Background: Methicillin-resistant Staphylococcus aureus (MRSA) generates NO through bacterial NO synthase (bNOS).
Results: Loss of bNOS increases MRSA sensitivity to host neutrophils, cathelicidin antimicrobial peptides, and cell envelope-active antibiotics.
Conclusion: bNOS influences MRSA disease pathology.
Significance: Future development of bNOS-specific inhibitors could provide dual activities to reduce MRSA pathology and increase antibiotic effectiveness.
Keywords: Antibiotics, Antimicrobial Peptides, Neutrophil, Nitric Oxide, Nitric-oxide Synthase, Staphylococcus aureus, Virulence Factors
Abstract
Staphylococcus aureus infections present an enormous global health concern complicated by an alarming increase in antibiotic resistance. S. aureus is among the few bacterial species that express nitric-oxide synthase (bNOS) and thus can catalyze NO production from l-arginine. Here we generate an isogenic bNOS-deficient mutant in the epidemic community-acquired methicillin-resistant S. aureus (MRSA) USA300 clone to study its contribution to virulence and antibiotic susceptibility. Loss of bNOS increased MRSA susceptibility to reactive oxygen species and host cathelicidin antimicrobial peptides, which correlated with increased MRSA killing by human neutrophils and within neutrophil extracellular traps. bNOS also promoted resistance to the pharmaceutical antibiotics that act on the cell envelope such as vancomycin and daptomycin. Surprisingly, bNOS-deficient strains gained resistance to aminoglycosides, suggesting that the role of bNOS in antibiotic susceptibility is more complex than previously observed in Bacillus species. Finally, the MRSA bNOS mutant showed reduced virulence with decreased survival and smaller abscess generation in a mouse subcutaneous infection model. Together, these data indicate that bNOS contributes to MRSA innate immune and antibiotic resistance phenotypes. Future development of specific bNOS inhibitors could be an attractive option to simultaneously reduce MRSA pathology and enhance its susceptibility to commonly used antibiotics.
Introduction
Mammalian NO production is regulated by the activity of NO synthase (NOS)4 enzymes, which catalyze the production of NO from l-arginine. The three mammalian NOS isoforms have two catalytic domains, an oxygenase domain and a reductase domain, responsible for binding the substrate arginine and co-factors, respectively. NO production has been implicated in many physiological and pathological processes, including host innate immune and inflammatory responses to pathogens (1). Paradoxically, genome sequencing has revealed the existence of mammalian NOS homologues in a limited number of prokaryotic organisms, principally Gram-positive bacteria (2). Structural analysis has shown that bacterial NOS (bNOS) enzymes are highly similar to the oxygenase domain of mammalian NOS (3–5) but lack a corresponding reductase domain. Nonetheless, bNOS can efficiently generate NO from l-arginine by hijacking alternate cellular redox partners (2).
Bacterial NO production contributes to Bacillus anthracis virulence by enhancing resistance to oxidative stress (6, 7) and to virulence of the plant pathogen Streptomyces turgidiscabies by nitrating and activating a phytotoxin (8). In addition, recent studies demonstrate a link between NO and bacterial antibiotic resistance. First, Gusarov et al. (9) found that endogenous NO production through bNOS protects the producing bacterium against various antibiotics, either through chemical modification of the agent or by counteracting the increased levels of intracellular oxidative stress induced by bactericidal drugs. In a complementary fashion, exposure to exogenous NO protects both Gram-positive and Gram-negative bacteria from aminoglycoside toxicity by limiting drug uptake (10).
Staphylococcus aureus is a Gram-positive human pathogen causing a wide spectrum of clinical disease, ranging from minor skin infections to severe invasive conditions such as pneumonia, sepsis, and endocarditis. Recently, marked increases in infections by antibiotic (methicillin)-resistant S. aureus (MRSA) have occurred in both hospital and community settings, posing a grave challenge to the public health. Epidemic strains of community-acquired MRSA (CA-MRSA) such as the USA300 clone can produce severe disease even in previously healthy individuals (11, 12), reflecting a formidable capacity of the pathogen to evade normal host innate immune clearance (13).
A key element of innate defense against invasive pathogens such as S. aureus is the function of neutrophils (14–16). Generation of reactive oxygen species (ROS) through respiratory burst is a central element of normal neutrophil killing activity, as evidenced by the marked susceptibility of patients with genetic defects in NADPH oxidase (chronic granulomatous disease) to recurrent severe S. aureus infections (17). In addition, inflammatory activation of leukocytes results in inducible NOS (iNOS) induction and release of NO, which has a direct inhibitory activity against several bacterial species, and participates in downstream reactions with different ROS to produce synergistic antibacterial effects (1). Nonoxidant mechanisms also contribute to bacterial killing, for example cationic antimicrobial peptide (AMPs) stored in neutrophil granules that target and disrupt bacterial membrane integrity (18).
S. aureus has evolved multifaceted resistance mechanisms to neutrophil killing that contribute to its invasive disease potential. For example, ROS activity is mitigated by the S. aureus enzyme catalase (19, 20) and the carotenoid pigment of the bacterium (21). S. aureus can also tolerate severe nitrosative stress and high NO concentrations, because of adaptive metabolic responses including induction of the flavohemoprotein Hmp (22) and l-lactate dehydrogenase (23). Relative insensitivity of S. aureus to cathelicidin action is promoted by cationic cell wall modifications (24, 25) or proteolytic cleavage (26, 27).
S. aureus expresses a bNOS enzyme, but its potential contribution to bacterial immune and antibiotic resistance has not been tested. To assess the role of MRSA bNOS in these processes, we couple targeted mutagenesis with in vitro and in vivo assays of bacterial-host interaction and antibiotic susceptibility. Our analysis was conducted using a representative isolate of the epidemic USA300 CA-MRSA strain, the most common agent of serious bacterial infections in recent epidemiologic surveillance in the United States (12).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
S. aureus strain UAMS1182 is a community-acquired USA300 MRSA strain. Bacteria were cultured in tryptic soy broth (TSB) under shaking conditions or on TSB agar at 37 °C. Plasmid expression of pNOS or empty vector control was achieved using 20 μg/ml chloramphenicol. All of the experiments were performed with USA300 MRSA WT and ΔNOS mutant strain containing the empty vector control as an appropriate comparison for experiments using the complemented strain. Overnight cultures were diluted 1:30 in fresh TSB and grown to exponential phase A600 = 0.4 (2 × 108 CFU/ml), unless otherwise indicated. Similar growth kinetics were observed for all strains under the assay conditions used (data not shown).
