Background: Renal cilia defects such as cilia disassembly cause polycystic kidney disease.
Results: Decrease of Mxi1 induces renal cilia disassembly via reduction in levels of Ift20 and activates p-ERK.
Conclusion: Inactivation of Mxi1 induces polycystic kidney through renal cilia disassembly.
Significance: This cilia disassembly mechanism induced by Mxi1 inactivation in polycystic kidney is expected to be new model for renal cystogenesis.
Keywords: Ets Family Transcription Factor, Gene Knockout, Gene Regulation, Kidney, Metabolic Regulation, Ets-1, IFT20, Mxi1, PKD, Renal Cilia
Abstract
Cilia in ciliated cells consist of protruding structures that sense mechanical and chemical signals from the extracellular environment. Cilia are assembled with variety molecules via a process known as intraflagellar transport (IFT). What controls the length of cilia in ciliated cells is critical to understand ciliary disease such as autosomal dominant polycystic kidney disease, which involves abnormally short cilia. But this control mechanism is not well understood. Previously, multiple tubular cysts have been observed in the kidneys of max-interacting protein 1 (Mxi1)-deficient mice aged 6 months or more. Here, we clarified the relationship between Mxi1 inactivation and cilia disassembly. Cilia phenotypes were observed in kidneys of Mxi1-deficient mice using scanning electron microscopy to elucidate the effect of Mxi1 on renal cilia phenotype, and cilia disassembly was observed in Mxi1-deficient kidney. In addition, genes related to cilia were validated in vitro and in vivo using quantitative PCR, and Ift20 was selected as a candidate gene in this study. The length of cilium decreased, and p-ERK level induced by a cilia defect increased in kidneys of Mxi1-deficient mice. Ciliogenesis of Mxi1-deficient mouse embryonic fibroblasts (MEFs) decreased, and this abnormality was restored by Mxi1 transfection in Mxi1-deficient MEFs. We confirmed that ciliogenesis and Ift20 expression were regulated by Mxi1 in vitro. We also determined that Mxi1 regulates Ift20 promoter activity via Ets-1 binding to the Ift20 promoter. These results indicate that inactivating Mxi1 induces ciliary defects in polycystic kidney.
Introduction
Most eukaryotic cells contain organelles known as cilia, which are antenna-like projections extending from the cell body. Nonmotile, primary cilia may be sensory cellular organelles that coordinate cellular signaling pathway including cell division and differentiation (1). A single primary cilium emanates from the basal body in kidney cells and exhibits a 9 + 0 (nine sets of microtubule doublets and no singlet microtubule in the center) axoneme microtubule structure (2). The process of intraflagellar transport (IFT)4 is responsible for building and maintaining the structure and function of primary cilia (3). Central to the functional basis of IFT is its ability to traffic various ciliary protein cargo (4). IFT particles are divided into two groups termed complex A and B. Components of IFT complex A function are related to retrograde IFT (Ift144/140/139/122), whereas components of IFT complex B function are related to anterograde IFT (Ift172/88/81/80/74/57/52/46/27/20) (5).
Recent research has demonstrated that cilia play a crucial role regulating vertebrate developmental pathways and tissue homeostasis, and defects in genes involved in primary cilia assembly or function are associated with diseases such as polycystic kidney disease (PKD) (6–10). Autosomal dominant PKD (ADPKD), which is one of the most common human monogenic diseases, is a life-threatening genetic disease in which epithelial-lined cysts develop extensively in the kidneys resulting in renal failure (11–15). PKD is related to abnormal proliferation of renal epithelial cells with ciliary defects. In addition, ADPKD is one of a number of human genetic diseases that are rooted in defects in cilia formation, maintenance, or function (16–18).
Max-interacting protein 1 (Mxi1), which is an antagonist of the c-Myc oncogene, is a transcription factor related to cellular growth and differentiation (19–21). The Mad2 protein encoded by the Mxi1 gene is a basic helix-loop-helix leucine zipper (22). Deletion or rearrangement of the Mxi1 gene causes several types of cancer in humans including prostate tumors, renal cell carcinoma, and endometrial cancers (22, 23). Mice lacking Mxi1 exhibit progressive, multisystem abnormalities including polycystic kidney (24). Previously, we reported multiple cysts in the kidneys of Mxi1-deficient mouse aged 6 months and older (25). Also, the common characteristics of polycystic kidney such as inflammation, fibrosis, and increase of renal cell proliferation were observed in Mxi1-deficient kidney (22, 26). Based on these previous studies, Mxi1-deficient mice are a suitable model to study PKD.
Here, we show that inactivating Mxi1 affects ciliogenesis in vivo and in vitro by down-regulating Ift20. Additionally, the p-ERK level was increased in cilia-defective models by the inactivation of Mxi1. To investigate the relationship between Mxi1 and Ift20, various transcription factors binding to the Ift20 promoter region were screened using the TRANSFAC database. The results confirmed that Ets-1 acts as a mediator involved in the relationship between Mxi1 and Ift20. These results suggest that inactivation of Mxi1 induces renal cilia disassembly through Ift20 reduction in polycystic kidneys.
EXPERIMENTAL PROCEDURES
Sample Preparation
Renal tissue was isolated from 9-month-old Mxi1-deficient mice and age-matched control mice. For histological analysis, tissues were fixed in freshly prepared 4% paraformaldehyde in PBS, dehydrated in graded ethanol, cleared in xylene, and embedded in paraffin. Kidney tissues were taken from sacrificed mice and immediately stored in 2.5% glutaraldehyde for scanning electron microscopy.
Cell Culture
Wild-type and Mxi1-deficient mouse embryonic fibroblasts (MEFs) were isolated from E13.5 mouse embryos. Briefly, embryos were mechanically fragmented and then incubated with trypsin (0.25% in PBS, pH 7.5) at 37 °C for 15–20 min with a magnetic stirrer. After 10 min of centrifugation at 1800 rpm, the pellets were resuspended in Dulbecco's modified Eagle's medium (DMEM) without fetal bovine serum (FBS) and centrifuged for 10 min at 1800 rpm. After three washes, the cell suspension was distributed in a culture dish containing complete DMEM with 10% FBS. Inner medullary collecting duct (mIMCD-3) cells were cultured in DMEM and Ham's F12 medium supplemented with 10% FBS. Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2. To induce ciliogenesis, mIMCD-3 cells were maintained at confluence for 5 days (27). MEFs were maintained at serum starvation for 48 h to induce ciliogenesis (28).
Mxi-1 and Ets-1 Expression Vector
The Mxi1 expression vector, pCMV Tag2B (Stratagene), was constructed by cloning Mxi1 cDNA isolated from mIMCD-3 cells. Mxi1 coding sequence constructs were made by PCR amplification with the following primers: upstream, 5′-GATCGGATCC (BamHI) ATGCCGAGCCCCCGG-3′ and downstream, 5′-TCGACTCGAG (XhoI) CTAGGACGCGAAGGAGG-3′. The Ets-1 expression vector, also pCMV Tag2B, was constructed by cloning Ets-1 cDNA isolated from mIMCD-3 cells. Ets-1 coding sequence constructs were made by PCR amplification with the following primers: upstream, 5′-ATTTGCGGCCGC (NotI) ATGAAGGCGGCCGTCGAT-3′ and downstream, 5′-CGCGGATCCCTA (BamHI) GTCAGCATCCGGCTTTACAT-3′.
