Background: As a phosphatase belonging to the PP2C family, ILKAP plays key roles in the regulation of cell survival and apoptosis.
Results: ILKAP interacts with importin α proteins; nuclear ILKAP interacts with RSK2 and induces apoptosis by inhibiting RSK2 activity.
Conclusion: ILKAP contains a functional NLS and induces apoptosis by regulating RSK2 signaling.
Significance: ILKAP may regulate cell survival and apoptosis through the activation of nuclear pathways.
Keywords: Apoptosis, Cell Cycle, Nuclear Transport, Phosphatase, Signal Peptidase, ILKAP, PP2C, RSK2
Abstract
Integrin-linked kinase-associated phosphatase (ILKAP) is a serine/threonine (S/T) phosphatase that belongs to the protein phosphatase 2C (PP2C) family. Many previous studies have demonstrated that ILKAP plays key roles in the regulation of cell survival and apoptosis. Researchers have thus far considered ILKAP a cytoplasmic protein that negatively regulates integrin signaling by interacting with and phosphorylating integrin-linked kinase 1 (ILK1). In this study, we found that both endogenous and tagged ILKAP mainly localize to the nucleus and that the nuclear transport of ILKAP is nuclear localization signal (NLS) importin-mediated. The ILKAP protein interacts directly with importin α1, α3, and α5. The NLS in ILKAP is located in the N-terminal region between amino acids 71 and 86, and the NLS-deleted ILKAP protein was distributed in the cytoplasm. In addition, we show that Lys-78 and Arg-79 are critical for the binding of ILKAP to importin α. We also found that nuclear ILKAP interacts with ribosomal protein S6 kinase-2 (RSK2) and induces apoptosis by inhibiting RSK2 activity and down-regulating the expression level of the RSK2 downstream substrate cyclin D1. These results indicate that ILKAP is a nuclear protein that regulates cell survival and apoptosis through the regulation of RSK2 signaling.
Introduction
ILKAP4 is a serine/threonine (S/T) phosphatase of the protein phosphatase 2C family, which was originally identified by Tong et al. (1) in 1998. Recent studies indicate that ILKAP plays key roles in the regulation of cell survival and apoptosis. ILKAP activates the apoptosis signal-regulating kinase 1 (ASK1) by enhancing the cellular phosphorylation of Thr-845 (2) and forms a complex with ILK1 to inhibit glycogen synthase kinase 3β-mediated integrin-ILK1 signaling in vivo, which ultimately inhibits cell cycle progression (3, 4).
The catalytic domain of ILKAP shares appreciable sequence homology with the protein phosphatase 2C subfamily; however, the sequence of the noncatalytic domain distinguishes this protein from the other family members because this sequence is absent in other mammalian protein phosphatase 2Cs and shares no homology with any known proteins. It is likely that this sequence may enable ILKAP to specifically interact with other proteins, such as its physiological substrates that are involved in the different cellular functions of ILKAP.
The importin α/β nuclear import pathway is one of the best understood nuclear trafficking systems in the cell (5). Importin α is an ∼60-kDa protein that contains two functional domains: a short basic N-terminal domain (the importin β-binding (IBB) domain) that is sufficient for importin β binding and an NLS-binding domain that is composed of armadillo (ARM) repeats. The latter domain binds to NLS-containing proteins and links these proteins to importin β, which is the karyopherin that ferries the ternary complex through the nuclear-pore complex (6). This transport process depends on the ability of importin α to recognize the specific NLSs that are presented by the cargo protein.
The classical NLSs typically contain either a single cluster of basic residues (monopartite NLS) or two clusters of basic amino acids that are separated by 10–12 variant residues (bipartite NLS) (7). The SV40 large T antigen (PKKKRKV) and nucleoplasmin (KRPAATKKAGQAKKKK) classical NLSs are prototypic mono- and bipartite classical NLSs (8, 9). Importin α has two NLS-binding sites that are formed by conserved residues in its ARM repeat domain; these are considered the major and the minor NLS-binding sites. The N- and C-terminal clusters of a bipartite NLS interact with the minor and the major NLS-binding sites, which correspond to ARM repeats 4–8 and 1–4, respectively. However, a monopartite NLS interacts primarily with the major binding site (10–12).
Although there are many studies that have investigated the anticancer function of ILKAP, the subcellular localization of ILKAP is not yet clearly understood. Researchers hypothesize that cytoplasmic ILKAP interacts with ILK1 to form a complex with integrins (3, 4, 13). Based on these results, the National Center for Biotechnology Information (NCBI), UniProt, and European Bioinformatics Institute (EBI) protein databases indicate that ILKAP is a cytoplasmic protein. In 2004, Doehn et al. (31) found that ILKAP (also called protein phosphatase 2Cδ) forms a complex with RSK2 in vivo. As a well known serine/threonine kinase, RSK2 plays a crucial role in oncogenesis and tumor progression. RSK2 has been reported to localize to the nucleus, and the nuclear localization of RSK2 is crucial for the regulation of cell proliferation and survival (14–16).
In this study, we found that ILKAP mainly localized to the nucleus and that the nuclear transport of ILKAP is NLS importin-mediated. The ILKAP protein interacts directly with importin α1, α3, and α5. We found that the NLS domain in ILKAP is located in its N-terminal region between amino acids 71–86 and that the Lys-78 and Arg-79 residues are critical for the binding of ILKAP to importin α. We also found that nuclear ILKAP interacts with RSK2 and induces apoptosis by inhibiting RSK2 activity and down-regulating the expression level of the RSK2 downstream substrate cyclin D1. These results classify ILKAP as a newly identified nuclear protein that regulates cell survival and apoptosis through the regulation of RSK2 signaling.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies against ILKAP (Abnova: B01P), importin α (Santa Cruz Biotechnology: sc-136204), importin β (Cell Signaling: 8673), RSK2 (Santa Cruz Biotechnology: sc-1430), cyclin D1 (Santa Cruz Biotechnology: sc-753), α-tubulin (Santa Cruz Biotechnology: sc-5286), H2A (Santa Cruz Biotechnology: sc-8648), GFP (Santa Cruz Biotechnology: sc-9996), Myc (Cell Signaling: 2272), GST (Santa Cruz Biotechnology: sc-138), HA (Santa Cruz Biotechnology: sc-805), FLAG (Santa Cruz Biotechnology: sc-807), and β-actin (Santa Cruz Biotechnology: sc-47778) were obtained.
Cell Culture and Transfections
All of the cell lines were maintained in DMEM with 10% FBS and were cultured at 37 °C in a humidified atmosphere with 5% CO2. The mitogenic stimulation involved the use of base medium containing 3 μg/ml insulin, 150 ng/ml epidermal growth factor, and 5% FBS. The transfections were performed using FuGENE HD (Roche Applied Science) according to the manufacturer's instructions.
Plasmid Constructs
cDNA encoding importin α1/3/5, importin β1, RSK2, ILKAP, or its fragments was amplified by PCR and subcloned into the pcDNA3.1/Myc-HisA, pcDNA3.1/Myc-His, pEGFP-C1, and pGEX-4T1 vectors for mammalian or Escherichia coli expression. The pEGFP-C1-ILKAP N-Δ71–87 and pEGFP-C1-ILKAPΔ71–87 constructs were generated by ligating the DNA synthesis fragments into the NheI and Bpu1102I sites of pEGFP-C1-ILKAP 1–107 and the pEGFP-C1-ILKAP vector, respectively. The construction of the ILKAP mutants was performed by PCR using the QuikChange site-directed mutagenesis kit from Qiagen.
Extraction of Nuclear, Cytoplasmic, and Whole-cell Proteins
The whole-cell protein extracts were obtained using cell lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm DTT, and 1× protease inhibitor mixture) according to the manufacturer's instructions. The cytoplasmic and nuclear protein extracts were prepared using the nuclear and cytoplasmic protein extraction kit (Beyotime Institute of Biotechnology) following the manufacturer's instructions. Briefly, the cells were washed with ice-cold PBS and then lysed in cell lysis buffer containing 10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.4% IGEPAL, and 1 mm phenylmethanesulfonyl fluoride (PMSF) for 20 min on ice. After centrifugation, the supernatants (corresponding to the cytoplasmic extracts) were collected, and the nuclei pellets were washed once with ice-cold cell lysis buffer and then resuspended in nuclear extraction buffer (0.4 m NaCl, 20 mm HEPES, pH 7.9, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF). After vigorous shaking for 30 min at 4 °C, the nuclear extracts were collected by centrifugation.
