Background: Lavandula accumulate irregular monoterpenes of unknown biosynthetic origin.
Results: We cloned a cis-prenyl diphosphate synthase (cis-PDPS) that produces precursor for irregular monoterpenes in lavenders.
Conclusion: Unlike other plants that utilize trans-PDPSs, Lavandula employ a cis-PDPS to initiate the biosynthesis of irregular monoterpenes.
Significance: This is the first report of the involvement of a cis-PDPS in irregular monoterpene biosynthesis.
Keywords: Biosynthesis, Genomics, Isoprenoid, Plant Biochemistry, Plant Molecular Biology, Lavandula, Irregular Monoterpenes, Lavandulol, Lavandulyl Diphosphate, Prenyltransferases
Abstract
Lavender essential oils are constituted predominantly of regular monoterpenes, for example linalool, 1,8-cineole, and camphor. However, they also contain irregular monoterpenes including lavandulol and lavandulyl acetate. Although the majority of genes responsible for the production of regular monoterpenes in lavenders are now known, enzymes (including lavandulyl diphosphate synthase (LPPS)) catalyzing the biosynthesis of irregular monoterpenes in these plants have not been described. Here, we report the isolation and functional characterization of a novel cis-prenyl diphosphate synthase cDNA, termed Lavandula x intermedia lavandulyl diphosphate synthase (LiLPPS), through a homology-based cloning strategy. The LiLPPS ORF, encoding for a 305-amino acid long protein, was expressed in Escherichia coli, and the recombinant protein was purified by nickel-nitrilotriacetic acid affinity chromatography. The approximately 34.5-kDa bacterially produced protein specifically catalyzed the head-to-middle condensation of two dimethylallyl diphosphate units to LPP in vitro with apparent Km and kcat values of 208 ± 12 μm and 0.1 s−1, respectively. LiLPPS is a homodimeric enzyme with a sigmoidal saturation curve and Hill coefficient of 2.7, suggesting a positive co-operative interaction among its catalytic sites. LiLPPS could be used to modulate the production of lavandulol and its derivatives in plants through metabolic engineering.
Introduction
Monoterpenes, the C10 class of the isoprenoids, are derived from the universal terpene building blocks isopentenyl diphosphate (IPP)3 and dimethylallyl diphosphate (DMAPP) predominantly synthesized through the 2-C-methyl-d-erythritol 4-phosphate metabolic pathway in plants (1). En route to monoterpene synthesis IPP and DMAPP, or two DMAPP units, can be condensed to form structurally diverse branch point C10 precursor molecules (2). The diversity of these molecules arises from the capability of prenyl diphosphate synthase enzymes (PDPSs) to condense isoprenes in head-to-tail or non–head-to-tail orientations (3, 4) and outcomes with cis or trans geometric configurations (5). For example, the head-to-tail coupling of two isoprene units to geranyl diphosphate (C10; GPP) or its cisoid isomer neryl diphosphate (C10; NPP) is catalyzed by geranyl diphosphate synthase (GPPS) and neryl diphosphate synthase (NPPS), respectively. The non–head-to-tail condensation of isoprene units to lavandulyl diphosphate (C10; LPP) and chrysanthemyl diphosphate (C10; CPP), on the other hand, is catalyzed by lavandulyl diphosphate synthase (LPPS) and chrysanthemyl diphosphate synthase (CPPS), respectively (Fig. 1). The final stage of monoterpene synthesis, transforming and elaborating the different precursor molecules to monoterpenes, is catalyzed by another group of enzymes called monoterpene synthases (mTPSs).
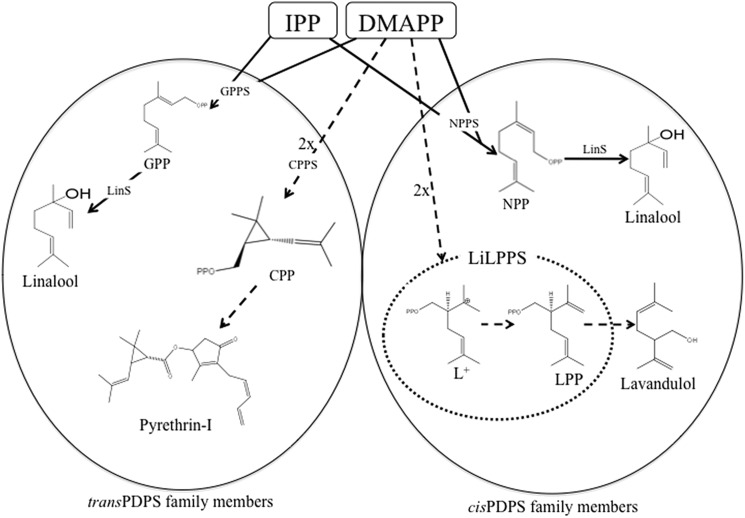
FIGURE 1.
Schematic representation of IPP and DMAPP coupling reactions catalyzed by typical trans- and cis-PDPS family members. The newly identified enzyme and the reaction it catalyzed are circled (adapted from Thulasiram et al. (3)). CPP, chrysanthemyl diphosphate; CPPS, chrysanthemyl diphosphate synthase; DMAPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; GPP, geranyl diphosphate; GPPS, geranyl diphosphate synthase; LPP, lavandulyl diphosphate; LiLPPS, L. x intermedia lavandulyl diphosphate synthase; LinS, linalool synthase; NPP, neryl diphosphate; and NPPS, neryl diphosphate synthase.
