Background: Endogenous functions of the recently identified membrane-associated RING-CH (MARCH) family of transmembrane ubiquitin ligases are undefined, except for MARCH-1.
Results: MARCH-8 interacts with death receptor TRAIL receptor (R) R1, ubiquitinates TRAIL-R1, and down-regulates TRAIL-R1 from the cell surface at steady state.
Conclusion: Steady-state cell surface levels of TRAIL-R1 are controlled by MARCH-8-mediated ubiquitination.
Significance: TRAIL-R1 is the first identified substrate of endogenous MARCH-8.
Keywords: Apoptosis, E3 Ubiquitin Ligase, Receptor Endocytosis, Receptor Modification, Ubiquitination
Abstract
The eleven members of the membrane-associated RING-CH (MARCH) ubiquitin ligase family are relatively unexplored. Upon exogenous (over)expression, a number of these ligases can affect the trafficking of membrane molecules. However, only for MARCH-1 endogenous functions have been demonstrated. For the other endogenous MARCH proteins, no functions or substrates are known. We report here that TRAIL-R1 is a physiological substrate of the endogenous MARCH-8 ligase. Human TRAIL-R1 and R2 play a role in immunosurveillance and are targets for cancer therapy, because they selectively induce apoptosis in tumor cells. We demonstrate that TRAIL-R1 is down-regulated from the cell surface, with great preference over TRAIL-R2, by exogenous expression of MARCH ligases that are implicated in endosomal trafficking, such as MARCH-1 and -8. MARCH-8 attenuated TRAIL-R1 cell surface expression and apoptosis signaling by virtue of its ligase activity. This suggested that ubiquitination of TRAIL-R1 was instrumental in its down-regulation by MARCH-8. Indeed, in cells with endogenous MARCH expression, TRAIL-R1 was ubiquitinated at steady-state, with the conserved membrane-proximal lysine 273 as one of the potential acceptor sites. This residue was also essential for the interaction of TRAIL-R1 with MARCH-1 and MARCH-8 and its down-regulation by these ligases. Gene silencing identified MARCH-8 as the endogenous ligase that ubiquitinates TRAIL-R1 and attenuates its cell surface expression. These findings reveal that endogenous MARCH-8 regulates the steady-state cell surface expression of TRAIL-R1.
Introduction
TNF-related apoptosis inducing ligand (TRAIL)4 is a death ligand expressed on natural killer cells that contributes to immune surveillance against infected and transformed cells (1, 2). TRAIL receptor signaling has also been implicated in suppression of cancer metastasis (3). TRAIL is of interest as an anti-cancer therapeutic, as it can selectively induce apoptosis in cancer cells, leaving normal cells unharmed. Recombinant TRAIL and agonistic antibodies that trigger TRAIL death receptor activity are under clinical evaluation in cancer patients (4). In the human, there are two closely related TRAIL death receptors: TRAIL-R1 and TRAIL-R2, also called DR4 and DR5, that are broadly expressed and are often present on the same cells (5). Death receptors have a cytoplasmic death domain that enables them to induce apoptosis. Upon ligand binding, they recruit and activate inducer caspase-8 and/or 10, which enables apoptotic execution (6, 7). Death receptors can also transduce anti-apoptotic signals, primarily via the NF-κB pathway (8). The subcellular localization of the receptor may determine the nature of the signal, because in certain tumor cells, death receptors TNF-R1 and CD95 activated NF-κB from the cell surface, but caspase-8 from endosomes (9, 10). TRAIL receptors, however, activated caspase-8 from the cell surface (11, 12). Therefore, the amount of TRAIL receptor at the cell surface may determine the strength of the apoptotic response to physiological TRAIL or TRAIL-R targeting therapeutics.
Generally, cell surface receptors are internalized from the plasma membrane at steady-state, as well as after ligand binding, potentially by distinct mechanisms (13). In the endocytic pathway, receptors are sorted and recycle back to the plasma membrane, or traffic to lysosomes for degradation (13). Ubiquitination is an important principle in regulating endocytosis and lysosomal transport of membrane receptors (14). Modification with monoubiquitin or K63-linked polyubiquitin directs receptor sorting by enabling interaction of the receptor with proteins that regulate endosomal trafficking (14).