Transient Transfection and siRNA Treatment
Transient transfection of Mxi1 and Ets-1 was performed with following materials. Cells were subcultured 1 day before and transfected using FuGENE 6 Transfection Reagent (Roche Applied Science). Cells were transfected with Mxi1 and Ets-1 small interfering RNA (siRNA; Santa Cruz Biotechnology) using Lipofectamine RNAi MAX (Invitrogen), according to the manufacturer's protocol.
Immunofluorescence Microscopy
Cells for immunofluorescence microscopy were grown on acid-washed glass coverslips. The cells were fixed for 10 min in 4% paraformaldehyde in PBS. Five- to 10-μm-thick paraffin sections were used for immunofluorescence microscopy and incubated with primary antibodies overnight at 4 °C. The primary antibodies used included acetylated anti-tubulin (Sigma-Aldrich). The cells were then washed three times and incubated with a 1:1000 dilution of FITC-conjugated anti-mouse IgG (Vector Laboratories) secondary antibody for 1 h at room temperature. The cells were then mounted with Vectashield (Vector Laboratories) and visualized by fluorescence microscopy. Counterstaining was carried out with DAPI.
Reporter Constructs and Luciferase Assay
Mouse genomic DNA was isolated from wild-type mouse embryos using BD Vacutainers. The 5′-flanking region of the mouse Ift20 gene was amplified by PCR using specific primers. The synthetic linkers and restriction sites for XhoI (CCGCTCGAG) and HindIII (GGGAAGCTT) are indicated in uppercase. The primers used were: 0.5 kb forward, 5′-CCGCTCGAGCACCCCTCCTCTTCTGAAAAACT-3′ and 1.0 kb forward, 5′-CCGCTCGAGAGAAACCCAAGTGACTAGGCTTG-3′; the reverse primer for all constructs except for 0.5kb-1.0kb, 5′-GGGAAGCTTAGCACCAGCATCCGCCCACCCTGCGG-3′ and the reverse primer for 0.5kb-1.0kb, 5′-GGGAAGCTTTGGCGGGAGAGCAGCCTACACT-3′. A series of 5′-deletion constructs were cloned into the pGL3-basic vector (Promega). For site-directed mutagenesis, the pGL3-basic/0.5-kb cloned vector was used as a template with the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The mutated region was the same as the Ets-1 mutant probe used in the shift assay. Luciferase plasmids and a pSV-β-galactosidase control vector were cotransfected into mIMCD-3 cells, Mxi1 transiently overexpressed, and Ets-1 transiently overexpressed mIMCD-3 cells with the FuGENE 6 Transfection Reagent. After 2 days, the cells were washed with PBS and harvested in 500 μl of 1× Passive Lysis Buffer (Promega). Supernatants were mixed with luciferase reagent (Promega), and luciferase activity was measured using a luminometer (Aureon Biosystems) as described in the manufacturer's protocol.
Quantitative PCR
Total RNA was extracted using a Nucleospin® Kit (Macherey-Nagel) according to the manufacturer's instructions, and cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's instructions. qPCR was conducted using the qPCR SYBR Green kit (Qiagen). Primer sequences for qPCR are presented in Table 1.
TABLE 1.
Primer sequences
| Primer (qPCR)a | 5′→ 3′ |
|---|---|
| β-Actin (F) | GACGATGCTCCCCGGGCTGTATTC |
| β-Actin (R) | TCTCTTGCTCTGGGCCTCGTCACC |
| Mxi1 (F) | CGGATTCAGAGCGAGAGGAGATT |
| Mxi1 (R) | AGGCTGCTGTGGTCGTCAAGGTC |
| Ift20 (F) | GGATGCTGGTGCTTCTGGACTC |
| Ift20 (R) | GCTCTGCGGCCCTGACGACTGT |
| Ift88 (F) | TGGTCAGCCCGCTCCTCCTC |
| Ift88 (R) | ACCCGTGTCATTCTCCAACTCCTC |
| Kif3α (F) | CCAGCCTCGCCCCCAAACC |
| Kif3α (R) | CCGGCACCTAACCACCACCTTC |
a F, forward; R, reverse.
Western Blotting
Proteins were separated on polyacrylamide gels and transferred to PVDF membranes (Amersham Biosciences). The primary antibodies used were anti-cytoskeletal actin (Bethyl Laboratories), anti-ERK, anti-p-ERK (Cell Signaling Technology), Mxi1 (Santa Cruz Biotechnology), Ets-1 (Novus), and Ift20 (Abcam). Blots were incubated with primary antibodies overnight at 4 °C. Blots were incubated with horseradish peroxidase-conjugated secondary antibody (Upstate Biotechnology, 1:5000) for 1 h at room temperature. Immunoreactive complexes were visualized using ECL reagents (Amersham Biosciences).
Supershift Assay and Isolation of Nuclear Extracts
Nuclear extracts of mIMCD-3 cells were prepared using a Nuclear Extraction Kit (Affymetrix) according to the manufacturer's protocol. The sequences of oligonucleotides used in the supershift assay were: Ets-1 wild-type probe, 5′-TCCTTCTACTTCCGGGTTCGG-3′ and Ets-1 mutant probe, 5′-TCCTTCTACAAAAGGGTTCGG-3′. Probes were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Takara), followed by purification on a Sephadex G-50 column (Roche Applied Science). Nuclear extracts were preincubated with unlabeled probe (competitor) for 90 min at room temperature for the competitive binding assay. Nuclear extracts were preincubated with antibodies against Ets-1 and IgG for 90 min at room temperature for the supershift assay. All antibodies for the supershift assay were purchased from Santa Cruz Biotechnology. Samples were loaded on polyacrylamide gel in 0.5× TBE buffer for 3 h at 180 V. The dried gels were exposed to film and detected with a FLA7000 (Fuji film).
Statistical Analysis
Data were described as mean ± S.D. and analyzed by a paired t test. The paired t test was performed by InStat software (GraphPad). For analysis, p value < 0.05 was considered statistically significant.