The head-to-tail condensation of IPP and DMAPP and the enzymes catalyzing these reactions are the most common in nature. Thus, monoterpenes derived from GPP and NPP such as linalool, 1,8-cineole, limonene, and so forth, are widely distributed and are referred to as “regular monoterpenes.” Subsequently, their biosynthetic route is well established, and cDNAs encoding for GPPS (6–10), NPPS (11), and mTPSs capable of transforming GPP and/or NPP into core monoterpenes have been described from diverse plant species (12, 13). GPPS, a trans-PDPS, and NPPS, a cis-PDPS, elongate the linear prenyl chain by coupling DMAPP with IPP through a chain elongation (c1′- 4) reaction to generate GPP and NPP, respectively (3). Despite catalyzing the same reaction, however, GPPSs and NPPS are more distantly related to one another than they are to PDPSs of similar geometric outcomes. GPPSs share high sequence similarity with trans-PDPSs catalyzing the higher order terpene precursors and are distinguished by two aspartate-rich conserved motifs, DDX2–4D and (N/D)DXXD. These motifs serve as a substrate and divalent metal ion cofactor, often Mg2+, binding sites for the carbocation rearrangement-mediated condensation reactions (2, 5). A third conserved motif, the CXXXC motif (where X is any hydrophobic residue), is present in heterodimeric PDPSs (14). Heterodimeric PDPSs, like the GPPSs cloned from Mentha x piperita (8), Salvia miltiorrhiza (15) etc., are enzymes constituted of two subunits (large and small) which interact through the CXXXC conserved motif to be catalytically active. NPPS and other cis-PDPSs, on the other hand, do not necessarily retain the above motifs but share five conserved regions designated as Regions I–V. In particular, the aspartate residue in Region IV and the glutamate residue of Region V are catalytically essential (16, 17).
PDPSs catalyzing non–head-to-tail coupling reactions, and monoterpenes derived from LPP and CPP such as lavandulol and pyrethrins, respectively, are encountered less frequently in nature (18–20). Irregular monoterpenes, like pyrethrins, are the major ingredients in leading botanical and EPA-certified insecticides (21, 22). Lavandulol and its ester derivative lavandulyl acetate were identified in pheromones of major insect pests and are subsequently used in artificial pheromone preparations to disrupt the mating behaviors of economically important pests (23, 24). Yet, little is known about the biosynthetic pathways leading to these monoterpenes and their derivatives. Only two cDNAs encoding for CPPSs, both trans-PDPS family members, have been isolated and functionally characterized from Chrysanthemum cinerariaefolium (20) and Artemisia tridentate ssp. spiciformis (25) so far. CPPSs catalyze predominantly the cyclopropanation (c1′-2-3) reaction in which the non–head-to-tail condensation of two DMAPP molecules generates the pyrethrin branch point intermediate, CPP. In addition to their major product, the A. tridentate CPPS and its chimeric derivatives were found to catalyze the non–head-to-tail coupling of two DMAPP molecules through branching (c1′-2) to generate LPP (3, 4). LPP is the branch point precursor of monoterpenes with head-to-middle condensed PDP backbones, such as lavandulol and lavandulyl acetate (19). It is also the source of the lavandulyl side group of sophoraflavanone G in Sophora flavescens Ait (26) that determines their antitumor (27) and phospholipase Cγ1 inhibition properties (28). To our knowledge a wild type LPPS gene has not been described from plants or other organisms.
Essential oils (EOs) of the genus Lavandula (lavenders) are constituted primarily of a few “regular” monoterpenes and their derivatives. For example, the economically important EOs derived from L. angustifolia and L. x intermedia species, contain large amounts of linalool, linalool acetate, and 1,8-cineole. Consequently, these oils are industrially utilized as ingredients of various cosmetic and antiseptic products (29). The biosynthetic pathway leading to these monoterpenes from IPP and DMAPP has been defined both experimentally (30–32) and through in silico analysis of Lavandula EST databases (30, 31, 33). In this regard, four cDNAs encoding for mTPSs that transform GPP and NPP into linalool, 1,8-cineole, limonene, and β-phellandrene in vitro have been cloned from L. angustifolia and L. x intermedia species (30–32). However, lavender EOs, particularly L. x intermedia species, also contain the irregular monoterpene lavandulol and its ester derivative lavandulyl acetate (34) whose biosynthetic origin has not yet been investigated. Here, we report the isolation and functional characterization of a novel cis-PDPS cDNA encoding for LPPS, an enzyme that condenses two DMAPP molecules to generate the lavandulol branch point precursor LPP in vitro, from a L. x intermedia oil gland cDNA library.
EXPERIMENTAL PROCEDURES
LiLPPS Candidate Selection
Our group recently reported the construction of a cDNA library and its corresponding annotated expressed sequence tag (EST) database from L. x intermedia cv. Grosso secretory cells of oil glands, tissues specialized for EO biosynthesis and secretion (31). All PDPS homologs in the database were retrieved by searching the EST database using the strings “diphosphate synthase” and “pyrophosphate synthase.” We then synchronized the search results and manually excluded EST homologs known to be involved in regular terpene biosynthesis including GPPS, trans-farnesyl diphosphate synthase (trans-FPPS), and trans-geranylgeranyl diphosphate synthase (trans-GGPPS), and PDPS homologs that are not involved in terpene biosynthesis. This led us to acquire a novel cis-PDPS homolog contig that was later determined to be L. x intermedia lavandulyl diphosphate synthase (LiLPPS). The transcriptional expression pattern of this contig and previously described mTPSs throughout L. x intermedia cv. Grosso flower developmental stages was assessed by microarray analysis using the Agilent oligonucleotide-based microarray technology, through services provided by the University Health Network Microarray Centre (Toronto, ON, Canada). The three floral developmental stages were unopened buds, anthesis, and mature flowers in which 30% of the buds were in bloom (for photographic description see Ref. 33). After validating the microarray data using standard PCR, we selected the contig for further detailed analysis.
Cloning, Protein Expression, and Enrichment of LiLPPS
Glandular trichome secretory cells were isolated from mature flowers of L. x intermedia cv. Grosso plants grown at the University of British Columbia, Okanagan campus lavender field following a previously described modified glass bead abrasion method (31). Total RNA was extracted from 100 mg of the tissue using an RNA extraction kit (OMEGA bio-tek) and reverse transcribed in a reaction containing the oligo(dT) primer (Fisher Scientific) and M-MuLV reverse transcriptase enzyme (New England Biolabs) following the manufacturer's directions. The putative LiLPPS ORF was amplified with the full-length cloning primer sets (Table 1) and iProof™ High-Fidelity DNA Polymerase (Bio-Rad). The PCR program used was 95 °C for 5 min, followed by 37 cycles of 95 °C for 1 min, 58 °C for 30 s, and 72 °C for 1 min, and a 5-min final extension at 72 °C. The amplified fragments were cloned into the NdeI/EcoRI sites of the pET41b(+) expression vector and expressed in Escherichia coli BL21(DE3) strain (EMD Chemicals, Darmstadt, Germany) following previously described procedures (30, 31). Except for the bind buffer that was slightly modified (0.5 m NaCl, 50 mm KH2PO4, pH 8.0) the procedure described in the aforementioned papers was followed to enrich the expressed protein. Sequence information for LiLPPS was deposited in the NCBI public data bank with the accession number JX985358.
