Background: Yeast App1p is a phosphatidate phosphatase (PAP) that interacts with endocytic proteins at cortical actin patches.
Results: App1p PAP was purified from yeast and characterized with respect to its enzymological, kinetic, and regulatory properties.
Conclusion: App1p PAP exhibits similar, but also distinct properties from other PAP enzymes.
Significance: App1p PAP may regulate the balance of phosphatidate and diacylglycerol for endocytosis.
Keywords: Diacylglycerol, Kinetics, Lipids, Phosphatase, Phosphatidate, Phospholipid, Phospholipid Metabolism, Yeast
Abstract
Yeast App1p is a phosphatidate phosphatase (PAP) that associates with endocytic proteins at cortical actin patches. App1p, which catalyzes the conversion of phosphatidate (PA) to diacylglycerol, is unique among Mg2+-dependent PAP enzymes in that its reaction is not involved with de novo lipid synthesis. Instead, App1p PAP is thought to play a role in endocytosis because its substrate and product facilitate membrane fission/fusion events and regulate enzymes that govern vesicular movement. App1p PAP was purified from yeast and characterized with respect to its enzymological, kinetic, and regulatory properties. Maximum PAP activity was dependent on Triton X-100 (20 mm), PA (2 mm), Mg2+ (0.5 mm), and 2-mercaptoethanol (10 mm) at pH 7.5 and 30 °C. Analysis of surface dilution kinetics with Triton X-100/PA-mixed micelles yielded constants for surface binding (KsA = 11 mm), interfacial PA binding (KmB = 4.2 mol %), and catalytic efficiency (Vmax = 557 μmol/min/mg). The activation energy, turnover number, and equilibrium constant were 16.5 kcal/mol, 406 s−1, and 16.2, respectively. PAP activity was stimulated by anionic lipids (cardiolipin, phosphatidylglycerol, phosphatidylserine, and CDP-diacylglycerol) and inhibited by zwitterionic (phosphatidylcholine and phosphatidylethanolamine) and cationic (sphinganine) lipids, nucleotides (ATP and CTP), N-ethylmaleimide, propranolol, phenylglyoxal, and divalent cations (Ca2+, Mn2+, and Zn2+). App1p also utilized diacylglycerol pyrophosphate and lyso-PA as substrates with specificity constants 4- and 7-fold lower, respectively, when compared with PA.
Introduction
PAP,2 the enzyme that catalyzes the dephosphorylation of PA to form DAG and Pi (1), has emerged as a key regulator of lipid homeostasis and cell physiology (2–7). PAP plays an important role in balancing the cellular amounts of PA and DAG, lipid intermediates that govern the synthesis of triacylglycerol and phospholipids, as well as other aspects of cell physiology that include transcription, lipid signaling, and vesicular trafficking (1, 8–27).
Two types of PAP activity are found in eukaryotic organisms; one type of PAP is dependent on Mg2+ for activity, and the other type of PAP has no divalent cation requirement for activity (2–4). A DXDX(T/V) catalytic motif directs the Mg2+-dependent activity (28, 29), whereas a three-domain catalytic motif consisting of the sequences KX6RP, PSGH, and SRX5HX3D directs the Mg2+-independent activity (30, 31). Mg2+-dependent PAP is a soluble enzyme that associates with the membrane to utilize its substrate PA (2–5, 32). In contrast, Mg2+-independent PAP is an intrinsic membrane enzyme that utilizes PA as well as other lipid phosphates (e.g. LPA and DGPP) as substrates (2–5, 32).
Insights into the roles of PAP enzymes in cell physiology have come from studies using the yeast Saccharomyces cerevisiae. In this model eukaryote, Mg2+-dependent Pah1p PAP activity is responsible for producing at the nuclear/ER membrane a pool of DAG used for the synthesis of triacylglycerol (2, 29, 33, 34). This activity also controls a pool of PA used for the synthesis of phospholipids via the liponucleotide intermediate CDP-DAG (2, 29, 33, 34). The PA content, as controlled by Pah1p PAP activity, also regulates the expression (e.g. transcriptional mechanism) of phospholipid synthesis enzymes (2, 26, 29, 33–35). Two membrane-associated Mg2+-independent PAP enzymes, Dpp1p and Lpp1p, are thought to control the signaling functions of PA, LPA, and DGPP in vacuole and Golgi membranes, respectively (2–4, 36–42).
The focus of the current work was to biochemically characterize the novel Mg2+-dependent PAP encoded by the APP1 gene (43). App1p (named for actin patch protein) is unique among Mg2+-dependent PAP enzymes in yeast and higher eukaryotes in that its reaction is not involved with de novo lipid synthesis (43). Instead, this PAP may regulate the local concentrations of PA and DAG at the membrane to control endocytic events (43). This assertion stems from the fact that App1p associates with endocytic proteins at cortical actin patches (2, 44–51), which are the sites of endocytosis in yeast (52). Indeed, the PAP substrate PA and product DAG are known to facilitate membrane fission/fusion events (53–58). Moreover, these lipids regulate enzymes (e.g. phosphatidylinositol 4-phosphate kinase) that govern vesicular movement (59–63). Detailed knowledge of the biochemical properties of App1p PAP is required to elucidate the mode of action and regulation of the enzyme at its site of action. Toward this end, we purified App1p PAP from S. cerevisiae and characterized its enzymological, kinetic, and regulatory properties. These studies revealed similarities but also distinct differences between App1p and other PAP enzymes with respect to substrate specificity and regulatory properties.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade. Growth medium supplies were purchased from Difco. Restriction endonucleases, modifying enzymes, and recombinant VentR DNA polymerase were purchased from New England Biolabs. Plasmid isolation and gel extraction kits, Ni2+-NTA-agarose resin, and spin columns were purchased from Qiagen. The Yeastmaker yeast transformation kit was purchased from Clontech. Invitrogen was the source of the DNA size standards and His6-tagged tobacco etch virus protease. Protease inhibitor mixture tablets were from Roche Applied Science. Sepharose Fast-Flow, IgG-Sepharose, protein A-Sepharose CL-4B, and the Superdex 200 column were purchased from GE Healthcare. Protein assay reagent, electrophoresis reagents, and Poly-Prep chromatography columns were purchased from Bio-Rad. Radiochemicals were purchased from PerkinElmer Life Sciences. Bovine serum albumin, nucleotides, oligonucleotides, Triton X-100, anti-protein A antibodies, and molecular mass markers for gel filtration chromatography were purchased from Sigma. Malachite green was from Fisher. Dioleoyl-PA, dioleoyl-DGPP, dioleoyl-DAG, and oleoyl-LPA, as well as the other lipids used in this study, were obtained from Avanti Polar Lipids. Scintillation counting supplies were purchased from National Diagnostics.
Plasmid Construction
The 1.8-kb APP1 coding sequence with a 30-bp linker at the 3′ end was amplified from pMC102 (43) (forward, 5′-TGGAGAGAGCTCAAAAAAATGAATAGTCAAGG-3′; reverse, 5′-ATACAGGTTTTCGGTCGTTGGGATGGATCCGTTTGAATACTTCTCCCTAATTCTGCG-3′), and the 0.45-kb Staphylococcus aureus spa coding sequence with a 30-bp linker at the 5′ end was amplified from YCplac111-PAH1-PtA (35) (forward, 5′-GGATCCATCCCAACGACCGAAAACCTGTAT-3′; reverse, 5′-ATAACACTCGAGTTAGCATGCTGAATTCGCG-3′). The APP1 and spa coding sequences were combined through the common 30-bp linker by overlap extension PCR. The 2.2-kb APP1-spa sequence was digested with SacI and XhoI and inserted into plasmid pYES2 at the same restriction sites. This construction added 12 amino acids to App1p and increased its subunit molecular mass from 66 to 67.5 kDa (minus the protein A tag). The recombinant plasmid for galactose-inducible expression of protein A-tagged App1p was named pMC104.
Strains and Growth Conditions
Escherichia coli strain DH5α (F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1) (64) was used for plasmid maintenance and amplification. S. cerevisiae strain W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) (65) was used for the galactose-inducible expression of protein A-tagged App1p PAP. Bacterial cells were grown at 37 °C in lysogeny broth medium (66). Ampicillin (100 μg/ml) was added to select for the cells carrying plasmid. Yeast cells were grown at 30 °C in standard synthetic complete medium (64, 67) containing 2% glucose. Uracil was omitted from the growth medium to select for cells carrying plasmid pMC104, and 2% agar was included for growth on plates. For App1p PAP purification, yeast cells (250 ml) were first grown to saturation in synthetic complete medium with 2% raffinose as the carbon source. The culture was then diluted into synthetic complete medium (2 liters) with 2% galactose as the carbon source to induce the expression of protein A-tagged App1p PAP. Maximum expression, as confirmed by immunoblot analysis (68, 69) using anti-protein A antibodies, was obtained after 6 h of growth (late exponential phase).