Targeted Mutagenesis and Complementation Vector Construction
Markerless precise allelic replacement of S. aureus bNOS was performed using PCR-based methods as described (28) with minor changes. Primer sequences were based on the published USA300 FPR3757 (29). Briefly, 828 bp of sequence immediately upstream of nos was amplified with primers NOS-UpF+attB1, 5′-ggggacaagtttgtacaaaaaagcaggctggcaacatattcgtctccataagt-3′, and NOS-UpR, 5′-gcatatgctacacctctactaacttacaacacctcgctttatattatag-3′, and 743 bp immediately downstream of nos amplified with the primers NOS-DownF, 5′-ctataatataaagcgaggtgttgttagttagtagaggtgtagcatatgc-3′, and NOS-DownR+attB2, 5′-ggggaccactttgtacaagaaagctgggtcggttgtaattgcagaatcagcctg-3′. The NOS-upR and NOS-downF primers were constructed with ∼25-bp 5′ overhangs for the opposite flanking region. The upstream and downstream PCR products were fused in a second round of PCR using primers NOS-UpF+attB1 + NOS-downR+attB2 and subcloned into the temperature-sensitive vector pKOR1 using BP clonase reaction (Invitrogen). The resulting plasmid pKOR1-ΔNOS was first passed through S. aureus RN4220, before electroporation into strain UAMS1182. Precise in-frame allelic replacement of the nos gene was established by a two-step process of temperature shifting and antisense counterselection (28) and confirmed by PCR and sequence analysis. For complementation analysis, bNOS was PCR-amplified from the UAMS1182 chromosome using NOS-FL-F, 5′-gcggaattcatgtaatttaaagaggctcaagc-3′, and NOS-FL-R, 5′-gcgggatccttaatgatggaaagggcagtgg-3′, and cloned into shuttle expression vector pDC123 (30), yielding plasmid pNOS.
Detection and Measurement of Bacterial NO Production/NOS Activity
Bacteria were grown to logarithmic phase in TSB and then transferred into phosphate buffer (45 mm, 80 mm K2HPO4, 4 mm Na3C6H5O7) with 5 mm Arg + 0.8% glucose and incubated for 10 min. A NO-reactive fluorescent probe (CuFL) (31) was prepared just prior to use by mixing FL and CuCl2 in a 1:1 ratio and then added to the cells to a final concentration of 20 μm (2). Twenty minutes later, fluorescence and bright field images of bacteria treated with the NO-detecting probe were taken by digital camera attached to an Axio microscope (Carl Zeiss MicroImaging Inc.).
Hydrogen Peroxide Killing Assay
Bacteria were grown to logarithmic phase in TSB, and 5 × 106 bacteria were incubated with 0.2% H2O2 at 37 °C in 250 μl in a 96-well microplate. Samples were diluted in PBS and plated on TSB agar for enumeration of surviving bacteria at the indicated time points. The first dilution contained 2,500 units/ml of catalase (Sigma) to quench residual H2O2. For exogenous NO rescue experiments, bacteria were pretreated with 500 μm of methylamine hexamethylene NONOate (Sigma) for 3 min before adding bacteria to wells that contained H2O2. The NO donor was present for the duration of the assay at 50 μm.
Antibiotic Susceptibility Assays
Minimum inhibitory concentration (MIC) assays were performed by broth microdilution. Vancomycin MIC and daptomycin MIC assays were performed in TSB and cation-supplemented Mueller-Hinton broth (50 mg/liter Ca2+ + 12.5 mg/liter Mg2+). Briefly, overnight cultures were diluted in fresh TSB and grown to logarithmic phase at 37 °C in a shaking incubator. The culture was pelleted and resuspended to A600 = 0.4 in PBS. The bacteria were diluted in the appropriate broth, and 5 × 105 CFU were added to a 96-well test plate in triplicate containing different concentration of vancomycin (Hospira, Lake Forest, IL) or daptomycin. The test plate containing the antibiotic dilutions and target bacteria was incubated at 37 °C for 18–20 h. MIC was determined to be the lowest concentration of antibiotic that inhibited bacterial growth as determined by turbidometric assessment at A600. Daptomycin time kill assays were similarly performed in triplicate in 96-well plates containing 5 × 105 bacteria and 2 μg/ml daptomycin in a total volume of 250 μl. Samples were taken every hour, and viability was determined by dilution plating. Growth curves in the presence of daptomycin were performed in cation-supplemented Mueller-Hinton broth and 2 μg/ml daptomycin at 37 °C under shaking conditions. Optical density was measured every 30 min at 600 nm. We also used growth inhibition (Bioscreen C MBR machine) and killing assays to determine sensitivity to several ribosomally active, generally bacteriostatic antibiotics, including the aminoglycosides gentamicin and streptomycin and the oxazolidinone linezolid. Briefly, bacteria were grown overnight in TSB containing 20 μg/ml chloramphenicol. The next day the bacteria were regrown to A600 = 0.4 in TSB, washed, and resuspended in cation-adjusted Mueller-Hinton broth. Bacteria (5 × 105 CFU) were added to Bioscreen Honeycomb plates in a total volume of 200 μl of cation-adjusted Mueller-Hinton broth containing different concentrations of the indicated antibiotic. Growth was determined by measuring A600 every 15 min for 18 h. As a measurement of antibiotic inhibition, the percentage of maximum growth compared with conditions without antibiotic was calculated using the following formula: A600 at 18 h with antibiotic/A600 at 18 h without antibiotic × 100%. Bacterial killing assays were performed to verify the antibiotic sensitivity by quantifying bacterial survival through serial dilution plating using a similar set up as described for daptomycin.
Measurement of SOD Activity
SOD activity was determined by negative staining on native polyacrylamide gels (32). Bacteria were grown in TSB, and samples were collected every hour. Pelleted cells were resuspended in lysis buffer (20 mm Tris, pH 7.9, 1 mm EDTA, and 25 μg/ml lysostaphin; Sigma), followed by repeated freeze thawing until complete lysis. The cell debris was separated from the soluble fraction by centrifugation (15 min, 14,000 rpm) at 4 °C. The samples were normalized according to the total protein concentration and resolved on 10% nondenaturing PAGE. The SOD activity was visualized by negative staining by the nitroblue tetrazolium method. Additionally, SOD enzymatic activity was measured using a colorimetric assay (Cayman Chemical).
Antimicrobial Peptide Killing and Degradation
Log phase grown bacteria were resuspended to 2 × 106 CFU/ml in assay buffer (PBS containing 20% TSB and 50 mm NaHCO3), and 25 μl of this suspension was added to 225 μl containing 16 μm murine cathelicidin antimicrobial peptide in assay buffer. The test plate was incubated at 37 °C, and samples were taken at 2, 4, 6, and 8 h to quantify surviving CFU by serial dilution plating. To test AMP degradation by S. aureus supernatants, S. aureus cultures were grown overnight in TSB containing 20 μg/ml chloramphenicol. Supernatants were collected by filter sterilization using a 0.22 μm filter. 18 μl of supernatant was incubated with 32 μm AMP for the indicated time points in a 37 °C water bath. The samples were run under reducing conditions on 12% NuPAGE Bis-Tris precast gels (Invitrogen) with MES running buffer (Invitrogen) and stained with SimplyBlue Safestain (Invitrogen) to visualize AMP degradation.
Neutrophil Killing Assays
Overnight bacterial cultures were grown to A600 = 0.4 in RPMI 1640 at 37 °C + 5% CO2 without shaking and diluted with assay medium (RPMI 1640 containing 2% nuclease-free heat-inactivated (70 °C, 30 min) FCS) (33) to the desired concentration. Human neutrophils were isolated from healthy donors using the PolymorphPrepTM system (Axis-Shield, Oslo, Norway) and resuspended in assay medium. The neutrophils were seeded in 48-well plates at 5 × 105 cells/250 μl of medium/well and prestimulated with 25 nm PMA for 20 min at 37 °C in the presence of 5% CO2. For extracellular killing, cytochalasin D was added together with PMA at a final concentration of 10 μg/ml. 250 μl of bacterial suspension was added directly to PMA-stimulated PMN at a multiplicity of infection of 2. The plates were centrifuged at 1200 rpm for 8 min and further incubated at 37 °C/5% CO2. After 30 or 90 min, neutrophils were lysed using 0.025% Triton X-100, and the surviving bacteria were quantified by serial dilution plating.