TABLE 1.
Oligonucleotides used in this study
| Primers | Sequences |
|---|---|
| LiLPPS set I | F: 5′-CACTCATATGGCATTCCTGCAGTTACC-3′ |
| R1: 5′-CTCTTCATGAATTCCTTTTCGTTCACCG-3′ | |
| LiLPPS set II | F: 5′-CACTCATATGGCATTCCTGCAGTTACC-3′ |
| R2: 5′-AATTCTCTTCATGAATTCCTTTTCGTTCACCG-3′ |
LiLPPS Product Assay and EO Constituent Identification
Initial in vitro enzyme activity assays were performed in a 500-μl reaction volume containing the assay buffer (50 mm Tris-HCl, 5% glycerol, 1 mm MnCl2, 1 mm MgCl2, pH 8.0), 1 mm DTT, 40 μm IPP, and 40 μm DMAPP (Echelon, Salt Lake City, UT), and 2.5–50 μg of LiLPPS. After a 2-h incubation at 30 °C, the reaction mix was heated at >80 °C for 10 min and kept on ice for 2 min. Then 30 units of calf intestinal alkaline phosphatase (CIP; New England Biolabs) were added to the reaction mix, overlaid by 500 μl of pentane and incubated overnight at 32.5 °C to hydrolyze the prenyl product. GPP and NPP standards (Echelon) were also hydrolyzed in the same reaction mix and conditions, as a control of the hydrolysis reaction. The reaction was stopped by vigorous vortexing followed by flash freezing in liquid nitrogen and stored in a −80 °C freezer until analyzed. Assay product identification and quantification, and EO extraction and quantification were performed by gas chromatography-mass spectrometry (GC-MS) following procedures described previously (32, 35). Purified protein extract of E. coli BL21(DE3) strain transformed with empty pET41b(+) expression vector, i.e. without insert, was also assayed under the same conditions as a negative control.
Biochemical Assay
Biochemical characterization of LiLPPS was performed following the procedure described above with slight modifications: the reaction volume was reduced to 125 μl, 2.5 μg of LiLPPS was used, and assays were incubated at 37 °C for 1 h after adding CIP. Six temperature levels (25, 27.5, 30, 32.5, 35, and 37.5 °C) were tested to determine LiLPPS optimum temperature, and the optimum pH was determined using MES and MOPS buffers at pH 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5. The kinetic properties of LiLPPS were deduced from LPP accumulation data in assays with increasing DMAPP concentrations (6.25, 12.5, 25, 50, 100, 150, 200, 300, 450, and 600 μm) at the optimal conditions. Prism software version 5.0d (GraphPad Software, Inc., La Jolla, CA) was used to fit the data into a sigmoidal substrate concentration-dependent enzyme response curve (y = a*bx/(cx + bx)), and to calculate the Vmax, Km, and kcat values. Substrate specificity of the enzyme was determined from the type and amount of reactants consumed versus product accumulated in assays containing a constant IPP concentration (150 μm) combined with various DMAPP levels (6.25–200 μm) under the optimized conditions. We also tested LPP as a potential substrate for L. x intermedia 1,8-cineole synthase (LiCINS), an enzyme that primarily produced 1,8-cineole from both GPP and NPP (31).
Phylogenetic Relationship Analysis
LiLPPS was aligned with cis-PDPSs isolated from tomato using the default parameters of the ClustalW alignment tool available at the EBI platform. The phylogenetic relation of LiLPPS with other PDPSs was constructed using the Jukes-Cantor genetic distance model and UPGMA tree building method of Geneious Tree Builder module (Geneious 5.0.3 software, Auckland, New Zealand).
RESULTS
Lavandula x intermedia cv. Grosso EO Composition
GC-MS analysis of EO distilled from L. x intermedia cv. Grosso floral tissues profiled the following terpenes in their respective abundance order: linalool, linalool acetate, 1,8-cineole, camphor, isoborneol, lavandulyl acetate, α-terpineol, α-bisabolol, α-cadinol, β-ocimen, β-caryophyllene, lavandulol, limonene, δ-carene, myrcene, geraniol, nerol, neryl acetate, and a few other minor products (supplemental Fig. S1). Except for the irregular monoterpenes lavandulol and lavandulyl acetate, which are derived from LPP, the remaining EO mono- and sesquiterpene constituents arise from the corresponding linear precursors GPP and trans-FPP, respectively.
PDPS Candidate Selection and Sequence Analysis
Searching our EST library using the string pyrophosphate synthase as a query identified 11 different prenyltransferase homolog unigenes whereas the string diphosphate synthase identified 31. Duplicate search results and prenyltransferases with no known involvement in terpene biosynthesis (e.g. ribose phosphate pyrophosphokinase and cytidine diphosphate diacylglycerol synthase) were disregarded to consolidate the search results. The final synchronized search result identified 25 unigenes that were homologous to known PDPSs involved in terpene biosynthesis (supplemental Table S1). From these sequences 12 were homologous to PDPSs involved in the 2-C-methyl-d-erythritol 4-phosphate pathway, eight were homologous to trans-PDPSs (GPPSs, FPPSs, GGPPSs), and one was homologous to bornyl diphosphate synthase. The remaining four candidates were homologous to cis-PDPSs. Two of these homologs corresponded to dehydrodolichol diphosphate synthase (dDPPS) genes, which catalyzes the prenyl chain elongation reaction to produce the polyprenyl backbone of dolichol. The other unigenes, one of which belonged to a contig with 18 EST members and the other was a singleton, exhibited significant homology to NPPS (FJ797956) and cis-FPPS (ACJ38408) from cultivated (Solanum lycopersicum) (11) and wild tomato (Solanum habrochaites) (17), respectively. The singleton was later determined to be a splice variant of the contig and was disregarded.