Purification of App1p PAP
Yeast cells (∼3 g wet weight) that expressed the protein A-tagged App1p PAP were disrupted with glass beads (0.5-mm diameter) using a Biospec Products Bead Beater in lysis buffer containing 20 mm Tris-HCl (pH 7.5), 150 mm KCl, 5 mm MgCl2, 1% Triton X-100, and a mixture of protease inhibitors. Unbroken cells and glass beads were removed by centrifugation at 1,500 × g for 10 min. The cell extract (supernatant) was precleared of contaminating proteins by incubation with 1 ml of Sepharose Fast-Flow resin in a centrifuge tube. The enzyme sample was obtained by centrifugation at 1,500 × g for 10 min, and then it was passed through 0.2 ml of IgG-Sepharose resin packed in a Bio-Rad Poly-Prep chromatography column. The column was washed with 10 ml of lysis buffer followed by 10 ml of wash buffer containing 50 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, 0.5 m NaCl. The resin was then transferred to a 1-ml spin column and incubated with an equal volume of elution buffer that contained 50 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, 0.5 m NaCl, 3 mm glutathione, 0.3 mm oxidized glutathione, and 100 units of tobacco etch virus protease. After incubation for 5 h, the column was subjected to centrifugation at 1,500 × g for 1 min, and the eluted enzyme (supernatant) was transferred to another 1-ml spin column containing a mixture of protein A-Sepharose resin (0.1 ml) and Ni2-NTA-agarose resin (0.1 ml) for the removal of contaminating IgG and tobacco etch virus protease. After a 30-min incubation, the spin column was centrifuged at 1,500 × g for 1 min to obtain the purified App1p PAP enzyme. The protein concentration of the purified enzyme preparation was estimated by the method of Bradford (70) using bovine serum albumin as the standard. ImageQuant analysis of Coomassie Blue-stained SDS-polyacrylamide gels containing purified App1p and known amounts of bovine serum albumin was also used to estimate the concentration of the App1p preparation. Both methods of protein estimation gave similar results. The protein concentration of the purified enzyme was 62.5 μg/ml.
SDS-PAGE and Liquid Chromatography/Tandem Mass Spectrometry
SDS-PAGE (71) was performed with 10% slab gels. Proteins in gels were visualized by staining with Coomassie Brilliant Blue R-250. An SDS-polyacrylamide gel slice containing purified App1p was subjected to trypsin digestion (72), followed by liquid chromatography/tandem mass spectrometry using a Thermo Fisher Scientific LTQ Obitrap Velos instrument (73). This work was performed at the Center for Advanced Proteomics Research facility of the University of Medicine and Dentistry of New Jersey (Newark, NJ).
Superdex 200 Chromatography
The native molecular mass of App1p PAP was estimated by gel filtration chromatography (74). A Superdex 200 column (1 × 30 cm) attached to a GE Pharmacia ÄKTA fast protein liquid chromatography system was equilibrated and eluted with 50 mm Tris-HCl (pH 7.5) and 0.15 m NaCl. The column was calibrated with blue dextran 2000 (for the void volume), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). Purified App1p was applied and eluted from the column at a flow rate of 40 ml/h. Fractions (0.5 ml) were collected and analyzed for App1p by measuring PAP activity. The molecular mass of App1p was calculated from a plot of log molecular mass versus the elution volume/void volume (V/V0) of the standards.
PAP Assay
PAP activity was measured for 20 min by following the release of water-soluble 32Pi from chloroform-soluble [32P]PA (500 cpm/nmol) at 30 °C (75). [32P]PA was synthesized enzymatically from dioleoyl-DAG and [γ-32P]ATP with E. coli DAG kinase (75). The radioactive PA was diluted with nonradioactive dioleoyl-PA to obtain the desired concentration of substrate used in the enzyme reactions. Alternatively, the formation of water-soluble Pi from chloroform-soluble non-radioactive lipid phosphate substrates was measured with the malachite green-molybdate reagent (76) as described previously (77). The reaction mixture contained 50 mm Tris-HCl buffer (pH 7.5), 1 mm MgCl2, 2 mm PA, 20 mm Triton X-100, 10 mm 2-mercaptoethanol, and 1 ng of enzyme protein in a total volume of 0.1 ml. Enzyme assays were conducted in triplicate, and the average S.D. of the assays was ±5%. All reactions were linear with time and protein concentration. A unit of enzyme activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of product/min.
Preparation of Triton X-100/Lipid-mixed Micelles
Lipid in chloroform was transferred to a test tube, and the solvent was evaporated with nitrogen followed by drying under vacuum. Triton X-100 was added to the dried lipid to prepare Triton X-100/lipid-mixed micelles. The mol fraction of lipid in a Triton X-100/lipid-mixed micelle was calculated using the formula, mol fraction = [lipid (molar)]/([lipid (molar)] + [Triton X-100 (molar)]). The mol % was the mol fraction × 100. The total lipid concentration in the Triton X-100/lipid-mixed micelles was kept below 15 mol % to ensure that the structure of the lipid-mixed micelles was similar to that of pure Triton X-100 micelles (78, 79).
Data Analyses
Kinetic data were analyzed according to the Michaelis-Menten and Hill equations using the SigmaPlot enzyme kinetics module. Student's t test (SigmaPlot software) was used to determine statistical significance, and p values of <0.05 were taken as a significant difference.
RESULTS
Purification of App1p PAP and Determination of Its Native Molecular Mass
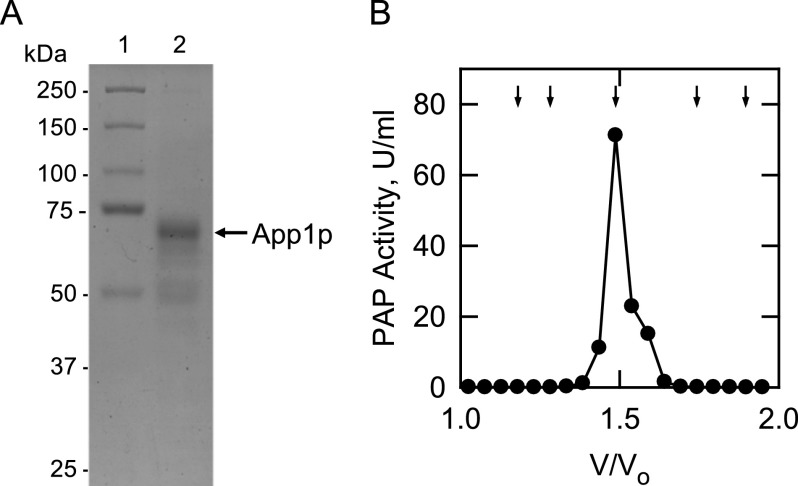
A purified preparation of App1p was required to examine the enzymological and regulatory properties of its PAP activity under well defined conditions. Due to its very low abundance and lability, it has not been possible to obtain pure native enzyme from yeast by classical protein purification methods (43). The identity of the APP1 gene encoding PAP (43) permitted the molecular construction and overexpression of protein A-tagged App1p to facilitate its purification from yeast. The protein A-tagged enzyme was purified by IgG-Sepharose affinity chromatography, followed by tobacco etch virus protease cleavage to remove the protein A tag. The procedure yielded a highly purified preparation of App1p that migrated upon SDS-PAGE at the expected minimum subunit molecular mass of ∼67 kDa (Fig. 1A). The identity of this protein as App1p was confirmed by trypsin digestion followed by analysis of peptides by liquid chromatography-tandem mass spectrometry. The purified enzyme was subjected to gel filtration chromatography with a Superdex 200 column to determine the native molecular mass (Fig. 1B). This analysis indicated a native size of ∼67 kDa, and thus, the active form of the enzyme was monomeric. The specific activity of purified App1p PAP was typically 370 ± 14 μmol/min/mg. This was equivalent to a 185,000-fold purification of App1p PAP relative to the activity (2 nmol/min/mg) in the cell extract of the pah1Δ dpp1Δ lpp1Δ triplet mutant devoid of other PAP enzyme activities.3
FIGURE 1.
SDS-PAGE analysis of purified App1p and the elution profile of PAP activity after chromatography with Superdex 200. A, App1p was expressed and purified from S. cerevisiae as described under “Experimental Procedures.” Lane 1, molecular mass standards; lane 2, a sample (1.25 μg) of the purified App1p (indicated by the arrow). The SDS-polyacrylamide gel was stained with Coomassie Brilliant Blue R-250. B, purified App1p was subjected to Superdex 200 chromatography. Fractions (0.5 ml) were collected and assayed for PAP activity. The molecular mass standards (indicated by arrows from left to right) were β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa).
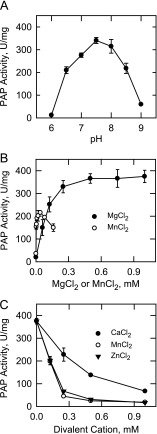
Effects of pH and Divalent Cations on App1p PAP Activity
The effect of pH on App1p PAP activity was examined using a Tris-maleate-glycine composite buffer ranging from pH 6 to 9. Maximum activity was found at pH 7.5 (Fig. 2A). Mg2+ (0.5–1 mm) was required for maximum PAP activity (Fig. 2B). Mn2+ (40 μm) could substitute for the Mg2+ ion requirement, but the maximum activity obtained with Mn2+ was nearly 2-fold lower than that obtained with Mg2+ (Fig. 2B). Moreover, concentrations of Mn2+ above 40 μm were inhibitory to the PAP activity. Under standard assay conditions (i.e. with 1 mm MgCl2), the addition of Ca2+, Mn2+, or Zn2+ inhibited PAP activity by 81, 95, and 96%, respectively (Fig. 2C).
FIGURE 2.