Quantification of NET-mediated Killing by Immunofluorescence
The neutrophils were seeded on poly-l-lysine-coated glass cover slides (5 × 105 cells/12 mm/1.5 glass slides), pretreated with PMA, and infected at an multiplicity of infection of 2 as described above. After 30 min of infection, a Live/Dead BacLightTM bacterial viability kit (Invitrogen) was used to determine the viability of NET-entrapped S. aureus entrapped by fluorescence microscopy. After staining as recommended by the manufacturer, the cells were washed three times with PBS, fixed with 1% paraformaldehyde for 5 min, washed three times, and mounted onto glass slides with ProlongGold with DAPI (Invitrogen). Mounted samples were examined using an inverted confocal laser scanning two-photon microscope Olympus Fluoview FV1000 with FluoviewTM spectral scanning technology (Olympus). Viable (green) versus dead (red) bacteria, which were entrapped by NETs (blue, DAPI-stained), were counted from a minimum of four images of three individual experiments.
Macrophage Survival Assay
Bone marrow cells were flushed from tibias and femurs of 6–10-week old C57bl6 mice (Charles River) in RPMI and cultured for 7 days at 37 °C and 5% CO2 in DMEM + 20% FBS + 20% L-929 cell conditioned medium. Adherent macrophages were lifted in ice-cold PBS containing 5 mm EDTA, pelleted, and seeded into 24-well plates at 4 × 105 cells/well in DMEM/10% FBS. 16 h after seeding, the wells were inoculated with 0.2 ml of DMEM/FBS containing PBS-washed bacteria grown to mid-log phase in TSB (A600 = 0.4), at multiplicity of infection of 1. The plates were centrifuged at 1,600 rpm for 5 min, incubated at 37 °C and 5% CO2 for 1 h, and treated with 100 μg/ml gentamicin (Invitrogen) for 30 min to kill noninternalized bacteria. The wells were then rinsed twice in DPBS and reconstituted with 0.9 ml of DMEM and 10% FBS. Enumeration of surviving bacteria was performed at 0 h (immediately post-gentamicin), 3 h, and 6 h, as follows. 0.25% Triton X-100 was added to wells to lyse macrophages by trituration, and serial dilutions were plated to enumerate CFU. Intracellular survival was calculated as the percentage of bacteria remaining at 3 or 6 h compared with 0 h CFU.
Murine Model of Subcutaneous Infection
MRSA were grown to mid-log phase in TSB (A600 = 0.4), washed twice in PBS, and resuspended in PBS. Shaved 7-week-old female C57bl6 mice were anesthetized using isoflurane and injected in the mid-left flank in 100 μl containing ∼1.3 × 107 CFU. Abscesses were photographed daily, and the area was quantified using Image J. Abscess area was defined as the zone of leukocytic infiltration including the contained zone of dermonecrosis. On day 3, abscesses were excised and homogenized in PBS prior to serial dilution and drop plating. Abscess burden was calculated as abscess area × CFU/g. In a co-infection experiment, equal numbers (8 × 106 CFU) of WT and ΔNOS mutant MRSA were injected into the same site on a single mouse; lesions were excised after 3 days, and the ratio of WT to mutant CFU recovered was calculated for each mouse.
Statistical Analysis
The data were analyzed with GraphPad Prism 5.03 (GraphPad Software). For H2O2, antibiotic, AMP, and neutrophil killing assays data were analyzed by one-way analysis of variance with Tukey's multiple comparison correction after log transformation. Analysis of the mouse in vivo challenge experiments was performed using the unpaired Student's t test. p < 0.05 was considered significant.
Assurances
Human blood collection and neutrophil isolation studies were performed under approved protocols of the University of California, San Diego Human Research Protection Program. The mouse infection studies were performed under the approved protocols of the University of California, San Diego Institutional Animal Use and Care Committee.
RESULTS
bNOS Protects MRSA from Oxidative Stress
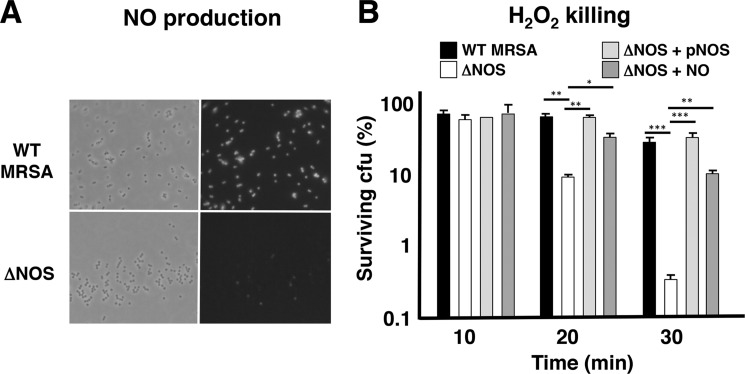
The nos gene is present in the genomes of all sequenced S. aureus strains and coagulase negative Staphylococcus strains showing high sequence identity (supplemental Table S1). Previous biochemical analysis showed that purified S. aureus bNOS exists as a dimer and generates NO from l-arginine (34). To characterize the biological function of bNOS in S. aureus, we generated an isogenic deletion mutant in a strain of the highly virulent CA-MRSA epidemic clone USA300 by allelic exchange mutagenesis. Deletion of the nos gene was confirmed by PCR analysis and did not affect growth rate in liquid culture nor bacterial density (data not shown). As assessed by a highly specific fluorescent probe (CuFL) (31), NO production was completely abrogated in the ΔNOS mutant compared with WT parent strain (Fig. 1A). These results confirm that S. aureus bNOS generates NO under laboratory growth conditions. Endogenous NO production was previously shown to protect B. anthracis against H2O2-mediated killing (6). Likewise, we found the ΔNOS mutant was much more sensitive to H2O2 killing, with 2 log fewer surviving bacteria after 30 min of H2O2 exposure compared with the WT MRSA strain (Fig. 1B). ΔNOS mutant H2O2 resistance could be completely restored to WT levels by in trans complementation of nos on a plasmid vector or by addition of exogenous NO (Fig. 1B), which the bacteria could encounter in activated phagocytes in the host.
FIGURE 1.