The trans-PDPS and dDPPS homologs were anticipated to be present in a database derived from L. x intermedia secretory cells because they catalyze reactions that are consistent with EO composition in this tissue. However, given the fact that GPP is the preferred substrate for regular monoterpenes (11, 36) and typical sesquiterpenes synthesized from cis-FPP (17) were not detected in the L. x intermedia EO (supplemental Fig. S1), the identification of NPPS and cis-FPPS homologs was unforeseen. In addition, the results of our transcript-profiling experiment indicated that the transcriptional expression of this EST paralleled those of other lavender mTPSs (31) and was developmentally regulated in L. x intermedia flowers (data not shown). Therefore, we decided to further investigate this contig which was later determined to be LiLPPS. The ORF of LiLPPS was expressed in E. coli BL21(DE3) cells for further analysis.
The ORF of LiLPPS is 918 nucleotides long encoding a 305-amino acid protein that includes a short N-terminal plastidial transit peptide (30 amino acids) and shares 66.4 and 66% sequence similarity with NPPS and cis-FPPS of tomato, respectively. Like many other cis-PDPSs, LiLPPS lacks the two aspartate rich trans-PDPS signature motifs DDX2–4D and (N/D)DXXD but maintains all five semiconserved regions (I–V) that define the cis-PDPSs (Fig. 2). In particular, the catalytically important aspartate and glutamate residues of Regions IV and V were conserved in LiLPPS at 201 and 276 positions from the N-terminal, respectively (Fig. 2). Multiple alignment of LiLPPS and its splice variant, both at the nucleotide and amino acid level, revealed that the splice variant lacks a stretch of 22 amino acids in the middle of its ORF that included a portion of the conserved region III (Fig. 2). Repeated attempts to amplify the splice variant from reverse transcribed total mRNA isolated from L. x intermedia glandular secretory cells failed possibly due to the low abundance of the corresponding cDNA. It is also possible that the splice variant singleton was an artifact that resulted from sequence assembly errors during database construction. Therefore, only the full-length contig was considered for further investigation.
FIGURE 2.
Multiple alignment of LiLPPS with NPPS and cis-FPPS of tomato. Bars indicate the five conserved regions (I–V), and the aspartate and glutamate residues in Regions IV and V, respectively, are boxed. Identical amino acid residues are represented by asterisks, conserved amino acid substitutions are represented by a semicolon, and semiconserved amino acid substitutions are represented by a period. LiLPPS (JX985358), L. x intermedia lavandulyl diphosphate synthase; zFPS_SOLHA (B8XA40.1), Z,Z-farnesyl diphosphate synthase from (S. habrochaites) and NPPS (NP_001234633.1), neryl diphosphate synthase from (S. lycopersicum).
Cloning, Expression, and Functional Characterization of LiLPPS
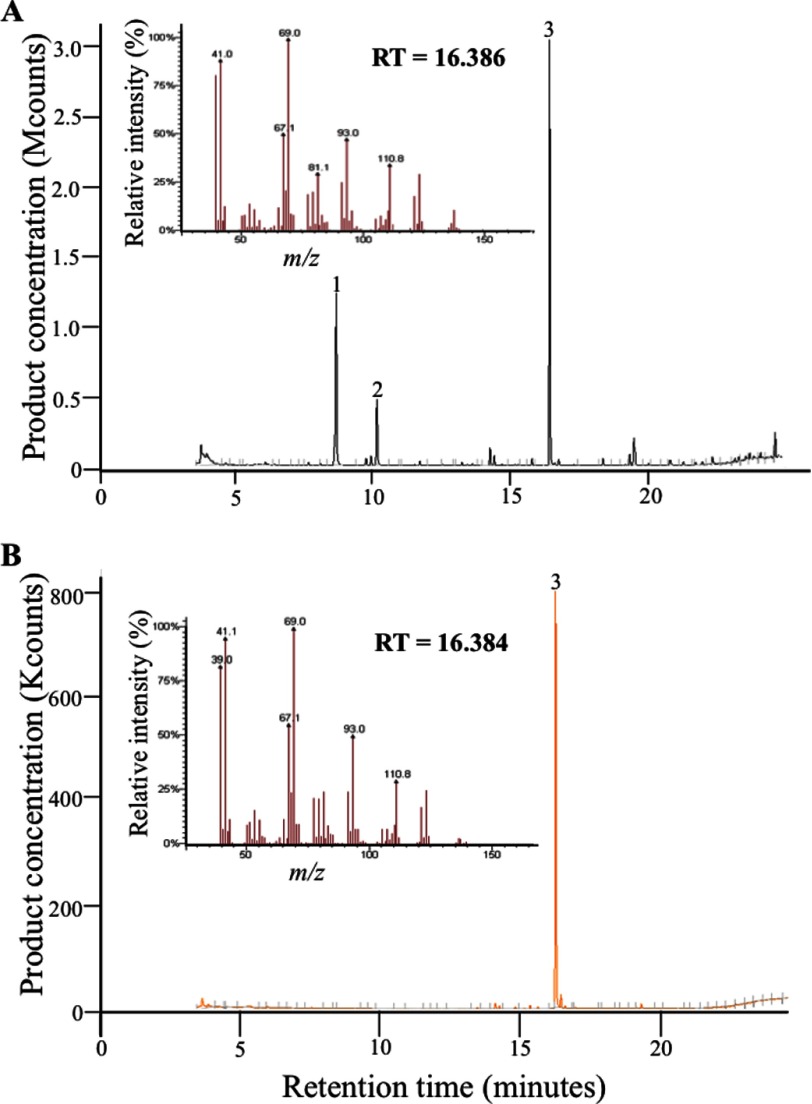
The approximately 34.5-kDa recombinant LiLPPS was successfully expressed in bacterial cells and enriched using the nickel-nitrilotriacetic acid-agarose affinity chromatography system. Incubation of the purified recombinant protein with IPP and DMAPP followed by the hydrolysis of the assay mix by CIP resulted in the production of lavandulol as the only product (Fig. 3A). The identity of the product was determined by comparing its mass spectrum and retention time with those of an authentic lavandulol standard (Sigma) (Fig. 3B). Given that the alkaline CIP-mediated hydrolysis of LPP is known to yield lavandulol (3, 4, 19), we concluded that LiLPPS catalyzes the synthesis of LPP from isoprene units. In this respect, the hydrolysis of GPP and NPP with CIP under the same conditions produced geraniol and nerol, respectively, as anticipated. Hydrolysis of the control assay mixes contained only the prenyl alcohol derivatives of IPP (buten-1-ol, <3-methyl-3) and DMAPP (buten-1-ol, <3-methyl-2) (Fig. S2, A and B).