RESULTS
Influence of Mxi1 Inactivation on Cilia Assembly
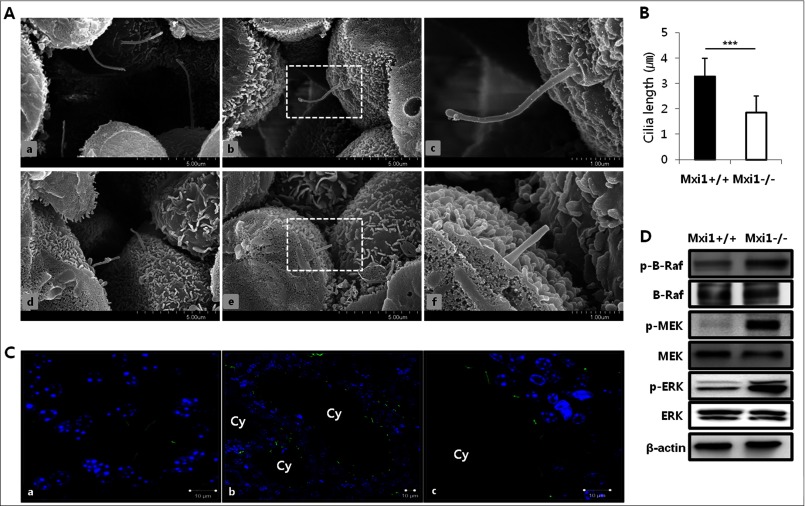
To confirm the effect of Mxi1 deletion on cilia assembly, we evaluated the length of the primary cilia in kidneys of Mxi1-deficient and wild-type mice by scanning electron microscopy (Fig. 1A). Cilia formation was normal in wild-type kidneys (Fig. 1A, a–c), whereas cells with defective and truncated cilia were observed in Mxi1-deficient mice (Fig. 1A, d–f). A comparison of the length of the cilium in Mxi1-deficient kidneys with that of the wild-type kidney (n = 20, 3.284 ± 0.697 μm) revealed that cilia in the Mxi1-deficient kidney (n = 15, 1.84 ± 0.663 μm) were much shorter than those in the wild-type (Fig. 1B). Next, we investigated the phenotype of cilia in cyst-lining cells of the Mxi1-deficient kidney compared with that in the wild-type (Fig. 1C). Results from immunofluorescence using an antibody against acetylated α-tubulin, known as a primary cilia-specific marker, showed that cilia still formed in the cyst-lining cells (Fig. 1C, b and c), but their phenotype was abnormal compared with that in wild-type kidney (Fig. 1C, a). Cilia formed in wild-type kidney were normally protruded to lumen and maintained normal cilia length. However, cilia formed in Mxi1-deficient kidney did not have extended form to lumen with short length. In addition, the B-Raf/MEK/ERK pathway was confirmed in kidney of Mxi1-deficient mice. Because decrease of intracellular calcium level in polycystic kidney with cilia defects induce cAMP level that results in activation of B-Raf/MEK/ERK pathway (29). The p-B-Raf, p-MEK, and p-ERK level increased in the cilia-defective kidney induced by Mxi1 inactivation compared with that in controls (Fig. 1D). These results show that inactivation of Mxi1 induces truncated cilia and activation of B-Raf/MEK/ERK pathway in the kidney.
FIGURE 1.
Identification of primary cilium in kidney of Mxi1-deficient mice. A, representative photographs of scanning electron micrographs of kidneys from wild-type (a–c), aged 9 months, and Mxi1-deficient kidney (d–f) as age-matched controls. B, comparison of the length of cilium in Mxi1-deficient kidney with wild-type kidney. The length of cilium was quantified by measuring from ciliary tip to basal body (basal body is bottom and protruding region of cilia) using the scale bar indicated in the figure. The graph shows mean ± S.D. (error bars) of three independent experiments. The one-tailed p value is <0.0001, considered extremely significant (***). C, representative photographs of anti-acetylated α-tubulin-stained Mxi1-deficient kidney (b and c) and wild-type kidney (a) (anti-acetylated α-tubulin staining (green), DAPI staining (blue), Cy, cyst. Original magnification, ×3000. Scale bars, 10 μm. D, Western blot of proteins related to B-Raf/MEK/ERK pathway in Mxi1-deficient kidney and wild-type. β-Actin was used as a loading control.
Influence of Mxi1 Inactivation on Ciliogenesis in MEFs
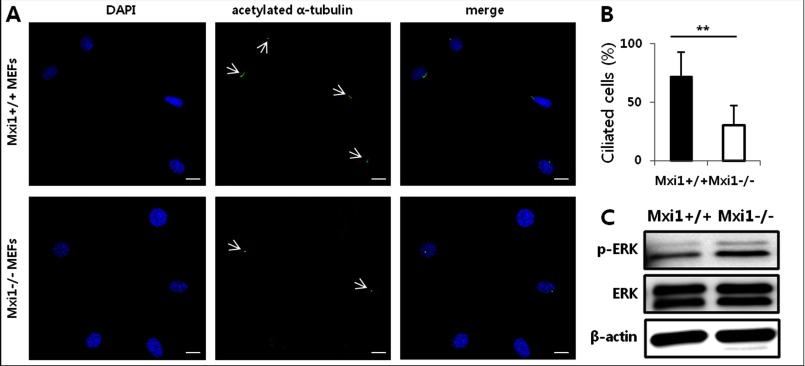
We imaged ciliated cells in Mxi1+/+ and Mxi1−/− MEFs to confirm whether ciliogenesis was affected by the Mxi1 gene in MEFs. In normal cells, the proper ciliogenesis occurred in one cell, so one cilium is localized to one cell. Consistent with this, many Mxi1+/+ MEFs have cilia in each cell (Fig. 2A). However, ciliated cells of Mxi1−/− MEFs was reduced compared with Mxi1+/+ MEFs (Fig. 2A). These results suggest that ciliogenesis was inhibited in Mxi1−/− MEFs compared with those in Mxi1+/+ MEFs. Quantification of ciliogenesis was indicated in Fig. 2B. The percentage of ciliated cells decreased in Mxi1−/− MEFs (n = 11, 30.151 ± 16.674%) compared with those in Mxi1+/+ MEFs (n = 11, 71.969 ± 20.84%). Additionally, the p-ERK level increased in Mxi1−/− MEFs compared with that in Mxi1+/+ MEFs (Fig. 2C). These results indicated that Mxi1 regulates ciliogenesis and the p-ERK level in MEFs.
FIGURE 2.
Effect of Mxi1 on ciliogenesis and p-ERK level in MEFs. A, representative photographs of anti-acetylated α-tubulin-stained Mxi1+/+, Mxi1−/− MEFs. White arrow indicates cilia stained with anti-acetylated α-tubulin. Anti-acetylated α-tubulin staining is shown in green, DAPI staining is blue. Original magnification, ×400. Scale bar, 20 μm. B, quantification of ciliated cells in Mxi1+/+, Mxi1−/− MEFs. To measure ciliated cells, a ratio of the number of cilia to the number of nuclei was calculated and multiplied by 100. The graph shows mean ± S.D. (error bars) of three independent experiments. The one-tailed p value is < 0.001, considered extremely significant (**). C, Western blot of p-ERK and total ERK in Mxi1+/+ and Mxi1−/− MEFs. β-Actin was used as a loading control.