MRSA NO production depends on bNOS and protects bacteria from oxidative stress. A, bNOS-dependent NO production in live bacteria. A representative fluorescent image of WT MRSA and ΔNOS mutant bacteria treated with Cu(II)-based NO-detecting probe (CuFL) is shown. B, bNOS protects MRSA from H2O2 toxicity. Log phase bacteria were exposed to 0.2% H2O2, and survival was monitored every 10 min by serial dilution plating. Survival of ΔNOS bacteria was rescued by complementation in trans or exposure to exogenous NO. One of three representative experiments is shown (means and S.D.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
bNOS Increases MRSA Antibiotic Resistance
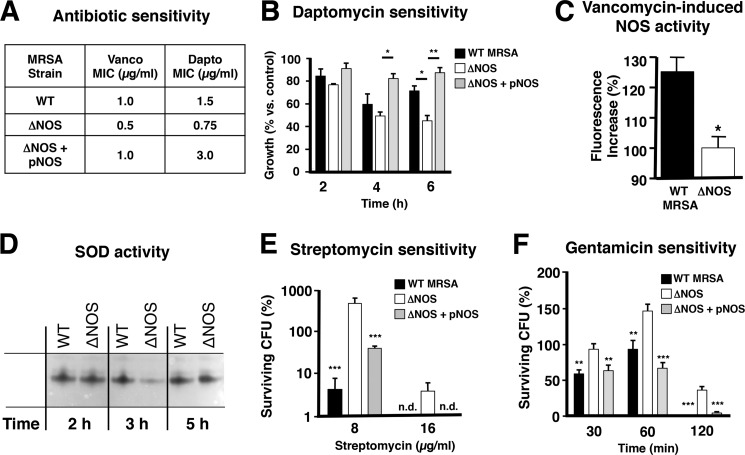
Different classes of bactericidal, but not bacteriostatic, antibiotics initiate the formation of intracellular hydroxyl radicals through the Fenton reaction resulting in bacterial cell death (35). In line with this finding, iron chelators like 2,2-dipyridyl or desferroxiamine that block Fenton chemistry decrease the bactericidal effects of antibiotics (35, 36). Similarly, H2O2-mediated toxicity involves Fenton chemistry (37). bNOS increases bacterial resistance to intracellular oxidative stress by inhibition of the damaging Fenton reaction (7). Correspondingly, production of endogenous NO was shown to protect Bacillus subtilis and the nonvirulent S. aureus laboratory strain RN4220 from killing by cefuroxime by elevating the threshold for oxidative stress (9). However, cefuroxime, like other cephalosporin antibiotics, is clinically ineffective against MRSA (38, 39). Therefore, we studied whether bNOS influenced S. aureus susceptibility to vancomycin and daptomycin, two front line agents in current pharmacotherapy of MRSA infections (40). Bactericidal concentrations of vancomycin are known to increase intracellular hydroxyl radical formation in S. aureus (35). We found that MICs for both vancomycin and daptomycin were 2-fold lower for ΔNOS bacteria compared with the WT parent strain (Fig. 2A). This difference in susceptibility was reversed upon complementation of the mutant with the nos expression plasmid, resulting in increased daptomycin-resistance (MIC = 4 μg/ml); Fig. 2A). Expression of bNOS also contributed to MRSA resistance to daptomycin activity in a kinetic assay, especially under conditions of overexpression, as is the case in the complemented mutant (Fig. 2B and supplemental Fig. S1). Despite the increased sensitivity of the ΔNOS strain to bactericidal antibiotics, growth of WT and mutant strains was not differentially affected by the addition of iron alone (supplemental Fig. S2).
FIGURE 2.
Deletion of bNOS affects MRSA antibiotic resistance. A, bNOS increases MRSA MIC for vancomycin (Vanco) and daptomycin (Dapto). MICs were measured in broth-based assays in triplicate. B, influence of daptomycin on MRSA growth. Growth curves were performed in the absence or presence of daptomycin (2 μg/ml), and A600 was recorded at the indicated time points. The data are presented as percentages of growth relative to control. Pooled data from two independent experiments in duplicate are shown (means and S.E.). C, vancomycin exposure increases bNOS activity. WT MRSA and ΔNOS mutant bacteria were treated with freshly prepared CuFL (20 μm) and vancomycin (20 μg/ml), and fluorescence was measured using real time fluorometer (PerkinElmer LS-55). The data were normalized to ΔNOS control cells that were grown, treated with CuFL and vancomycin, and processed the same way. D, bNOS controls SodA expression. WT MRSA and ΔNOS mutant bacteria were grown in TSB at 37 °C. The samples were collected at the indicated time points, and the cell extracts were analyzed by native PAAG and staining for SOD activity. E and F, loss of bNOS increases resistance to the aminoglycosides streptomycin (E, 2-h time point) and gentamicin (F, 0.25 μg/ml). Pooled data of two independent experiments are shown (means and S.E.). n.d., not detected. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In addition to blocking the Fenton reaction, endogenous NO can also protect bacteria from oxidative stress by increasing SOD expression or catalase activity (7, 9). Indeed, vancomycin treatment increased NO levels (Fig. 2C), which correlated to a temporary increased SOD activity (Fig. 2D and supplemental Fig. S3) in the WT but not ΔNOS strain. bNOS may help MRSA counteract the oxidative stress imposed by bactericidal antibiotics both through Fenton reaction inhibition and through increased SOD activity. A significant role for catalase appears less likely, because we observed no difference in vancomycin (MIC = 1 μg/ml) or daptomycin (MIC = 2 μg/ml) susceptibility between a WT S. aureus strain (Newman) and its isogenic catalase-deficient mutant (20).
A recent paper by McCollister et al. (10) demonstrates that exposure to exogenous NO can also protect bacteria, including S. aureus, from aminoglycoside toxicity by blocking drug uptake. In contrast, we observe that endogenous NO production renders the bacteria more susceptible to the antibiotic effect, with ΔNOS bacteria displaying 10–50-fold increased resistance to gentamicin and streptomycin (Fig. 2, E and F). This effect seems specific to this particular class of antibiotics because strains were equally susceptible to linezolid (supplemental Fig. S4), which also inhibits bacterial protein synthesis initiation but by acting on the 50 S rather than the 30 S subunit of bacterial ribosomes. Therefore, MRSA bNOS seems to affect antibiotic sensitivity in a distinct and more complex manner when compared with Bacillus.
bNOS Promotes MRSA Cathelicidin Resistance
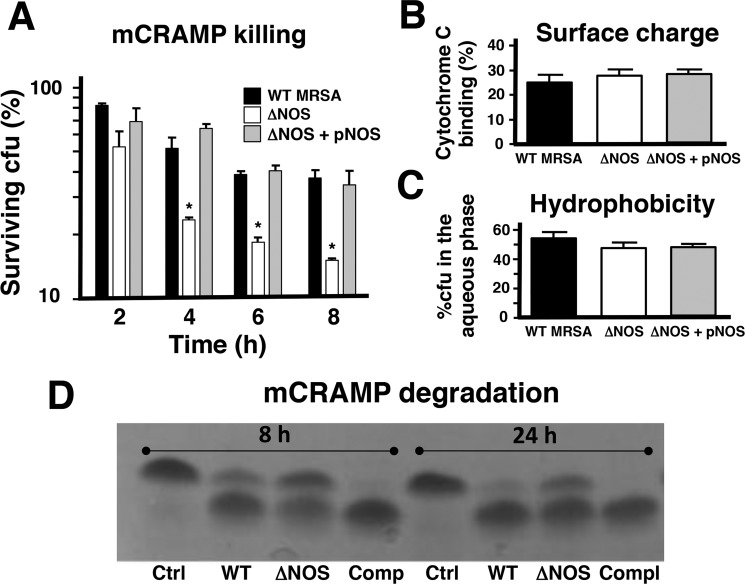
Vancomycin and daptomycin both act on the cell envelope and bear relative positive charge (41, 42). Similarly, host cathelicidins produced during the innate immune response are α-helical peptides that are cationic in nature and exert direct antimicrobial activity by pore formation in the target bacterial membrane (43, 44). Abundantly present in neutrophils, cathelicidins can enhance ROS production by host phagocytes (45, 46). Because of these similarities with vancomycin and daptomycin, we hypothesized that bNOS could contribute to MRSA cathelicidin resistance. We found that loss of bNOS sensitized MRSA to killing by the murine cathelicidin murine cathelicidin antimicrobial peptide (mCRAMP) (Fig. 3A). Previously, staphylococcal resistance to cathelicidin killing has been associated with surface factors or biochemical modifications that increase positive charge or decrease hydrophobicity of the bacterial cell wall (24, 25, 47). However, we did not observe a difference between the WT MRSA, ΔNOS, and complemented strains in surface charge, as measured by cytochrome c binding assay (Fig. 3B), nor hydrophobicity, as measured by N-hexadecane partition assay (Fig. 3C). Cathelicidin resistance has also been associated with S. aureus protease expression, resulting in inactivation of the AMP by degradation (26, 27). We observed a decrease in mCRAMP degradation by ΔNOS supernatant compared with the WT MRSA and complemented strains (Fig. 3D), suggesting a potential linkage to the AMP susceptibility phenotype. Thus the contribution of bNOS to MRSA resistance to cathelicidin antimicrobial peptides is likely multifaceted, including resistance to ROS induced by AMP exposure (48) and diminished expression of proteolytic activity(s) against the peptides.