FIGURE 3.
GC chromatogram and mass spectrum of LiLPPS product from DMAPP after calf intestinal alkaline phosphatase hydrolysis (A) and authentic lavandulol standard (B). Peaks corresponds to: IPP prenyl alcohol (1), DMAPP prenyl alcohol (2), and lavandulol (3).
Substrate Specificity and Kinetic Properties of LiLPPS
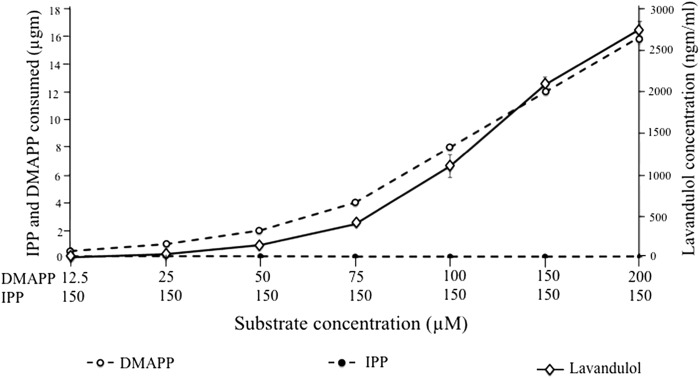
When LiLPPS was incubated with IPP alone, detectable amounts of lavandulol or any other terpene prenyl alcohols were not identified after the alkaline hydrolysis step. We also observed that assay reactions containing 5 μm each IPP and DMAPP, or 5 μm DMAPP alone produced equivalent amounts of LPP. Furthermore, lowering the concentration of DMAPP in the assay resulted in a parallel reduction in LPP production, although reducing the concentration or excluding IPP from the assay had no effect. When LiLPPS was incubated with a constant IPP concentration (150 μm) combined with an increasing DMAPP amount (6.25–200 μm), the amount of LPP accumulated correlated (R2 = 0.99; p < 0.0001) with the amount of DMAPP consumed whereas the IPP consumption remained close to zero at all combinations (Fig. 4).
FIGURE 4.
LPP accumulation versus DMAPP and IPP consumption (R2 = 0.99, p < 0.0001, n = 2). Error bars indicate S.D.
The optimum temperature and pH of LiLPPS were found to be 30 °C and 8.0 (supplemental Fig. S3, A and B), respectively. Unlike other PDPSs cloned from related plant species, the substrate concentration-dependent saturation curve of LiLPPS did not follow the standard Michaelis-Menten kinetics for single substrate enzymes. Instead, the amount of LPP accumulated in response to increasing DMAPP concentration levels (6.25–600 μm) fitted a sigmoidal saturation curve (Fig. 5), typical of two substrate enzymes. The nonlinear regression equation Vo = Vmax*[S]H/(kmH + [S]H) was used to calculate the Km and Vmax of LiLPPS which were determined to be 208 ± 12 μm and 448 ± 22 pmol/min, respectively. The Hill coefficient (H) value of LiLPPS was 2.7 ± 0.3 whereas the kcat (Vmax/[E]) and catalytic efficiency (kcat/Km) of the enzyme were calculated as 0.1 s−1 and 5 × 10−10 m−1 s−1, respectively.
FIGURE 5.
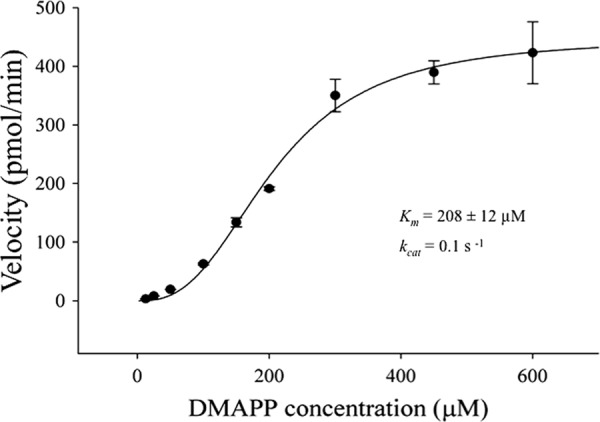
Kinetic properties of LiLPPS at increasing DMAPP concentrations. The nonlinear regression equation used to fit LPP accumulation against DMAPP concentrations was V = Vmax*[S]H/(kmmH + [S]H) (n = 2). Error bars indicate S.D.
LPP as a Substrate for Regular mTPSs
mTPSs involved in the biosynthesis of regular monoterpenes in lavenders accept both GPP and NPP as in vitro substrates. We thus examined the ability of the recombinant LiCINS to utilize LPP as a substrate in standard assay reactions. As anticipated, LiCINS, which produced 1,8-cineole as a major product upon incubation with GPP and NPP (32), did not produce detectable quantities of a product from LPP.
DISCUSSION
Like those of many other plants, the EOs of lavenders are dominated by regular monoterpenes synthesized through the head-to-tail condensation of isoprene units. It was therefore not surprising that the EST database derived from L. x intermedia secretory cells, tissues specialized for EO biosynthesis, contained a substantial number of ESTs corresponding to enzymes involved in the biosynthesis of regular monoterpenes (supplemental Table S1). Given that L. x intermedia plants also accumulate irregular monoterpenes, particularly the LPP derivatives lavandulol and lavandulyl acetate (19) (supplemental Fig. S1), it was also not surprising to find ESTs related to irregular monoterpene biosynthesis, including the cloned LiLPPS, in our database. However, it was somewhat unexpected to find a cis-PDPS in L. x intermedia glandular trichomes because (i) all known Lamiaceae PDPSs (6–10), including those of Lavandula (30–32), are of the trans-PDPS type. (ii) The only reported PDPSs with confirmed role in irregular monoterpene biosynthesis, CPPSs (3, 4, 20), also belong to the trans-PDPSs family. The previously reported cis-PDPSs, NPPS and cis-FPPS, catalyzed the biosynthesis of regular (and not irregular) mono- and sesquiterpene precursors, respectively, in tomato glandular trichomes.
