells. The MS/MS spectra revealed that SIK2 undergoes acetylation on Lys-53 (Fig. 1A) as well as phosphorylation on Ser-358 and Ser-587 (supplemental Fig. S1). As the phosphorylation sites were previously reported (11, 14), we thus focused on further characterizing the acetylation modification. Next, to pinpoint the acetyltransferase responsible for this acetylation, we co-expressed p300, CBP, or Tat-interactive protein 60 (Tip60) with SIK2 and examined the acetylation level of SIK2. We discovered that SIK2 acetylation was enhanced by ectopic expression of p300 or CBP but remained unaltered in the presence of Tip60 (Fig. 1B), indicating their possible regulatory roles. To verify whether the p300/CBP-mediated acetylation is specific to the Lys-53 residue of SIK2, we co-expressed SIK2-WT or the nonacetylatable mutant (K53R) with p300. Immunoblotting analysis showed that the acetylation level of the SIK2-K53R mutant protein was abolished (Fig. 1C), demonstrating that Lys-53 is a predominant acetylation site on SIK2 targeted by p300. Similar results were obtained when the experiment was performed with CBP (data not shown).
FIGURE 1.
SIK2 is acetylated on Lys-53 by p300/CBP. A, FLAG-SIK2 was overexpressed and purified from HEK293T, and subjected to mass spectrometry-based analysis for post-translational modifications (see ”Experimental Procedures“). MS/MS spectra of signal m/z 815.0 that corresponds to a trypsinized SIK2 fragment containing Lys-53 acetylation. B, HEK293T cells were transiently transfected with the indicated expression constructs. Recombinant SIK2 was immunoprecipitated and subsequently probed using antibodies against acetylated lysine or FLAG. Expression of HATs was monitored by anti-HA antibody. C, wild-type (WT) or the K53R mutant of SIK2 was overexpressed in HEK293T cells, along with HA-p300. Immunoprecipitated recombinant SIK2 as well as lysate input were probed with the indicated antibodies.
Lys-53 Acetylation Inhibits the ATP-binding Ability and Kinase Activity of SIK2
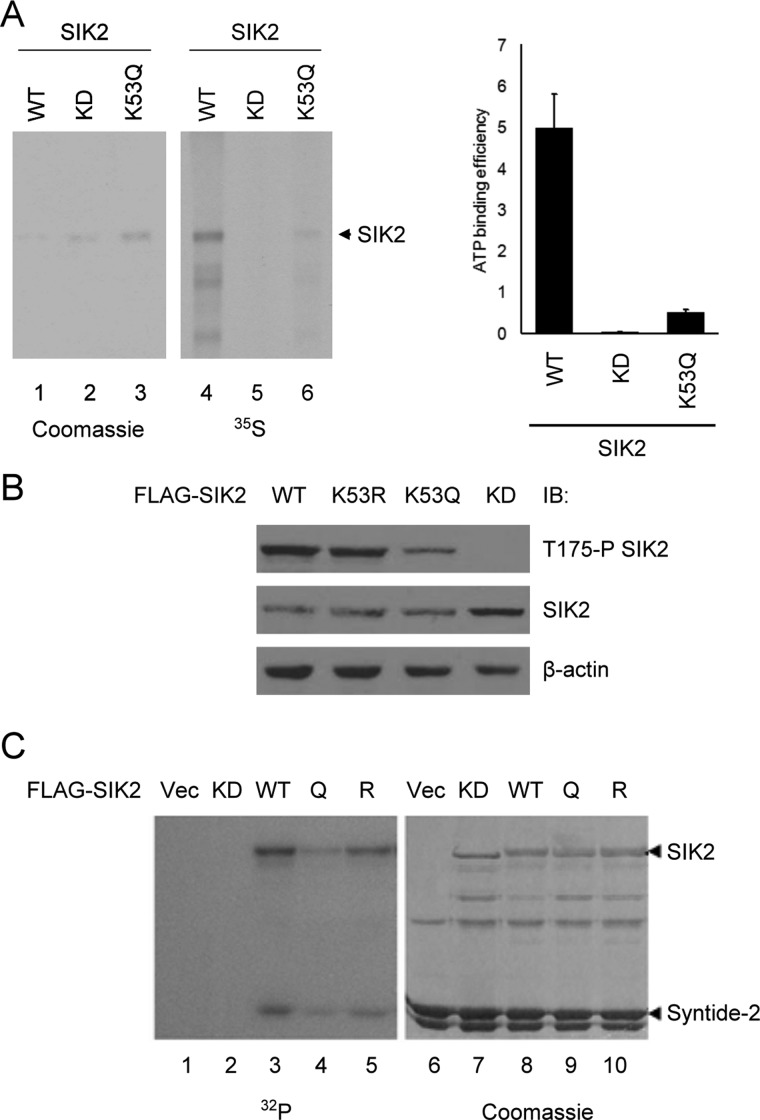
Because Lys-53 of SIK2 is a major acetylation site and located 4 residues C-terminal to the known ATP-binding residue Lys-49, we thus hypothesized that acetylation on this residue might affect the ATP-binding ability and kinase activity of SIK2 due to charge alteration. To address this issue, we performed an in vitro ATP-binding assay on SIK2-KD (a K49M, kinase-dead mutant) and the acetylation-mimetic SIK2-K53Q variant. The K53Q mutant clearly exhibited a deficiency in ATP binding comparable with that of the KD mutant (Fig. 2A), implying that a neutralization of charge by acetylation affects ATP binding. Next, we examined kinase activation by monitoring the phosphorylation of Thr-175 in the activation loop on both wild-type and mutant SIK2s (Fig. 2B). The immunoblotting results first indicated that Thr-175 phosphorylation of SIK2-KD was abolished, suggesting that this phosphorylation is mediated by autophosphorylation. Furthermore, Thr-175 phosphorylation of the K53Q variant was reduced, whereas the K53R form exhibited comparable phosphorylation as SIK2-WT, indicating that the kinase activity is indeed impaired by Lys-53 acetylation. As a means to further assess the kinase activity of the acetylation mutants, immunoprecipitated recombinant SIK2 was subjected to an in vitro kinase assay with GST-Syntide2 as the substrate. As shown in Fig. 2C, Syntide-2 was phosphorylated by the SIK2-K53Q mutant to a significantly lesser extent than by SIK2-WT or SIK2-K53R, providing further evidence that Lys-53 acetylation suppresses SIK2 kinase activity. Consistent with the above observation (Fig. 2B), autophosphorylation of SIK2 correlated with its activity toward Syntide-2, as indicated by the extent of phosphorylation. Taken together, these results indicate that SIK2 is inactivated by p300/CBP-mediated acetylation of Lys-53 in the vicinity of the ATP-binding pocket.