A recently discovered group of ubiquitin ligases that targets membrane molecules is the MARCH family. These mammalian membrane-associated RING-CH ligases have been identified on the basis of their homology to ligases of murine and human herpesviruses (15–17). The MARCH proteins have an amino-terminal cytoplasmic ligase domain, with characteristically spaced cysteine and histidine residues (C4HC3) (18–20). This typical E2-binding RING finger domain is generally followed by two transmembrane segments and a cytoplasmic carboxyl-terminal region (18–20).
The different MARCH family members reside in different compartments within the cell (18–20). The closely related MARCH-1 and MARCH-8 are located on endosomes and the plasma membrane and are both implicated in regulating cell surface expression of their substrates (15, 16, 21–23). MARCH-1 is the only family member for which genetically deficient mice have been studied. This work has established that endogenous MARCH-1 regulates antigen presentation and T-cell costimulatory functions of dendritic cells by attenuating cell surface expression of its substrates MHC class II and CD86 (22, 24, 25). Although MARCH-1 is mainly expressed in cells of the immune system, expression of MARCH-8 is broader (15, 16). For MARCH-8 and a number of its relatives, ubiquitination substrates have been reported, but these were all identified by exogenous MARCH (over)expression, except for MARCH-1 (18–20). The functions of endogenous MARCH ligases are therefore largely unknown.
Here, we identify TRAIL-R1 as a substrate for ubiquitination by endogenous MARCH-8 in breast cancer cells. TRAIL-R1 was targeted with preference over TRAIL-R2 for down-regulation from the cell surface by various exogenously expressed MARCH ligases. In this way, the MARCH ligases attenuated apoptosis signaling in response to TRAIL. We identified a unique membrane-proximal lysine in the cytoplasmic tail of TRAIL-R1 that is important for its ubiquitination, its interaction with MARCH-1 and -8, and its down-regulation by these ligases. RNA interference identified MARCH-8 as the endogenous ligase that ubiquitinates TRAIL-R1 and attenuates its steady-state cell surface expression. These findings identify MARCH-8 as a regulator of TRAIL-R1 signaling and a potential determinant for tumor cell sensitivity to TRAIL receptor-targeted therapy.
EXPERIMENTAL PROCEDURES
Cells and Reagents
MCF-7Casp-3 breast cancer cells (26), Mel Juso melanoma cells, and retrovirus packaging cells were cultured in Dulbecco's modified Eagle's medium (DMEM). H358 lung cancer cells were cultured in Roswell Park Memorial Institute medium (RPMI). Both media were supplemented with 8% fetal bovine serum and antibiotics. MG132 was from Calbiochem and bafilomycin A1 from Santa Cruz Biotechnology. Soluble recombinant human IZ-TRAIL was kindly provided by Dr. Henning Walczak (Division of Medicine, Imperial College London, United Kingdom). Transferrin (Sigma) was conjugated to FITC by a standard procedure and FITC-transferrin was purified by Sephadex G-25 gel filtration. Mouse monoclonal antibodies used were: biotin-conjugated anti-human TRAIL-R1 mAb DJR1 and anti-human TRAIL-R2 mAb DJR2–4 (eBioscience); HRP-conjugated anti-HA mAb clone HA-7 (Sigma) and HRP-conjugated anti-FLAG mAb M2 (Sigma); anti-Dynamin-1 mAb 41 (BD Biosciences) and anti-Actin MAB1501R (Millipore). Rabbit polyclonal antibodies used were: anti-TRAIL-R1 AB16955 (Millipore), anti-mRFP and anti-GFP (made in house) (27) and anti-active Caspase-3 (BD Biosciences). Allophycocyanine-conjugated streptavidin was from BD Biosciences and HRP-conjugated swine anti-rabbit Ig was from DAKO A/S. Secondary polyclonal antibodies conjugated with Alexa Fluor 568 and Alexa Fluor 647 were from Molecular Probes. Goat anti-rabbit IgG or goat anti-mouse IgG, conjugated with IRDye 682 or -800 were from LI-COR (Lincoln NE).