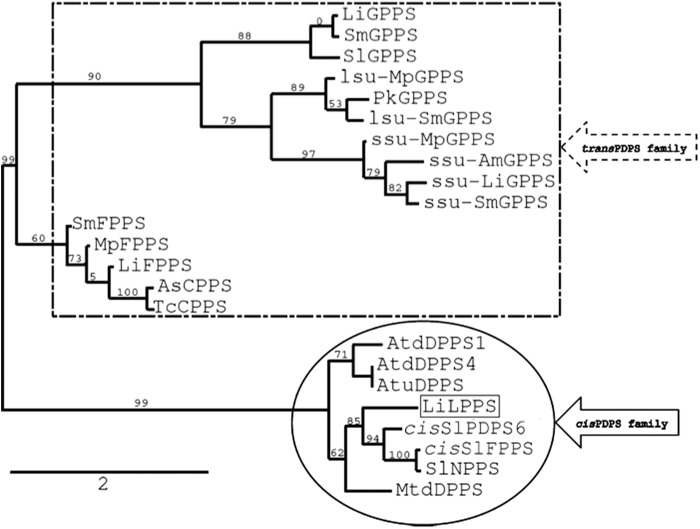
The cis and trans PDPS family members often share higher sequence similarity with each other irrespective of the genetic relatedness among donor organisms (5, 16). This is contrary to some other enzymes involved in terpene biosynthesis, particularly terpene synthases. Typically, terpene synthases of a given species are more related to one another than to those of distantly related species, even if they catalyze the same reaction. For example β-phellandrene synthase of L. angustifolia shares 48 and 47% identity with linalool and limonene synthases of the same species, respectively. However, the gene only shares 31 and 25% similarity with β-phellandrene synthases of grand fir and tomato, respectively (30). In this study, we observed that L. x intermedia trans-PDPS homologs (GPPS, FPPS, and GGPPS) share a higher sequence similarity among each other and with other trans-PDPSs cloned from genetically unrelated species than they do with LiLPPS. LiLPPS is closely related to NPPS and cis-FPPS of cultivated (S. lycopersicum) (11) and wild (S. habrochaites) (17) tomato, respectively. The phylogenetic tree presented in Fig. 6 clearly suggests that LiLPPS together with NPPS and cis-FPPS diverged very early from trans-PDPSs.
FIGURE 6.
Phylogenetic relationship and classification of prenyltransferases. Prenyltransferases within the same class share a minimum of 50% amino acid identity. The trans-PDPS family members are boxed with a broken line, cis-PDPS family members are circled with a solid line, and the prenyltransferase described in this article is boxed with a solid line. The scale bar represents amino acid substitutions per site, and numbers represent the branch support values in percentage. Accession numbers of prenyltransferases used to generate the phylogenetic tree are: AsCPPS, AY308478.1; AtdDPPS1, NP_565551.1; AtdDPPS4, NP_568883.1; AtuDPPS, AAM67372.1; cis-SlFPPS, B8XA40.1; cis-SlPDPS6, AFW98430.1; LiLPPS, JX985358; lsu-MpGPPS, AAF08793.1; lsu-SmGPPS, AEZ55681.1; MpFPPS, AAK63847.1; MtdDPPS, XP_003615624.1; PkGPPS, AAW66658.1; SmFPPS, ABV08819.1; SmGPPS, ACR19637.1; SlGPPS, NP_001234302.1; SlNPPS, NP_001234633.1; ssu-AmGPPS, AAS82859.1; ssu-LiGPPS, JX985359; ssu-MpGPPS, AAF08792.1; ssu-SmGPPS, AEZ55678.1; and TcCPPS, HQ235057.1.
LiLPPS catalyzes the head-to-middle condensation of two DMAPP molecules to synthesize the linear lavandulol branch point precursor, LPP (C10). As demonstrated by Thulasiram et al. (3, 4), LPP synthesis proceeds via rearrangement of the double bond position to create the highly reactive carbocation intermediate lavandulyl cation (L+). L+ will eventually be transformed to LPP after a mandatory proton loss (Fig. 1). LiLPPS catalyzes these reactions with an apparent Km and kcat of 208 ± 12 μm and 0.1 s−1, respectively. These catalytic properties are within the range of previously reported cis-PDPSs including NPPS, which has a Km and kcat of 177 μm and 0.2 s−1, respectively (11). The Km and kcat values of the aromatic prenyltransferase cloned from fungi were 325 μm and 0.03 s−1, respectively (39). Unlike other PDPSs, LiLPPS displayed a sigmoidal saturation curve (Fig. 5). This is a typical feature of enzymes with multiple substrates, in which the binding of the first substrate affects the affinity of the enzyme for the second substrate through conformational changes or stabilization of the active pocket environment (40). LiLPPS, like any other typical PDPS enzyme, has binding sites for a divalent metal ion cofactor and two substrates (5, 16). PDPSs accepting IPP and DMAPP (e.g. GPPS or NPPS) can be saturated with one of the substrates and forced to follow Michaelis-Menten kinetics typical of single substrate enzymes for the other substrate. Because DMAPP was the only substrate for LiLPPS, saturating one site was not an option. Thus, plotting LiLPPS product (LPP) accumulation against increasing substrate (DMAPP) concentrations assumed a sigmoidal saturation curve with a Hill coefficient (H) value of 2.7. The positive Hill coefficient value (2.7) indicates a positive co-operativity among the binding pockets.
If LPP was a condensation result of IPP and DMAPP, an equimolar consumption of the two isoprenes would be expected in our assays. However, LPP production was independent of IPP and required only DMAPP. In our assays, a reduction in DMAPP supply resulted in an equivalent reduction in LPP accumulation. However, reducing the concentration or excluding IPP from the reaction did not alter LPP production. This result was also confirmed when IPP was provided at a constant high concentration level (150 μm) while the amount of DMAPP was increased progressively (6.25–200 μm), and the reaction was allowed to run until LPP synthesis seized or DMAPP molecules were nearly consumed. At all IPP and DMAPP concentration combinations assayed, IPP consumption remained very close to 0, whereas that of DMAPP increased in parallel to the amount of LPP synthesized (Fig. 4). This outcome indicated that, like the CPPS enzymes reported by Thulasiram et al. (4) and Rivera et al. (20), LiLPPS utilizes DMAPP as the only substrate to synthesize LPP. The structural features of LiLPPS underlying this catalytic property or residues involved in DMAPP recognition and c1′-2 coupling reaction catalysis are yet to be determined. Our results, however, suggest that the active sites of LiLPPS selectively bind two DMAPP (and not IPP) units and position them in such a way that the first carbon atom of one unit is in close proximity to the second carbon of the other to facilitate the c1′-2 bond formation. One possibility is that LiLPPS preferentially recognizes DMAPP by identifying its double bond position. It is also possible that the position of the double bond in IPP is not favorable for head-to-middle condensation.