FIGURE 2.
The ATP-binding ability and kinase activity of SIK2 are both impaired by Lys-53 acetylation. A, HEK293T cells were transfected with the indicated wild-type and mutant SIK2 expression constructs. The recombinant proteins were immunoprecipitated for [γ-35S]ATP binding assay. Quantitative results of the relative ATP binding are presented in the right panel, with the intensity of [γ-35S]ATP being normalized to the corresponding protein level. Bar graph shows mean ± S.D. of two experimental replicates. B, wild-type (WT) or mutant (KD, K53R, and K53Q) SIK2 overexpressing HEK293T cells were harvested and the level of phosphorylated SIK2/Thr-175 was analyzed by Western blot with Thr-175 phosphospecific antibody. C, HEK293T cells were transiently transfected with the indicated constructs. The recombinant SIK2 proteins were immunoprecipitated and subjected to in vitro kinase reaction with GST-Syntide-2 as the substrate. Vec, vector control; Q, the K53Q mutant; R, the K53R mutant.
HDAC6 Activates SIK2 by Reversing Acetylation at Lys-53
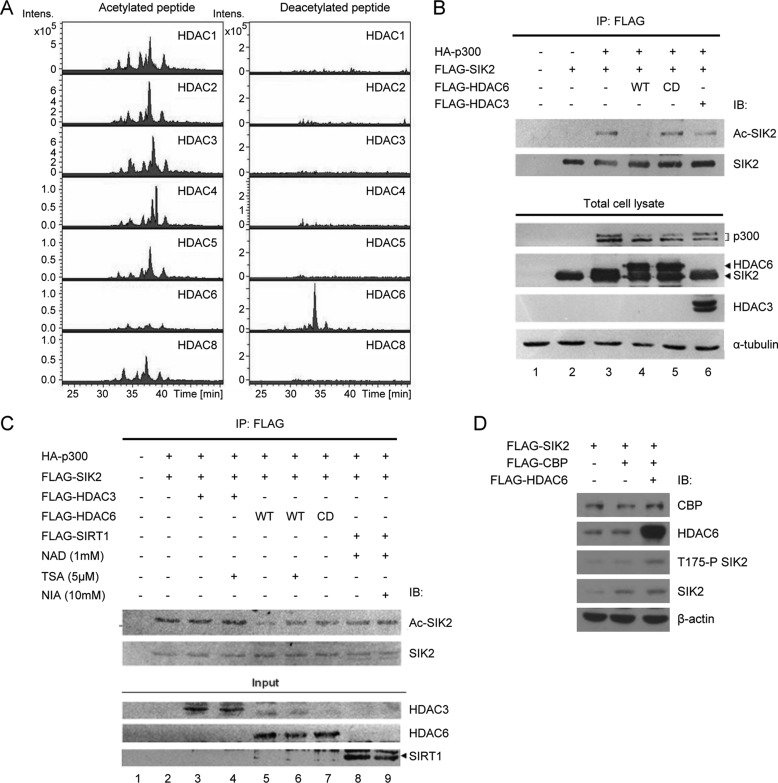
In an attempt to search for the enzyme that deacetylates SIK2 on Lys-53, a peptide containing an acetylated Lys-53 (KIIDK*SQLDAC) was used for the in vitro deacetylation assay in conjunction with purified recombinant HDAC1, -2, -3, -4, -5, -6, and -8 proteins. Upon performing LC-MS/MS analyses, we found that only HDAC6 could deacetylate the synthetic acetylpeptide (Fig. 3A). Furthermore, the acetylation level of SIK2 was significantly reduced when HDAC6 was overexpressed in the cells, indicating that HDAC6 deacetylates SIK2 in vivo (Fig. 3B). Such decline in SIK2 acetylation was not observed when cells expressed a catalytically inactive HDAC6 mutant or HDAC3 (Fig. 3B, lanes 5 and 6), further supporting a specific requirement for the deacetylase activity of HDAC6. To further clarify whether HDAC6 counterbalances p300-mediated SIK2 acetylation, we performed an in vitro deacetylation assay of acetylated-SIK2. Acetylated-SIK2 was first immunoprecipitated from cells co-expressing p300 and SIK2 in the presence of trichostatin A. The in vitro deacetylation reaction was then performed with recombinant HDAC6, HDAC3, or SIRT1 in the presence or absence of inhibitors. As shown in Fig. 3C, the SIK2 acetylation level was decreased when incubated with HDAC6 (lane 5), whereas it remained unchanged in the presence of a catalytically inactivated HDAC6 mutant (HDAC6-CD) or the HDAC inhibitor trichostatin A (lanes 6 and 7). Furthermore, HDAC3 or SIRT1 did not affect the acetylation level of SIK2 (Fig. 3C, lanes 3–4 and 8–9). Similar data were obtained when CBP was expressed (data not shown). Collectively, these data suggest that p300/CBP and HDAC6 specifically and reciprocally regulate the acetylation level of SIK2/Lys-53.
FIGURE 3.