Constructs
The TRAIL-R1.mRFP fusion was created by PCR/restriction enzyme-based cloning of TRAIL-R1 cDNA from pcDNA3-TRAIL-R1 (kindly provided by Dr. H. Walczak) into pmRFP-N1 (28), resulting in carboxyl-terminal mRFP tagging of the type I TRAIL receptor. Point and truncation mutants of TRAIL-R1.mRFP were generated by PCR-based mutagenesis. The TRAIL-R1.mRFP cDNA was subcloned from pmRFP-N1 into the retroviral vector pMXIRESBlasticidin to allow for stable expression by gene transduction in MCF-7Casp-3 cells. Plasmids pUHD10–1-MARCH-1, pUHD10–1-MARCH-2, pUHD10–1-MARCH-8 (kindly provided by Dr. Klaus Früh, Vaccine and Gene Therapy Institute, Oregon Health and Science University, Beaverton OR), and pOTB7-MARCH-4 and pOTB7-MARCH-9 (Geneservice) served as PCR templates for MARCH cDNAs, which were cloned into pEGFP-N1 (Clontech) to create MARCH.GFP fusions. MARCH-1.HA and MARCH-8.HA cDNA was generated by subcloning MARCH cDNAs into pHAN1, which is peGFPN1 (Clontech) in which the eGFP coding sequence is replaced by a double HA tag with the sequence YPYDVPDYA (kindly provided by L. Janssen, The Netherlands Cancer Institute, Amsterdam). The pcDNA3.1-Dynamin-1 WT and K44A plasmids were derived from the pMT2-Dynamin-1 WT and K44A plasmids (29). Plasmid pcDNA3.1 encoding FLAG-tagged human ubiquitin was a gift from Dr. Simon Cook (Babraham Institute, Cambridge, UK). Point and truncation mutants of TRAIL-R1 and MARCH-8 were generated by PCR-based mutagenesis. The pLKO.1 empty plasmid or plasmids containing shRNA hairpins targeting MARCH-8 (CCTCCTTCTCTCGCACTTCTA, nucleotide 194–214), or MARCH-1 mRNA (1b GAGAAGAACTTCTCATGTAAT, nucleotide 745–765; 1c GTACAGTGTAAAGTCTATGTT, nucleotide 649–669) were purified from bacterial glycerol stocks (Open Biosystems, Thermo Scientific). All vector construction was done using standard cloning and PCR techniques. All constructs were verified by dideoxynucleotide sequencing.
Transfection and Retroviral Transduction
Cells were transfected with FuGENE 6 according to the manufacturer's instructions (Roche Applied Science). Cells were used for assays at 24 h after transfection. When shRNAs were transfected, cells were used 48–72 h after transfection. Retroviral transduction was done as described (30). In brief, for production of amphotropic retrovirus carrying pMXIRESBlasticidin TRAIL-R1.mRFP WT or -K/A constructs were transfected into the HT1080-derived packaging cell line FLY. MCF-7Casp-3 cells were transduced with freshly harvested virus-containing FLY cell supernatant and cells were selected after 3 days with 10 μg/ml of Blasticidin (Sigma).
Apoptosis Assay and Flow Cytometry
For apoptosis induction, cells were stimulated with the indicated concentrations of recombinant soluble TRAIL in culture medium for the indicated periods of time at 37 °C in 5% CO2. Active caspase-3 content was determined by flow cytometry as described (26). Cells were stained with propidium iodide (1 μg/ml) in PBS with BSA for 5 min at room temperature to detect membrane-permeable dead cells. For flow cytometric detection of endogenous TRAIL receptors, cells were harvested in PBS/EDTA followed by staining with biotinylated anti-TRAIL-R1 mAb DJR1 and anti-TRAIL-R2 mAb DJR2–4 (1:250 and 1:500) combined with allophycocyanine-conjugated streptavidin. The mean fluorescence intensity (MFI) obtained upon staining with secondary reagent only was subtracted from the MFI obtained after TRAIL receptor staining to yield the TRAIL receptor MFI expressed in the figures. Samples were gated on live cells. Data were analyzed using FCS Express (De Novo Software, Thornhill, Canada) or FlowJo (Tree Star, Ashland, OR).