With >55,000 members, isoprenoids are the most structurally and stereochemically diverse biochemical compounds known to mankind (42). Much of this diversity has been attributed to the astounding mechanistic heterogeneity and promiscuity of terpene synthases. PDPSs, cis or trans, also play a major role in the structural diversity of isoprenoids by providing intermediate precursor molecules of varying chain length (C10, C15, C20, C30, etc.) destined for different isoprenoid groups. In addition, PDPSs generate the structurally distinct C10 precursor molecules GPP/NPP, LPP, CPP, maconelliyl diphosphate (C10) or planococcyl diphosphate (C10) by simply changing the position of the carbon-carbon bond (3,4, 20). These linear precursors are then elaborated upon by mTPSs to create various monoterpenes (42, 43). In Lavandula, GPP and LPP are the linear precursors for the biosynthesis of regular and irregular monoterpenes, respectively (30–32).
In conclusion, through the identification and functional characterization of LiLPPS, a novel cis-PDPS, we have elucidated the biosynthetic origin of irregular monoterpene constituents of Lavandula EOs. To our knowledge, this is the first report of a cis-PDPS involved in the biosynthesis of irregular monoterpenes. In addition, LiLPPS is the first wild type gene that catalyzes the unusual head-to-middle condensation of two DMAPP molecules to synthesize LPP (C10) as its primary function. The elucidation of this pathway enables researchers to further investigate the biosynthesis of irregular monoterpenes in lavenders and other plants. Further, the cloned gene could be used to modulate the accumulation of lavandulol, lavandulyl acetate, and prenylated metabolites with a lavandulyl group to a desired level through metabolic engineering (44).
Supplementary Material
Acknowledgment
We thank Mariana Galata for help with manuscript preparation.
This work was supported through grants or in-kind contributions by the University of British Columbia Okanagan, Investment Agriculture Foundation of British Columbia, National Research Council Plant Biotechnology Institute through the NAPGEN program, and Genome British Columbia (to S. S. M.) and by the Natural Sciences and Engineering Research Council of Canada (to S. S. M. and M. R. R.).

This article contains supplemental Figs. S1–S3 and Table S1.
- IPP
- isopentenyl diphosphate
- CIP
- calf intestinal alkaline phosphatase
- CPP
- chrysanthemyl diphosphate
- CPPS
- CPP synthase
- dDPPS
- dehydrodolichol diphosphate synthase
- DMAPP
- dimethylallyl diphosphate
- EO
- essential oil
- EST
- expressed sequence tag
- FPP
- farnesyl diphosphate
- FPPS
- FPP synthase
- GGPPS
- geranylgeranyl diphosphate synthase
- GPP
- geranyl diphosphate
- GPPS
- GPP synthase
- LiCINS
- L. x intermedia 1,8-cineole synthase
- LiLPPS
- L. x intermedia LPPS
- LPP
- lavandulyl diphosphate
- LPPS
- LPP synthase
- mTPS
- monoterpene synthase
- NPP
- neryl diphosphate
- NPPS
- NPP synthase
- PDP
- prenyl diphosphate
- PDPS
- PDP synthase.
REFERENCES
- 1. Croteau R., Kutchan T., Lewis N. (2000) in Biochemistry and Molecular Biology of Plants (Gruissem W., Jones R., eds) pp. 1250–1318, American Society of Plant Biologists, Rockville, MD [Google Scholar]
- 2. Walsh C. T. (2007) Revealing coupling patterns in isoprenoid alkylation biocatalysis. ACS Chem. Biol. 2, 296–298 [DOI] [PubMed] [Google Scholar]
- 3. Thulasiram H. V., Erickson H. K., Poulter C. D. (2008) A common mechanism for branching, cyclopropanation, and cyclobutanation reactions in the isoprenoid biosynthetic pathway. J. Am. Chem. Soc. 130, 1966–1971 [DOI] [PubMed] [Google Scholar]
- 4. Thulasiram H. V., Erickson H. K., Poulter C. D. (2007) Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science 316, 73–76 [DOI] [PubMed] [Google Scholar]
- 5. Liang P. H., Ko T. P., Wang A. H. (2002) Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269, 3339–3354 [DOI] [PubMed] [Google Scholar]
- 6. Bouvier F., Suire C., d'Harlingue A., Backhaus R. A., Camara B. (2000) Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24, 241–252 [DOI] [PubMed] [Google Scholar]
- 7. Burke C., Croteau R. (2002) Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch. Biochem. Biophys. 405, 130–136 [DOI] [PubMed] [Google Scholar]
- 8. Burke C. C., Wildung M. R., Croteau R. (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc. Natl. Acad. Sci. U.S.A. 96, 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt A., Gershenzon J. (2008) Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69, 49–57 [DOI] [PubMed] [Google Scholar]
- 10. Tholl D., Kish C. M., Orlova I., Sherman D., Gershenzon J., Pichersky E., Dudareva N. (2004) Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16, 977–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schilmiller A. L., Schauvinhold I., Larson M., Xu R., Charbonneau A. L., Schmidt A., Wilkerson C., Last R. L., Pichersky E. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. U.S.A. 106, 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen F., Tholl D., Bohlmann J., Pichersky E. (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229 [DOI] [PubMed] [Google Scholar]
- 13. Degenhardt J., Köllner T. G., Gershenzon J. (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70, 1621–1637 [DOI] [PubMed] [Google Scholar]
- 14. Wang G., Dixon R. A. (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 106, 9914–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma Y., Yuan L., Wu B., Li X., Chen S., Lu S. (2012) Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63, 2809–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kharel Y., Koyama T. (2003) Molecular analysis of cis-prenyl chain elongating enzymes. Nat. Prod. Rep. 20, 111–118 [DOI] [PubMed] [Google Scholar]