Acetylation-dependent inhibition of SIK2 is reversed by HDAC6-mediated deacetylation. A, the synthetic SIK2 peptide containing a Lys-53 acetylation (KIIDacKSQLDAC) was incubated with recombinant HDAC proteins (HDAC1, -2, -3, -4, -5, -6, and -8). Following the reaction, the mixtures were subjected to mass spectrometric analysis to monitor any changes in the level of peptide acetylation. Extracted ion chromatography of acetylated peptides (left panel) and nonacetylated peptides (right) is shown, illustrating removal of the acetyl group in the HDAC6-containing reaction. B, HEK293T cells were transiently transfected with FLAG-SIK2-WT, p300, or HDAC6 in various combinations, as indicated (CD, catalytically deficient). Recombinant FLAG-SIK2 was immunoprecipitated and the acetylation level of SIK2 was probed with antibody against acetylated lysine. C, acetylated SIK2 protein was immunoprecipitated from the lysates of HEK293T cells ectopically expressing FLAG-SIK2 and HA-p300. The immunoprecipitated SIK2 proteins were then incubated with the indicated deacetylases, and in the absence or presence of the deacetylase inhibitors. Immunoblotting analysis was performed as above. D, HEK293T cells were transfected with plasmids encoding FLAG-SIK2, FLAG-CBP, and FLAG-HDAC6 in the indicated combinations. Phosphorylation level of SIK2-Thr-175 as well as the expression of the recombinant proteins were probed with the denoted antibodies.
For further confirmation of the role of acetylation/deacetylation in SIK2 activity regulation, we probed the phosphorylation level of SIK2/Thr-175 in the context of CBP and HDAC6 overexpression. The results showed that Thr-175 phosphorylation was inhibited by CBP, whereas overexpression of HDAC6 restored the CBP-mediated inhibition of Thr-175 phosphorylation (Fig. 3D). These findings thus imply a reversible SIK2 activity regulatory switch that hinges on Lys-53 acetylation/deacetylation.
Acetylation Elicits SIK2 Sequestration to Autophagosomes
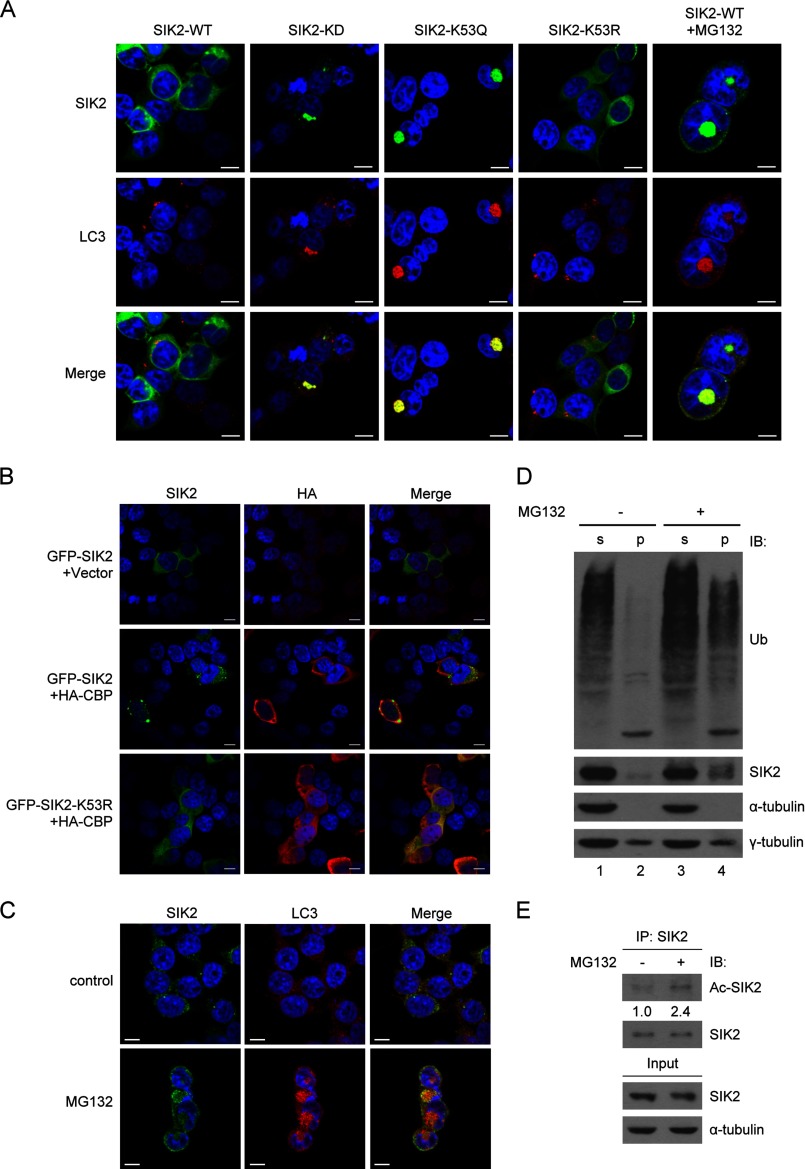
Having demonstrated the kinase inhibitory effect of Lys-53 acetylation on SIK2, we then sought to assess whether this modification alters cellular attributes of SIK2 such as subcellular localization. To this end, immunofluorescence staining analysis of SIK2-WT, SIK2-KD, or the acetylation site mutants was performed. In contrast to the wild-type SIK2 and K53R mutant, which exhibited diffused patterns of cytoplasmic distribution, the kinase-deficient mutants, KD and K53Q, were found sequestered in autophagosome structures characterized by LC3 (Fig. 4A). This observation suggests that the subcellular localization of SIK2 relies on its Lys-53 acetylation as well as kinase activity. To further corroborate the role of acetylation on the spatial co-localization of SIK2 with autophagosomes, we performed additional experiments on cells co-transfected with CBP and SIK2-WT or SIK2-K53R. Confocal microscopy analysis illustrated that co-expression of CBP indeed promoted accumulation of SIK2 in aggresomes, whereas the nonacetylatable K53R remained in the cytosol (Fig. 4B), supporting the notion that acetylation of SIK2 underlies such a unique distribution.
FIGURE 4.