Validation of Gene Expression Related to Ciliogenesis in Mxi1-deficient Kidneys and MEFs
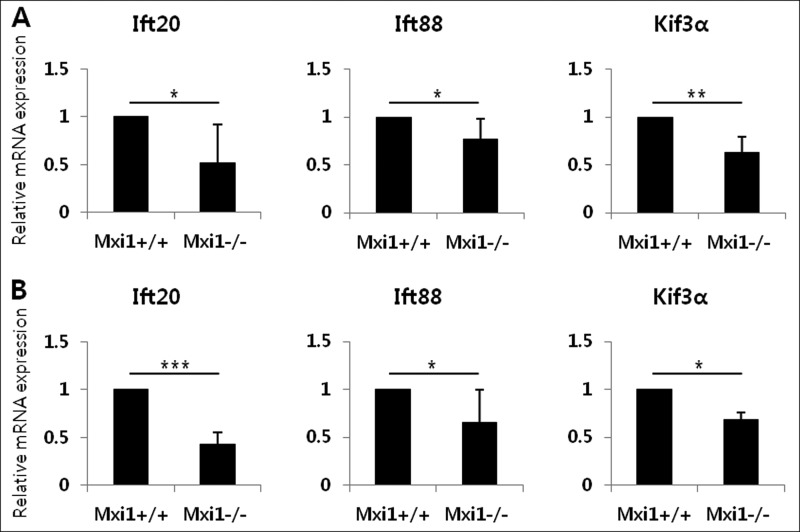
To identify the expression pattern of the IFT family involved in ciliogenesis, we evaluated the expression of Ift20, Ift88, and Kif3α in Mxi1-deficient kidneys using qPCR (Fig. 3A). Ift20 (0.521 ± 0.394), Ift88 (0.771 ± 0.212), and Kif3α (0.633 ± 0.159) were down-regulated in Mxi1-deficient mice with polycystic kidneys. We also confirmed expression of these genes in MEFs (Fig. 3B). Ift20 (0.428 ± 0.125), Ift88 (0.658 ± 0.337), and Kif3α (0.684 ± 0.078) were down-regulated in Mxi1-deficient MEFs compared with that in controls. Among these genes, Ift20 expression decreased significantly in the Mxi1-deficient kidney and MEFs, so Ift20 was selected for our subsequent experiments.
FIGURE 3.
Influence of Mxi1 inactivation on IFT-related gene expression. A, verification of Ift20, Ift88, and Kif3α mRNA expression in Mxi1-deficient mice. The graphs show mean ± S.D. (error bars) of three independent experiments. *, p < 0.05; **, p < 0.01. B, verification of Ift20, Ift88, and Kif3α mRNA expression in Mxi1-deficient MEFs. The graphs show mean ± S.D. of three independent experiments. *, p < 0.05; ***, p < 0.005.
Influence of Mxi1 Transient Overexpression on Ift20 Expression and Ciliogenesis in Mxi1−/− MEFs
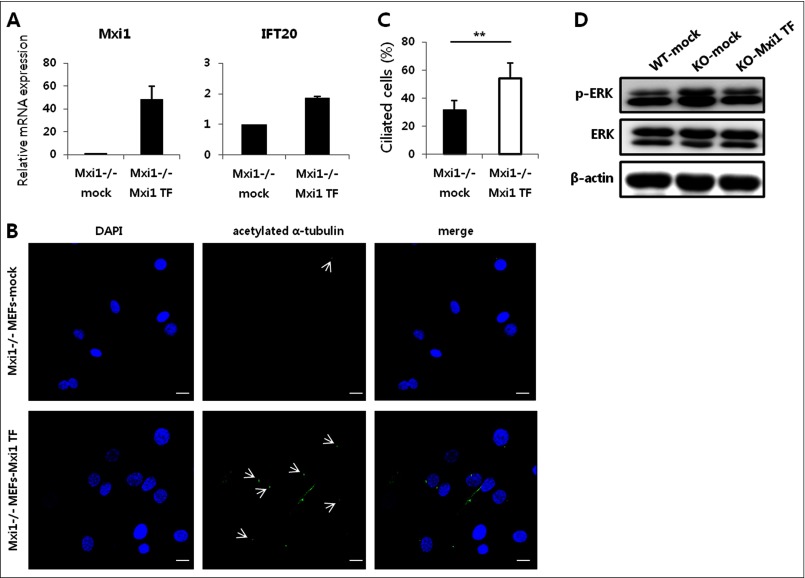
To confirm that Mxi1 regulates Ift20 expression, we compared Ift20 expression in Mxi1 transiently overexpressed Mxi1−/− MEFs and Mxi1−/− MEFs (Fig. 4A). When Mxi1 was transfected into Mxi1−/− MEFs, Mxi1 (48.721 ± 11.301) and Ift20 (1.867 ± 0.056) mRNA levels were increased compared with that in Mxi1−/− MEFs. These results indicate that Mxi1 is a positive regulator of Ift20. When Mxi1 was transfected into Mxi1−/− MEFs, ciliated cells of Mxi1−/− MEFs were increased as indicated by arrows compared with Mxi1−/− MEFs, suggesting that Mxi1 transfection increased ciliogenesis of Mxi1−/− MEFs (Fig. 4B). Quantification of ciliated cells in Mxi1−/− (n = 20, 31.909 ± 6.421%) and Mxi1 transiently overexpressed Mxi1−/− MEFs (n = 13, 54.047 ± 11.16%) is described in Fig. 4C. Additionally, p-ERK level was measured in Mxi1+/+ MEFs, Mxi1−/− MEFs, and Mxi1 transiently overexpressed Mxi1−/− MEFs (Fig. 4D). When Mxi1 was transfected into Mxi1−/− MEFs, p-ERK level was decreased compared with Mxi1−/− MEFs and similar to that in Mxi1+/+ MEFs. These results indicated that Mxi1 overexpression can rescue Mxi1−/− MEFs from the ciliary-defective phenotype, such as abnormal ciliogenesis and the p-ERK level.
FIGURE 4.
Transient overexpression of Mxi1 has an effect on ciliary phenotype of Mxi1−/− MEFs. A, verification of Mxi1 and Ift20 mRNA expression in Mxi1−/− and Mxi1 transiently overexpressed Mxi1−/− MEFs. The graphs show mean ± S.D. (error bars) in triplicate. B, representative photographs of anti-acetylated α-tubulin-stained Mxi1−/− and Mxi1 transiently overexpressed Mxi1−/− MEFs. White arrows indicates cilia stained with anti-acetylated α-tubulin. Original magnification, ×400. Scale bars, 20 μm. C, quantification of ciliated cells in Mxi1−/− and Mxi1 transiently overexpressed Mxi1−/− MEFs. To measure ciliated cells, the ratio of the number of cilia to the number of nuclei was calculated and multiplied by 100. The graph shows mean ± S.D. of three independent experiments. The one-tailed p value is 0.007, considered significant (**). D, Western blot of p-ERK and total ERK in Mxi1+/+, Mxi1−/−, and Mxi1 transiently overexpressed Mxi1−/− MEFs. β-Actin was used as a loading control.