Effects of pH and divalent cations on App1p PAP activity. A, PAP activity was measured at the indicated pH values with 50 mm Tris-maleate-glycine buffer. B, PAP activity was measured in the absence and presence of the indicated concentrations of MgCl2 or MnCl2. C, PAP activity was measured with 1 mm MgCl2 in the absence and presence of the indicated concentrations of CaCl2, MnCl2, or ZnCl2. The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
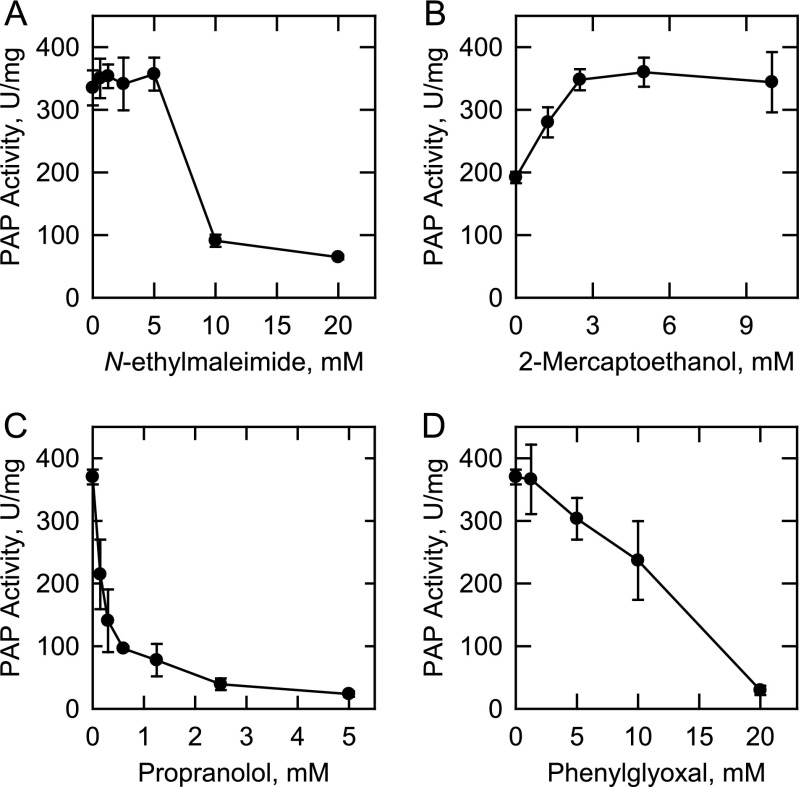
Effects of N-Ethylmaleimide, 2-Mercaptoethanol, Propranolol, and Phenylglyoxal on App1p PAP Activity
The alkylating reagent N-ethylmaleimide (80) has been used to differentiate Mg2+-dependent and Mg2+-independent PAP enzymes (18, 81). App1p PAP activity was insensitive to inhibition by N-ethylmaleimide at concentrations up to 5 mm, but the activity was inhibited (∼80%) at concentrations of 10 mm and above (Fig. 3A). In addition, the PAP activity was stimulated (87%) by the addition of 2-mercaptoethanol to the assay systems that did not contain N-ethylmaleimide (Fig. 3B). Thus, 2-mercaptoethanol was included in the standard reaction for the enzyme. Propranolol (a non-selective β-adrenergic receptor antagonist (80)) and phenylglyoxal (an arginine-reactive compound (80)) have been shown to inhibit Mg2+-dependent and Mg2+-independent forms of PAP (41, 82, 83). Both of these compounds inhibited App1p PAP activity; propranolol (IC50 = 0.3 mm) was a more potent inhibitor when compared with phenylglyoxal (IC50 = 12 mm) (Fig. 3, C and D).
FIGURE 3.
Effects of N-ethylmaleimide, 2-mercaptoethanol, propranolol, and phenylglyoxal on App1p PAP activity. PAP activity was measured under standard assay conditions in the absence and presence of the indicated concentrations of N-ethylmaleimide (A) 2-mercaptoethanol (B), propranolol (C), or phenylglyoxal (D). The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
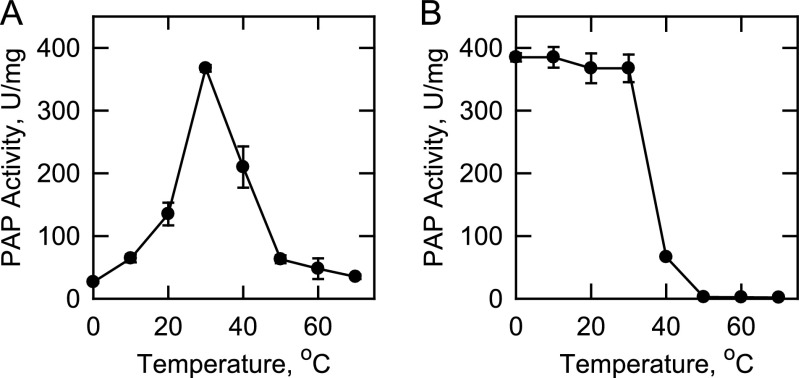
Effects of Temperature on App1p PAP Activity and Stability
The effects of temperature on App1p PAP activity and stability were examined. Maximum PAP activity was observed at 30 °C, but the activity was reduced at temperatures higher than 30 °C (Fig. 4A). An Arrhenius plot analysis of the data from 0 to 30 °C yielded an activation energy for the reaction of 16.5 kcal/mol. The thermal stability of App1p PAP was examined by incubating the enzyme at temperatures ranging from 0 to 70 °C for 20 min. Following the incubation, the samples were incubated on ice to allow for renaturation and then assayed for PAP activity at 30 °C. The enzyme was labile above 30 °C, with total inactivation at 50 °C (Fig. 4B). The purified enzyme was completely stable for at least 3 months of storage at −80 °C and stable to three cycles of freeze-thawing.
FIGURE 4.
Effects of temperature on App1p PAP activity and stability. A, PAP activity was measured at the indicated temperatures for 20 min in a temperature-controlled water bath. B, the enzyme samples were first incubated for 20 min at the indicated temperatures. After incubation, the samples were cooled in an ice bath for 10 min to allow for enzyme renaturation, and PAP activity was then measured for 20 min at 30 °C. The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
Equilibrium Constant for App1p PAP
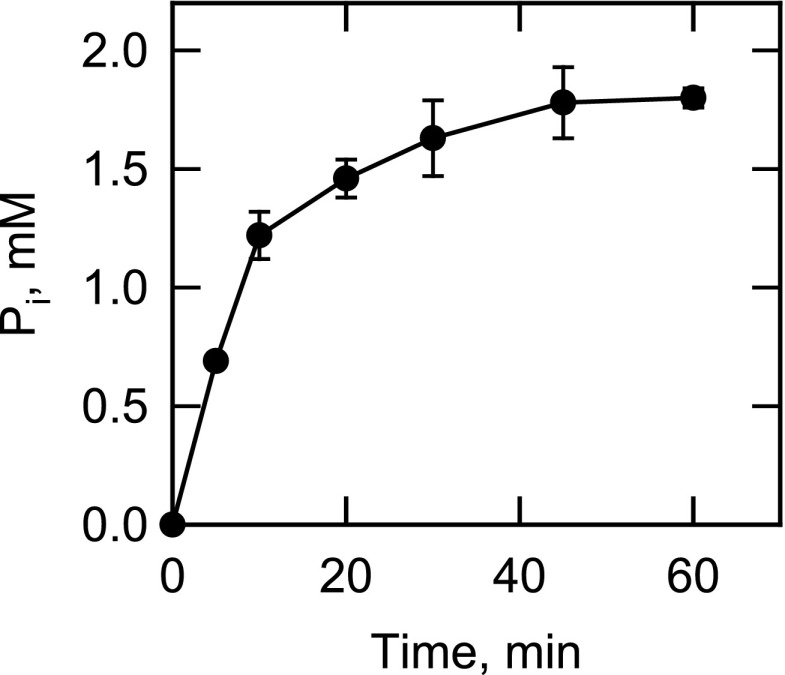
The PAP reaction was carried out to equilibrium using a 10-fold excess of App1p and an initial PA concentration of 2 mm (Fig. 5). At equilibrium, the concentrations of DAG and Pi (as determined by the generation of 32Pi from [32P]PA) were 1.8 mm each, and the equilibrium constant was calculated to be 16.2. This indicated that the forward reaction was favored in vitro.
FIGURE 5.
Time dependence of the App1p PAP reaction. PAP activity was measured with 2 mm [32P]PA (500 cpm/nmol) and 10 ng enzyme for the indicated time intervals. Following the incubations, the water-soluble 32Pi hydrolyzed from the substrate was analyzed by scintillation counting. The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
Effects of PA and Triton X-100 on App1p PAP Activity
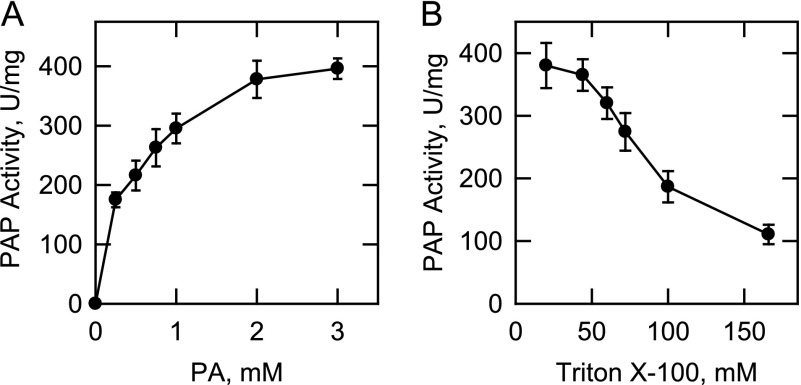
PAP activity was measured as a function of the molar concentration of PA (Fig. 6A). The water-insoluble PA substrate was solubilized with the nonionic detergent Triton X-100, which is known to form uniform Triton X-100/PA-mixed micelles (84). In this experiment, the molar ratio of Triton X-100 to PA was maintained at 10:1 (e.g. 9.1 mol % PA) because this ratio resulted in maximum PAP activity. With respect to PA, the maximum activity was obtained at a molar concentration of 2–3 mm. Analysis of the data according to the Michaelis-Menten equation yielded a Vmax of 463 μmol/min/mg and a Km value for PA of 0.52 mm. When the concentration of PA was held constant at 2 mm and the concentration of Triton X-100 was varied from 20 to 170 mm (e.g. diluting the surface concentration of PA), there was a dose-dependent reduction in the PAP activity (Fig. 6B). These data indicated that App1p PAP activity was responsive to both the molar and surface concentrations of PA, kinetic properties that are characteristic of surface dilution kinetics (85, 86).