FIGURE 3.
bNOS protects MRSA from killing by mCRAMP by increasing proteolytic activity. A, killing kinetics of MRSA by the mCRAMP. Bacteria were incubated with 16 μm mCRAMP. Survival was assessed by dilution plating at indicated time points. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Combined results from three independent experiments are shown. The error bars represent S.E. B and C, deletion of bNOS does not affect surface charge as measured by cytochrome c (B) or hydrophobicity as measured by n-hexadecane separation (C). The error bars represent S.D. D, Overnight supernatants of MRSA WT and complemented strains degrade mCRAMP more efficiently than supernatant of the ΔNOS mutant. Ctrl, control; Comp or Compl, complemented.
bNOS Promotes MRSA Neutrophil Resistance
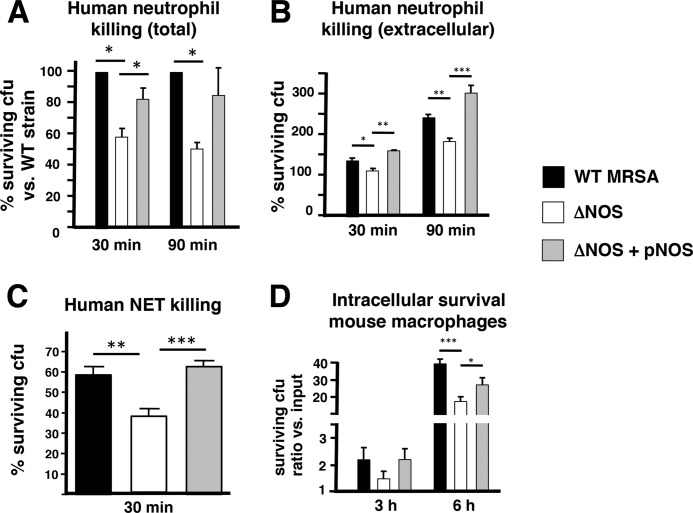
Neutrophils are crucial immune cells in host defense against S. aureus (14–16) and contribute to antibacterial defense through oxidative burst, antimicrobial peptides (including cathelidicin), granule proteases, and the elaboration of DNA-based extracellular traps (NETs). Recent studies have revealed that community-acquired MRSA strains are highly resistant to neutrophil killing compared with hospital-acquired MRSA strains (49). Because bNOS-deficient bacteria were more susceptible to oxidative stress and AMP in isolation, we determined bacterial survival in the presence of human neutrophils. We found that the ΔNOS mutant was more susceptible than the WT MRSA parent strain to killing by freshly isolated human neutrophils (Fig. 4A); this defect could be corrected by complementation of the nos gene on a plasmid vector (Fig. 4A). Differential neutrophil survival was not due to differential induction of ROS, NO production, or cell lysis (supplemental Figs. S5–S7). Interestingly, significant differences in neutrophil survival were maintained in killing assays performed in the presence of cytochalasin D, an actin inhibitor that blocks phagocytic uptake but not NET formation (Fig. 4B). Because extracellular killing is independent of oxidants, these results indicate that bNOS also contributes to S. aureus resistance to extracellular oxidant-independent killing mechanisms. NETs entrap and kill microbes through the action of cationic antimicrobials such as cathelicidins and histone proteins (33), a phenomenon that contributes to S. aureus killing (50, 51). We used PMA to induce complete extracellular trap formation, a so-called “total NET assay” (52), and tested their antibacterial activity against the strains. Once again, WT MRSA and complemented strains survived better than the isogenic ΔNOS mutant (Fig. 4C). NO production by S. aureus did not alter the amount of NETs produced by neutrophils (supplemental Fig. S8). Thus bNOS likely contributes to the CA-MRSA neutrophil resistance phenotype by enhancing resistance to both intracellular (e.g., oxidative burst) and extracellular (NETs including cathelicidin, oxidant-independent) killing mechanisms. Finally, comparable with previous findings on B. anthracis bNOS (6), bNOS also contributed to MRSA intracellular replication in murine macrophages (Fig. 4D).
FIGURE 4.
bNOS-mediated protection from neutrophil and macrophage killing. A–C, deletion of bNOS in MRSA increases total human neutrophil killing (A), extracellular neutrophil killing (B), and NET-mediated killing (C). For total killing and extracellular killing, freshly isolated human neutrophils were prestimulated with PMA and incubated with MRSA WT, ΔNOS, or complemented strains for 30 and 90 min. Bacterial survival (CFU) was assessed by plating serial dilution. NET-mediated killing was quantified by counting live and dead bacteria within NET structures. D, intracellular survival in mouse macrophages. Pooled data from three independent experiments are shown (means and S.E.). For A, WT was set at 100%. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
bNOS Contributes to S. aureus Skin Infection
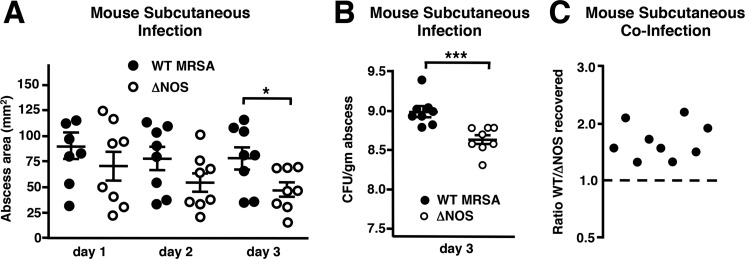
S. aureus ranks first as bacterial cause for skin and soft tissue infections worldwide (53), with CA-MRSA strains currently accounting for the majority of these presentations (54). Cathelicidin peptides and neutrophils are crucial aspects of host innate defense against invasive skin infection (14, 55). We therefore assessed whether bNOS contributed to MRSA skin pathology in vivo, using a well established murine subcutaneous infection model (21). C57bl6 mice were injected with either WT MRSA bacteria or the isogenic ΔNOS mutant bacteria. The size of the resulting abscesses was measured every 24 h. On day 3, we euthanized the mice and harvested skin for bacterial quantification. WT MRSA bacteria produced significantly larger lesions by the third day (Fig. 5A), and increased bacterial numbers were recovered from the WT-induced abscesses compared with the ΔNOS-induced abscesses (Fig. 5B). To verify that enhanced WT survival in vivo contributed per se to the observed findings (rather than solely differences in the induced host response), a co-infection experiment was performed showing that when injected together into the same mouse, more WT than bNOS mutant bacteria were recovered from the lesion (Fig. 5C). A similar effect of bNOS in promoting MRSA subcutaneous lesion development was observed upon infection of outbred CD-1 mice (supplemental Fig. S9).