GST Pulldown Experiments
The GST and GST fusion proteins were incubated with glutathione-Sepharose 4B (GE) for 2 h at 4 °C, extensively washed with PBS, immobilized on the beads, and then incubated with the cell lysates (5 mg) overnight at 4 °C with gentle agitation. The beads were collected by centrifugation and washed five times. After the supernatants were removed in the final wash, the samples were separated by SDS-PAGE and analyzed through immunoblotting.
Co-immunoprecipitation Assay
The primary antibodies were added to 50 μl of protein G beads (Roche Applied Science) and incubated at 4 °C for 2 h. The corresponding cell lysates or nuclear protein extracts were incubated with the antibody-beads complex at 4 °C for 4 h. The beads were washed five times with 1:10 diluted lysis buffer and then eluted by boiling in Laemmli sample buffer. The precipitated proteins were subjected to immunoblot analysis with the corresponding antibodies.
siRNA Knockdown of ILKAP
The pGenesil RNAi system (shRNA) was used to reduce the expression of ILKAP in cells through RNA interference technology according to the manufacturer's protocols. Forward and reverse oligonucleotides encoding the anti-ILKAP short hairpin RNA (shRNA) sequence were used.
Homology Modeling and Molecular Docking
The sequence of ILKAP was searched against the Protein Data Bank using the NCBI-BLAST search tool to identify a related protein structure that could be used as a template. The MODELLER program was used to build the three-dimensional structure of ILKAP. The model with the highest score was chosen for further refinement through energy minimization. The energy minimization was performed using the NAMD package. The popularity of protein-protein docking as a powerful method for the prediction of the structures of protein complexes is growing almost daily (17, 18). In this study, ZDOCK was used to perform the automated molecular docking. ZDOCK is an initial stage rigid body protein-docking algorithm that explicitly searches the rotational space and uses a fast Fourier transform algorithm to significantly speed up the search of the translational space (19). The rotational sampling interval was set to 6°, and all of the default parameters were used. The best docking conformations were selected for optimization by the RDOCK program, which is a three-stage energy minimization algorithm that was designed as a refinement and re-ranking tool for the top predictions obtained by ZDOCK.
RSK2 Kinase Assays
The RSK2 kinase assay was performed as described previously (20). Briefly, agarose beads with the immunoprecipitated kinase were drained with a syringe and resuspended in 20 μl of 1.5× kinase assay buffer. The kinase reaction was initiated by the addition of 10 μl of the substrate mixture containing ATP (300 μm, 0.2 μCi of [γ-32P]ATP) and the S6 peptide (800 μm). After 10 min at 30 °C, 20 μl of the supernatant was removed and spotted onto phosphocellulose paper (Whatman). The paper was washed five times with 150 mm orthophosphoric acid. The [32P]phosphate that was incorporated into the protein substrate was quantified on a VersaDocTM MP5000 imager using the Quantity One software (Bio-Rad). To quantify the amount of RSK2 in the immunoprecipitates, the remaining reaction mixture (minus the 20 μl of the supernatant) was solubilized and analyzed by immunoblotting.
Annexin V/Propidium Iodide Apoptosis Assay
The cells were plated onto 24-well glass-bottomed dishes, and the assays were performed using the annexin V/propidium iodide apoptosis assay kit (Sigma) according to the manufacturer's instructions. The images were collected using a BX51 microscope (Olympus).
Statistical Analyses
All of the measurements were collected in triplicate for each independent preparation. The results were statistically analyzed using Student's t test and analysis of variance. The SPSS software, version 16.0, was used for all of the statistical analyses, and differences with a p value less than 0.05 were considered statistically significant.
RESULTS
Both Endogenous and Tagged ILKAP Were Detected in the Nucleus
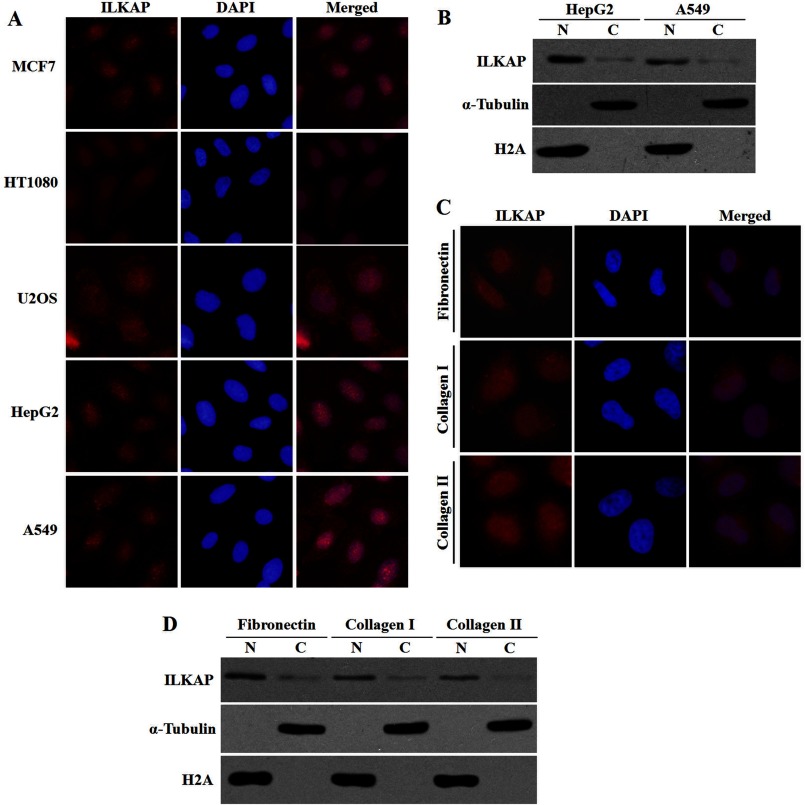
We examined the localization of endogenous ILKAP in different cell lines using the anti-ILKAP antibody. As shown in Fig. 1A, the endogenous ILKAP protein was detected in the nucleus, although weak cytoplasmic staining was also observed. To further confirm the subcellular localization of the endogenous ILKAP protein, the subcellular fractions that were prepared from HepG2 and A549 cells were analyzed by immunoblotting.
FIGURE 1.
Subcellular distribution of endogenous ILKAP. A, cells grown on coverslips were fixed and immunostained with anti-ILKAP (red) antibody. DAPI (blue) staining of the nuclei is shown. B, subcellular localization of endogenous ILKAP examined by cell fractionation and immunoblotting. HepG2 and A549 cells were separated into nuclear (N) and cytoplasmic (C) fractions. Equal protein amounts (80 μg) were loaded onto SDS-PAGE (10%) gels, and the protein expression was analyzed with anti-ILKAP, anti-H2A, and anti-α-tubulin antibodies. C, A549 cells grown on coverslips coated with fibronectin (10 mg/ml), collagen type I (5 mg/ml), or collagen type IV (5 mg/ml) were fixed and immunostained with anti-ILKAP (red) antibody. DAPI (blue) staining of the nuclei is shown. D, the A549 cells grown on the different extracellular matrices were examined by cell fractionation and immunoblotting.
We found that the expression level of endogenous ILKAP was rather low in the tested cells. In addition, ILKAP was mainly detected in the nuclear fraction (Fig. 1B), which is in agreement with the immunofluorescence results. The purity of the nuclear and cytosolic fractions was demonstrated by probing with antibodies against H2A and α-tubulin.
ILKAP was first found as a regulator of integrin signaling. Thus, we examined the subcellular localization of ILKAP in A549 cells that were cultured on different types of extracellular matrices, including fibronectin, collagen type I, and collagen type IV, to activate integrin signaling. We found that the endogenous ILKAP protein was mainly detected in the nuclear fraction in the cells that were cultured in the different extracellular matrices (Fig. 1, C and D).