Western Blotting and Immunoprecipitation
Cells were harvested and lysed in Nonidet P-40 buffer consisting of 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 150 mm NaCl, 1 mm PMSF and Complete Protease Inhibitors (Roche). For the experiment depicted in Fig. 4C, cells were lysed in 50 mm Tris-HCl, pH 8.0, 1% SDS, 10 mm DTT, 0.5 mm EDTA, for 10 min at 95 °C. The SDS was quenched by addition of 9 volumes of Nonidet P-40 buffer. Cell lysates were clarified by centrifugation for 10 min at 13,000 × g and protein content was measured by Bio-Rad protein assay. Immunoprecipitation was performed with antibody to mRFP, followed by Protein G-Sepharose beads (GE Healthcare). Immunoprecipitates were washed, resuspended in reducing NuPAGE sample buffer (with 0.1 m DTT), and heated for 10 min at 95 °C. SDS-PAGE was done on pre-cast 4–12% NuPAGE minigels, according to the manufacturer's protocol (Invitrogen). Total cell lysate (taken prior to immunoprecipitation) was run at 30 μg of protein per lane, as determined by Bio-Rad protein assay. Proteins were transferred to nitrocellulose membranes by wet blotting for 90 min at 70 V. Membranes were blocked for 1 h at room temperature with 5% (w/v) skim milk (Oxoid) in Tris-buffered saline (TBS). Antibody probing was performed in TBS with 1% (w/v) skim milk and 0.05% (v/v) Tween 20. For detection by ECL (Pierce Biotechnology), blots were incubated with HRP-conjugated anti-HA or anti-FLAG mAb, or with rabbit anti-mRFP followed by HRP-conjugated swine anti-rabbit Ig. Alternatively, blots were incubated with unconjugated primary antibody, followed by IRDye-conjugated second step antibody and proteins were detected on the Odyssey infrared imager (LI-COR). Quantification of signals was done using ImageLab software (Bio-Rad) or Odyssey software (LI-COR), respectively.
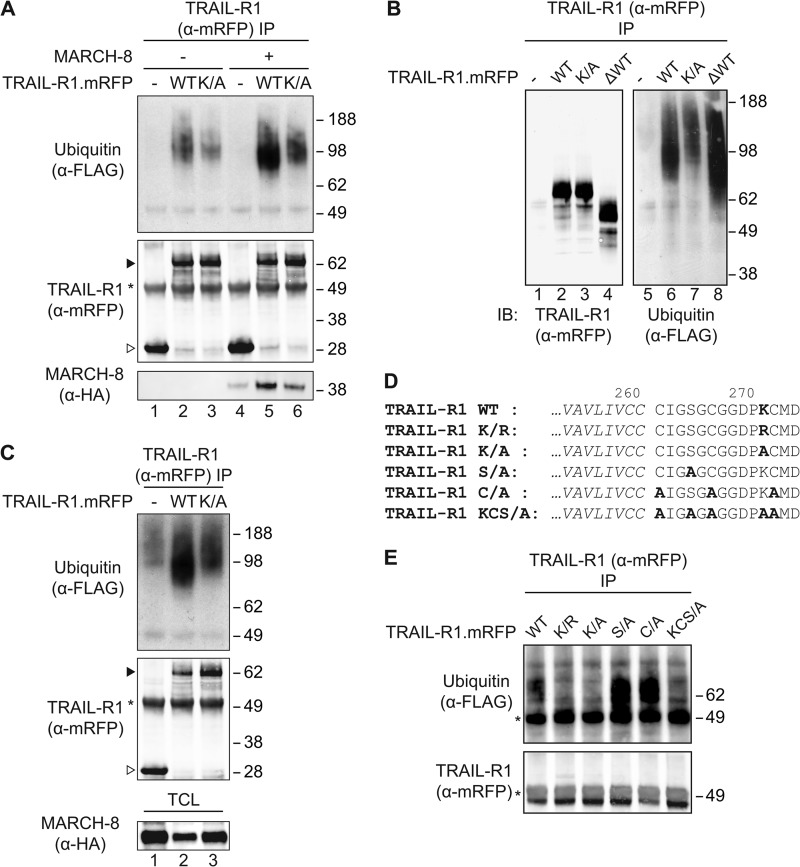
FIGURE 4.