FIGURE 5.
bNOS contributes to MRSA virulence in a murine skin infection model. Groups of shaved 7-week-old female C57bl6 mice were challenged with an equivalent dose (∼1.3 × 107 CFU) of either WT or ΔNOS MRSA. A, abscesses were photographed and quantified using Image J. Abscess area was defined as the zone of leukocytic infiltration including the contained zone of dermonecrosis. B, on day 3, abscesses were excised and homogenized to quantify surviving CFU per gm abscess weight. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, in a co-infection experiment, equal numbers (8 × 106 CFU) of WT and ΔNOS mutant MRSA were injected into the same site on a single mouse; lesions were excised after 3 days, and the ratio of WT to mutant CFU recovered was calculated for each mouse.
DISCUSSION
In mammals, NO is an important regulator of diverse biological functions. Of the three described NOS isoforms (NOS1–3) that generate NO from l-arginine, NOS2 or iNOS is mainly responsible for functions related to immune regulation and host defense against pathogens (56). Activation of leukocytes by inflammatory stimuli results in local release of ROS and NO as a result of iNOS induction. Studies in knock-out animals support a critical role for phagocyte oxidative burst in controlling S. aureus (57). In contrast, studies of iNOS knock-out mice have yielded conflicting results, with one study showing increased susceptibility to S. aureus infection (58) and a second showing no difference compared with WT animals (59).
S. aureus is among a limited number of Gram-positive bacterial species possessing an iNOS homologue (2). In addition, S. aureus is relatively resistant to nitrosative stress, i.e., exogenous NO, through transcriptional adaptation that is essential for virulence (22, 23). In this paper, we describe that deletion of bNOS in a virulent strain of community-acquired MRSA increased bacterial susceptibility to oxidative stress, similar to earlier reports in Bacillus species (2, 6, 7). Oxidant resistance of the ΔNOS mutant could be restored to WT levels by plasmid complementation of the nos gene or short term exposure of the mutant to exogenous NO. Such short term NO exposure does not involve transcriptional adaptation but rather increases activity of oxidant resistance proteins like catalase or blocking Fenton-induced cytotoxicity (7). Because NO is freely diffusible across cell membranes and S. aureus is intrinsically resistant to nitrosative stress, these results may indicate that S. aureus can co-opt host-derived NO to supplement bNOS-derived NO in mitigating ROS stress during encounters with activated phagocytes. In line with this hypothesis, it was recently shown that the pathogen Salmonella typhimurium, which lacks bNOS, utilizes macrophage-derived NO to protect itself from the activity of aminoglycosides by reducing drug uptake (10).
Expression of bNOS increased resistance to two front-line pharmaceutical antibiotics used in treatment of serious MRSA infections: vancomycin and daptomycin. In addition, we show for the first time that bNOS can contribute to defense against so-called “natural antibiotics” such as host-derived AMP (cathelicidin). In particular, overexpression of bNOS on a plasmid significantly increased MRSA resistance to these antimicrobial agents, resulting in the generation of a daptomycin-resistant MRSA strain. This indicates that the ability to further up-regulate NO levels could aid the bacterium in dealing with antibiotic-induced (oxidative) stress. Consistent with this observation, we observed that MRSA bNOS activity increased following vancomycin exposure and subsequently boosted activity of SOD, an ROS detoxifying enzyme. How bNOS increases SOD activity is currently not known, but similar experiments in B. subtilis previously demonstrated increased bNOS-dependent Sod transcription (9). Overall, we suggest that bNOS increases MRSA resistance to antibiotics that act on the cell envelope at least in part by increasing resistance to antibiotic-induced Fenton-generated oxidative stress and through induction of oxidant neutralizing enzymes such as SOD.
In contrast to previous observations in B. subtilis and B. anthracis (9), endogenous NO actually sensitizes MRSA to the toxic effect of aminoglycosides including gentamicin and streptomycin. Interestingly, bNOS does not affect susceptibility to linezolid, representative of the oxazolidinone class of protein synthesis inhibitors. This indicates that the spectrum of effects of S. aureus bNOS on antibiotic resistance are distinct from those observed in Bacillus (9). The underlying mechanism for this divergent effect is interesting because it may hint toward a biological function of bNOS to influence protein synthesis through the 30 S ribosomal subunit.
Neutrophils are crucial effector cells for host containment of S. aureus infection (14, 15, 59), deploying both ROS and cathelicidin as key components of their bactericidal arsenal. Recent studies have revealed that community-acquired MRSA strains are highly resistant to neutrophil killing compared with hospital-acquired MRSA strains (13, 49). We demonstrate here that endogenous NO production contributes to the CA-MRSA neutrophil resistance phenotype. Moreover, because addition of cytochalasin D did not abolish this difference, bNOS contributes as well to protection against killing by NETs. Susceptibility of the MRSA ΔNOS mutant to NET killing is likely linked to increased sensitivity to cathelicidin AMPs that are present in abundance in these structures (33, 52).
Overall, the effects of endogenous NO production on S. aureus antibiotic susceptibility and immune survival are subtler and more specific than previously reported in Bacillus species (6, 7, 9). MRSA possesses a large repertoire of established ROS and AMP resistance mechanisms (49, 60, 61), and this may provide functional redundancy that allows greater compensation for loss of bNOS expression. The regulatory effects of endogenous NO are also likely to differ among species, just as differential adaptive responses of S. aureus and B. anthracis to exogenous NO have been described (22, 62). Finally, despite the overall structural similarity between S. aureus and B. anthracis bNOS and mammalian iNOS (3), biochemical differences have been described for the two bNOS enzymes (63). This is reflected by the observation that commonly used mammalian NOS inhibitors fail to inhibit MRSA bNOS (data not shown), in contrast to B. anthracis, where mammalian NOS inhibitors can reproduce the ΔNOS mutant phenotype (6). Paradoxically, certain iNOS inhibitors have even been reported to increase S. aureus bNOS activity (64), emphasizing the need for additional studies on the function and regulation of the MRSA bNOS enzyme.
Using a mouse model of S. aureus skin infection, we demonstrate that MRSA bNOS contributes to lesion development and bacterial survival in vivo. Enhanced survival of the WT versus ΔNOS mutant in a co-infection experiment suggests that increased resistance to killing by host defenses (e.g., AMPs, ROS, and neutrophils) is a contributor to the bNOS-associated virulence phenotype. However, future work could also explore whether bNOS regulates expression of MRSA cytotoxins or induces increased host inflammatory response pathways that also contribute to larger lesion sizes.
As an alternative to traditional antibiotics in an era of increasing bacterial resistance, new attention has focused upon development of agents that target bacterial virulence factors, effectively disarming the pathogen to allow clearance by the host (61, 65, 66). Our observations suggest that S. aureus bNOS deserves consideration as a therapeutic target, particularly because inhibition of bNOS would not only reduce MRSA pathology but may in specific cases may have the dual benefit of increasing bacterial susceptibility to commonly used antibiotics including vancomycin and daptomycin.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI077780, HD071600, and AI057153 (to V. N.). This work was also supported by a National Institutes of Health Director's Pioneer award (to E. N.).