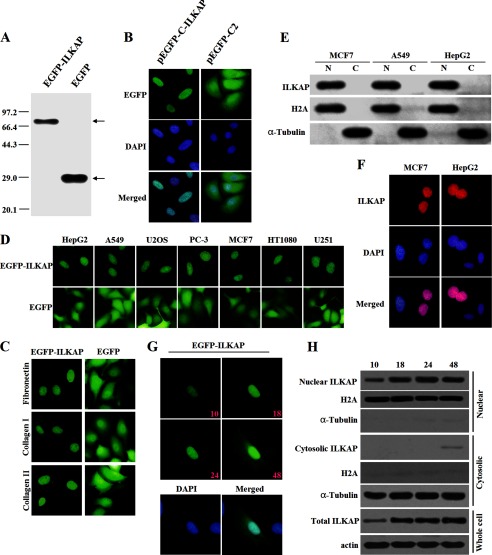
We next examined the subcellular localization of the tagged ILKAP protein. The immunoblotting analysis with anti-EGFP antibody indicated that the EGFP-ILKAP fusion protein was properly expressed in the transiently transfected MCF7 cells (Fig. 2A). EGFP-ILKAP was detected exclusively in the nucleus (Fig. 2B, left panel), whereas EGFP alone was diffusely distributed in both the cytoplasm and the nucleus (Fig. 2B, right panel). Similar results were obtained with MCF7 cells that were cultured in different extracellular matrices (Fig. 2C) and with other cell lines (Fig. 2D). Additional immunoblotting analyses were performed using the subcellular fractions prepared from EGFP-ILKAP-expressing HepG2, A549, and MCF7 cells. In agreement with the immunofluorescence results, EGFP-ILKAP was predominantly found in the nuclear fraction (Fig. 2E).
FIGURE 2.
Subcellular distribution of tagged ILKAP. A, detection of the protein expression of the full-length EGFP-ILKAP and the EGFP empty constructs. MCF7 cells were transiently transfected with each construct and allowed to recover for 24 h. The protein expression was confirmed by immunoblotting using an anti-EGFP antibody. B, localization of EGFP-ILKAP and EGFP in MCF7 cells. MCF7 cells were transiently transfected with the respective constructs and grown on coverslips for 24 h after transfection. The cells were then fixed and counterstained with DAPI for nuclear staining (blue). C, localization of EGFP-ILKAP and EGFP in MCF7 cells grown on coverslips coated with fibronectin (10 mg/ml), collagen type I (5 mg/ml), or collagen type IV (5 mg/ml). D, localization of EGFP-ILKAP and EGFP in different cell lines. E, the subcellular localization of EGFP-ILKAP was examined by cell fractionation and immunoblotting. HepG2, A549, and MCF7 cells were separated into nuclear (N) and cytoplasmic (C) fractions, and the protein expression levels were analyzed with anti-EGFP, anti-H2A, and anti-α-tubulin antibodies. F, distribution of the Myc-ILKAP fusion protein in MCF7 and HepG2 cells. MCF7 and HepG2 cells were immunostained with anti-Myc antibody. The protein localization was detected using a Cy3-conjugated secondary antibody. G, time-lapse images of EGFP-ILKAP in A549 cells after transfection. The protein was detected through the microscopic observation of EGFP (green) over a period of 48 h. The merged image was formed by merging the images that were obtained 48 h after transfection. H, immunoblotting analysis of EGFP-ILKAP by cell fractionation.
To exclude the effect of the position of the EGFP on the nuclear localization of the ILKAP fusion protein, we also examined the subcellular localization of the Myc-ILKAP fusion protein, in which the tagged protein was fused to the C-terminal end of ILKAP, in MCF7 and HepG2 cells using the anti-Myc antibody. No staining was detected when the cells were transfected with pcDNA3.1/Myc-HisA or when the secondary antibody was used alone, which demonstrates the specificity of the immunofluorescence staining of the Myc fusion protein. Similarly to EGFP-ILKAP, the Myc-ILKAP fusion protein was predominantly distributed in the nucleus of the different cell lines that were examined (Fig. 2F).
We also examined the localization of EGFP-ILKAP over time. A series of time-lapse images of EGFP-ILKAP was captured 10, 18, 24, and 48 h after transfection. In addition, immunoblotting analyses were also performed to examine the localization of EGFP-ILKAP at these time points. The real-time imaging clearly demonstrated that EGFP-ILKAP was mainly localized in the nucleus. As shown in Fig. 2, G and H, both the real-time imaging and the immunoblotting results clearly showed that EGFP-ILKAP was mainly localized in the nucleus. We also found weak cytosolic ILKAP localization 48 h after transfection. These results indicated that both the endogenous and the tagged ILKAP were mainly localized in the nucleus.
Modeling and Docking Study
The human phosphatase 2C (Protein Data Bank (PDB) id: 1A6Q) has a high level of sequence identity with ILKAP; therefore, a model of ILKAP was built using the MODELLER software. The structure with the minimum free energy and the highest score was used as the initial model. Energy minimization, which comprised 2000 steps of steepest decent followed by 2000 steps of conjugate gradient, was then performed to optimize the ILKAP model.
The crystal structure of importin α isoform 1 (karyopherin α2, PDB code 3FEX C-chain) was used for the docking simulations with ILKAP. The resultant docked structure was a 2.2 Å resolution x-ray structure for the cap-binding complex-importin α complex, which provides insight into the binding of importin α to the CBP80 subunit of the cap-binding complex (21). The importin α structure from this complex contains 10 ARM structural repeats and lacks the first 69 amino acid residues (importin β-binding domain).
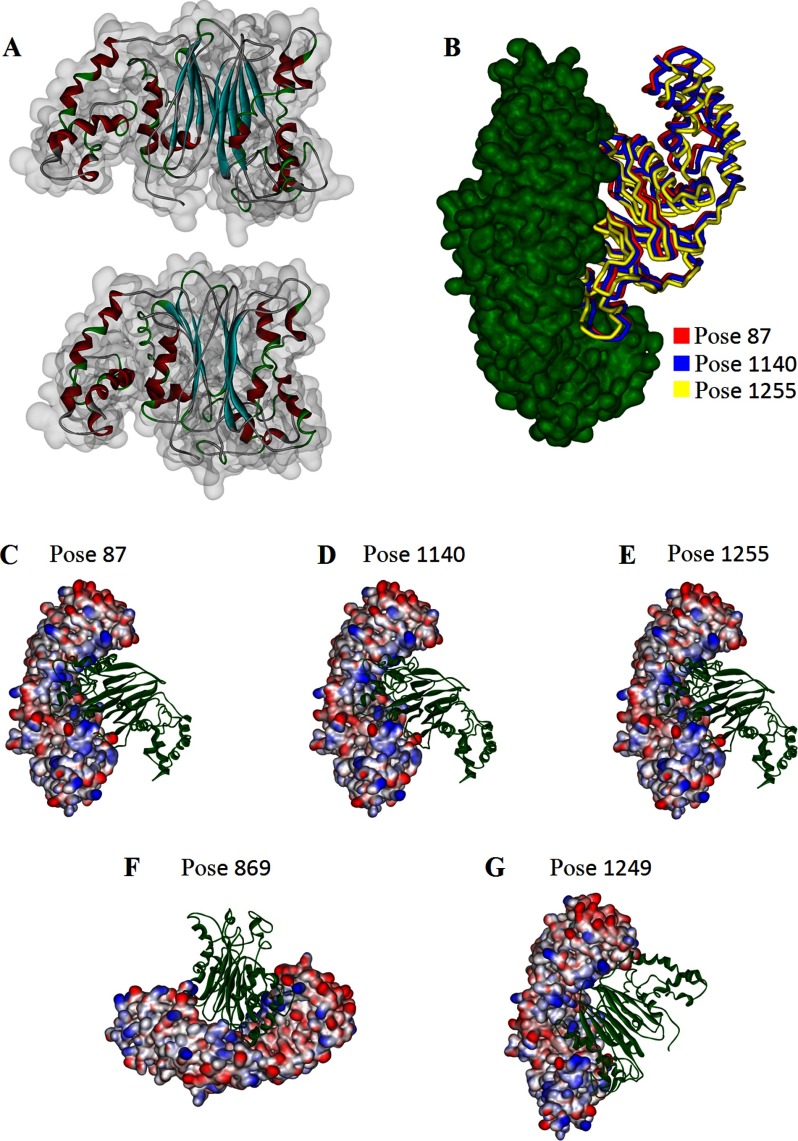
A total of 60 clusters and 2000 complexes were obtained after the ZDOCK calculation. Of these, the best 15 docking poses were selected based on the top scores and were re-ranked by the ZRANK score, which is a more detailed energetics-based scoring function that has been developed and tested for the re-ranking of docked protein poses (22). Based on the scoring methods, the best five possible structures (poses 87, 869, 1140, 1249, and 1255) were selected for RDOCK refinement. The models of the five top binding complexes are shown in Fig. 3. In addition, root mean square deviation analysis revealed that poses 87, 1140, and 1255 were very similar (Fig. 3).