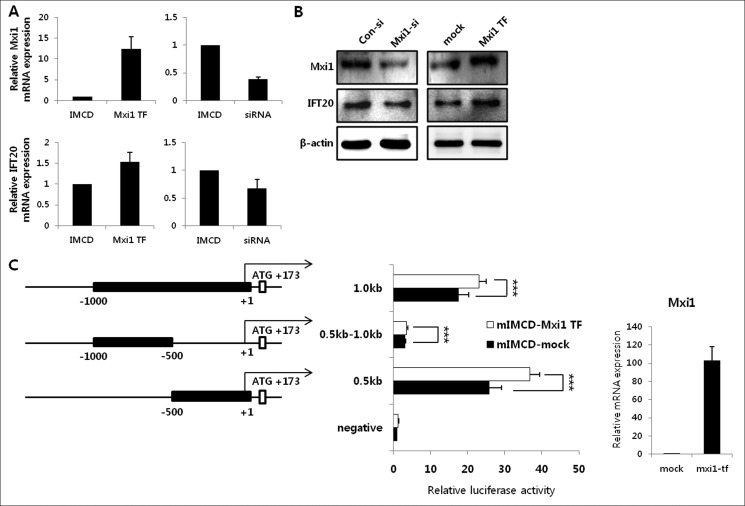
Mxi1 Regulates Ift20 Expression and Its Promoter Activity in mIMCD-3 Cells
Next, we used mIMCD-3 cells to identify the effect of Mxi1 on cilia assembly in renal epithelial cells. We verified the expression of Mxi1 and Ift20 in mIMCD-3 cells, Mxi1 transiently overexpressed mIMCD-3 cells, and Mxi1 siRNA-treated mIMCD-3 cells. Mxi1 mRNA expression was up-regulated in Mxi1 transiently overexpressed mIMCD-3 cells (12.397 ± 2.923) and down-regulated in Mxi1 siRNA-treated mIMCD-3 cells (0.381 ± 0.046) compared with each control. Ift20 mRNA expression was increased in Mxi1 transiently overexpressed mIMCD-3 cells (1.532 ± 0.236) and decreased in Mxi1 siRNA-treated mIMCD-3 cells (0.670 ± 0.170) compared with each control (Fig. 5A). Consistent with this, Ift20 protein level was regulated by Mxi1 protein level (Fig. 5B). Therefore, we found that Ift20 expression was regulated by Mxi1 expression. Next, we confirmed whether Mxi1 regulates Ift20 promoter activity because Mxi1 is a basic helix-loop-helix leucine zipper transcription factor. We performed a transient transfection assay, in which luciferase reporter constructs driven by various lengths of the Ift20 5′-flanking region (−0.5 kb, −1.0 kb, −1.5 kb, and −2.0 kb) in mIMCD-3 cells and Mxi1 transiently overexpressed mIMCD-3 cells (data not shown). Promoter activity of the −0.5-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (6.563 ± 1.198) compared with mIMCD-3 cells (2.935 ± 0.553). Promoter activity of the −1.0-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (5.0223 ± 1.026) compared with mIMCD-3 cells (1.7823 ± 0.303). Promoter activity of the −1.5-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (3.041 ± 0.475) compared with mIMCD-3 cells (0.902 ± 0.103). Promoter activity of the −2.0-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (2.933 ± 0.664) than mIMCD-3 cells (0.903 ± 0.276). This luciferase assay shows that promoter activities with the −0.5 kb, −1.0 kb, −1.5 kb, and −2.0 kb were increased in Mxi1 transiently overexpressed mIMCD-3 cells compared with mIMCD-3 cells. Interestingly, the shortest fragment of −0.5 kb displayed the highest luciferase activity among various fragments, which suggested the presence of positive regulatory elements in the −0.5-kb region of the Ift20 promoter. From these reasons, −0.5-kb fragment of Ift20 promoter was selected for our subsequent experiments. Mxi1 mRNA level of mIMCD-3 cells used in this promoter activity measurement was described in the right part of Fig. 5C as a qPCR graph. To confirm whether the −0.5-kb region was critical for Ift20 promoter regulation related to Mxi1, we designed the +1 to −0.5-kb deleted luciferase reporter (0.5kb-1.0kb) construct and checked activity with −0.5 kb and −1.0 kb (Fig. 5C). As a result, Ift20 promoter activity of the 0.5-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (36.747 ± 2.681) compared with mIMCD-3 (25.773 ± 3.39). Promoter activity of the 1.0-kb construct increased in Mxi1 transiently overexpressed mIMCD-3 cells (23.131 ± 1.94) compared with mIMCD-3 cells (17.583 ± 2.772). However, Ift20 promoter activity in each cell line using 0.5kb-1.0kb was significantly low. These results indicated that Mxi1 may play a positive regulator in the transcriptional regulation of the −0.5-kb region of Ift20 promoter.
FIGURE 5.
Influence of Mxi1 on Ift20 expression and its promoter activity. A, verification of Mxi1 and Ift20 mRNA expression in mIMCD-3 cells (controls), Mxi1 transiently overexpressed mIMCD-3 cells, and Mxi1 siRNA-treated mIMCD-3 cells. The graphs show mean ± S.D. (error bars) in triplicate. B, Western blot of Ift20 and Mxi1 level in Mxi1 siRNA-treated mIMCD-3 cells and Mxi1 transiently overexpressed mIMCD-3 cells compared with that in their respective controls. β-Actin was used as a loading control. C, deletion constructs of the Ift20 5′-flanking region and luciferase activity in mIMCD-3 and Mxi1 transiently overexpressed mIMCD-3 cells. The representative graph shows mean ± S.D. of three independent assays. ***, p < 0.005. The right part of the luciferase assay shows Mxi1 mRNA expression in Mxi1 transiently overexpressed mIMCD-3 cells compared with that in control used in this luciferase assay. The graph shows mean ± S.D. in triplicate.
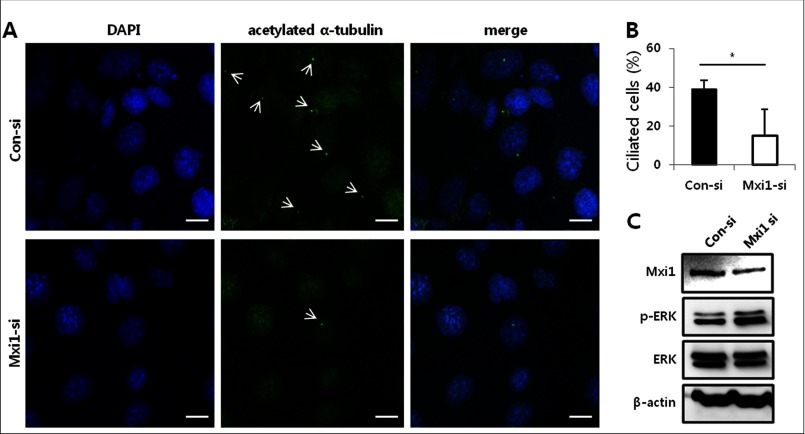
Ciliogenesis Is Regulated by Mxi1 in mIMCD-3 Cells
We imaged ciliated cells in mIMCD-3 and Mxi1 siRNA-treated mIMCD-3 to confirm whether Ift20 regulated by Mxi1 has an effect on mIMCD-3 ciliogenesis. mIMCD-3 cells treated with control siRNA have many ciliated cells, as indicated by arrows in Fig. 6A. However, knockdown of Mxi1 using siRNA reduced the number of ciliated cells (Fig. 6A). Quantification of ciliated cells in Mxi1 siRNA-treated mIMCD-3 (n = 12, 15.050 ± 13.799%) and control siRNA-treated mIMCD-3 cells (n = 12, 39.026 ± 4.541%) is depicted in Fig. 6B. In addition, the p-ERK level increased in Mxi1 siRNA-treated mIMCD-3 compared with that in controls (Fig. 6C). These results show that ciliogenesis defects affected by inactivating Mxi1 induce p-ERK in mIMCD-3 cells.