Steady-state ubiquitination of TRAIL-R1 on lysine residue 273 by an endogenous machinery. A, MCF-7Casp-3 cells were transfected to express FLAG-ubiquitin, together with mRFP only (−), or with mRFP-chimeras of WT TRAIL-R1 (WT) or its K273A lysine mutant (K/A). MARCH-8.HA cDNA (+) or an empty control vector (−) were additionally transfected as indicated. Cells were lysed in Nonidet P-40 buffer, TRAIL-R1 was isolated with anti(α)-mRFP antibody and immunoprecipitates (IP) were analyzed by immunoblotting (IB) with α-mRFP antibody to detect TRAIL-R1, α-FLAG antibody to detect ubiquitin and α-HA antibody to detect MARCH-8. Asterisk denotes the heavy chain of the antibody used for IP. Solid and open arrowheads indicate, respectively, TRAIL-R1.mRFP and mRFP only. Blot is representative of 4 independent experiments. B, MCF-7Casp-3 cells were transfected to express FLAG-ubiquitin, together with either mRFP only (−), with mRFP chimeras of WT TRAIL-R1 (WT) or the K273A TRAIL-R1 mutant (K/A), or with a truncated TRAIL-R1 lacking the C-terminal 116 residues (ΔWT). TRAIL-R1 was isolated with α-mRFP antibody and immunoprecipitates were analyzed by immunoblotting with α-mRFP antibody to detect TRAIL-R1 and with α-FLAG antibody to detect ubiquitin. Data shown are representative of two independent experiments. C, MCF-7Casp-3 cells were transfected to express FLAG-ubiquitin, together with mRFP only (−), or with mRFP-chimeras of WT TRAIL-R1 (WT) or its K273A lysine mutant (K/A). Cells were lysed by boiling in SDS, Nonidet P-40 buffer was added in excess and immunoprecipitation of TRAIL-R1 and analysis were performed as outlined for panel A. Asterisk denotes the heavy chain of the antibody used for IP. Solid and open arrowheads indicate, respectively, TRAIL-R1.mRFP and mRFP only. The blot is representative of 2 independent experiments. D, alignment of primary amino acid sequence of part of the transmembrane segment (italic) and the remaining 14 residues of the cytoplasmic tail of the truncated TRAIL-R1 mutants used in E. Relevant potential ubiquitination sites are shown in bold. E, MCF-7Casp-3 cells were transfected to express FLAG-ubiquitin, together with mRFP-tagged TRAIL-R1 WT or mutants shown in D. TRAIL-R1 was isolated with α-mRFP antibody and immunoprecipitates were analyzed by immunoblotting with α-mRFP antibody to detect TRAIL-R1 and α-FLAG antibody to detect ubiquitin. The blot is representative of 2 independent experiments. Asterisk denotes the heavy chain of the antibody used for IP.
RT-PCR
RNA was isolated according to the manufacturer's protocol (RNeasy mini kit; QiaGen). Copy-DNA (cDNA) was generated from the RNA using SuperScript II RT (Invitrogen). Quantitative RT-PCR was performed using FAST SYBR Green master mix (Applied Biosystems).
Statistics
Statistical analyses were performed using GraphPad Prism version 4 for Windows (Graph Pad Software). The tests employed and the criteria for significance are indicated in the figure legends.