Sequestration of acetylated-SIK2 to autophagosomes. A, HEK293T cells overexpressing the WT, KD, K53Q, or K53R variant of GFP-SIK2 were analyzed by immunofluorescence staining. Endogenous LC3 (red) was detected by anti-LC3B antibody. The rightmost panel shows GFP-SIK2-WT cells with MG132 (5 μm) treatment for 16 h. Scale bar, 10 μm. B, HEK293T cells were co-transfected with HA-CBP- (or the vector control) and GFP-SIK2-encoding plasmids, followed by immunofluorescence staining of the overexpressed HA-CBP (red) using anti-HA antibody. Scale bar, 10 μm. C-E, HEK293T cells were treated with or without MG132 (5 μm) for 16 h. They were then subjected to immunofluorescence staining (C), separated into soluble (s) and insoluble (p) fractions for protein expression analysis (D), or extracted for anti-SIK2 immunoprecipitation (E). The scale bar in C is equivalent to 10 μm. Western blots in D and E were done using the indicated antibodies. Relative levels of SIK2 acetylation were determined by normalizing the levels of acetylated SIK2 signals to those of total SIK2 protein in the respective samples, with the control sample being represented as 1.
Notably, in response to MG132-mediated proteasome inhibition, even the wild-type form of SIK2 was sequestered to autophagosomes (Fig. 4A, rightmost panel). The endogenous SIK2 protein also exhibited this spatial overlap with autophagosome upon proteasome blockade (Fig. 4C). We further confirmed this observation by analyzing protein expression in the soluble and insoluble fractions of MG132-treated cell extracts (Fig. 4D). In this assay, no contamination of insoluble fraction with soluble constituents was evidenced by the presence of γ-tubulin and absence of α-tubulin. In line with the regulated subcellular distribution, the protein level of endogenous SIK2, similarly to the ubiquitinated protein levels, was elevated in the insoluble fraction after MG132 treatment (Fig. 4D, compare lanes 2 and 4). Additionally, congruent with the above observations that linked the acetylation state of SIK2 to its stress-associated distribution, the acetylation level of SIK2 was elevated after MG132 treatment (Fig. 4E). Together, these findings therefore suggest that stalled protein turnover may trigger a dynamic, acetylation-dependent distribution of SIK2, and further signify its link to aggregated protein removal.
SIK2 Is Indispensible for the Processing of Autophagosomes
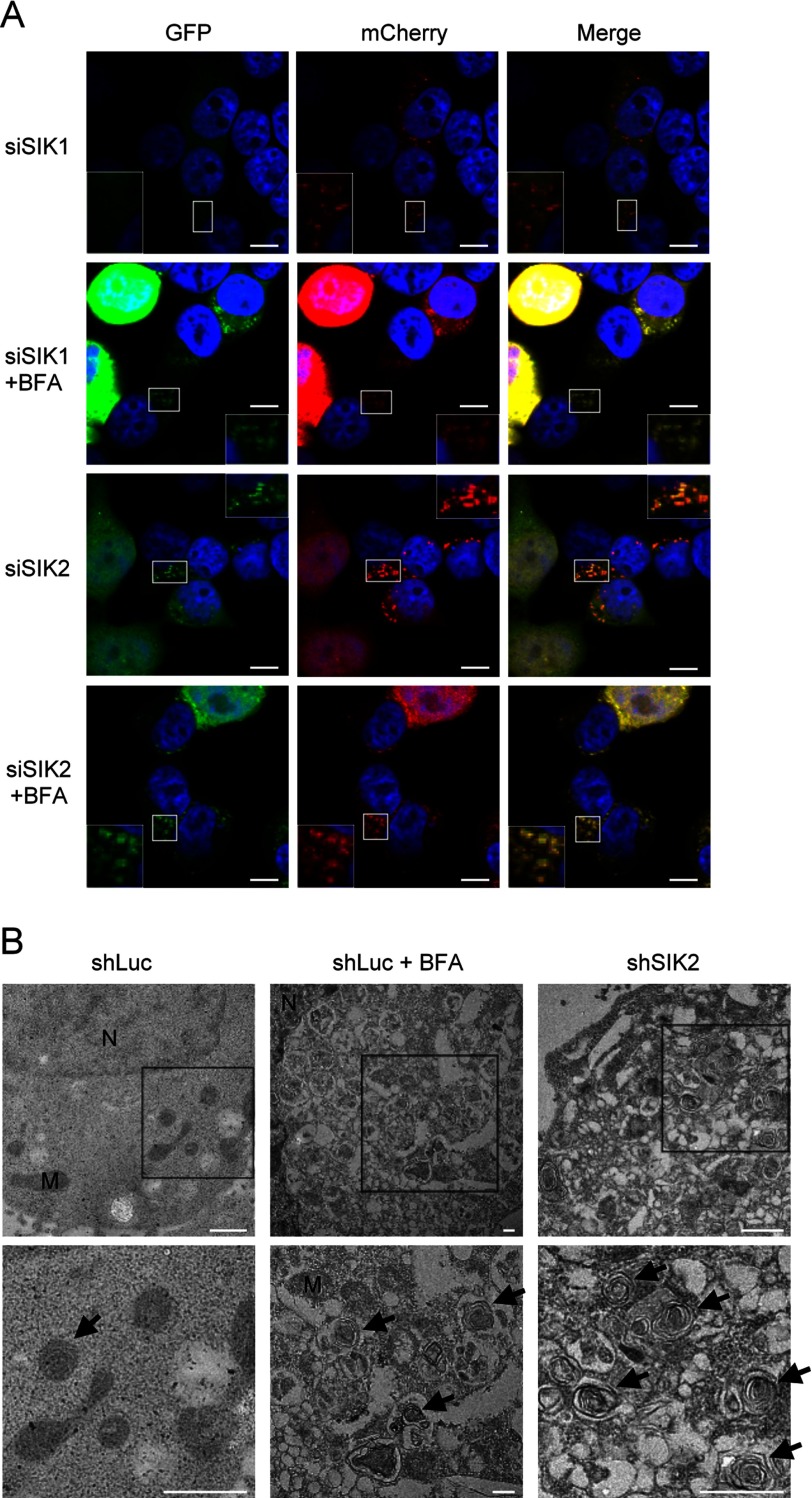
Abnormal accumulation of autophagosomes thus implies that the kinase activity of SIK2 may be required for facilitating the clearance of aggregated proteins. To next explore this possibility, we utilized a pH-sensitive mCherry-GFP-LC3, whose GFP ceases to fluoresce once fused with lysosome (15), to examine autophagosome-lysosome fusion efficiency in SIK2-knockdown HEK293T cells. The immunofluorescence staining data subsequently revealed that knockdown of SIK2, but not SIK1, led to persisted GFP signals, thus indicating a disrupted autophagosome-lysosome fusion (Fig. 5A). Correspondingly, fine structure analysis using transmission electron microscopy revealed marked accumulation of autophagosome structures in the absence of SIK2 (Fig. 5B), suggesting a positive role of SIK2 in the processing of autophagosomes.