This article contains supplemental text, Table S1, and Figs. S1–S9.
- NOS
- nitric-oxide synthase
- bNOS
- bacterial NOS
- iNOS
- inducible NOS
- MRSA
- methicillin-resistant S. aureus
- CA-MRSA
- community-acquired MRSA
- CFU
- colony forming unit
- NET
- neutrophil extracellular trap
- AMP
- antimicrobial peptide
- ROS
- reactive oxygen species
- MIC
- mean inhibitory concentration
- SOD
- superoxide dismutase
- TSB
- tryptic soy broth
- mCRAMP
- murine cathelicidin antimicrobial peptide.
REFERENCES
- 1. Wink D. A., Hines H. B., Cheng R. Y., Switzer C. H., Flores-Santana W., Vitek M. P., Ridnour L. A., Colton C. A. (2011) Nitric oxide and redox mechanisms in the immune response. J. Leukocyte Biol. 89, 873–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gusarov I., Starodubtseva M., Wang Z. Q., McQuade L., Lippard S. J., Stuehr D. J., Nudler E. (2008) Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283, 13140–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bird L. E., Ren J., Zhang J., Foxwell N., Hawkins A. R., Charles I. G., Stammers D. K. (2002) Crystal structure of SANOS, a bacterial nitric oxide synthase oxygenase protein from Staphylococcus aureus. Structure 10, 1687–1696 [DOI] [PubMed] [Google Scholar]
- 4. Pant K., Bilwes A. M., Adak S., Stuehr D. J., Crane B. R. (2002) Structure of a nitric oxide synthase heme protein from Bacillus subtilis. Biochemistry 41, 11071–11079 [DOI] [PubMed] [Google Scholar]
- 5. Pant K., Crane B. R. (2006) Nitrosyl-heme structures of Bacillus subtilis nitric oxide synthase have implications for understanding substrate oxidation. Biochemistry 45, 2537–2544 [DOI] [PubMed] [Google Scholar]
- 6. Shatalin K., Gusarov I., Avetissova E., Shatalina Y., McQuade L. E., Lippard S. J., Nudler E. (2008) Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 105, 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gusarov I., Nudler E. (2005) NO-mediated cytoprotection. Instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 13855–13860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kers J. A., Wach M. J., Krasnoff S. B., Widom J., Cameron K. D., Bukhalid R. A., Gibson D. M., Crane B. R., Loria R. (2004) Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429, 79–82 [DOI] [PubMed] [Google Scholar]
- 9. Gusarov I., Shatalin K., Starodubtseva M., Nudler E. (2009) Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325, 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCollister B. D., Hoffman M., Husain M., Vázquez-Torres A. (2011) Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. 55, 2189–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeLeo F. R., Otto M., Kreiswirth B. N., Chambers H. F. (2010) Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tenover F. C., Goering R. V. (2009) Methicillin-resistant Staphylococcus aureus strain USA300. Origin and epidemiology. J. Antimicrob. Chemother. 64, 441–446 [DOI] [PubMed] [Google Scholar]
- 13. Voyich J. M., Braughton K. R., Sturdevant D. E., Whitney A. R., Saïd-Salim B., Porcella S. F., Long R. D., Dorward D. W., Gardner D. J., Kreiswirth B. N., Musser J. M., DeLeo F. R. (2005) Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175, 3907–3919 [DOI] [PubMed] [Google Scholar]
- 14. Mölne L., Verdrengh M., Tarkowski A. (2000) Role of neutrophil leukocytes in cutaneous infection caused by Staphylococcus aureus. Infect. Immun. 68, 6162–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Köckritz-Blickwede M., Rohde M., Oehmcke S., Miller L. S., Cheung A. L., Herwald H., Foster S., Medina E. (2008) Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am. J. Pathol. 173, 1657–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robertson C. M., Perrone E. E., McConnell K. W., Dunne W. M., Boody B., Brahmbhatt T., Diacovo M. J., Van Rooijen N., Hogue L. A., Cannon C. L., Buchman T. G., Hotchkiss R. S., Coopersmith C. M. (2008) Neutrophil depletion causes a fatal defect in murine pulmonary Staphylococcus aureus clearance. J. Surg. Res. 150, 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van den Berg J. M., van Koppen E., Ahlin A., Belohradsky B. H., Bernatowska E., Corbeel L., Español T., Fischer A., Kurenko-Deptuch M., Mouy R., Petropoulou T., Roesler J., Seger R., Stasia M. J., Valerius N. H., Weening R. S., Wolach B., Roos D., Kuijpers T. W. (2009) Chronic granulomatous disease. The European experience. PLoS One 4, e5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soehnlein O. (2009) Direct and alternative antimicrobial mechanisms of neutrophil-derived granule proteins. J. Mol. Med. 87, 1157–1164 [DOI] [PubMed] [Google Scholar]
- 19. Mandell G. L. (1975) Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal-leukocyte interaction. J. Clin. Invest. 55, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park B., Nizet V., Liu G. Y. (2008) Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J. Bacteriol. 190, 2275–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu G. Y., Essex A., Buchanan J. T., Datta V., Hoffman H. M., Bastian J. F., Fierer J., Nizet V. (2005) Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson A. R., Dunman P. M., Fang F. C. (2006) The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939 [DOI] [PubMed] [Google Scholar]
- 23. Richardson A. R., Libby S. J., Fang F. C. (2008) A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319, 1672–1676 [DOI] [PubMed] [Google Scholar]
- 24. Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., Götz F. (1999) Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 [DOI] [PubMed] [Google Scholar]
- 25. Kristian S. A., Lauth X., Nizet V., Goetz F., Neumeister B., Peschel A., Landmann R. (2003) Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J. Infect. Dis. 188, 414–423 [DOI] [PubMed] [Google Scholar]
- 26. Jin T., Bokarewa M., Foster T., Mitchell J., Higgins J., Tarkowski A. (2004) Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 27. Sieprawska-Lupa M., Mydel P., Krawczyk K., Wójcik K., Puklo M., Lupa B., Suder P., Silberring J., Reed M., Pohl J., Shafer W., McAleese F., Foster T., Travis J., Potempa J. (2004) Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48, 4673–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bae T., Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 [DOI] [PubMed] [Google Scholar]
- 29. Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A., Mongodin E. F., Sensabaugh G. F., Perdreau-Remington F. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 [DOI] [PubMed] [Google Scholar]
- 30. Chaffin D. O., Rubens C. E. (1998) Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219, 91–99 [DOI] [PubMed] [Google Scholar]
- 31. Lim M. H., Lippard S. J. (2006) Fluorescent nitric oxide detection by copper complexes bearing anthracenyl and dansyl fluorophore ligands. Inorg. Chem. 45, 8980–8989 [DOI] [PubMed] [Google Scholar]
- 32. Beauchamp C., Fridovich I. (1971) Superoxide dismutase. Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287 [DOI] [PubMed] [Google Scholar]
- 33. von Köckritz-Blickwede M., Nizet V. (2009) Innate immunity turned inside-out. Antimicrobial defense by phagocyte extracellular traps. J. Mol. Med. 87, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong I. S., Kim Y. K., Choi W. S., Seo D. W., Yoon J. W., Han J. W., Lee H. Y., Lee H. W. (2003) Purification and characterization of nitric oxide synthase from Staphylococcus aureus. FEMS Microbiol. Lett. 222, 177–182 [DOI] [PubMed] [Google Scholar]