FIGURE 6.
Effects of PA and Triton X-100 on App1p PAP activity. A, PAP activity was measured as a function of the molar concentration of PA. The molar ratio of Triton X-100 to PA was maintained at 10:1 (9.1 mol % PA). B, PAP activity was measured with the indicated concentrations of Triton X-100. The molar concentration of PA was maintained at 2 mm. The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
App1p PAP Activity Follows Surface Dilution Kinetics with Triton X-100/PA-mixed Micelles
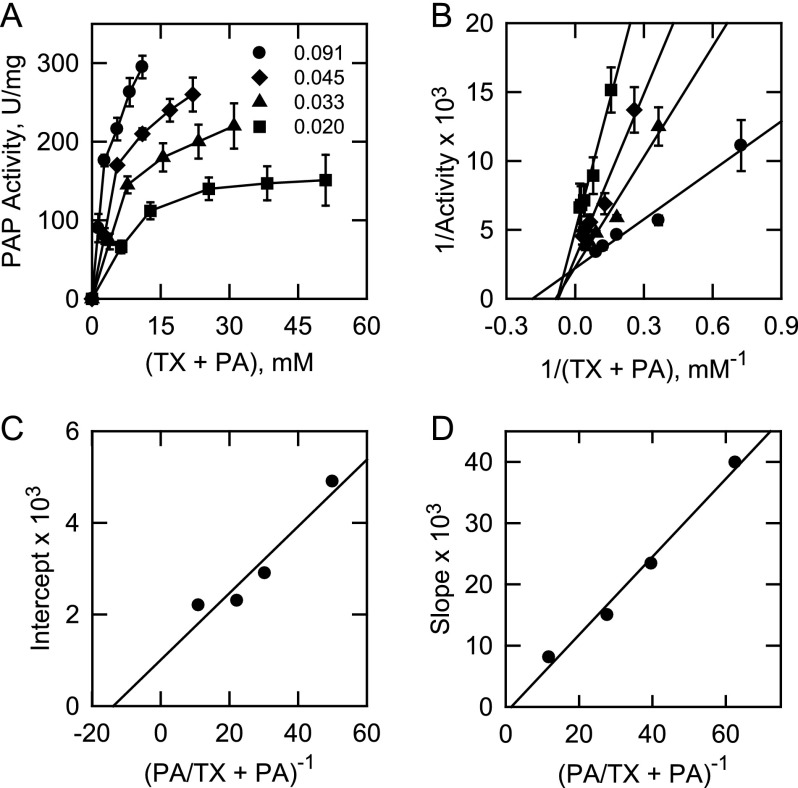
A kinetic analysis of App1p PAP was performed according to the surface dilution kinetic model (85, 86). In the bulk step (Reaction 1), the enzyme E (e.g. PAP) first associates with the mixed micelle A (e.g. Triton X-100 + PA) to form an enzyme-mixed micelle complex EA. In the surface step, the enzyme associates with the mixed micelle and then interacts with the phospholipid substrate B (e.g. mol fraction of PA in the mixed micelle) to form the EAB complex. Catalysis occurs, the products Q (e.g. DAG and Pi) are formed, and EA is regenerated. The kinetic equation for the model is shown in Equation 1 (85), where KsA = k−1/k1 and KmB = (k−2 + k3)/k2. The Vmax is the true Vmax at an infinite mol fraction and an infinite molar concentration of phospholipid substrate, KmB is the interfacial Michaelis constant (expressed in surface concentration), and KsA is the dissociation constant (expressed in molar concentration) for the mixed micelle binding site (85). The following assumptions are made with this model (85). The size and aggregation number of mixed micelles remain constant over the concentration range employed as the molar concentration or surface concentration of substrate is varied; the average surface area per Triton X-100 molecule is within the same range as the lipid substrate; both remain constant as the molar concentration or the surface concentration of substrate is varied; the micellar surface area per enzyme binding site remains constant; and the enzyme has access to the entire lipid pool.
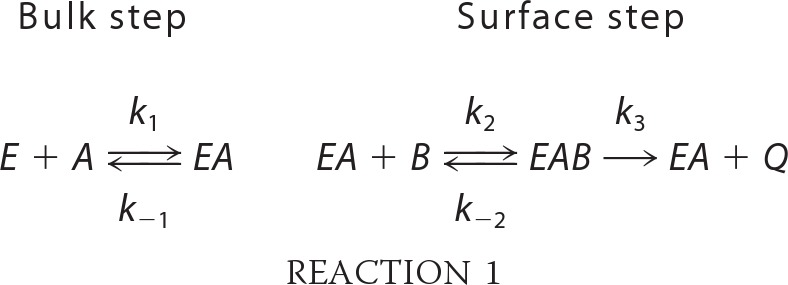 |
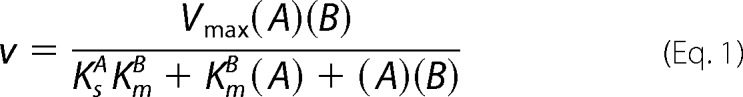 |
App1p PAP activity was measured as a function of the sum of the molar concentrations of Triton X-100 + PA (e.g. A of Equation 1) at a series of set mol fractions of PA (e.g. B of Equation 1). The PAP activity was dependent on the sum of molar concentrations of Triton X-100 + PA at each surface concentration of PA (Fig. 7A). As the surface concentration of PA in the mixed micelle increased, there was an increase in the PAP activity (Fig. 7A). A double reciprocal plot of the data indicated that the enzyme exhibited saturation kinetics with respect to the molar concentrations of Triton X-100 + PA at each surface concentration of PA (Fig. 7B). The family of lines shown in Fig. 7B was consistent with an equilibrium-ordered step, as predicted by Equation 1. A replot of the 1/activity intercepts versus 1/B (e.g. PA/Triton X-100 + PA) (Fig. 7C) and a replot of the slopes versus 1/B (Fig. 7D) were linear, as predicted by Equation 1 (85, 86). The intercept of the intercept axis and the intercept of the 1/B axis in Fig. 7C are the 1/Vmax and −1/KmB values, respectively, and the slope in Fig. 7D is equal to KsAKmB/Vmax (85, 86). The Vmax, KmB, and KsA values were calculated to be 557 μmol/min/mg, 4.2 mol %, and 11 mm, respectively.
FIGURE 7.
App1p PAP activity follows surface dilution kinetics with Triton X-100/PA-mixed micelles. A, PAP activity was measured as a function of the sum of the molar concentrations of Triton X-100 (TX) + PA at the indicated set mol fractions of PA. B, double reciprocal plot of the data in A. C, replot of the 1/activity intercepts obtained in B versus the reciprocal of the mol fraction of PA. D, replot of slopes obtained in B versus the reciprocal of the mol fraction of PA. The data shown in A are means ± S.D. (error bars) from triplicate enzyme determinations. The lines drawn in B–D are a result of a least-squares analysis of the data.
Substrate Specificity of App1p PAP
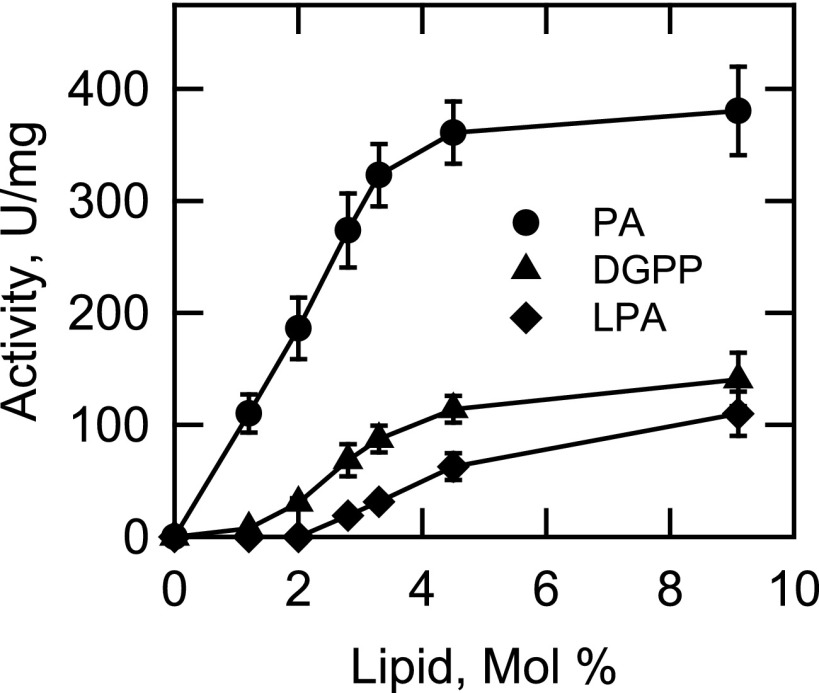
App1p was examined for its ability to utilize a variety of lipid phosphates as substrates. These included DGPP, LPA, ceramide 1-phosphate, sphinganine 1-phosphate, and sphingosine 1-phosphate. The PAP activity was also measured as a control. The lipids were delivered to the assay as uniform Triton X-100/lipid-mixed micelles, and the release of Pi from the substrates was measured with the malachite green-molybdate reagent. Of these molecules, PA, DGPP, and LPA served as substrates. The kinetic analyses for these substrates were simplified by measuring the activities as a function of their surface concentrations (mol %) and maintaining their molar concentrations at a saturating level of 2 mm (Fig. 8). Under these conditions, the enzyme followed positive cooperative kinetics (Hill numbers of 2.3, 3.3, and 3.4, respectively) with respect to the surface concentrations of PA (Vmax = 400 μmol/min/mg, Km = 2 mol %), DGPP (Vmax = 143 μmol/min/mg, Km = 2.9 mol %), and LPA (Vmax = 139 μmol/min/mg, Km = 4.8 mol %). PA (200 μmol/min/mg/mol %) was the preferred substrate of App1p with a specificity constant 4-fold and 7-fold greater, respectively, when compared with DGPP (49 μmol/min/mg/mol %) and LPA (29 μmol/min/mg/mol %).