FIGURE 5.
SIK2 is essential for autophagosome processing. A, HEK293T cells were co-expressed mCherry-GFP-LC3 with siRNA targeting SIK1 or SIK2. After treatment with or without 0.2 μm bafilomycin A1 (BFA) for 16 h, cells were fixed and analyzed by immunofluorescence and confocal microscopy (scale bar, 10 μm). B, electron microscopy images of SIK2 knockdown cells. HEK293T cells expressing control (shLuc) or SIK2-targeting (shSIK2) shRNA were subjected to transmission electron microscopy analysis. Control cells were also treated with (shLuc + BFA) or without (shLuc) 0.2 μm bafilomycin A1 for 16 h. Bottom figures represent enlarged images of the boxed regions in the top figures. Arrow, autophagosome; M, mitochondria; N, nucleus. Scale bar, 1 μm.
SIK2 Activity Is Required for the Clearance of TDP-43Δ Inclusion Bodies
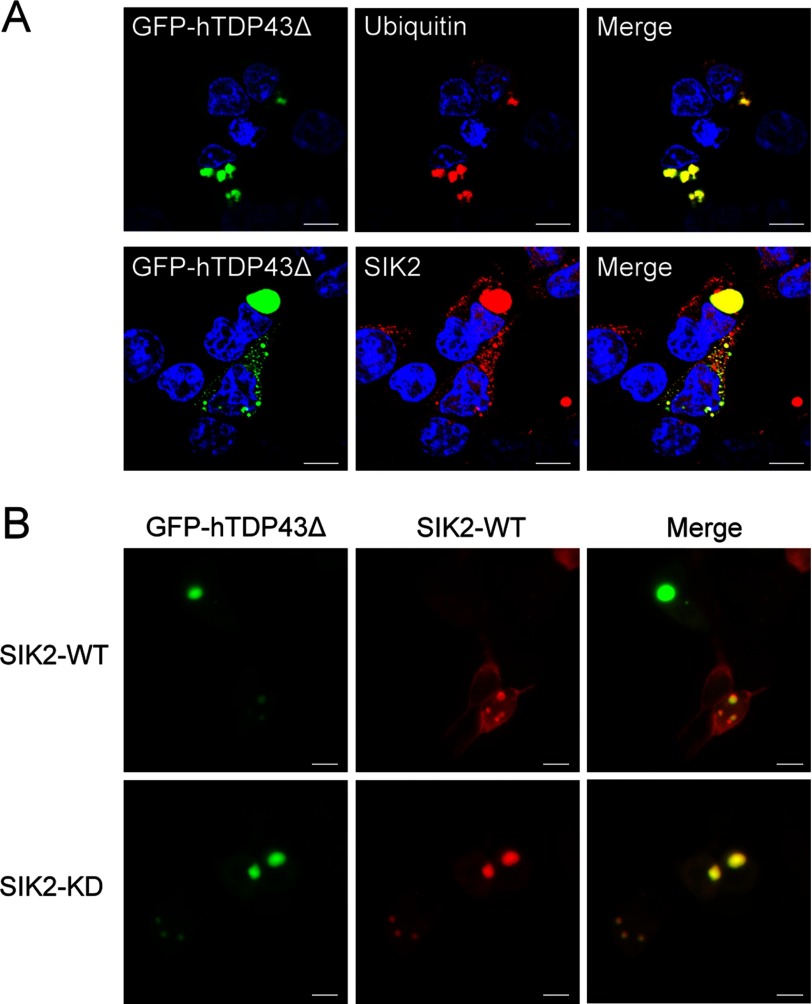
Owning to the possible role of SIK2 in mediating removal of aggregated proteins, we next set out to address whether SIK2 is associated with aggregated proteins in inclusion bodies. To this end, we transfected a 25-kDa truncated form of TDP-43, a pathological signature of neurodegeneration (16–18), into HEK293T cells, and examined the subcellular localization of relevant proteins. Immunofluorescence staining analysis demonstrated partial co-localization of endogenous SIK2 with the ubiquitin-positive TDP-43Δ inclusion bodies in the perinuclear region (Fig. 6A). Interestingly, SIK2-KD completely coincided with TDP-43Δ inclusion bodies as compared with the limited co-localization displayed by SIK2-WT (Fig. 6B). Together, these results suggest that SIK2 could be sequestrated to TDP-43Δ-associated inclusion bodies, and may further imply a functional relevance.
FIGURE 6.
SIK2 co-localizes with the TDP-43Δ inclusion bodies. A, HEK293T cells ectopically expressing GFP-hTDP43Δ were fixed and immunostained with anti-ubiquitin (upper) or anti-SIK2 (lower) antibodies. Scale bar, 10 μm. B, cells were co-transfected with GFP-hTDP43Δ- and FLAG-SIK2-encoding plasmids, followed by immunofluorescence staining using anti-FLAG antibody. Single-section images of confocal microscopy were acquired and shown (scale bar, 10 μm).