Confocal Laser Scanning Microscopy
See supplemental Methods.
RESULTS
MARCH Family Ligases Down-regulate Cell Surface Levels of TRAIL-R1 at Steady-state
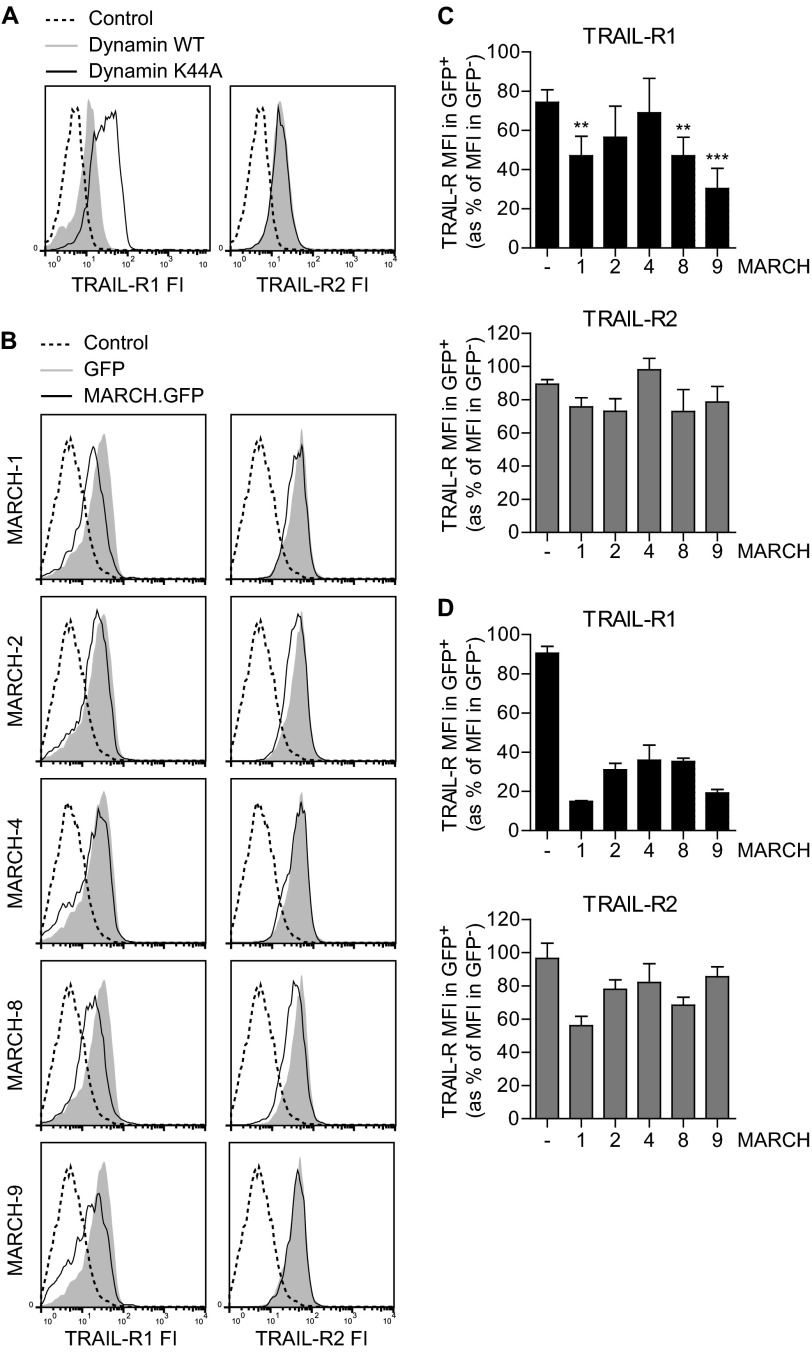
To study how cell surface expression of TRAIL receptors is regulated, we blocked receptor internalization in MCF-7 breast carcinoma cells, engineered to stably express caspase-3 (MCF-7Casp-3). These cells have endogenous TRAIL-R1 and -R2 and effectively undergo apoptosis upon TRAIL treatment (26). Receptor internalization was blocked by dominant-negative dynamin-1 (K44A), which inhibits endosome formation (14, 31). K44A dynamin-1 inhibited transferrin uptake (Fig. S1), confirming the inhibitory effect of this mutant on receptor endocytosis.
Interestingly, this experiment revealed a differential impact of K44A dynamin-1 on the steady-state cell surface expression of TRAIL-R1 versus TRAIL-R2. In cells that expressed high levels of dynamin, as revealed by high GFP expression, the K44A mutant specifically up-regulated cell surface expression of TRAIL-R1, whereas it did not affect TRAIL-R2 expression (Fig. 1A). This indicated that, at steady-state, TRAIL-R1 has a higher turnover by dynamin-dependent endocytosis than TRAIL-R2.
FIGURE 1.
MARCH proteins preferentially down-regulate TRAIL-R1 cell surface levels in MCF-7Casp-3 cells. A, MCF-7Casp-3 breast cancer cells were transfected to express WT or K44A dynamin together with GFP. To detect endogenous TRAIL-R1 or TRAIL-R2 at the cell surface, the cells were stained with specific antibodies and a second step reagent, or with second step reagent only (Control) followed by flow cytometric analysis. Histograms of TRAIL-R fluorescence intensity (FI) in the GFP positive populations are shown. B and C, MCF-7Casp-3 cells were transfected to express GFP-tagged MARCH-1, -2, -4, -8, -9, or GFP only (Control) and cell surface levels of TRAIL-R1 (left) and TRAIL-R2 (right) were determined by flow cytometry. Representative histograms of TRAIL-R intensity in the GFP positive populations are shown in B. Panel C shows the quantification of TRAIL-R1 and -R2 expression in MCF-7Casp-3 cells expressing the indicated MARCH-GFP proteins or GFP only (−). The MFI, denoting TRAIL-R cell surface levels in GFP+ cells expressing MARCH-GFP or GFP only is expressed as percentage of the MFI in untransfected GFP− cells in the same cell population. Data represent mean ± S.D. of values from at least 3 independent experiments. Asterisks indicate statistically significant differences between MARCH-transfected and GFP-transfected control cells (one-way analysis of variance, Bonferroni correction; *, p < 0.05; **, p < 0.01; ***, p < 0.001). D, this experiment was performed and quantified as outlined in B and C, but in this case using the melanoma cell line Mel JuSo. Data represent mean ± S.D. of values from 2 independent experiments.