FIGURE 3.
Modeling results of ILKAP structure and docking of ILKAP with importin α. A, optimal model of ILKAP. The structure is shown as a ribbon display model. The α-helices, the β-sheets, and the β-turns are shown in red, cyan, and green, respectively. B, docking complex of importin α (shown in green) with ILKAP in different poses (poses 87, 1140, and 1255 are shown in red, blue, and yellow, respectively) obtained after root mean square deviation analysis. The best five possible structures of the complex (poses 87, 1140, 1255, 869, and 1249) are shown in C–G.
Initial Prediction of the NLS by Analyzing the Interface Residues of the Complexes
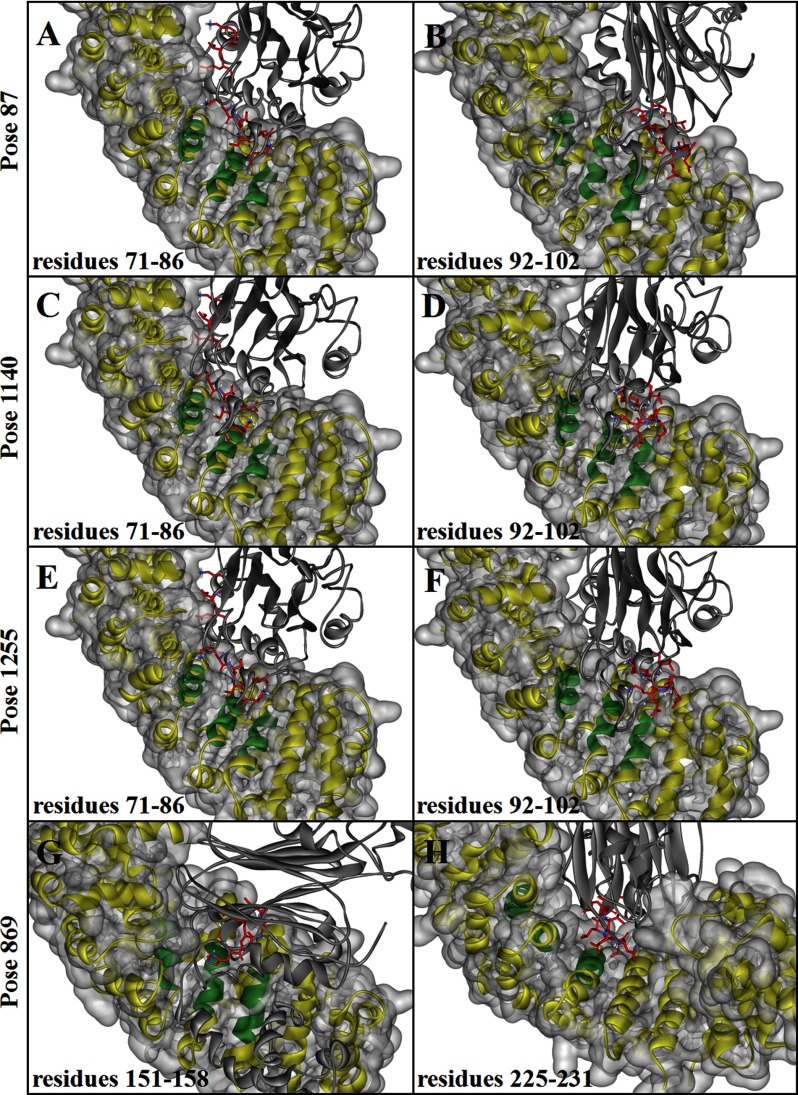
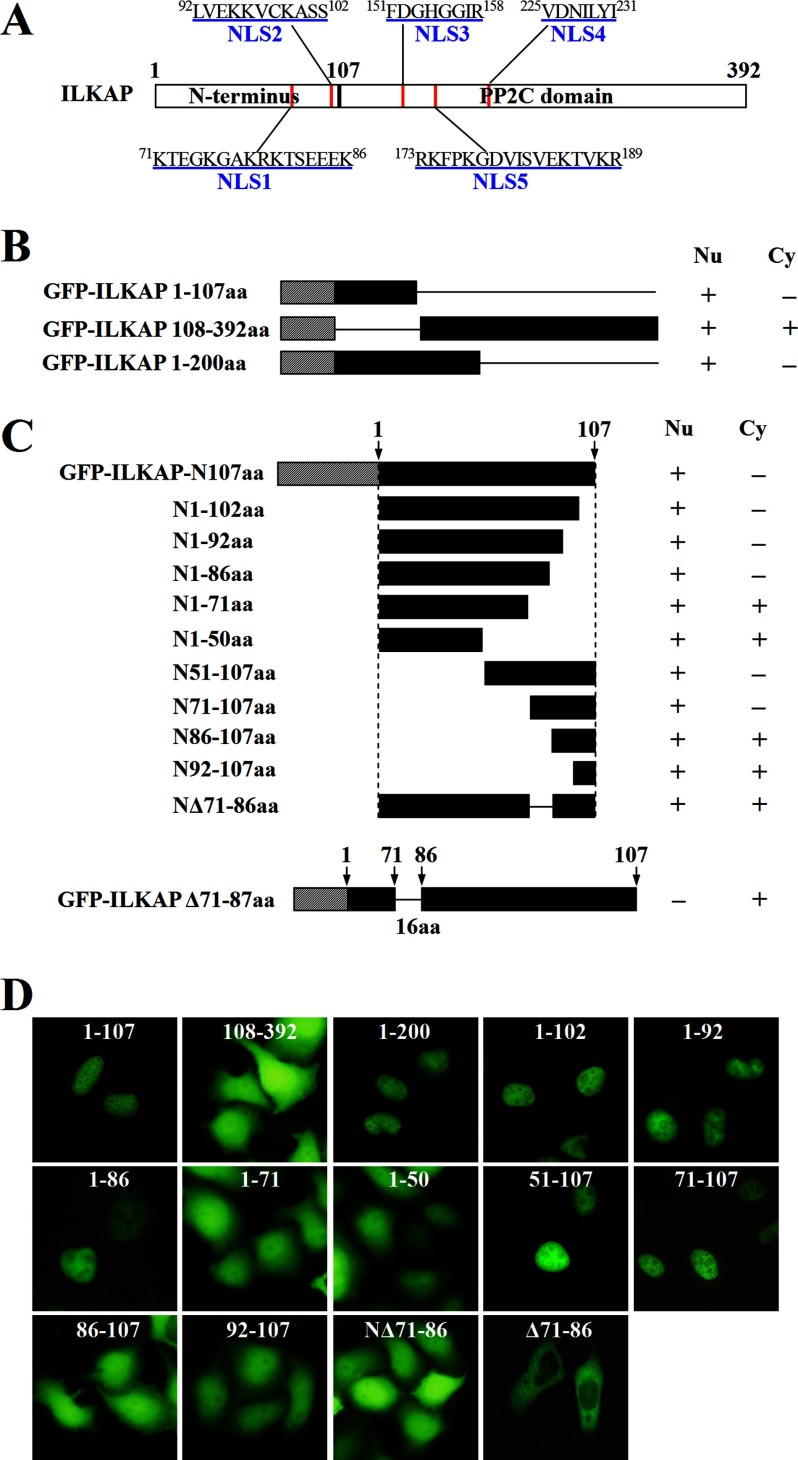
The NLS-binding sites are located in the concave face of the protein near regions of invariant Trp and Asn arrays (23, 24). We defined importin α as the receptor and found two groups of interface residues (residues 71–86 and 92–102 in the ILKAP model face) that face the 6–7 ARM repeats of the receptor model in poses 87, 1140, and 1255 by analyzing the interface residues of the predicted structures (Fig. 4, A–F). Two groups of interface residues, containing amino acids 151–158 and 225–231, face the 6–7 ARM repeats of the receptor model in pose 869 (Fig. 4, G and H); no residues face the appropriate site in pose 1249. According to these results, we identified four putative NLS regions that contain multiple positively charged amino acid residues; these four regions are located at amino acids 71–86, 92–102, 151–158, and 225–231 and are designated NLS1, NLS2, NLS3, and NLS4, respectively (Fig. 5A). The interaction energy of each group with importin α was then calculated (Table 1). We determined that residues 71–86 in the predominant model of ILKAP (poses 87, 1140, and 1255) have a higher interaction energy and were thus most likely to contain the NLS sequence.