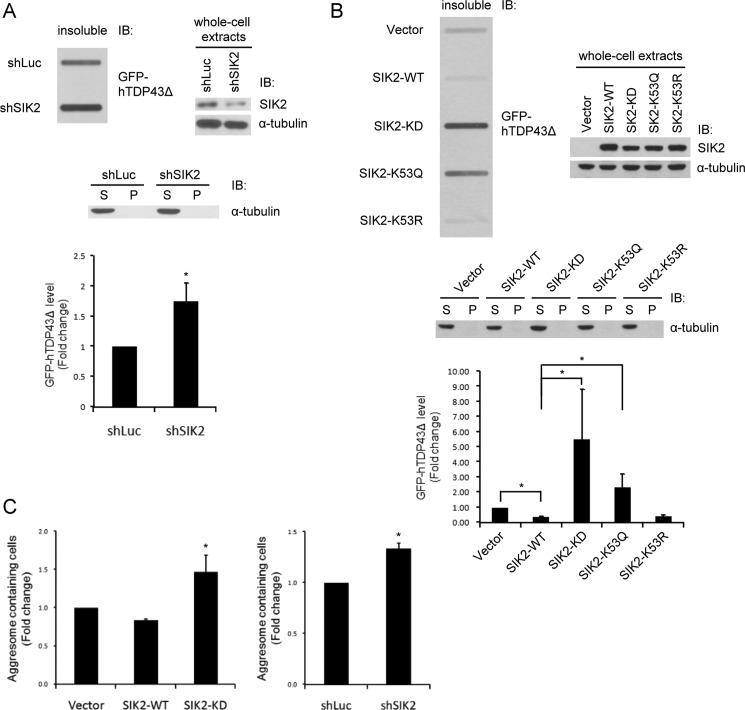
To next investigate whether SIK2 participates in the removal of aggregated proteins, slot-blot was performed to analyze the level of TDP-43Δ in insoluble fractions of SIK2-depleted cells. As illustrated in Fig. 7A, TDP-43Δ accumulated in insoluble fraction when SIK2 was down-regulated. Similarly, overexpression of SIK2-WT and SIK2-K53R resulted in lower levels of TDP-43Δ in the insoluble fraction as compared with the vector control, whereas SIK2-KD and SIK2-K53Q led to TDP-43Δ accumulation in the insoluble fraction (Fig. 7B). Next, we quantified the number of inclusion bodies containing cells in the presence of SIK2-WT/KD or SIK2-targeting shRNA, and subsequently found increased occurrence of such aberrant cells upon SIK2-KD overexpression (Fig. 7C, left) or SIK2 ablation by RNAi (Fig. 7C, right). These results are consistent with the above observations of autophagosome accumulation (Figs. 4A and 5B), and recapitulate the notion that the kinase activity of SIK2 represents a critical determinant for autophagosome maturation and aggresome removal.
FIGURE 7.
SIK2 is required for the processing/removal of TDP-43Δ inclusion bodies. A and B, HEK293T cells were transfected with plasmids expressing luciferase- or SIK2-targeting shRNAs (A), or the WT, KD, K53Q, or K53R variants of SIK2 (B). GFP-hTDP43Δ was also co-expressed. Cell extracts were separated into soluble (S) and insoluble (P) fractions. Insoluble fractions were analyzed by slot-blot assay using anti-GFP antibody (top left panel). Soluble and insoluble fractions (middle) and whole cell extracts (top right panel) were probed with the indicated antibodies to show the expression levels of various proteins. Bar graph in the bottom panel shows the quantification of GFP-hTDP43Δ intensity on the slot-blot. Mean ± S.D. were calculated from 3 independent experiments. *, p < 0.05. C, HEK293T cells were transiently transfected plasmids for the co-expression of: GFP-hTDP43Δ with FLAG-SIK2 WT or KD (left panel) or GFP-hTDP43Δ with control shRNA or SIK2-targeting shRNA (right panel). Forty-eight h after transfection, cells were fixed and analyzed for the proportion of aggresome-containing cells. Quantitative data are mean ± S.D. of two independent experiments (*, p < 0.05).
DISCUSSION
In this article, we uncovered a two-component regulatory switch for the SIK2 kinase activity and function, kinase autoactivation and the acetylation/deacetylation-mediated inactivation/activation. p300/CBP-mediated Lys-53 acetylation inhibited SIK2 kinase activity, whereas HDAC6-mediated deacetylation restored the activity. Importantly, alterations in the ATP-binding pocket, exhibited by the kinase-dead (K49M) and acetylation-mimetic (K53Q) variants, were shown to impede the phosphorylation of Thr-175 (Fig. 2B). Therefore, as a single-polypeptide member of the AMPK family, autophosphorylation of SIK2 on the activation loop (Thr-175) may represent a distinct feature of its kinase activation. Intriguingly, SIK2 was previously shown to target p300 HAT through inhibitory phosphorylation on Ser-89 (4). Our present study thus raises the possibility of a feedback mechanism that cross-regulates these two enzymatic activities, which may have significant functional implications in processes such as gene expression and autophagy.