FIGURE 6.
Effect of Mxi1 on ciliogenesis and p-ERK level in mIMCD-3 cells. A, representative photographs of anti-acetylated α-tubulin-stained mIMCD-3 cells and Mxi1 siRNA-treated mIMCD-3 cells. White arrows indicate cilia stained with anti-acetylated α-tubulin. Original magnification, ×1200. Scale bars, 10 μm. B, quantification of ciliated cells in control siRNA-treated and Mxi1 siRNA-treated mIMCD-3 cells. To measure ciliated cells, a ratio of the number of cilia to the number of nuclei was calculated and multiplied by 100. The graph shows mean ± S.D. (error bars) of three independent experiments. The one-tailed p value is 0.0263, which was considered significant (*). C, Mxi1, p-ERK, and total ERK protein levels in mIMCD-3 and Mxi1 siRNA-treated mIMCD-3 cells. β-Actin was used as the loading control.
Mxi1 Regulates the Ift20 Promoter via Ets-1
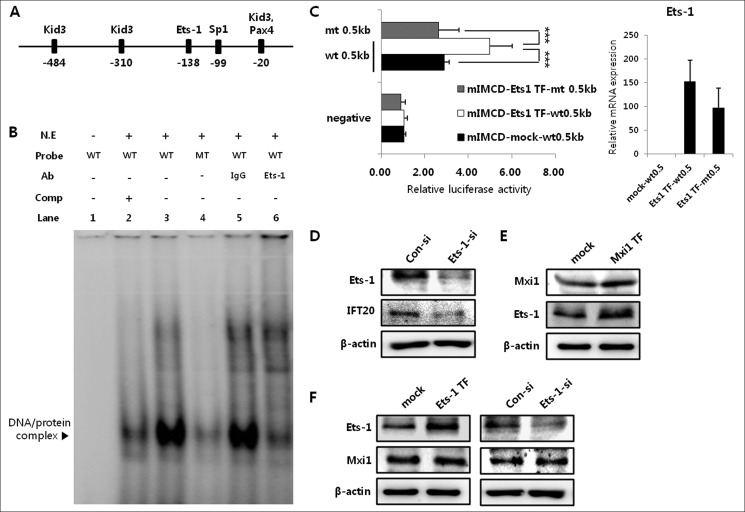
Given the previous demonstration that Mxi1 regulates Ift20 promoter activity, we tested whether Mxi1 had a direct or indirect effect on Ift20 promoter regulation. To identify the transcription factors that bind to the Ift20 promoter, we screened the consensus binding site for known transcription factors using the TRANSFAC database within the +1 to −0.5-kb region. Transcription factors with high scores were selected and described in Fig. 7A. No consensus binding site for Mxi1 was found in the Ift20 promoter region (−0.5 kb), prompting our prediction that Mxi1 does not directly bind but affects Ift20 promoter activity indirectly. We performed an electrophoretic mobility shift assay (EMSA) of the consensus binding sites obtained from the TRANSFAC database (data not shown) to identify candidate proteins because we expected that Mxi1 affects Ift20 promoter activity through a candidate protein that binds to the Ift20 promoter. Among various transcription factors, Ets-1 was selected as a candidate for binding to the Ift20 promoter, −135 to −138. A strong shifted band (DNA-protein complex) was detected in the wild-type probe (lane 3) containing the Ets-1 binding site (Fig. 7B). In addition, the density of this complex decreased dramatically following incubation with the mutant probe (lane 4) containing the Ets-1 binding site mutant sequence. A supershift assay was carried out using the Ets-1 antibody to confirm whether this DNA-protein complex contained the Ets-1 protein (Fig. 7B). The DNA-protein complex band was diminished (lane 6) compared with that in IgG-incubated samples used as a negative control (lane 5). Next, we checked Ift20 promoter activity in Ets-1 transiently overexpressed mIMCD-3 cells to identify whether Ets-1 had an effect on Ift20 promoter regulation. Ets-1 expression level of mIMCD-3 used in this promoter activity measurement is described in the right part of the luciferase assay graph in Fig. 7C. Luciferase activity increased in Ets-1 transiently overexpressed mIMCD-3 cells (4.995 ± 1.017) compared with that in controls (2.9 ± 0.205) (Fig. 7C), and this increased activity decreased in mutant luciferase constructs (2.644 ± 0.919) containing the mutant Ets-1 binding site sequence despite Ets-1 overexpression (Fig. 7C). Consistent with this promoter regulation, the Ift20 protein level decreased in Ets-1 siRNA-treated mIMCD-3 cells (Fig. 7D) compared with that in controls. These results show that Ets-1 binds to −135 to −138 of the Ift20 promoter and regulates Ift20 expression and promoter activity in mIMCD-3 cells.
FIGURE 7.
Ets-1 is a mediator of the regulatory mechanism between Mxi1 and Ift20. A, screening of known transcription factors that bind to the Ift20 promoter using TRANSFAC database. B, lanes 2–4, gel shift assay. Lane 6, supershift assay. Lane 1, negative control for the gel shift assay. Lane 5, negative control for the supershift assay. N.E., nuclear extracts; Comp, competitor. C, luciferase assay with construct of 0.5-kb wt (wild-type) and 0.5-kb mt (mutant) containing mutant sequence for the Ets-1 binding site in mIMCD-3 and Ets-1 transiently overexpressed mIMCD-3. The representative graph shows mean ± S.D. (error bars) of three independent assays. ***, p < 0.005. The right part of the luciferase assay graph indicates Ets-1 mRNA expression in mock (0.5-kb wt), Ets-1 transiently overexpressed (TF) 0.5-kb wt, and Ets-1 transiently overexpressed (TF) 0.5-kb mt transfected mIMCD-3 cells used in this luciferase assay. The graph shows mean ± S.D. in triplicate. D, Western blot of Ets-1 and Ift20 protein levels in mIMCD-3 and Ets-1 siRNA-treated mIMCD-3 cells. β-Actin was used as a loading control. E, Mxi1 and Ets-1 protein levels in Mxi1 transiently overexpressed mIMCD-3 and mIMCD-3 cells. β-Actin was used as a loading control. F, Ets-1 and Mxi1 protein levels in mIMCD-3 cells, Ets-1 transiently overexpressed mIMCD-3 cells, and Ets-1 siRNA-treated mIMCD-3 cells.
From these results, we predicted that the indirect regulatory mechanism of Ift20 promoter involved in Mxi1 occurred through Ets-1, so we identified the relationship between Mxi1 and Ets-1. No studies have been conducted on this relationship, so we tested whether Mxi1 could be an upstream regulator of Ets-1. The Ets-1 level increased in Mxi1 transiently overexpressed mIMCD-3 cells compared with that in controls (Fig. 7E), but the Mxi1 level did not change in Ets-1 transiently overexpressed mIMCD-3 and Ets-1 siRNA-treated mIMCD-3 cells compared with that in each controls (Fig. 7F), suggesting that Mxi1 acted as an upstream regulator of Ets-1. The collective results support the suggestion that Mxi1 regulates Ift20 promoter activity via Ets-1, which binds to the Ift20 promoter at −135 to −138.