FIGURE 8.
Dependence of App1p PAP activity on the surface concentrations of PA, DGPP, and LPA. Activity was measured as a function of the indicated surface concentration of PA, DGPP, or LPA. The molar concentrations of the substrates were held constant at 2 mm, and the Triton X-100 concentrations were varied to obtain the indicated surface concentrations. The amount of phosphate released from the substrates in the enzyme reactions was determined by the colorimetric assay using the malachite green-molybdate reagent. The data shown are means ± S.D. (error bars) from triplicate determinations.
Regulatory Effects of Lipids and Nucleotides on App1p PAP Activity
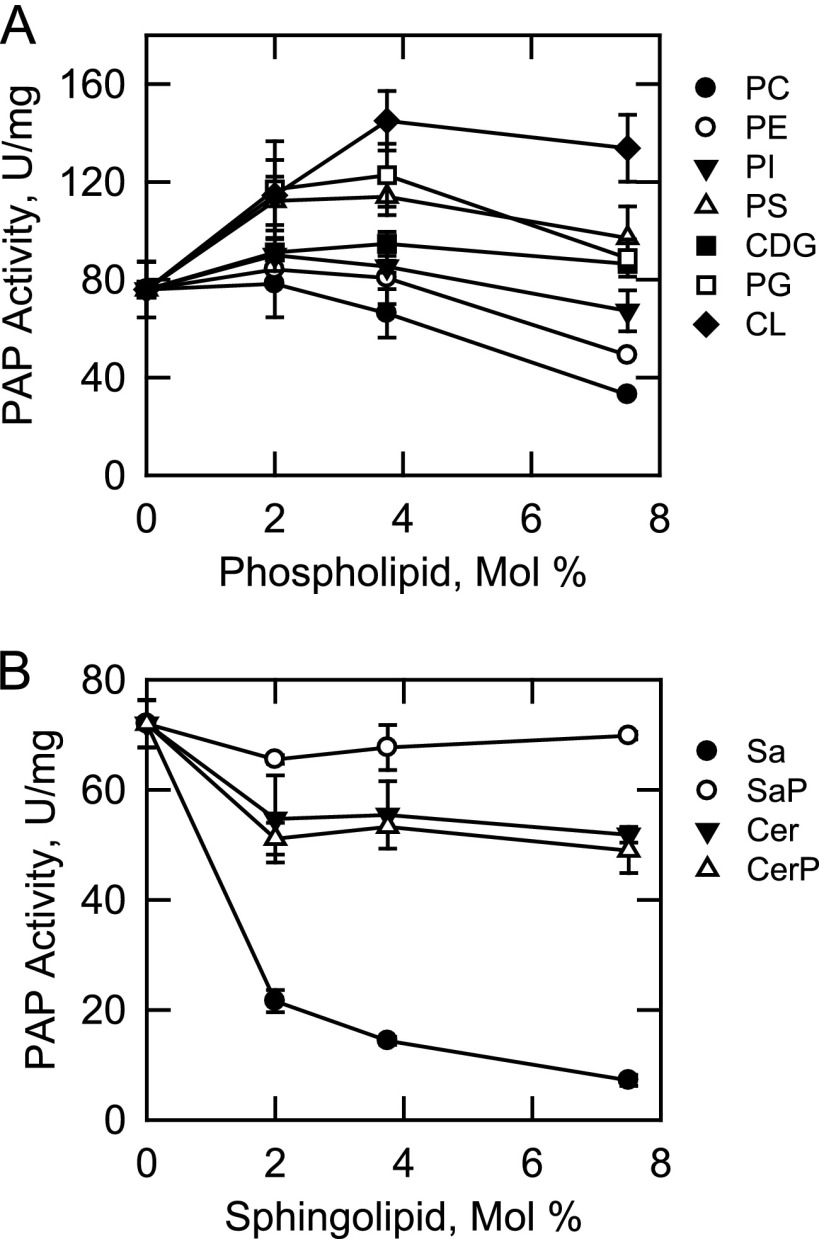
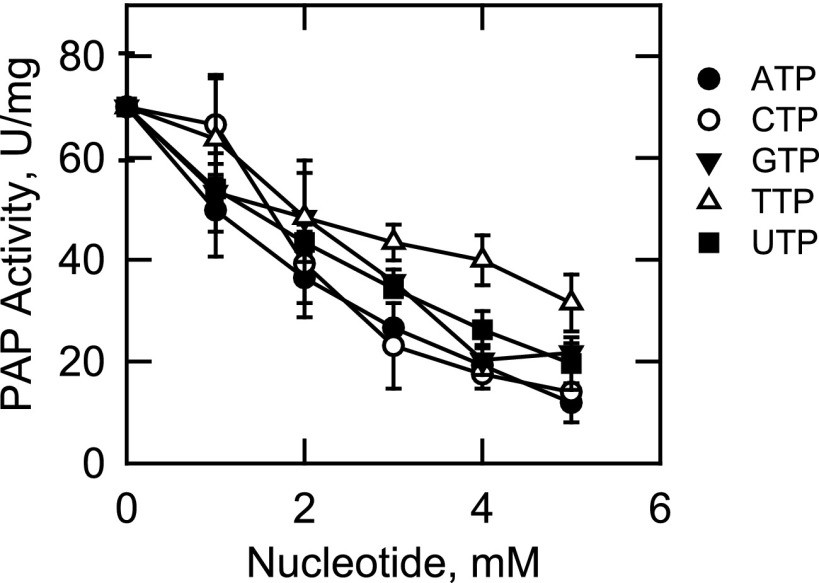
In addition to their roles as structural components of membranes, phospholipids and sphingolipids function as cofactors, inhibitors, and activators of several lipid metabolic enzymes (87–94), including Mg2+-dependent Pah1p PAP (95–97). The nucleotides ATP and CTP are substrates in pathways leading to the synthesis of the lipid intermediates (e.g. CDP-DAG) (98, 99). We questioned if these molecules had regulatory effects on App1p PAP activity. In these experiments, a surface concentration of PA (2.5 mol %) near its Km value was used to simultaneously observe stimulatory or inhibitory effects of lipids and nucleotides on PAP activity. Fig. 9 shows the effects of phospholipids (A) and sphingolipids (B) on PAP activity. At a surface concentration of 3.75 mol %, cardiolipin (90%), phosphatidylglycerol (62%), phosphatidylserine (50%), and CDP-diacylglycerol (25%) stimulated PAP activity. At 7.5 mol %, phosphatidylcholine (57%) and phosphatidylethanolamine (36%) inhibited PAP activity. The other phospholipids tested did not have a major effect on the enzyme. Of the sphingolipids examined, sphinganine (dihydrosphingosine) had the greatest effect on PAP activity. This sphingoid base inhibited activity in a dose-dependent manner (IC50 = 1.5 mol %) with 90% inhibition at a concentration of 7.5 mol %. The PAP activity was inhibited by ATP (IC50 = 3.1 mm), CTP (IC50 = 3.3 mm), GTP (IC50 = 3.8 mm), TTP (IC50 = 5.5 mm), and UTP (IC50 = 3.8 mm) in dose-dependent manners (Fig. 10). ATP and CTP were the most potent inhibitors of the enzyme, with ∼80% inhibition at a concentration of 5 mm.
FIGURE 9.
Effect of lipids on App1p PAP activity. PAP activity was measured in the absence and presence of the indicated surface concentrations of phospholipids (A) and sphingolipids (B). The surface concentration of PA was 2.5 mol % (molar concentration of 0.2 mm). The data shown are means ± S.D. (error bars) from triplicate enzyme determinations. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; CDG, CDP-diacylglycerol; PG, phosphatidylglycerol; CL, cardiolipin; Sa, sphinganine; SaP, sphinganine 1-phosphate; Cer, ceramide; CerP, ceramide 1-phosphate.
FIGURE 10.
Effect of nucleotides on App1p activity. PAP activity was measured in the absence and presence of the indicated concentrations of nucleotides. The surface concentration of PA was 2.5 mol % (molar concentration of 0.2 mm). The data shown are means ± S.D. (error bars) from triplicate enzyme determinations.
DISCUSSION
App1p is a novel Mg2+-dependent PAP enzyme that associates with cortical actin patches and is postulated to play a role in endocytosis/vesicle movement in S. cerevisiae (43, 50). Little is known about the mode of action of App1p PAP, and thus, the aim of this work was to gain information on its enzymological, kinetic, and regulatory properties. Our studies were performed under well defined conditions devoid of competing and/or modifying enzymes or molecules that might affect the interpretation of the results. IgG-Sepharose affinity chromatography of protein A-tagged App1p expressed in yeast afforded a nearly homogenous enzyme preparation. The purified enzyme was confirmed by mass spectrometry to be App1p. This further established that APP1 encodes a PAP enzyme in yeast (43). That the pure enzyme had such a high specific activity when compared with the activity in cell extracts indicated that App1p is a very low abundance PAP enzyme in yeast. In contrast, the other yeast Mg2+-dependent PAP (i.e. Pah1p) appears to be a much more abundant protein (29, 100). Although App1p was less abundant than Pah1p, the turnover number (kcat = 406 s−1) for its PAP activity was 34-fold greater than that (kcat = 12 s−1) of Pah1p PAP activity (101). The importance of this relatively high turnover number might be related to App1p function at cortical actin patches.