In addition to the inhibition of SIK2 activity, acetylation also led to sequestration of SIK2 in autophagosomes. Remarkably, proteasome inhibition by MG132 resulted in sequestration of SIK2 to autophagosomes as well as elevation of its acetylation level (Fig. 4). Interestingly, we also found that the stability of SIK2 is dependent on the heat-shock protein HSP90 and negatively regulated by proteasome-mediated turnover.4 These observations prompted us to assume that SIK2 might play a regulatory role in autophagy induced by proteasome inhibition. Supporting this assumption, we further demonstrated that SIK2 is indispensible for autophagosome processing (Fig. 5). Unexpectedly, our present results have shown some similarities as well as discrepancies in the phenotypes between knockdown of SIK2 and overexpression of SIK2-KD or SIK2-K53Q, whereas knockdown of SIK2 resulted in compromised autophagosome maturation (Fig. 5), overexpression of SIK2-KD or SIK2-K53Q triggered a more severe extent of aggresome accumulation than SIK2 knockdown (Fig. 4A). This unique phenotype may be attributed to a forced sequestering of aggregated proteins to the aggresomes by the overexpressed SIK2-K53Q or SIK2-KD. In support of this premise, we have observed that highly elevated levels of polyubiquitinated proteins were immunoprecipitated with overexpressed SIK2-KD as compared with SIK2-WT.5
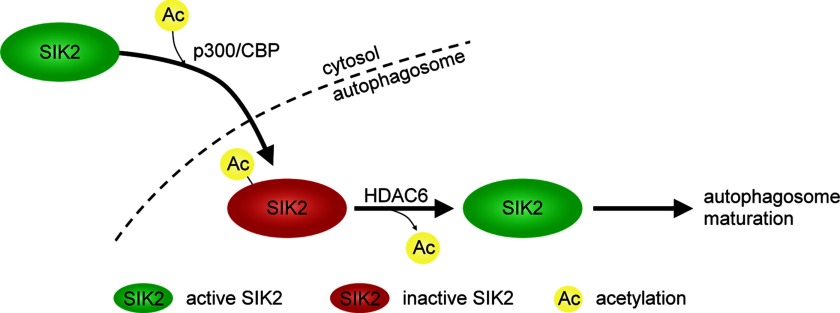
Furthermore, there is emerging evidence suggesting the critical role of acetylation in autophagy regulation (19–21). Both p300 and HDAC6, the enzymes controlling SIK2 acetylation, were previously reported as key regulators of autophagy. p300 modulates autophagy through augmentation of Atg5, Atg7, Atg8, and Atg12 acetylation (19), and HDAC6 is required for the formation and maturation of autophagy (22, 23). Therefore, it seems very likely that the function of SIK2 in autophagy is regulated through reversible modification controlled by p300 and HDAC6. Our results are consistent with the scenario that, upon inactivation of SIK2 by p300/CBP-mediated Lys-53 acetylation and its subsequent sequestration to autophagosomes, SIK2 may undergo deacetylation at Lys-53 by HDAC6 and regain its kinase activity required for the subsequent maturation of autophagosome (Fig. 8). Thus, the present findings provide a physiological explanation for how SIK2 is functioning in autophagy in a kinase activity-dependent manner.
FIGURE 8.
SIK2 kinase activity, controlled by an acetylation-based regulatory switch, contributes to the progression of autophagy. Upon proteasome inhibition, SIK2 is inhibited by p300/CBP-mediated Lys-53 acetylation and sequestrated to autophagosomes. Deacetylation of SIK2 by HDAC6 in autophagosomes likely reactivates SIK2 and promotes autophagosome processing. (See text for discussion.)
The physiological as well pathophysiological significance of autophagy is evidenced by its link to many diseases including cancer and neurodegeneration (24, 25). Autophagy is intricately linked to cancer, it may serve to inhibit continuous cell growth at the early stage, whereas promote transformed cells survival by recycling damaged organelles (24). In contrast, autophagy is thought to protect cells against neurodegenerative disorders (26, 27). Notably, the anomalous autophagic structures in the SIK2 knockdown cells (Fig. 5B) were also observed in dystrophic neuritis of Alzheimer disease patients (28). More importantly, we demonstrated that the kinase activity of SIK2 is required for the clearance of the TDP-43 truncated fragment aggregates, which are the hallmark of several neurodegenerative diseases, such as frontotemporal lobar degeneration and amyotrophic lateral sclerosis (29–31). Given that SIK2 was recently found highly expressed in neurons (8), our observations of its involvement in autophagosome processing and TDP-43Δ inclusion body removal strongly implies a protective role of this kinase in neurons. Hence, the development of agonists for boosting SIK2 activity or inhibitors of SIK2 Lys-53 acetylation, both of which presumably would facilitate autophagy, might be an effective pharmacological strategy in treating neurodegenerative or protein aggregate disorders.
Supplementary Material
Acknowledgments
We thank Drs. Tso-Pang Yao for HDAC6 and Mitsuhiro Okamoto for SIK2 and GST-Syntide plasmids. We are indebted to Dr. Yeu-Kuang Hwu of Academia Sinica and the National Synchrotron Radiation Research Center for kindly providing a Jeol 2100F EM, to the Institute of Molecular Biology, Academia Sinica, for the FEI Tecnai G2 EM service, and Hsiao-Hsuian Shen, Shu-Ping Tsai, Wen-Li Peng, Sue-Ping Lee, and Yi-Yun Chen for EM technical assistance. We appreciate Yi-Li Liu and I-Ching Huang for their technical expertise on DNA sequencing, supported in part by the Department of Medical Research, National Taiwan University Hospital.
This work was supported by National Science Council Grants NSC96-2321-B-002-008 and NSC100-2320-B-002-008 (to S.-C. L.), NSC99-2632-B-182-011-MY3, NSC101-2320-B-182-036-MY3, and NSC101-2321-B-182-009 (to B. C-M. T.), and grants from the Academia Sinica (to M.-D. T.) and the Institute of Biological Chemistry, Academia Sinica (to S.-C. L).

This article contains supplemental Fig. S1.
Y. H. Lin, unpublished data.
Y. H. Lin and C. T. Chuang, unpublished data.
- SIK2
- salt-inducible kinase 2
- AMPK
- AMP-activated protein kinase
- CREB
- cAMP-response element-binding protein
- KD
- kinase dead.