FIGURE 4.
Interactions between the binding groove of importin α with ILKAP. Interactions between the binding groove (in green; the other parts of importin α are shown as a yellow and/or gray surface) of importin α with ILKAP (shown as a gray ribbon) are shown. The interface residues (NLS-binding sites) in ILKAP are shown in red: residues 71–86 (A) and 92–102 (B) in pose 87, residues 71–86 (C) and 92–102 (D) in pose 1140, residues 71–86 (E) and 92–102 (F) in pose 1255, and residues 151–158 (G) and 225–231 (H) in pose 869.
FIGURE 5.
A 16-amino acid sequence in the N terminus of ILKAP is necessary to mediate the nuclear localization of EGFP-ILKAP. A, schematic representation of ILKAP and its five putative NLSs (red). B, the N-terminal and C-terminal deletion mutants derived from pEGFP-C1-ILKAP are depicted schematically. The different constructs were introduced into A549 cells, and the subcellular distributions of the fusion proteins were visualized through fluorescence microscopy. Nu, nucleus; Cy, cytoplasm. C, nuclear localization of the deletion mutants of EGFP-ILKAP-N107 in transfected A549 cells. The EGFP-ILKAP-N107 constructs were generated with deletions at either the N or the C terminus of the N-terminal domain of ILKAP, as indicated. The cells were viewed directly by immunofluorescence 24 h after transfection. aa, amino acids. D, the localization of the different constructs in A549 cells was visualized through fluorescence microscopy.
TABLE 1.
The interaction energies of the four putative NLS regions (NLS1, NLS2, NLS3, and NLS4) with importin-α in various poses
The interaction energy (Einter) includes the electrostatic energy (Eelec) and the van der Waals interactions (EvdW).
| Pose | Residues | NLS | Eelec | EvdW | Einter |
|---|---|---|---|---|---|
| 87 | 71–86 | NLS1 | −211.80114 | −24.35565 | −236.15679 |
| 87 | 92–102 | NLS2 | 34.17237 | 45.07577 | 79.24794 |
| 1140 | 71–86 | NLS1 | −138.39299 | 29.87968 | −108.51331 |
| 1140 | 92–102 | NLS2 | −44.79956 | −6.13201 | −50.93157 |
| 1255 | 71–86 | NLS1 | −73.16285 | 60.00461 | 42.00687 |
| 1255 | 92–102 | NLS2 | −17.99775 | 60.00461 | 42.00687 |
| 869 | 151–158 | NLS3 | −15.23122 | −15.59997 | −30.83199 |
| 869 | 225–231 | NLS4 | −65.59090 | 10.08778 | −55.50312 |
Identification of a 16-Amino Acid Sequence in the N Terminus of ILKAP That Is Necessary for Nuclear Targeting
To confirm the hypothesis and identify the NLS sequence of ILKAP, we investigated the subcellular localization of several ILKAP deletion mutants based on the docking results. We also used a web-based computer program called PSORT II to search the functional NLS in ILKAP and found only one putative NLS region, which was located at amino acids 173–189 and is designated NLS5 (Fig. 5A). We fused a fragment of 200 amino acids in the N terminus of ILKAP to the C terminus of EGFP and examined the subcellular distribution of this fusion protein in A549 cells. As shown in Fig. 5B, the N-terminal fragment (pEGFP-C1-ILKAP 1–200) of ILKAP was capable of directing EGFP to the nucleus (Fig. 5, B and D).
Next, we determined whether the unique N terminus of ILKAP was required for its nuclear localization. As shown in Fig. 5, B and D, the N-terminal fragment (pEGFP-C1-ILKAP 1–107) of ILKAP was capable of directing EGFP to the nucleus.
Following this procedure, the C-terminal deletion construct pEGFP-C1-ILKAP 108–392 was generated and transiently expressed in A549 cells. However, this C-terminal fragment (pEGFP-C1-ILKAP 108–392) of ILKAP was diffusely distributed in both the cytoplasm and the nucleus. These results suggested that the N terminus of ILKAP is necessary and sufficient for nuclear targeting. We concluded that the nuclear importation of the ILKAP protein is distinctly regulated and that the functional NLS is located in its unique N terminus, which contains NLS1 and NLS2.
To identify the minimal sequence within the N terminus of ILKAP that was capable of localizing to the nucleus, a series of truncation constructs, in which different fragments of the N terminus of ILKAP (1–107) were fused to the C terminus of EGFP, was generated and transiently expressed in A549 cells. As shown in Fig. 5C, the construct containing the first 86 amino acids of the N terminus was predominantly localized in the nucleus, but the first 71 amino acids failed to accumulate in the nucleus (Fig. 5, C and D).
Based on the results obtained with the EGFP-ILKAP-N107aa deletion constructs, it is apparent that the EGFP fusion proteins with either 70 residues deleted from the N terminus or 21 residues deleted from the C terminus of ILKAP-N107aa were capable of being targeted to the nucleus. These results suggest that NLS1 (a minimum stretch of 16 amino acids) is required for nuclear targeting, whereas the construct lacking this region was not capable of translocating EGFP to the nucleus.
The ILKAP Protein Interacts Directly with Importin Proteins
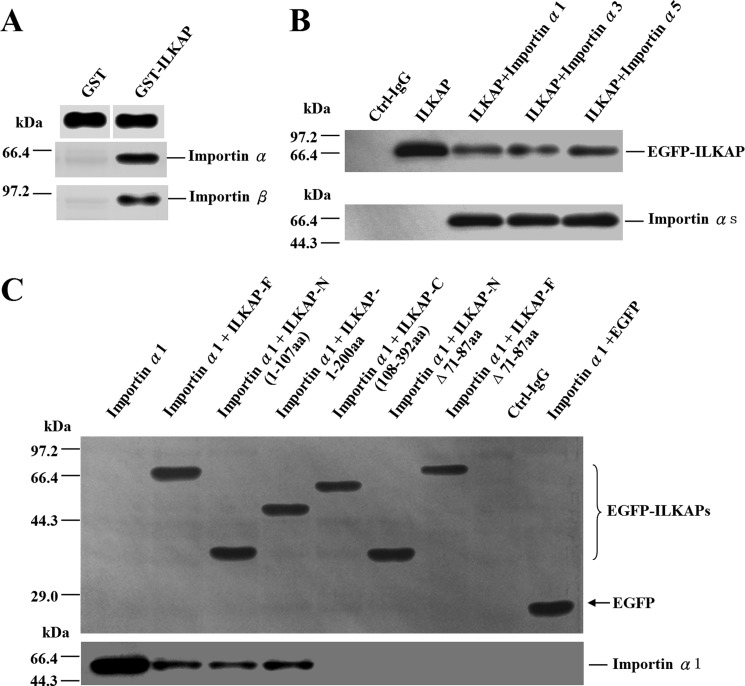
To test the hypothesis that the nuclear transport of the ILKAP protein is NLS-mediated, we performed an in vitro GST pulldown assay to test the ability of ILKAP to interact with importin α and importin β. ILKAP was expressed as a fusion with GST in bacteria. The GST protein alone was utilized as a negative control. As shown in Fig. 6A, ILKAP was able to interact with both importin α and importin β (Fig. 6A).
FIGURE 6.
Co-immunoprecipitation analysis of ILKAP-importin interactions. A, importin α and importin β were found to interact with the GST-ILKAP fusion proteins through a GST pulldown assay. The GST and GST fusion proteins expressed in E. coli were immobilized on glutathione-Sepharose beads. After extensive washing, the beads were incubated with equal amounts of protein extracts from untransfected A549 cells. The retained proteins were analyzed by immunoblotting with anti-GST, anti-importin α, or anti-importin β antibodies. B, the lysates of COS-7 cells expressing Myc-importin α proteins (importin α1, α3, and α5) and EGFP-ILKAP were immunoprecipitated with an anti-Myc antibody. Each precipitate sample was blotted with anti-Myc and anti-EGFP antibodies to detect importin α (Myc) and ILKAP (EGFP). Ctrl-IgG, mouse IgG and beads (negative control). C, the lysates of COS-7 cells expressing various EGFP-ILKAP constructs (residues 1–107, 1–200, 108–392, NΔ71–86, FΔ71–86, or full-length protein) and Myc-importin α1 were immunoprecipitated with an anti-EGFP antibody. Each precipitate sample was blotted with anti-Myc and anti-EGFP antibodies to detect both importin α1 (Myc) and ILKAP (EGFP).