Because certain MARCH ligases were previously shown to down-regulate various molecules, including CD95 (15), from the cell surface upon exogenous expression, we hypothesized that MARCH ligases might also regulate TRAIL receptor plasma membrane levels. To test this, we focused on MARCH-1, -2, -4, -8 and -9, because these locate to the plasma membrane and/or endosomal compartments and were already implicated in modulation of cell surface expression of several molecules (18–20). The MARCH proteins were transiently expressed as GFP-chimeras in MCF-7Casp-3 cells and their impact on endogenous TRAIL-R1 and TRAIL-R2 cell surface levels was determined by flow cytometry. Several MARCH proteins reduced the cell surface level of TRAIL-R1, as revealed by reduced TRAIL-R1 fluorescence intensity of MARCH-transfected cells compared with control vector (GFP)-transfected cells (Fig. 1B). Quantification of the results from multiple independent experiments showed that in MCF-7Casp-3 cells, expression of MARCH-1, -8, and -9 significantly reduced TRAIL-R1 cell surface levels (Fig. 1C). TRAIL-R2 cell surface levels, on the other hand, were only slightly affected by the MARCH proteins in MCF-7Casp-3 cells and the down-regulation was not statistically significant (Fig. 1, B and C). In the same cells, CD95 was most prominently down-regulated by MARCH-1 and MARCH-8 (supplemental Fig. S2), confirming published data (15). In Mel JuSo melanoma cells, exogenous expression of the different MARCH proteins also resulted in a more prominent down-regulation of TRAIL-R1 than TRAIL-R2 cell surface expression (Fig. 1D) and similar results were obtained in HeLa cervix carcinoma cells (data not shown). We conclude that MARCH family members preferentially target TRAIL-R1 over TRAIL-R2 and down-regulate its cell surface expression at steady-state.
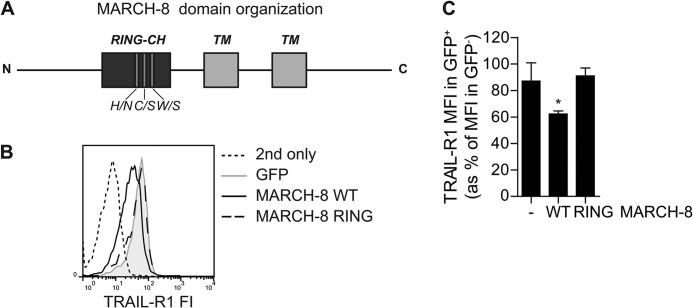
Down-regulating TRAIL-R1 Cell Surface Expression Requires Ligase-competent MARCH-8
As MARCH proteins are ubiquitin ligases, we hypothesized that they down-regulate TRAIL-R1 cell surface expression by virtue of their ligase activity. To test this, we created a ligase-dead MARCH-8 variant (MARCH-8 RING) by mutating three conserved residues in its RING-CH domain (H107N, C110S, W114S; Fig. 2A). The impact of WT MARCH-8 versus the MARCH-8 RING mutant on TRAIL-R1 cell surface levels in MCF-7Casp-3 cells was analyzed by flow cytometry, as outlined above for Fig. 1. A representative histogram is shown in Fig. 2B. Quantification of multiple experiments demonstrated that in contrast to WT MARCH-8, the MARCH-8 RING mutant did not down-regulate TRAIL-R1 cell surface levels (Fig. 2C). Thus, MARCH-8 requires an intact RING domain to reduce TRAIL-R1 cell surface expression. This indicates that the ligase activity of MARCH-8 is essential for TRAIL-R1 down-regulation.
FIGURE 2.
MARCH-8 requires a functional RING domain to down-regulate TRAIL-R1 cell surface expression. MCF-7Casp-3 cells were transfected to express GFP only (−), or GFP-tagged WT MARCH-8, or a MARCH-8 variant carrying ligase-inactivating mutations in its RING domain (MARCH-8 RING). Cell surface levels of TRAIL-R1 were determined by antibody staining, followed by flow cytometric analysis. A, schematic depiction of MARCH-8, on a relative scale, with indication of the RING-CH domain and transmembrane (TM) segments, as well as the three point mutations. B, primary data from a representative experiment, showing histograms of TRAIL-R1 cell surface expression (fluorescence intensity, FI) in GFP+ (MARCH-transfected) cells. C, TRAIL-R1 cell surface expression was quantified and statistically analyzed as described in the legend to Fig. 1C. Data represent mean ± S.D. of values from 3 independent experiments. Asterisk indicates statistically significant differences between cells with WT MARCH-8 versus control or the RING mutant (one-way analysis of variance, Bonferroni correction; *, p < 0.05).