- 35. Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 [DOI] [PubMed] [Google Scholar]
- 36. Yeom J., Imlay J. A., Park W. (2010) Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J. Biol. Chem. 285, 22689–22695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Repine J. E., Fox R. B., Berger E. M. (1981) Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J. Biol. Chem. 256, 7094–7096 [PubMed] [Google Scholar]
- 38. Chambers H. F., Hackbarth C. J., Drake T. A., Rusnak M. G., Sande M. A. (1984) Endocarditis due to methicillin-resistant Staphylococcus aureus in rabbits. Expression of resistance to β-lactam antibiotics in vivo and in vitro. J. Infect. Dis. 149, 894–903 [DOI] [PubMed] [Google Scholar]
- 39. Lemaire S., Glupczynski Y., Duval V., Joris B., Tulkens P. M., Van Bambeke F. (2009) Activities of ceftobiprole and other cephalosporins against extracellular and intracellular (THP-1 macrophages and keratinocytes) forms of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 2289–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckmann C., Dryden M. (2010) Treatment of complicated skin and soft-tissue infections caused by resistant bacteria. Value of linezolid, tigecycline, daptomycin and vancomycin. Eur. J. Med. Res. 15, 554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Best G. K., Durham N. N. (1965) Vancomycin adsorption to Bacillus subtilis cell walls. Arch. Biochem. Biophys. 111, 685–691 [DOI] [PubMed] [Google Scholar]
- 42. Hachmann A. B., Angert E. R., Helmann J. D. (2009) Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53, 1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nizet V., Gallo R. L. (2003) Cathelicidins and innate defense against invasive bacterial infection. Scand. J. Infect. Dis. 35, 670–676 [DOI] [PubMed] [Google Scholar]
- 44. Tomasinsig L., Zanetti M. (2005) The cathelicidins. Structure, function and evolution. Curr. Protein Pept. Sci. 6, 23–34 [DOI] [PubMed] [Google Scholar]
- 45. Alalwani S. M., Sierigk J., Herr C., Pinkenburg O., Gallo R., Vogelmeier C., Bals R. (2010) The antimicrobial peptide LL-37 modulates the inflammatory and host defense response of human neutrophils. Eur. J. Immunol. 40, 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng Y., Niyonsaba F., Ushio H., Nagaoka I., Ikeda S., Okumura K., Ogawa H. (2007) Cathelicidin LL-37 induces the generation of reactive oxygen species and release of human alpha-defensins from neutrophils. Br. J. Dermatol. 157, 1124–1131 [DOI] [PubMed] [Google Scholar]
- 47. Clarke S. R., Mohamed R., Bian L., Routh A. F., Kokai-Kun J. F., Mond J. J., Tarkowski A., Foster S. J. (2007) The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1, 199–212 [DOI] [PubMed] [Google Scholar]
- 48. Peters B. M., Shirtliff M. E., Jabra-Rizk M. A. (2010) Antimicrobial peptides. Primeval molecules or future drugs? PLoS Pathog. 6, e1001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palazzolo-Ballance A. M., Reniere M. L., Braughton K. R., Sturdevant D. E., Otto M., Kreiswirth B. N., Skaar E. P., DeLeo F. R. (2008) Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180, 500–509 [DOI] [PubMed] [Google Scholar]
- 50. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 [DOI] [PubMed] [Google Scholar]
- 51. Pilsczek F. H., Salina D., Poon K. K., Fahey C., Yipp B. G., Sibley C. D., Robbins S. M., Green F. H., Surette M. G., Sugai M., Bowden M. G., Hussain M., Zhang K., Kubes P. (2010) A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 185, 7413–7425 [DOI] [PubMed] [Google Scholar]
- 52. Lauth X., von Köckritz-Blickwede M., McNamara C. W., Myskowski S., Zinkernagel A. S., Beall B., Ghosh P., Gallo R. L., Nizet V. (2009) M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J. Innate Immun. 1, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moet G. J., Jones R. N., Biedenbach D. J., Stilwell M. G., Fritsche T. R. (2007) Contemporary causes of skin and soft tissue infections in North America, Latin America, and Europe. Report from the SENTRY Antimicrobial Surveillance Program (1998–2004). Diagn. Microbiol. Infect. Dis. 57, 7–13 [DOI] [PubMed] [Google Scholar]
- 54. Moran G. J., Krishnadasan A., Gorwitz R. J., Fosheim G. E., McDougal L. K., Carey R. B., Talan D. A. (2006) Methicillin-resistant S. aureus infections among patients in the emergency department. New Eng. J. Med. 355, 666–674 [DOI] [PubMed] [Google Scholar]
- 55. Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R. A., Pestonjamasp V., Piraino J., Huttner K., Gallo R. L. (2001) Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457 [DOI] [PubMed] [Google Scholar]
- 56. Chakravortty D., Hensel M. (2003) Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect. 5, 621–627 [DOI] [PubMed] [Google Scholar]
- 57. Pollock J. D., Williams D. A., Gifford M. A., Li L. L., Du X., Fisherman J., Orkin S. H., Doerschuk C. M., Dinauer M. C. (1995) Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9, 202–209 [DOI] [PubMed] [Google Scholar]
- 58. Sasaki S., Miura T., Nishikawa S., Yamada K., Hirasue M., Nakane A. (1998) Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect. Immun. 66, 1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Köhler J., Breitbach K., Renner C., Heitsch A. K., Bast A., van Rooijen N., Vogelgesang S., Steinmetz I. (2011) NADPH-oxidase but not inducible nitric oxide synthase contributes to resistance in a murine Staphylococcus aureus Newman pneumonia model. Microbes Infect. 13, 914–922 [DOI] [PubMed] [Google Scholar]
- 60. Ouhara K., Komatsuzawa H., Kawai T., Nishi H., Fujiwara T., Fujiue Y., Kuwabara M., Sayama K., Hashimoto K., Sugai M. (2008) Increased resistance to cationic antimicrobial peptide LL-37 in methicillin-resistant strains of Staphylococcus aureus. J. Antimicrob. Chemother. 61, 1266–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nizet V. (2007) Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 120, 13–22 [DOI] [PubMed] [Google Scholar]
- 62. Hochgräfe F., Wolf C., Fuchs S., Liebeke M., Lalk M., Engelmann S., Hecker M. (2008) Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 190, 4997–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Salard I., Mercey E., Rekka E., Boucher J. L., Nioche P., Mikula I., Martasek P., Raman C. S., Mansuy D. (2006) Analogies and surprising differences between recombinant nitric oxide synthase-like proteins from Staphylococcus aureus and Bacillus anthracis in their interactions with l-arginine analogs and iron ligands. J. Inorg. Biochem. 100, 2024–2033 [DOI] [PubMed] [Google Scholar]
- 64. Choi W. S., Chang M. S., Han J. W., Hong S. Y., Lee H. W. (1997) Identification of nitric oxide synthase in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 237, 554–558 [DOI] [PubMed] [Google Scholar]
- 65. Escaich S. (2010) Novel agents to inhibit microbial virulence and pathogenicity. Expert Opin. Ther. Pat. 20, 1401–1418 [DOI] [PubMed] [Google Scholar]
- 66. Clatworthy A. E., Pierson E., Hung D. T. (2007) Targeting virulence. A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.