The binding of ILKAP to three importin α proteins was also confirmed using an immunoprecipitation assay with COS-7 cell extracts. First, COS-7 cells were co-transfected with EGFP-tagged full-length ILKAP (EGFP-ILKAP) and various Myc-tagged importins (importin α1, α3, and α5). Importin α1, α3, and α5 represent each of the subfamilies of importin α proteins. Twenty-four hours after transfection, the cells were lysed, and the Myc-importin complexes were precipitated with anti-Myc antibody and protein G-Sepharose beads. The precipitates were immunoblotted for Myc and EGFP. In this assay, EGFP-ILKAP was able to interact with all of the tested importin α proteins (Fig. 6B). Then, COS-7 cells were co-transfected with Myc-tagged importin α1 and the various EGFP-tagged ILKAP constructs (residues 1–107, 1–210, 108–392, 1–107-Δ71–87, full-length-Δ71–87, and full-length protein). The immunoprecipitation results are shown in Fig. 6C; the EGFP-ILKAP full-length, 1–107, and 1–200 fusion proteins were recognized by importin α1. EGFP ILKAP 108–392, 1–107-Δ71–86, and full-length-Δ71–87, however, did not bind these importins. Thus, the nuclear transport of ILKAP is mediated by importin α via its binding to the 71–86 amino acid sequence.
Lys-78 and Arg-79 Are the Most Crucial Residues That Mediate the Interaction with Importin α and the Nuclear Localization of ILKAP
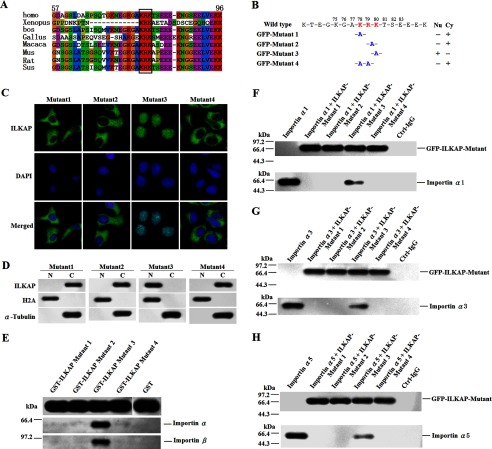
We have confirmed that the nuclear transport of ILKAP is mediated by importin α via its binding to the 71–86 amino acid sequence. To determine the contribution of the specific amino acids within this region, we aligned human ILKAP with orthologous proteins from other species and showed that 78KRK80 is highly conserved among all of the species that were examined (Fig. 7A). This result suggested that 78KRK80 likely acts as a functional NLS in ILKAP. To test this hypothesis, site-directed mutagenesis was performed to generate four mutant constructs (Fig. 7B). We examined the subcellular localization of the four ILKAP mutants. As shown in Fig. 7C, the K78A, R79A, and K78A/R79R substitutions disrupted the nuclear localization of EGFP-ILKAP. The K80A substitution did not impair nuclear localization. Immunoblotting analysis was performed using the subcellular fractions prepared from A549 cells that express each of four ILKAP mutants. In agreement with the immunofluorescence results, the mutated proteins with the K78A, R79A, or K78A/R79A substitutions were predominantly found in the cytosolic fraction, whereas the mutant with the K80A substitution was predominantly located in the nuclear fraction (Fig. 7D).
FIGURE 7.
Lys-78 and Arg-79 are the most crucial residues for mediating the interaction with importin α and the nuclear localization of ILKAP. A, the residues from 78 to 80 are highly conserved. The sequence alignment of the human ILKAP with orthologous proteins in nine species was performed using the ClustalW alignment program. The missing amino acids are indicated with hyphens. The identical and similar residues are highlighted. The highly conserved residues in the NLS1 of ILKAP are boxed, and the residue numbers corresponding to the human sequence are shown. homo, Homo sapiens; bos, Bos taurus; Gallus, Gallus gallus domesticus; Mus, Mus musculus; Sus, Sus scrofa domestica. B–D, determination of the essential amino acids of NLS by mutational analysis. A549 cells were transiently transfected with the four mutant constructs that were derived from pEGFP-C1-ILKAP through site-directed mutagenesis (B). Nu, nucleus; Cy, cytoplasm. The transfected proteins were then examined by fluorescence microscopy (C) and immunoblotting (D). N, nucleus; C, cytoplasm. E, importin α and importin β were found to interact with the GST-ILKAP mutant (mutants 1, 2, 3, and 4) fusion proteins through a GST pulldown assay. The GST and GST fusion proteins expressed in E. coli were immobilized on glutathione-Sepharose beads. After extensive washing, the beads were incubated with equal amounts of protein extracts from the untransfected A549 cells. The retained proteins were analyzed by immunoblotting with anti-GST, anti-importin α, or anti-importin β antibodies. F–H, the lysates of COS-7 cells expressing Myc-importin α proteins and the EGFP-ILKAP mutants (mutants 1, 2, 3, and 4) were immunoprecipitated with an anti-Myc antibody. Each precipitate sample was blotted with anti-Myc and anti-EGFP antibodies to detect importin α (Myc) and ILKAP (EGFP). Ctrl, mouse IgG and beads (negative control).
We then performed GST pulldown assays to test the ability of the ILKAP mutants to interact with importin α and β proteins. ILKAP mutants were expressed in fusion with GST in bacteria. The GST protein alone was utilized as a negative control. As shown in Fig. 7E, Mutant 3 (K80A substitution) was able to interact with both importin α and importin β. However, both the K78A and the R79A single substitutions disrupted the interaction with importin α and importin β. As expected, the K78A/R79A double mutant disrupted the interaction (Fig. 7E).
We also used immunoprecipitation assays with the COS-7 cell extracts to test the ability of the ILKAP mutants to interact with the importin α proteins. As shown in Fig. 7, F–H, the K80A substitution did not impair the interaction with any of the three importin α proteins. However, both the K78A and the R79A single substitutions disrupted the interaction with the importin α proteins. As expected, the K78A/R79A double mutant also disrupted the interaction. These results indicate that Lys-78 and Arg-79 are the most crucial residues that mediate the interaction with importin α and thus the nuclear localization of ILKAP.
ILKAP Interacts with RSK2 in the Nucleus and Reduces RSK2 Activity
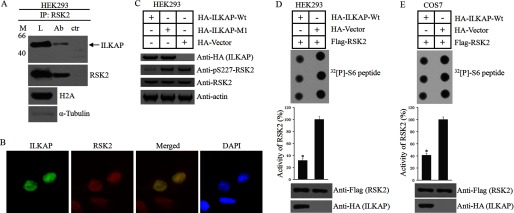
RSK2 is hypothesized to play key roles in the regulation of cell proliferation and survival (25). T. S. Eisinger et al. found that mitogen induced the accumulation of RSK2 in the nucleus and enhanced proliferation through the induction of cyclin D1. The nuclear localization of RSK2 is crucial for the regulation of cell proliferation and survival (25). In 2004, Doehn et al. (31) found that ILKAP forms a complex with RSK2 in vivo, but these researchers did not discuss the subcellular localization of these two proteins (15). In this study, we asked whether ILKAP interacts with RSK2 in the nucleus and whether ILKAP negatively regulates cell proliferation by affecting RSK2 signaling. We first performed co-immunoprecipitation assays using the nuclear fractions of HEK293 cells to investigate whether ILKAP could interact with RSK2 in the nucleus. HEK293 cells were treated with mitogen for 8 h. The nuclear fractions were isolated and lysed, and RSK2 was precipitated with anti-RSK2 antibody and protein G-Sepharose beads. The precipitates were immunoblotted for RSK2 and ILKAP antibody. We found that ILKAP could be detected in the precipitates (Fig. 8A). Then, we used immunofluorescence assays to investigate the co-localization of ILKAP and RSK2. After mitogen treatment, HA-ILKAP co-localized with RSK2 in the nucleus (Fig. 8B). All of these results demonstrated that ILKAP can interact with RSK2 in the nucleus.
FIGURE 8.