The basic enzymological and regulatory properties for App1p were determined, and this information will be useful in future studies when the effects of PAP activity on membranes at cortical actin patches are examined. The pH optimum, Mg2+ dependence, and sensitivities to divalent cations, propranolol, and phenylglyoxal of App1p PAP were generally similar to those shown for Pah1p PAP (29, 100, 102). However, there were some differences in enzymological properties that might be exploited to differentiate these enzymes in yeast extracts. Although the Mg2+ requirement for App1p PAP activity could be partially compensated by Mn2+, the Mg2+ requirement for Pah1p PAP activity is absolute (29, 100). In addition, App1p PAP activity was sensitive to inhibition by N-ethylmaleimide, but this reagent has no effect on Pah1p PAP activity (29). Differential effects of N-ethylmaleimide on Lpp1p (sensitive to inhibition) and Dpp1p (insensitive to inhibition) PAP activities might also be used to differentiate these Mg2+-independent enzymes in yeast extracts (36, 41), yet the sensitivity to N-ethylmaleimide inhibition cannot be used to differentiate Mg2+-dependent and Mg2+-independent PAP activities in yeast. Likewise, inhibition by propranolol, a nonspecific β-blocker (80) that is commonly used to establish roles of PAP enzymes in various physiological processes (63, 103, 104), cannot be used to distinguish between App1p and Pah1p PAP enzymes or to distinguish between Mg2+-dependent and Mg2+-independent PAP enzymes because these PAP enzymes are sensitive to this reagent (41, 77, 82, 105).
The kinetic analyses of enzymes using phospholipid substrates are conveniently carried out with uniform detergent/phospholipid-mixed micelles (84–86, 88, 92, 106–109). This system provides an environment that resembles the physiological surface of the membrane (86) where the substrate PA resides. The kinetic analysis according to the surface dilution model of Deems et al. (85) was applicable to App1p PAP activity and yielded constants for micelle surface binding (KsA), interfacial PA binding (KmB), and catalytic efficiency (Vmax). Notwithstanding the numerical values obtained from this analysis, the kinetic behavior can be extended to the in vivo condition, where soluble App1p PAP must first associate with the membrane surface at cortical actin patches followed by catalytically productive binding to PA at the membrane surface for DAG formation.
The kinetic analysis for App1p PAP was simplified by varying the PA surface concentration in Triton X-100 micelles at a set saturating molar concentration of PA. Although this approach has the limitation of not yielding a constant for micelle surface binding (KsA), it has been useful for examining the kinetic properties of other PAP enzymes from yeast and mammalian cells and, in particular, has facilitated studies to examine the substrate specificities of these enzymes (29, 36, 41, 77, 110). Moreover, the Km (2 mol %) and Vmax (400 μmol/min/mg) values determined for App1p PAP by the simplified kinetic approach are in the range of those values (Km = 4.2 mol % and Vmax = 557 μmol/min/mg) obtained according to the more complicated surface dilution kinetic scheme. In addition, this kinetic analysis showed that App1p PAP activity followed positive cooperative kinetics with respect to the surface concentration of PA. We posit that this kinetic behavior is a reflection of cooperative substrate binding as opposed to a reflection of enzyme oligomerization. The gel filtration analysis of the purified enzyme indicates that native App1p is monomeric in nature.
Mutants defective in the synthesis/turnover of anionic (e.g. phosphatidylserine and phosphatidylinositol 4,5-bisphosphate) and cationic (e.g. sphinganine) lipids exhibit defects in endocytosis (74, 111–113). Lipids might contribute to the endocytic process by governing membrane structural properties (e.g. curvature) and/or recruiting endocytic proteins to the membrane. That anionic (stimulation) and cationic (inhibition) membrane lipids modulated App1p PAP activity, which by the nature of its reaction controls the balance of PA and DAG, indicated another level by which lipids may play a role in endocytosis. An explanation for the lipid-mediated regulation of PAP activity is that charge plays a role in enzyme interaction with the membrane surface and/or interaction with PA. Alternatively, the lipid effector molecules might allosterically interact with App1p to regulate its PAP activity. Regardless of the mechanism, the local concentrations of the lipid effector molecules might be expected to fluctuate during the endocytic process. A lipidomics analysis of the membrane at cortical actin patches would shed light on this notion. Nucleotides (e.g. ATP and CTP) were also shown to negatively affect App1p PAP activity. They might interact directly with App1p to allosterically regulate activity, or as in the case of Pah1p PAP (114), they might inhibit activity by simply chelating the enzyme cofactor Mg2+. The cellular concentrations of ATP (1–2 mm) and CTP (0.2–0.8 mm) (115–117) are within the range of the IC50 values for PAP inhibition, and thus, this inhibition may be physiologically relevant. Whether or not these nucleotides fluctuate during the endocytic process to regulate PAP activity is also unknown.
An unexpected finding of this work was that App1p utilized DGPP and LPA as substrates. Substrate specificity is a major criterion for differentiating Mg2+-dependent and Mg2+-independent PAP enzymes (3, 4, 32, 118). The Mg2+-dependent Pah1p PAP (29, 100) and its mammalian counterpart lipin 1 PAP (77) are specific for PA. Differences in substrate specificity between the Mg2+-dependent and Mg2+-independent PAP enzymes have been ascribed to their distinct catalytic motifs (3, 4, 32, 118). Although App1p and Pah1p PAP activities are governed by the DXDX(T/V) catalytic motif (43, 119), this sequence in Pah1p is contained within a conserved haloacid dehalogenase-like domain (29) that is not found in App1p (43). The DXDX(T/V) motif in App1p is contained in a conserved domain found only in fungi that has overlapping regions with only weak sequence similarity to the haloacid dehalogenase-like domain (120). That the haloacid dehalogenase-like domain found in Pah1p and its mammalian counterpart Mg2+-dependent PAP enzymes (e.g. lipins) is the basis of strict specificity for PA has not been established. Nonetheless, PA was clearly the preferred substrate for App1p with a specificity constant 4–7-fold greater than that of DGPP and LPA, respectively. In contrast, the yeast Mg2+-independent Dpp1p (36, 121) and Lpp1p PAP (41) enzymes exhibit a much higher specificity for DGPP and LPA when compared with the App1p PAP enzyme. The lipidomics analysis of membranes at cortical actin patches might also shed light on whether App1p utilization of DGPP and LPA as substrates is physiologically relevant.
Acknowledgments
We thank Gil-Soo Han for helpful comments during the course of this work and in the preparation of the manuscript. The mass spectrometry data were obtained from an Orbitrap instrument funded in part by National Institutes of Health Grant NS046593, for the support of the University of Medicine and Dentistry of New Jersey Neuroproteomics Core Facility.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140.
The specific activity (0.7 nmol/min/mg) of App1p PAP in the pah1Δ dpp1Δ lpp1Δ mutant has been determined with a subsaturating concentration (0.2 mm) of PA (43). However, when measured with a saturating concentration (2 mm) of PA, the PAP activity in the triple mutant was 2 nmol/min/mg.
- PAP
- phosphatidate phosphatase
- PA
- phosphatidate
- DAG
- diacylglycerol
- LPA
- lysophosphatidate
- DGPP
- diacylglycerol pyrophosphate.