REFERENCES
- 1. Horike N., Takemori H., Katoh Y., Doi J., Min L., Asano T., Sun X. J., Yamamoto H., Kasayama S., Muraoka M., Nonaka Y., Okamoto M. (2003) Adipose-specific expression, phosphorylation of Ser-794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 278, 18440–18447 [DOI] [PubMed] [Google Scholar]
- 2. Dentin R., Liu Y., Koo S. H., Hedrick S., Vargas T., Heredia J., Yates J., 3rd, Montminy M. (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 [DOI] [PubMed] [Google Scholar]
- 3. Muraoka M., Fukushima A., Viengchareun S., Lombès M., Kishi F., Miyauchi A., Kanematsu M., Doi J., Kajimura J., Nakai R., Uebi T., Okamoto M., Takemori H. (2009) Involvement of SIK2/TORC2 signaling cascade in the regulation of insulin-induced PGC-1α and UCP-1 gene expression in brown adipocytes. Am. J. Physiol. Endocrinol. Metab. 296, E1430–1439 [DOI] [PubMed] [Google Scholar]
- 4. Bricambert J., Miranda J., Benhamed F., Girard J., Postic C., Dentin R. (2010) Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 120, 4316–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed A. A., Lu Z., Jennings N. B., Etemadmoghadam D., Capalbo L., Jacamo R. O., Barbosa-Morais N., Le X. F., Australian Ovarian Cancer Study Group, Vivas-Mejia P., Lopez-Berestein G., Grandjean G., Bartholomeusz G., Liao W., Andreeff M., Bowtell D., Glover D. M., Sood A. K., Bast R. C., Jr. (2010) SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell 18, 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horike N., Kumagai A., Shimono Y., Onishi T., Itoh Y., Sasaki T., Kitagawa K., Hatano O., Takagi H., Susumu T., Teraoka H., Kusano K., Nagaoka Y., Kawahara H., Takemori H. (2010) Down-regulation of SIK2 expression promotes the melanogenic program in mice. Pigment Cell Melanoma Res. 23, 809–819 [DOI] [PubMed] [Google Scholar]
- 7. Kumagai A., Horike N., Satoh Y., Uebi T., Sasaki T., Itoh Y., Hirata Y., Uchio-Yamada K., Kitagawa K., Uesato S., Kawahara H., Takemori H., Nagaoka Y. (2011) A potent inhibitor of SIK2, 3,3′,7-trihydroxy-4′-methoxyflavon (4′-O-methylfisetin), promotes melanogenesis in B16F10 melanoma cells. PLoS One 6, e26148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sasaki T., Takemori H., Yagita Y., Terasaki Y., Uebi T., Horike N., Takagi H., Susumu T., Teraoka H., Kusano K., Hatano O., Oyama N., Sugiyama Y., Sakoda S., Kitagawa K. (2011) SIK2 is a key regulator for neuronal survival after ischemia via TORC1-CREB. Neuron 69, 106–119 [DOI] [PubMed] [Google Scholar]
- 9. Liu Y., Poon V., Sanchez-Watts G., Watts A. G., Takemori H., Aguilera G. (2012) Salt-inducible kinase is involved in the regulation of corticotropin-releasing hormone transcription in hypothalamic neurons in rats. Endocrinology 153, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lizcano J. M., Göransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Mäkelä T. P., Hardie D. G., Alessi D. R. (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henriksson E., Jones H. A., Patel K., Peggie M., Morrice N., Sakamoto K., Göransson O. (2012) The AMPK-related kinase SIK2 is regulated by cAMP via phosphorylation at Ser358 in adipocytes. Biochem. J. 444, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J. Y., Chen W. H., Chang Y. L., Wang H. T., Chuang W. T., Lee S. C. (2006) Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 34, 2398–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsay Y. G., Wang Y. H., Chiu C. M., Shen B. J., Lee S. C. (2000) A strategy for identification and quantitation of phosphopeptides by liquid chromatography/tandem mass spectrometry. Anal. Biochem. 287, 55–64 [DOI] [PubMed] [Google Scholar]
- 14. Screaton R. A., Conkright M. D., Katoh Y., Best J. L., Canettieri G., Jeffries S., Guzman E., Niessen S., Yates J. R., 3rd, Takemori H., Okamoto M., Montminy M. (2004) The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74 [DOI] [PubMed] [Google Scholar]
- 15. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 16. Wang I. F., Wu L. S., Shen C. K. (2008) TDP-43. An emerging new player in neurodegenerative diseases. Trends Mol. Med. 14, 479–485 [DOI] [PubMed] [Google Scholar]
- 17. Lee E. B., Lee V. M., Trojanowski J. Q. (2012) Gains or losses. Molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 13, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amador-Ortiz C., Lin W. L., Ahmed Z., Personett D., Davies P., Duara R., Graff-Radford N. R., Hutton M. L., Dickson D. W. (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann. Neurol. 61, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee I. H., Finkel T. (2009) Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 284, 6322–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yi C., Ma M., Ran L., Zheng J., Tong J., Zhu J., Ma C., Sun Y., Zhang S., Feng W., Zhu L., Le Y., Gong X., Yan X., Hong B., Jiang F. J., Xie Z., Miao D., Deng H., Yu L. (2012) Function and molecular mechanism of acetylation in autophagy regulation. Science 336, 474–477 [DOI] [PubMed] [Google Scholar]
- 21. Yi C., Yu L. (2012) How does acetylation regulate autophagy? Autophagy 8, 1529–1530 [DOI] [PubMed] [Google Scholar]
- 22. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 23. Lee J. Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y. S., Pandey U. B., Kaushik S., Tresse E., Lu J., Taylor J. P., Cuervo A. M., Yao T. P. (2010) HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shintani T., Klionsky D. J. (2004) Autophagy in health and disease. A double-edged sword. Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knaevelsrud H., Simonsen A. (2010) Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 584, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 26. Ravikumar B., Duden R., Rubinsztein D. C. (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 27. Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 28. Nixon R. A., Wegiel J., Kumar A., Yu W. H., Peterhoff C., Cataldo A., Cuervo A. M. (2005) Extensive involvement of autophagy in Alzheimer disease. An immunoelectron microscopy study. J. Neuropathol. Exp. Neurol. 64, 113–122 [DOI] [PubMed] [Google Scholar]
- 29. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. (2007) Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 27, 10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohn T. T. (2008) Caspase-cleaved TAR DNA-binding protein-43 is a major pathological finding in Alzheimer's disease. Brain Res. 1228, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.