ILKAP interacts with RSK2 in the nucleus and inhibits RSK2 kinase activity. A, HEK293 cells were treated with mitogen (M) for 8 h. The nuclear fractions were isolated and lysed (L). RSK2 was precipitated with anti-RSK2 antibody (Ab) and protein G-Sepharose beads. The precipitates were immunoblotted for RSK2 and ILKAP antibody. ctr, control. B, A549 cells were transiently transfected with EGFP-ILKAP for 16 h and treated with mitogen for 8 h. The cells were then stained with anti-RSK2 (red) antibody. DAPI (blue) staining of the nuclei is shown. C, HEK293 cells were transfected with an empty vector or a plasmid expressing either HA-ILKAP or HA-ILKAP Mutant 1 for 16 h. The cells were then treated with mitogen for 8 h, lysed, subjected to SDS-PAGE, and immunoblotted using phosphospecific antibodies to RSK2 phosphorylated at Ser-227, non-phosphospecific antibodies to RSK2 or anti-HA antibody. D and E, HEK293 (D) and COS-7 (E) cells were co-transfected with plasmids expressing HA-ILKAP and FLAG-RSK2. Twenty-four hours after transfection, the cells were serum-starved for 4 h, exposed to 1 ng/ml EGF for 30 min, and lysed. RSK2 was precipitated from the cell lysates with antibody to the FLAG tag, and its kinase activity was determined and expressed as a percentage of the maximal value obtained. * indicates a significant difference (p < 0.05) when compared with the control. Error bars indicate S.D.
The phosphorylation of RSK2 at Ser-227 directly activates RSK2 activity. Thus, we examined the phosphorylation levels of RSK2 at Ser-227 in HA-ILKAP- and HA-ILKAP Mutant 1(K78A substitution)-expressing HEK293 cells. The results in Fig. 8C show that the expression of WT or mutant ILKAP did not affect the expression level of RSK2. We found that HA-ILKAP but not HA-ILKAP Mutant 1 or the control decreased the phosphorylation level of Ser(P)-227 (Fig. 8C).
We also examined whether ILKAP affects RSK2 kinase activity. FLAG-tagged RSK2 was co-expressed with HA-ILKAP in COS-7 and HEK293 cells. The cells were exposed to EGF prior to lysis. After the precipitation of RSK2 with an antibody to the FLAG tag, the kinase activity of RSK2 was determined. The RKS2 that was co-expressed with active ILKAP was found to be nearly inactive, whereas RSK2 alone showed high kinase activity (Fig. 8, D and E).
ILKAP Induces Apoptosis by Affecting the RSK2 Downstream Effector Cyclin D1
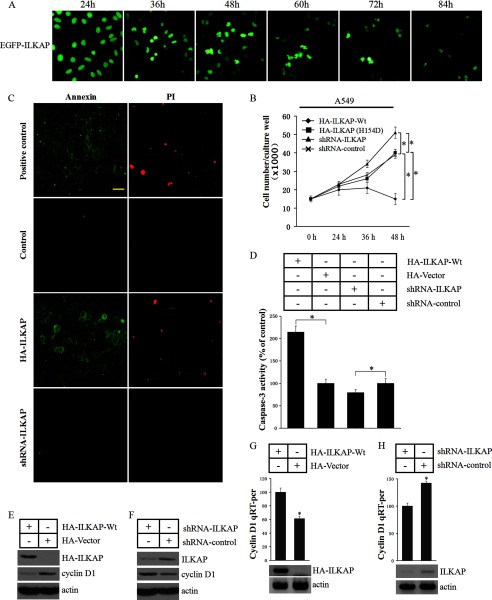
ILKAP is an apoptosis-associated phosphatase. Thus, we investigated the effect of ILKAP expression in A549 cells. We found no noticeable nuclear morphological changes in the EGFP-ILKAP-positive cells 24 h after the transfection of EGFP-ILKAP. However, 36–48 h after transfection, the nucleus of 35–50% of the EGFP-ILKAP-positive cells became irregular, and 5–10% of the positive cells showed nuclear condensation and fragmentation. After 48 h, the number of positive cells significantly decreased, and most of the positive cells exhibited nuclear condensation and fragmentation (Fig. 9A). We exogenously expressed HA-ILKAP, ILKAP-shRNA, and a loading control in A549 cells for 48 h. The cell survival assay showed that HA-ILKAP expression significantly reduced the cell viability (∼78%). In the ILKAP-shRNA-expressing cells, the cell viability was significantly higher (∼38.2%) when compared with the control (Fig. 9B). We next investigated whether ILKAP induced apoptosis in A549 cells using the annexin V/propidium iodide assay. We found that the expression of ILKAP could induce apoptosis in A549 cells when compared with the control (Fig. 9C). We also measured the caspase-3 activity in the tested cells. The caspase-3 activity of the ILKAP-expressing A549 cells was significantly higher than that in the control cells. In the ILKAP-shRNA-expressing cells, the caspase-3 activity was lower when compared with that in the shRNA control cells (Fig. 9D).
FIGURE 9.
ILKAP regulates cyclin D1 protein expression and induces apoptosis. A, HEK293 cells were transfected with the EGFP-ILKAP construct, and the expressed EGFP-ILKAP in the cells was visualized by UV microscopy at different times. B, A549 cells were transfected with HA-ILKAP, empty vector, shRNA-ILKAP or shRNA-control. The 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide assay was performed to examine the cell proliferation rate 24, 36, and 48 h after the cells were transfected with each construct. C, the cells were assayed using the annexin V/propidium iodide (PI) apoptosis assay kit (Sigma-Aldrich) 48 h after the cells were transfected with each construct. D. The caspase-3 activity in the transfected cells was determined. E and F, immunoblots were performed to examine the cyclin D1 protein levels in the transfected cells. G and H, the RNA was extracted, and a quantitative RT-PCR assay was performed. * indicates a significant difference (p < 0.05) when compared with the control. Error bars indicate S.D.
Nuclear RSK2 enhances proliferation through the induction of cyclin D1 expression (15). Thus, we next investigated whether ILKAP regulates the RSK2 downstream protein factor cyclin D1. As shown in Fig. 9E, the expression level of cyclin D1 was markedly reduced in the tested cells after the expression of WT-ILKAP. In the ILKAP-silenced A549 cells, the expression level of cyclin D1 was increased (Fig. 9F). We also used quantitative RT-PCR assays to examine whether ILKAP affects the cyclin D1 mRNA level. We found that the overexpression of ILKAP decreases the cyclin D1 mRNA level by ∼38.2% when compared with the control. The knockdown of ILKAP increases the cyclin D1 mRNA level by ∼41.2% when compared with the siRNA control (Fig. 9, G and H).
All of the above results demonstrate that nuclear ILKAP can interact with RSK2 and inhibit RSK2 activity. ILKAP also controls cell proliferation and survival by regulating the expression of the RSK2 downstream substrate cyclin D1.
DISCUSSION
ILKAP was initially identified in a yeast two-hybrid screen baited with ILK and was considered a cytoplasmic protein associated with the down-regulation of integrin signaling (3, 4, 13). ILK has been shown to act as a serine/threonine kinase and to directly activate several signaling pathways downstream of integrins. ILK localizes to focal adhesions and binds directly to the cytoplasmic domain of the integrin β1 and β3 subunits (13).
ILK is normally found in complex with two other proteins: particularly interesting new cysteinehisticline rich protein (PINCH) and parvin. The heterotrimeric complex between ILK, PINCH, and parvin, which is termed the IPP complex, is an essential signaling platform that regulates cell adhesion, spreading, and migration (26). The IPP complex is a central constituent of at least β1- and β3-integrin-containing adhesion sites, which bind other regulatory factors to influence multiple cellular processes. Without the formation of the IPP complex, integrin signaling cannot be regulated (27). Leung-Hagesteijn and co-workers (3, 4) showed that the ILKAP inhibition of ILK activity selectively inhibited the S9 phosphorylation of glycogen synthase kinase 3β without affecting the Ser-473 phosphorylation of PKB. These results indicated that ILKAP links to the IPP complex by interacting with ILK and is localized in β1- and β3-integrin-containing focal adhesions.