REFERENCES
- 1. Smith S. W., Weiss S. B., Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 2. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carman G. M., Han G.-S. (2006) Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carman G. M., Han G.-S. (2009) Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Csaki L. S., Reue K. (2010) Lipins. Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 30, 257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reue K., Dwyer J. R. (2009) Lipin proteins and metabolic homeostasis. J. Lipid Res. 50, S109–S114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reue K., Brindley D. N. (2008) Multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J. Lipid Res. 49, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kennedy E. P., Weiss S. B. (1956) The function of cytidine coenzyme in the biosynthesis of phospholipids. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 9. Borkenhagen L. F., Kennedy E. P. (1957) The enzymatic synthesis of cytidine diphosphate choline. J. Biol. Chem. 227, 951–962 [PubMed] [Google Scholar]
- 10. Weiss S. B., Smith S. W., Kennedy E. P. (1958) The enzymatic formation of lecithin from cytidine diphosphate choline and d-1,2-diglyceride. J. Biol. Chem. 231, 53–64 [PubMed] [Google Scholar]
- 11. Kennedy E. P. (1956) The synthesis of cytidine diphosphate choline, cytidine diphosphate ethanolamine, and related compounds. J. Biol. Chem. 222, 185–191 [PubMed] [Google Scholar]
- 12. Weiss S. B., Kennedy E. P., Kiyasu J. Y. (1960) The enzymatic synthesis of triglycerides. J. Biol. Chem. 235, 40–44 [PubMed] [Google Scholar]
- 13. Paulus H., Kennedy E. P. (1960) The enzymatic synthesis of inositol monophosphatide. J. Biol. Chem. 235, 1303–1311 [PubMed] [Google Scholar]
- 14. Kiyasu J. Y., Pieringer R. A., Paulus H., Kennedy E. P. (1963) The biosynthesis of phosphatidylglycerol. J. Biol. Chem. 238, 2293–2298 [PubMed] [Google Scholar]
- 15. Davidson J. B., Stanacev N. Z. (1971) Biosynthesis of cardiolipin in mitochondria isolated from guinea pig liver. Biochem. Biophys. Res. Commun. 42, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 16. Bishop W. R., Ganong B. R., Bell R. M. (1986) Attenuation of sn-1,2-diacylglycerol second messengers by diacylglycerol kinase. J. Biol. Chem. 261, 6993–7000 [PubMed] [Google Scholar]
- 17. Kearns B. G., McGee T. P., Mayinger P., Gedvilaite A., Phillips S. E., Kagiwada S., Bankaitis V. A. (1997) Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature 387, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waggoner D. W., Xu J., Singh I., Jasinska R., Zhang Q. X., Brindley D. N. (1999) Structural organization of mammalian lipid phosphate phosphatases. Implications for signal transduction. Biochim. Biophys. Acta 1439, 299–316 [DOI] [PubMed] [Google Scholar]
- 19. Sciorra V. A., Morris A. J. (2002) Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim. Biophys. Acta 1582, 45–51 [DOI] [PubMed] [Google Scholar]
- 20. Testerink C., Munnik T. (2005) Phosphatidic acid. A multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375 [DOI] [PubMed] [Google Scholar]
- 21. Exton J. H. (1990) Signaling through phosphatidylcholine breakdown. J. Biol. Chem. 265, 1–4 [PubMed] [Google Scholar]
- 22. Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 23. Brindley D. N. (2004) Lipid phosphate phosphatases and related proteins. Signaling functions in development, cell division, and cancer. J. Cell. Biochem. 92, 900–912 [DOI] [PubMed] [Google Scholar]
- 24. Howe A. G., McMaster C. R. (2006) Regulation of phosphatidylcholine homeostasis by Sec14. Can. J. Physiol. Pharmacol. 84, 29–38 [DOI] [PubMed] [Google Scholar]
- 25. Foster D. A. (2007) Regulation of mTOR by phosphatidic acid? Cancer Res. 67, 1–4 [DOI] [PubMed] [Google Scholar]
- 26. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrasco S., Mérida I. (2007) Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32, 27–36 [DOI] [PubMed] [Google Scholar]
- 28. Madera M., Vogel C., Kummerfeld S. K., Chothia C., Gough J. (2004) The SUPERFAMILY database in 2004. Additions and improvements. Nucleic Acids Res. 32, D235–D239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han G.-S., Wu W.-I., Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stukey J., Carman G. M. (1997) Identification of a novel phosphatase sequence motif. Protein Sci. 6, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Toke D. A., McClintick M. L., Carman G. M. (1999) Mutagenesis of the phosphatase sequence motif in diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. Biochemistry 38, 14606–14613 [DOI] [PubMed] [Google Scholar]
- 32. Brindley D. N., Pilquil C., Sariahmetoglu M., Reue K. (2009) Phosphatidate degradation. Phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim. Biophys. Acta 1791, 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carman G. M., Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henry S. A., Kohlwein S. D., Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu W.-I., Liu Y., Riedel B., Wissing J. B., Fischl A. S., Carman G. M. (1996) Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 271, 1868–1876 [DOI] [PubMed] [Google Scholar]
- 37. Toke D. A., Bennett W. L., Dillon D. A., Wu W.-I., Chen X., Ostrander D. B., Oshiro J., Cremesti A., Voelker D. R., Fischl A. S., Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 38. Han G.-S., Johnston C. N., Chen X., Athenstaedt K., Daum G., Carman G. M. (2001) Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J. Biol. Chem. 276, 10126–10133 [DOI] [PubMed] [Google Scholar]
- 39. Han G.-S., Johnston C. N., Carman G. M. (2004) Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J. Biol. Chem. 279, 5338–5345 [DOI] [PubMed] [Google Scholar]
- 40. Toke D. A., Bennett W. L., Oshiro J., Wu W.-I., Voelker D. R., Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273, 14331–14338 [DOI] [PubMed] [Google Scholar]
- 41. Furneisen J. M., Carman G. M. (2000) Enzymological properties of the LPP1-encoded lipid phosphatase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1484, 71–82 [DOI] [PubMed] [Google Scholar]
- 42. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 43. Chae M., Han G.-S., Carman G. M. (2012) The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J. Biol. Chem. 287, 40186–40196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bon E., Recordon-Navarro P., Durrens P., Iwase M., Toh-E A., Aigle M. (2000) A network of proteins around Rvs167p and Rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast 16, 1229–1241 [DOI] [PubMed] [Google Scholar]
- 45. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 46. Tong A. H., Drees B., Nardelli G., Bader G. D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., Quondam M., Zucconi A., Hogue C. W., Fields S., Boone C., Cesareni G. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321–324 [DOI] [PubMed] [Google Scholar]
- 47. Fazi B., Cope M. J., Douangamath A., Ferracuti S., Schirwitz K., Zucconi A., Drubin D. G., Wilmanns M., Cesareni G., Castagnoli L. (2002) Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1. Structural and functional analysis. J. Biol. Chem. 277, 5290–5298 [DOI] [PubMed] [Google Scholar]
- 48. Landgraf C., Panni S., Montecchi-Palazzi L., Castagnoli L., Schneider-Mergener J., Volkmer-Engert R., Cesareni G. (2004) Protein interaction networks by proteome peptide scanning. PLoS Biol. 2, E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tonikian R., Xin X., Toret C. P., Gfeller D., Landgraf C., Panni S., Paoluzi S., Castagnoli L., Currell B., Seshagiri S., Yu H., Winsor B., Vidal M., Gerstein M. B., Bader G. D., Volkmer R., Cesareni G., Drubin D. G., Kim P. M., Sidhu S. S., Boone C. (2009) Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 7, e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drees B. L., Sundin B., Brazeau E., Caviston J. P., Chen G. C., Guo W., Kozminski K. G., Lau M. W., Moskow J. J., Tong A., Schenkman L. R., McKenzie A., 3rd, Brennwald P., Longtine M., Bi E., Chan C., Novick P., Boone C., Pringle J. R., Davis T. N., Fields S., Drubin D. G. (2001) A protein interaction map for cell polarity development. J. Cell Biol. 154, 549–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michelot A., Costanzo M., Sarkeshik A., Boone C., Yates J. R., 3rd, Drubin D. G. (2010) Reconstitution and protein composition analysis of endocytic actin patches. Curr. Biol. 20, 1890–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaksonen M., Sun Y., Drubin D. G. (2003) A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487 [DOI] [PubMed] [Google Scholar]
- 53. Liao M. J., Prestegard J. H. (1979) Fusion of phosphatidic acid-phosphatidylcholine mixed lipid vesicles. Biochim. Biophys. Acta 550, 157–173 [DOI] [PubMed] [Google Scholar]
- 54. Koter M., de Kruijff B., van Deenen L. L. (1978) Calcium-induced aggregation and fusion of mixed phosphatidylcholine-phosphatidic acid vesicles as studied by 31P NMR. Biochim. Biophys. Acta 514, 255–263 [DOI] [PubMed] [Google Scholar]
- 55. Blackwood R. A., Smolen J. E., Transue A., Hessler R. J., Harsh D. M., Brower R. C., French S. (1997) Phospholipase D activity facilitates Ca2+-induced aggregation and fusion of complex liposomes. Am. J. Physiol. 272, C1279–C1285 [DOI] [PubMed] [Google Scholar]
- 56. Weigert R., Silletta M. G., Spanò S., Turacchio G., Cericola C., Colanzi A., Senatore S., Mancini R., Polishchuk E. V., Salmona M., Facchiano F., Burger K. N., Mironov A., Luini A., Corda D. (1999) CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature 402, 429–433 [DOI] [PubMed] [Google Scholar]
- 57. Goñi F. M., Alonso A. (1999) Structure and functional properties of diacylglycerols in membranes. Prog. Lipid Res. 38, 1–48 [DOI] [PubMed] [Google Scholar]
- 58. Chernomordik L., Kozlov M. M., Zimmerberg J. (1995) Lipids in biological membrane fusion. J. Membr. Biol. 146, 1–14 [DOI] [PubMed] [Google Scholar]
- 59. Roth M. G. (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 60. Morris A. J. (2007) Regulation of phospholipase D activity, membrane targeting and intracellular trafficking by phosphoinositides. Biochem. Soc. Symp., 247–257 [DOI] [PubMed] [Google Scholar]
- 61. Maissel A., Marom M., Shtutman M., Shahaf G., Livneh E. (2006) PKCeta is localized in the Golgi, ER and nuclear envelope and translocates to the nuclear envelope upon PMA activation and serum-starvation. C1b domain and the pseudosubstrate containing fragment target PKCeta to the Golgi and the nuclear envelope. Cell. Signal. 18, 1127–1139 [DOI] [PubMed] [Google Scholar]