DISCUSSION
In this study, we have shown that the nonciliary protein Mxi1 is a regulator of primary cilia assembly. Many studies have been conducted about the regulatory mechanism between ciliary proteins and primary cilia (2, 30–33), but these studies did not discuss the relationship between nonciliary proteins and primary cilia. From this aspect, our study is significant because it provides new evidence for the function of a nonciliary protein. Mxi1-deficient mice have been considered a PKD animal model, but no sufficient disease-inducing mechanisms have been identified. We identify the mechanism in this study.
Defects in renal primary cilia induce polycystic kidneys via abnormal cell proliferation, and aberrant ciliary gene expression has been observed in cystic kidneys. In addition, proteins encoded by Pkd1 and Pkd2 are main targets of polycystic kidney (34), and these proteins localize to renal cilia to regulate intracellular signaling pathways. Many proteins whose functions are disrupted in cystic kidney disease have been localized to the cilium (5), so cilia and ciliary proteins are considered key components of polycystic kidney. The first discoveries of the relationship between primary cilia and polycystic kidney were demonstrated in Oak Ridge Polycystic Kidney (Orpk) mice, which is mutated in the Tg737 gene and is a known ortholog of Chlamydomonas Ift88 (33). Indeed, malformation of primary cilia and abnormalities are observed in Orpk mice. Based on these findings, renal cystic phenotypes are observed in various ciliary gene-targeted mice, so these mice are considered as PKD model. In previous work from our laboratory, we found that inactivating Mxi1 leads to the formation of numerous renal cysts (25). Additionally, inflammation and abnormal cell proliferation accompanied by renal cysts are observed in the Mxi1-deficient kidney (22, 26).
Here, we explain the relationship between Mxi1 inactivation and cilia disassembly in Mxi1-deficient mice, which occurs in renal cysts, because the relevance between cilia defects and ADPKD was recently reported. We observed ciliary defects in Mxi1-deficient kidneys and MEFs, so we assumed that Mxi1 had an effect on renal cilia formation. Furthermore, Ift20, a complex B subunit, was the focus of this study to identify the relationship between the cilia defect and Mxi1 inactivation, because Ift20 expression is reduced in Mxi1-deficient mice and MEFs. Ift20 is required for embryonic viability, and Ift20 deletion in kidney collecting ducts leads to cystic kidney disease (31). Based on this reference, we expected that abnormal Ift20 expression would play a critical role in renal cyst formation through ciliary defects. Various promoter experiments were performed to demonstrate the mechanism related to Mxi1 and Ift20 regulation, and we found that Ets-1, which binds to the Ift20 promoter, is a mediator of this mechanism.
Ets-1 is a member of the Ets family of transcription factors and regulates various cellular signaling pathways related to proliferation, differentiation, and apoptosis (35). Few studies have been conducted about the relationship between Ets-1 and polycystic kidney or renal cilia, but it has been reported that Ets factors regulate the Pkd1 gene promoter (35) localized in renal cilia. We provide the new possibility that Ets-1 is a regulator of the Ift20 promoter in this study. From these findings, we expect that Ets-1 is related to polycystic kidney, but further studies are needed.
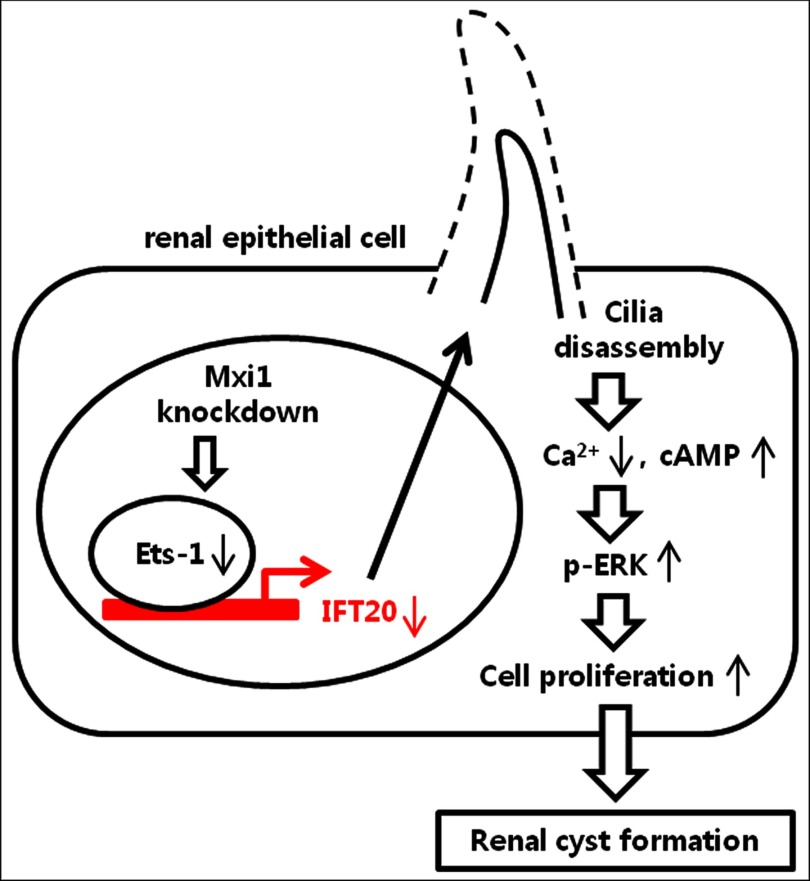
The collective data support a new model of renal cilia formation regulated by Mxi1 (Fig. 8). Our study is meaningful in revealing a new mechanism from three aspects. First, the renal cilia abnormality is caused by inactivation of the Mxi1. Second, Ets-1 acts as an Ift20 regulator and is associated with polycystic kidney. Third, Mxi1 is an upstream regulator of Ets-1. Therefore, we suggest that Mxi1 regulates Ift20 expression via Ets-1, which binds to the Ift20 promoter and has an effect on renal ciliogenesis.
FIGURE 8.
Proposed model illustrating links between inactivation of Mxi1 and renal cyst formation induced by cilia disassembly. Inactivation of Mxi1 decreases the level of Ets-1, which binds to the Ift20 promoter. Down-regulation of Ets-1 reduces Ift20 expression and induces cilia disassembly in renal epithelial cells. This abnormal cilia phenotype increases the p-ERK level and causes renal cyst formation.
This work was supported by Grants 2011-0031220 and 2011-0019452 from the Bio and Medical Technology Development Program of the National Research Foundation of Korea.
- IFT
- intraflagellar transport
- ADPKD
- autosomal dominant PKD
- Ets-1
- E26 avian leukemia oncogene 1
- MEF
- mouse embryonic fibroblast
- mIMCD-3
- mouse inner medullary collecting duct
- Mxi1
- max-interacting protein 1
- PKD
- polycystic kidney disease.