However, ILK lacks well conserved motifs that are required for eukaryotic protein kinase activity (28). The GXGXXG consensus sequence of the kinase subdomain I, which is required for covering and anchoring the nontransferable phosphates of ATP, is not conserved in ILK proteins from different species. ILK-deficient fibroblasts, chondrocytes, and keratinocytes do not show changes in Akt or glycogen synthase kinase 3β phosphorylation (23, 24, 29). Knock-in mice with mutations in the putative pleckstrin homology (PH) domain (R211A) or in the autophosphorylation site (S343A or S343D) are completely normal and do not show changes in Akt or glycogen synthase kinase 3β phosphorylation or actin organization downstream of integrins (30). Recently, Wickström et al. (27) indicated that ILK is a pseudokinase that functions as a scaffold protein and not as a kinase in invertebrates. The regulatory role of integrin signaling was not associated with the kinase activity of ILK. Our results showed a weak cytosolic localization of ILKAP. Thus, we hypothesize that ILKAP regulates integrin signaling by catalyzing some key downstream factors but not by directly phosphorylating ILK.
Interestingly, although our results showed a weak cytosolic localization of ILKAP, we did not find any significant focal adhesion localization. Moreover, both the endogenous and the tagged ILKAP proteins were strongly detected in the nucleus. We found that the nuclear import of ILKAP is specifically dependent on a short, poorly basic NLS that is located at the N terminus of the protein between residues 71 and 86. This NLS could not be predicted using conventional NLS prediction algorithms. We also found that the nuclear transport of ILKAP was mediated by the importin α/β nuclear transport pathway. We know that the NLS-binding sites are located in a concave groove on the surface of importin α. The major binding site is composed mainly of the H3 helices of ARM repeats 1–4, whereas the minor site is located at ARM repeats 6–8. The protein docking results suggested that ILKAP may bind to the minor binding site of importin α.
In 2004, Doehn et al. (31) found that ILKAP forms a complex with RSK2 in vivo. As a well known serine/threonine kinase, RSK2 plays a crucial role in oncogenesis and tumor progression. RSK2 has been reported to localize to the nucleus (14–16, 25). Thus, we hypothesized that ILKAP may interact with RSK2 in the nucleus and regulate cell proliferation and apoptosis by affecting RSK2 signaling. We found that nuclear ILKAP interacts with RSK2 and that ILKAP induces apoptosis by inhibiting RSK2 activity and down-regulating the expression level of the RSK2 downstream substrate cyclin D1.
In summary, we found that ILKAP is a nuclear protein and that the nuclear transport of ILKAP is NLS importin-mediated. The ILKAP protein interacts directly with importins α1, α3, and α5. We also found that ILKAP induces apoptosis by inhibiting RSK2 activity and down-regulating the expression level of the RSK2 downstream substrate cyclin D1. These results classify ILKAP as a newly identified nuclear protein that regulates cell survival and apoptosis through the regulation of RSK2 signaling.
This work was supported by the National Natural Science Foundation of China (Grant 81201556) and the Department of Health of the National Natural Science Foundation of China (Grant 2012159).
- ILKAP
- integrin-linked kinase-associated phosphatase
- ILK
- integrin-linked kinase
- RSK2
- ribosomal protein S6 kinase 2
- NLS
- nuclear localization signal
- EGFP
- enhanced green fluorescent protein
- ARM
- armadillo.
REFERENCES
- 1. Tong Y., Quirion R., Shen S. H. (1998) Cloning and characterization of a novel mammalian PP2C isozyme. J. Biol. Chem. 273, 35282–35290 [DOI] [PubMed] [Google Scholar]
- 2. Tamura S., Toriumi S., Saito J., Awano K., Kudo T. A., Kobayashi T. (2006) PP2C family members play key roles in regulation of cell survival and apoptosis. Cancer Sci. 97, 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar A. S., Naruszewicz I., Wang P., Leung-Hagesteijn C., Hannigan G. E. (2004) ILKAP regulates ILK signaling and inhibits anchorage-independent growth. Oncogene 23, 3454–3461 [DOI] [PubMed] [Google Scholar]
- 4. Leung-Hagesteijn C., Mahendra A., Naruszewicz I., Hannigan G. E. (2001) Modulation of integrin signal transduction by ILKAP, a protein phosphatase 2C associating with the integrin-linked kinase, ILK1. EMBO J. 20, 2160–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 6. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 7. Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. (2003) Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-α. J. Biol. Chem. 278, 27981–27987 [DOI] [PubMed] [Google Scholar]
- 8. Dingwall C., Laskey R. A. (1991) Nuclear targeting sequences – a consensus? Trends Biochem. Sci. 16, 478–481 [DOI] [PubMed] [Google Scholar]
- 9. Robbins J. (1991) Two interdependent basic domains in nucleoplasm in nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 [DOI] [PubMed] [Google Scholar]
- 10. Conti E., Kuriyan J. (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 11. Fontes M. R., Teh T., Kobe B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 12. Rexach M., Blobel G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692 [DOI] [PubMed] [Google Scholar]
- 13. Hehlgans S., Haase M., Cordes N. (2007) Signalling via integrins: implications for cell survival and anticancer strategies. Biochim. Biophys. Acta 1775, 163–180 [DOI] [PubMed] [Google Scholar]
- 14. Chen R. H., Sarnecki C., Blenis J. (1992) Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eisinger-Mathason T. S., Andrade J., Groehler A. L., Clark D. E., Muratore-Schroeder T. L., Pasic L., Smith J. A., Shabanowitz J., Hunt D. F., Macara I. G., Lannigan D. A. (2008) Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell 31, 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willard F. S., Crouch M. F. (2001) MEK, ERK, and p90RSK are present on mitotic tubulin in Swiss 3T3 cells: a role for the MAP kinase pathway in regulating mitotic exit. Cell. Signal. 13, 653–664 [DOI] [PubMed] [Google Scholar]
- 17. Gray J.J. (2006) High-resolution protein-protein docking. Curr. Opin. Struct. Biol. 16, 183–193 [DOI] [PubMed] [Google Scholar]
- 18. Sivasubramanian A., Chao G., Pressler H. M., Wittrup K. D., Gray J. J. (2006) Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure 14, 401–414 [DOI] [PubMed] [Google Scholar]
- 19. Wiehe K., Pierce B., Mintseris J., Tong W. W., Anderson R., Chen R., Weng Z. (2005) ZDOCK and RDOCK performance in CAPRI rounds 3, 4, and 5. Proteins 60, 207–213 [DOI] [PubMed] [Google Scholar]
- 20. Jensen C. J., Buch M. B., Krag T. O., Hemmings B. A., Gammeltoft S., Frödin M. (1999) 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem. 274, 27168–27176 [DOI] [PubMed] [Google Scholar]
- 21. Dias S. M., Wilson K. F., Rojas K. S., Ambrosio A. L., Cerione R. A. (2009) The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 16, 930–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pierce B., Weng Z. (2007) ZRANK: reranking protein docking predictions with an optimized energy function. Proteins 67, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 23. Lorenz K., Grashoff C., Torka R., Sakai T., Langbein L., Bloch W., Aumailley M., Fässler R. (2007) Integrin-linked kinase is required for epidermal and hair follicle morphogenesis. J. Cell Biol. 177, 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakai T., Li S., Docheva D., Grashoff C., Sakai K., Kostka G., Braun A., Pfeifer A., Yurchenco P. D., Fässler R. (2003) Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 17, 926–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang S., Chen J. (2011) Targeting RSK2 in human malignancies. Expert Opin. Ther. Targets 15, 11–20 [DOI] [PubMed] [Google Scholar]
- 26. Legate K. R., Montañez E., Kudlacek O., Fässler R. (2006) ILK, PINCH, and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 7, 20–31 [DOI] [PubMed] [Google Scholar]
- 27. Wickström S. A., Lange A., Montanez E., Fässler R. (2010) The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J. 29, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanks S. K., Quinn A. M., Hunter T. (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52 [DOI] [PubMed] [Google Scholar]
- 29. Grashoff C., Aszódi A., Sakai T., Hunziker E. B., Fässler R. (2003) Integrin-linked kinase regulates chondrocyte shape and proliferation. EMBO Rep. 4, 432–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lange A., Wickström S. A., Jakobson M., Zent R., Sainio K., Fässler R. (2009) Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature 461, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 31. Doehn U., Gammeltoft S., Shen S. H., Jensen C. J. (2004) p90 ribosomal S6 kinase 2 is associated with and dephosphorylated by protein phosphatase 2Cδ. Biochem. J. 382, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]