- 62. Lehel C., Oláh Z., Jakab G., Szállási Z., Petrovics G., Harta G., Blumberg P. M., Anderson W. B. (1995) Protein kinase C ϵ subcellular localization domains and proteolytic degradation sites. A model for protein kinase C conformational changes. J. Biol. Chem. 270, 19651–19658 [DOI] [PubMed] [Google Scholar]
- 63. Baron C. L., Malhotra V. (2002) Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295, 325–328 [DOI] [PubMed] [Google Scholar]
- 64. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 65. Thomas B., Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 66. Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rose M. D., Winston F., Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 68. Burnette W. (1981) Western blotting. Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 69. Haid A., Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 70. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 71. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 72. Das A., Li H., Liu T., Bellofatto V. (2006) Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150, 201–210 [DOI] [PubMed] [Google Scholar]
- 73. Jain M. R., Li Q., Liu T., Rinaggio J., Ketkar A., Tournier V., Madura K., Elkabes S., Li H. (2012) Proteomic identification of immunoproteasome accumulation in formalin-fixed rodent spinal cords with experimental autoimmune encephalomyelitis. J. Proteome Res. 11, 1791–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun Y., Carroll S., Kaksonen M., Toshima J. Y., Drubin D. G. (2007) PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J. Cell Biol. 177, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carman G. M., Lin Y.-P. (1991) Phosphatidate phosphatase from yeast. Methods Enzymol. 197, 548–553 [DOI] [PubMed] [Google Scholar]
- 76. Mahuren J. D., Coburn S. P., Slominski A., Wortsman J. (2001) Microassay of phosphate provides a general method for measuring the activity of phosphatases using physiological, nonchromogenic substrates, such as lysophosphatidic acid. Anal. Biochem. 298, 241–245 [DOI] [PubMed] [Google Scholar]
- 77. Han G.-S., Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Robson R. J., Dennis E. A. (1977) The size, shape, and hydration of nonionic surfactant micelles. Triton X-100. J. Phys. Chem. 81, 1075–1078 [Google Scholar]
- 79. Lichtenberg D., Robson R. J., Dennis E. A. (1983) Solubilization of phospholipids by detergents. Structural and kinetic aspects. Biochim. Biophys. Acta 737, 285–304 [DOI] [PubMed] [Google Scholar]
- 80. Dawson R. M. C., Elliott D. C., Elliott W. H., Jones K. M. (1986) Data for Biochemical Research, 3rd Ed., Oxford University Press, Oxford [Google Scholar]
- 81. Carman G. M. (1997) Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim. Biophys. Acta 1348, 45–55 [DOI] [PubMed] [Google Scholar]
- 82. Jamal Z., Martin A., Gomez-Muñoz A., Brindley D. N. (1991) Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J. Biol. Chem. 266, 2988–2996 [PubMed] [Google Scholar]
- 83. Kocsis M. G., Weselake R. J. (1996) Phosphatidate phosphatases of mammals, yeast, and higher plants. Lipids 31, 785–802 [DOI] [PubMed] [Google Scholar]
- 84. Lin Y.-P., Carman G. M. (1990) Kinetic analysis of yeast phosphatidate phosphatase toward Triton X-100/phosphatidate mixed micelles. J. Biol. Chem. 265, 166–170 [PubMed] [Google Scholar]
- 85. Deems R. A., Eaton B. R., Dennis E. A. (1975) Kinetic analysis of phospholipase A2 activity toward mixed micelles and its implications for the study of lipolytic enzymes. J. Biol. Chem. 250, 9013–9020 [PubMed] [Google Scholar]
- 86. Carman G. M., Deems R. A., Dennis E. A. (1995) Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 270, 18711–18714 [DOI] [PubMed] [Google Scholar]
- 87. Hjelmstad R. H., Bell R. M. (1991) Molecular insights into enzymes of membrane bilayer assembly. Biochemistry 30, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 88. Bae-Lee M., Carman G. M. (1990) Regulation of yeast phosphatidylserine synthase and phosphatidylinositol synthase activities by phospholipids in Triton X-100/phospholipid mixed micelles. J. Biol. Chem. 265, 7221–7226 [PubMed] [Google Scholar]
- 89. Nickels J. T., Jr., Buxeda R. J., Carman G. M. (1994) Regulation of phosphatidylinositol 4-kinase from the yeast Saccharomyces cerevisiae by CDP-diacylglycerol. J. Biol. Chem. 269, 11018–11024 [PubMed] [Google Scholar]
- 90. Moritz A., De Graan P. N., Gispen W. H., Wirtz K. W. (1992) Phoshatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J. Biol. Chem. 267, 7207–7210 [PubMed] [Google Scholar]
- 91. Bhat B. G., Wang P., Coleman R. A. (1994) Hepatic monoacylglycerol acyltransferase is regulated by sn-1,2- diacylglycerol and by specific lipids in Triton X-100/phospholipid-mixed micelles. J. Biol. Chem. 269, 13172–13178 [PubMed] [Google Scholar]
- 92. Walsh J. P., Bell R. M. (1986) sn-1,2-Diacylglycerol kinase of Escherichia coli. Mixed micellar analysis of the phospholipid cofactor requirement and divalent cation dependence. J. Biol. Chem. 261, 6239–6247 [PubMed] [Google Scholar]
- 93. Walsh J. P., Bell R. M. (1986) sn-1,2-Diacylglycerol kinase of Escherichia coli. Structural and kinetic analysis of the lipid cofactor dependence. J. Biol. Chem. 261, 15062–15069 [PubMed] [Google Scholar]
- 94. Jones G. A., Carpenter G. (1993) The regulation of phospholipase C-g1 by phosphatidic acid. Assessment of kinetic parameters. J. Biol. Chem. 268, 20845–20850 [PubMed] [Google Scholar]
- 95. Wu W.-I., Carman G. M. (1996) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by phospholipids. Biochemistry 35, 3790–3796 [DOI] [PubMed] [Google Scholar]
- 96. Wu W.-I., Lin Y.-P., Wang E., Merrill A. H., Jr., Carman G. M. (1993) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J. Biol. Chem. 268, 13830–13837 [PubMed] [Google Scholar]
- 97. Mullmann T. J., Siegel M. I., Egan R. W., Billah M. M. (1991) Sphingosine inhibits phosphatidate phosphohydrolase in human neutrophils by a protein kinase C-independent mechanism. J. Biol. Chem. 266, 2013–2016 [PubMed] [Google Scholar]
- 98. Kent C., Carman G. M. (1999) Interactions of pathways for phosphatidylcholine metabolism, CTP synthesis, and secretion through the Golgi apparatus. Trends Biochem. Sci. 24, 146–150 [DOI] [PubMed] [Google Scholar]
- 99. Carman G. M., Han G.-S. (2007) Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim. Biophys. Acta 1771, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lin Y.-P., Carman G. M. (1989) Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264, 8641–8645 [PubMed] [Google Scholar]
- 101. Karanasios E., Han G.-S., Xu Z., Carman G. M., Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Morlock K. R., McLaughlin J. J., Lin Y.-P., Carman G. M. (1991) Phosphatidate phosphatase from Saccharomyces cerevisiae. Isolation of 45-kDa and 104-kDa forms of the enzyme that are differentially regulated by inositol. J. Biol. Chem. 266, 3586–3593 [PubMed] [Google Scholar]
- 103. Asp L., Kartberg F., Fernandez-Rodriguez J., Smedh M., Elsner M., Laporte F., Bárcena M., Jansen K. A., Valentijn J. A., Koster A. J., Bergeron J. J., Nilsson T. (2009) Early stages of Golgi vesicle and tubule formation require diacylglycerol. Mol. Biol. Cell 20, 780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Havriluk T., Lozy F., Siniossoglou S., Carman G. M. (2008) Colorimetric determination of pure Mg2+-dependent phosphatidate phosphatase activity. Anal. Biochem. 373, 392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Carman G. M., Dowhan W. (1979) Phosphatidylserine synthase from Escherichia coli. The role of Triton X-100 in catalysis. J. Biol. Chem. 254, 8391–8397 [PubMed] [Google Scholar]
- 107. Hannun Y. A., Loomis C. R., Bell R. M. (1985) Activation of protein kinase C by Triton X-100 mixed micelles containing diacylglycerol and phosphatidylserine. J. Biol. Chem. 260, 10039–10043 [PubMed] [Google Scholar]
- 108. Warner T. G., Dennis E. A. (1975) Action of the highly purified, membrane-bound enzyme phosphatidylserine decarboxylase from Escherichia coli toward phosphatidylserine in mixed micelles and erythrocyte ghosts in the presence of surfactant. J. Biol. Chem. 250, 8004–8009 [PubMed] [Google Scholar]
- 109. Buxeda R. J., Nickels J. T., Jr., Belunis C. J., Carman G. M. (1991) Phosphatidylinositol 4-kinase from Saccharomyces cerevisiae. Kinetic analysis using Triton X-100/phosphatidylinositol-mixed micelles. J. Biol. Chem. 266, 13859–13865 [PubMed] [Google Scholar]
- 110. Dillon D. A., Chen X., Zeimetz G. M., Wu W.-I., Waggoner D. W., Dewald J., Brindley D. N., Carman G. M. (1997) Mammalian Mg2+-independent phosphatidate phosphatase (PAP2) displays diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 272, 10361–10366 [DOI] [PubMed] [Google Scholar]
- 111. Sun Y., Drubin D. G. (2012) The functions of anionic phospholipids during clathrin-mediated endocytosis site initiation and vesicle formation. J. Cell Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Singer-Krüger B., Nemoto Y., Daniell L., Ferro-Novick S., De Camilli P. (1998) Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J. Cell Sci. 111, 3347–3356 [DOI] [PubMed] [Google Scholar]
- 113. Zanolari B., Friant S., Funato K., Sütterlin C., Stevenson B. J., Riezman H. (2000) Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19, 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wu W.-I., Carman G. M. (1994) Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J. Biol. Chem. 269, 29495–29501 [PubMed] [Google Scholar]
- 115. Talwalkar R. T., Lester R. L. (1973) The response of diphosphoinositide and triphosphoinositide to perturbations of the adenylate energy charge in cells of Saccharomyces cerevisiae. Biochim. Biophys. Acta 306, 412–421 [DOI] [PubMed] [Google Scholar]
- 116. Ozier-Kalogeropoulos O., Fasiolo F., Adeline M.-T., Collin J., Lacroute F. (1991) Cloning, sequencing and characterization of the Saccharomyces cerevisiae URA7 gene encoding CTP synthetase. Mol. Gen. Genet. 231, 7–16 [DOI] [PubMed] [Google Scholar]
- 117. Ostrander D. B., O'Brien D. J., Gorman J. A., Carman G. M. (1998) Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 273, 18992–19001 [DOI] [PubMed] [Google Scholar]
- 118. Brindley D. N., Waggoner D. W. (1998) Mammalian lipid phosphate phosphohydrolases. J. Biol. Chem. 273, 24281–24284 [DOI] [PubMed] [Google Scholar]
- 119. Han G.-S., Siniossoglou S., Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 [DOI] [PubMed] [Google Scholar]
- 120. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Dillon D. A., Wu W.-I., Riedel B., Wissing J. B., Dowhan W., Carman G. M. (1996) The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 271, 30548–30553 [DOI] [PubMed] [Google Scholar]