REFERENCES
- 1. Satir P., Christensen S. T. (2008) Structure and function of mammalian cilia. Histochem. Cell Biol. 129, 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuo X., Fogelgren B., Lipschutz J. H. (2011) The small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubular epithelial cells. J. Biol. Chem. 286, 22469–22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Serra R. (2008) Role of intraflagellar transport and primary cilia in skeletal development. Anat. Rec. 291, 1049–1061 [DOI] [PubMed] [Google Scholar]
- 4. Blacque O. E., Cevik S., Kaplan O. I. (2008) Intraflagellar transport: from molecular characterisation to mechanism. Front. Biosci. 13, 2633–2652 [DOI] [PubMed] [Google Scholar]
- 5. Veland I. R., Awan A., Pedersen L. B., Yoder B. K., Christensen S. T. (2009) Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 111, p39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pedersen L. B., Veland I. R., Schrøder J. M., Christensen S. T. (2008) Assembly of primary cilia. Dev. Dyn. 237, 1993–2006 [DOI] [PubMed] [Google Scholar]
- 7. Wagner C. A. (2008) News from the cyst: insights into polycystic kidney disease. J. Nephrol. 21, 14–16 [PubMed] [Google Scholar]
- 8. Brueckner M. (2007) Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation 115, 2793–2795 [DOI] [PubMed] [Google Scholar]
- 9. Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006) The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 10. Kennedy M. P., Omran H., Leigh M. W., Dell S., Morgan L., Molina P. L., Robinson B. V., Minnix S. L., Olbrich H., Severin T., Ahrens P., Lange L., Morillas H. N., Noone P. G., Zariwala M. A., Knowles M. R. (2007) Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 115, 2814–2821 [DOI] [PubMed] [Google Scholar]
- 11. Ong A. C., Harris P. C. (2005) Molecular pathogenesis of ADPKD: the polycystin complex gets complex. Kidney Int. 67, 1234–1247 [DOI] [PubMed] [Google Scholar]
- 12. Al-Bhalal L., Akhtar M. (2005) Molecular basis of autosomal dominant polycystic kidney disease. Adv. Anat. Pathol. 12, 126–133 [DOI] [PubMed] [Google Scholar]
- 13. Reeders S. T. (1992) Multilocus polycystic disease. Nat. Genet. 1, 235–237 [DOI] [PubMed] [Google Scholar]
- 14. Torres V. E., Harris P. C. (2006) Mechanisms of disease: autosomal dominant and recessive polycystic kidney diseases. Nat. Clin. Pract. Nephrol. 2, 40–55 [DOI] [PubMed] [Google Scholar]
- 15. Park E. Y., Woo Y. M., Park J. H. (2011) Polycystic kidney disease and therapeutic approaches. BMB Rep. 44, 359–368 [DOI] [PubMed] [Google Scholar]
- 16. Pazour G. J., Rosenbaum J. L. (2002) Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 12, 551–555 [DOI] [PubMed] [Google Scholar]
- 17. Watnick T., Germino G. (2003) From cilia to cyst. Nat. Genet. 34, 355–356 [DOI] [PubMed] [Google Scholar]
- 18. Pazour G. J. (2004) Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J. Am. Soc. Nephrol. 15, 2528–2536 [DOI] [PubMed] [Google Scholar]
- 19. Kawamata N., Sugimoto K. J., Sakajiri S., Oshimi K., Koeffler H. P. (2005) Mxi1 isoforms are expressed in hematological cell lines and normal bone marrow. Int. J. Oncol. 26, 1369–1375 [PubMed] [Google Scholar]
- 20. Schreiber-Agus N., DePinho R. A. (1998) Repression by the Mad(Mxi1)-Sin3 complex. Bioessays 20, 808–818 [DOI] [PubMed] [Google Scholar]
- 21. Henriksson M., Lüscher B. (1996) Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv. Cancer Res. 68, 109–182 [DOI] [PubMed] [Google Scholar]
- 22. Ko J. Y., Yoo K. H., Lee H. W., Park J. H. (2011) Mxi1 regulates cell proliferation through insulin-like growth factor binding protein-3. Biochem. Biophys. Res. Commun. 415, 36–41 [DOI] [PubMed] [Google Scholar]
- 23. Wang D. Y., Xiang Y. Y., Li X. J., Hashimoto M., Tanaka M., Sugimura H. (2000) Mxi1 is a potential cellular target of carcinogens and frequently mutated in experimental rat tumors and tumor cell lines. Pathol. Int. 50, 373–383 [DOI] [PubMed] [Google Scholar]
- 24. Schreiber-Agus N., Meng Y., Hoang T., Hou H., Jr., Chen K., Greenberg R., Cordon-Cardo C., Lee H. W., DePinho R. A. (1998) Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393, 483–487 [DOI] [PubMed] [Google Scholar]
- 25. Yoo K. H., Sung Y. H., Yang M. H., Jeon J. O., Yook Y. J., Woo Y. M., Lee H. W., Park J. H. (2007) Inactivation of Mxi1 induces Il-8 secretion activation in polycystic kidney. Biochem. Biophys. Res. Commun. 356, 85–90 [DOI] [PubMed] [Google Scholar]
- 26. Yoo K. H., Kim Y. N., Lee M. J., Seong J. K., Park J. H. (2009) Identification of apolipoprotein A1 reduction in the polycystic kidney by proteomics analysis of the Mxi1-deficient mouse. Proteomics 9, 3824–3832 [DOI] [PubMed] [Google Scholar]
- 27. Zaucke F., Boehnlein J. M., Steffens S., Polishchuk R. S., Rampoldi L., Fischer A., Pasch A., Boehm C. W., Baasner A., Attanasio M., Hoppe B., Hopfer H., Beck B. B., Sayer J. A., Hildebrandt F., Wolf M. T. (2010) Uromodulin is expressed in renal primary cilia, and UMOD mutations result in decreased ciliary uromodulin expression. Hum. Mol. Genet. 19, 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., Motro B. (2010) Nek7 kinase targeting leads to early mortality, cytokinesis disturbance, and polyploidy. Oncogene 29, 4046–4057 [DOI] [PubMed] [Google Scholar]
- 29. Cowley B. D., Jr. (2008) Calcium, cyclic AMP, and MAP kinases: dysregulation in polycystic kidney disease. Kidney Int. 73, 251–253 [DOI] [PubMed] [Google Scholar]
- 30. Keady B. T., Samtani R., Tobita K., Tsuchya M., San Agustin J. T., Follit J. A., Jonassen J. A., Subramanian R., Lo C. W., Pazour G. J. (2012) IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell 22, 940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jonassen J. A., San Agustin J., Follit J. A., Pazour G. J. (2008) Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jonassen J. A., SanAgustin J., Baker S. P., Pazour G. J. (2012) Disruption of IFT complex A causes cystic kidneys without mitotic spindle misorientation. J. Am. Soc. Nephrol. 23, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takiar V., Caplan M. J. (2011) Polycystic kidney disease: pathogenesis and potential therapies. Biochim. Biophys. Acta 1812, 1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., Zhou J. (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 35. Puri S., Rodova M., Islam M. R., Magenheimer B. S., Maser R. L., Calvet J. P. (2006) Ets factors regulate the polycystic kidney disease-1 promoter. Biochem. Biophys. Res. Commun. 342, 1005–1013 [DOI] [PubMed] [Google Scholar]