- 17. Sallaud C., Rontein D., Onillon S., Jabès F., Duffé P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissier A. (2009) A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21, 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dewick P. M. (2009) Medicinal Natural Products: A Biosynthetic Approach, 2nd Ed., John Wiley and Sons, West Sussex, England [Google Scholar]
- 19. Epstein W. W., Klobus M. A., Edison A. S. (1991) Irregular monoterpene constituents of Artemisia tridentata cana: the isolation, characterization, and synthesis of two new chrysanthemyl derivatives. J. Org. Chem. 56, 4451–4456 [Google Scholar]
- 20. Rivera S. B., Swedlund B. D., King G. J., Bell R. N., Hussey C. E., Jr., Shattuck-Eidens D. M., Wrobel W. M., Peiser G. D., Poulter C. D. (2001) Chrysanthemyl diphosphate synthase: isolation of the gene and characterization of the recombinant non–head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. Proc. Natl. Acad. Sci. U.S.A. 98, 4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuda K., Kikuta Y., Haba A., Nakayama K., Katsuda Y., Hatanaka A., Komai K. (2005) Biosynthesis of pyrethrin I in seedlings of Chrysanthemum cinerariaefolium. Phytochemistry 66, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 22. Matsuda K. (2012) Pyrethrin biosynthesis and its regulation in Chrysanthemum cinerariaefolium. Top. Curr. Chem. 314, 73–81 [DOI] [PubMed] [Google Scholar]
- 23. Franco J. C., Zada A., Mendel Z. (2009) in Biorational Control of Arthropod Pests: Application and Resistance Management (Ishaaya I., Horowitz A. R., eds) pp 233–278, Springer, New York [Google Scholar]
- 24. Walton V. M., Daane K. M., Bentley W. J., Millar J. G., Larsen T. E., Malakar-Kuenen R. (2006) Pheromone-based mating disruption of Planococcus ficus (Hemiptera: Pseudococcidae) in California vineyards. J. Econ. Entomol. 99, 1280–1290 [DOI] [PubMed] [Google Scholar]
- 25. Hemmerlin A., Rivera S. B., Erickson H. K., Poulter C. D. (2003) Enzymes encoded by the farnesyl diphosphate synthase gene family in the big sagebrush Artemisia tridentata ssp. spiciformis. J. Biol. Chem. 278, 32132–32140 [DOI] [PubMed] [Google Scholar]
- 26. Zhao P., Inoue K., Kouno I., Yamamoto H. (2003) Characterization of Leachianone G 2″-dimethylallyltransferase, a novel prenyl side-chain elongation enzyme for the formation of the lavandulyl group of sophoraflavanone G in Sophora flavescens Ait. cell suspension cultures. Plant Physiol. 133, 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ko W. G., Kang T. H., Kim N. Y., Lee S. J., Kim Y. C., Ko G. I., Ryu S. Y., Lee B. H. (2000) Lavandulylflavonoids: a new class of in vitro apoptogenic agents from Sophora flavescens. Toxicol. In Vitro 14, 429–433 [DOI] [PubMed] [Google Scholar]
- 28. Lee H. S., Ko H. R., Ryu S. Y., Oh W. K., Kim B. Y., Ahn S. C., Mheen T. I., Ahn J. S. (1997) Inhibition of phospholipase Cγ1 by the prenylated flavonoids from Sophora flavescens. Planta Med. 63, 266–268 [DOI] [PubMed] [Google Scholar]
- 29. Woronuk G., Demissie Z., Rheault M., Mahmoud S. (2011) Biosynthesis and therapeutic properties of Lavandula essential oil constituents. Planta Med. 77, 7–15 [DOI] [PubMed] [Google Scholar]
- 30. Demissie Z. A., Sarker L. S., Mahmoud S. S. (2011) Cloning and functional characterization of β-phellandrene synthase from Lavandula angustifolia. Planta 233, 685–696 [DOI] [PubMed] [Google Scholar]
- 31. Demissie Z. A., Cella M. A., Sarker L. S., Thompson T. J., Rheault M. R., Mahmoud S. S. (2012) Cloning, functional characterization and genomic organization of 1,8-cineole synthases from Lavandula. Plant Mol. Biol. 79, 393–411 [DOI] [PubMed] [Google Scholar]
- 32. Landmann C., Fink B., Festner M., Dregus M., Engel K. H., Schwab W. (2007) Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Arch. Biochem. Biophys 465, 417–429 [DOI] [PubMed] [Google Scholar]
- 33. Lane A., Boecklemann A., Woronuk G. N., Sarker L., Mahmoud S. S. (2010) in Lavender: the Genus Lavandula (Lis-Balchin M., ed) pp. 117–123, Taylor & Francis, London [Google Scholar]
- 34. Lis-Balchin M. (2002) Lavender essential oil: standardisation, ISO, adulteration and its detection using GC, enantiomeric columns and bioactivity. Lavender: The Genus Lavandula 117–123 [Google Scholar]
- 35. Falk L., Biswas K., Boeckelmann A., Lane A., Mahmoud S. S. (2009) An efficient method for the micropropagation of lavenders: regeneration of a unique mutant. J. Essent. Oil Res. 21, 225–228 [Google Scholar]
- 36. Croteau R., Karp F. (1979) Biosynthesis of monoterpenes: preliminary characterization of bornyl pyrophosphate synthetase from sage (Salvia officinalis) and demonstration that geranyl pyrophosphate is the preferred substrate for cyclization. Arch. Biochem. Biophys. 198, 512–522 [DOI] [PubMed] [Google Scholar]
- 37.Deleted in proof [Google Scholar]
- 38.Deleted in proof [Google Scholar]
- 39. Haug-Schifferdecker E., Arican D., Brückner R., Heide L. (2010) A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyltranferase reaction. J. Biol. Chem. 285, 16487–16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deleted in proof [Google Scholar]
- 41. Chang T. H., Hsieh F. L., Ko T. P., Teng K. H., Liang P. H., Wang A. H. (2010) Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation. Plant Cell 22, 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Christianson D. W. (2008) Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao Y., Honzatko R. B., Peters R. J. (2012) Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat. Prod. Rep. 29, 1153–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sugiyama A., Linley P. J., Sasaki K., Kumano T., Yamamoto H., Shitan N., Ohara K., Takanashi K., Harada E., Hasegawa H., Terakawa T., Kuzuyama T., Yazaki K. (2011) Metabolic engineering for the production of prenylated polyphenols in transgenic legume plants using bacterial and plant prenyltransferases. Metab. Eng. 13, 629–637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.