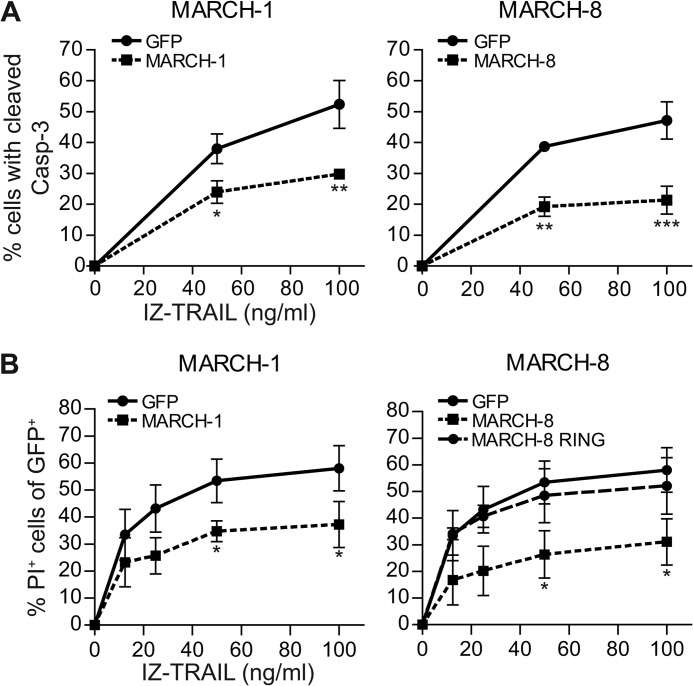
Overexpression of MARCH-1 or -8 Confers Resistance to TRAIL-induced Apoptosis
To evaluate the possible implications of our findings for tumor therapy with TRAIL receptor agonists, we tested whether MARCH ligases altered the sensitivity of MCF-7Casp-3 cells to TRAIL-induced apoptosis. We focused on MARCH-1 and -8, because they are closely related (20), they are most clearly implicated in the endocytic trafficking of membrane proteins and consistently affected TRAIL-R1 cell surface expression in MCF-7Casp-3 and Mel JuSo cells. The cells were transiently transfected to express GFP-tagged MARCH-1 or MARCH-8, or GFP alone (control) and treated with soluble recombinant TRAIL, at different doses. Apoptosis signaling was read out at 5 h after TRAIL treatment, by flow cytometric detection of cleaved caspase-3 in the GFP-positive (MARCH- or control GFP expressing) cell population. In addition, cell death was read out by staining with propidium iodide at 14 h after TRAIL treatment, followed by flow cytometry. Both MARCH-1 and MARCH-8 expressing cells were significantly less sensitive to TRAIL-induced apoptosis than the control GFP expressing cells (Fig. 3, A and B). Data depicted here were obtained with isoleucine-zippered (IZ) TRAIL (26, 32), but similar results were obtained with FLAG-tagged TRAIL (supplemental Fig. S3). Expression of the MARCH-8 RING mutant did not confer resistance to TRAIL-induced cell death (Fig. 3B). These data indicate that MARCH ligase activity can determine the sensitivity of tumor cells to TRAIL-induced apoptosis.
FIGURE 3.
MARCH overexpression inhibits apoptosis induction by TRAIL. A, MCF-7Casp-3 cells were transfected to express either GFP only, or GFP-tagged MARCH-1, or -8. At 24 h after transfection, cells were stimulated with IZ-TRAIL for 5 h and caspase-3 cleavage was determined by flow cytometry in the GFP positive cell populations. B, as in A, with the following adaptations. Cells were transfected with WT MARCH-1 or -8, or with the MARCH-8 RING mutant described in the legend to Fig. 2. Cells were stimulated with TRAIL for 14 h, and cell death was read out by propidium iodide (PI) uptake. Data in A and B represent mean ± S.D. of values from 3 to 4 independent experiments. The percentage of cells with cleaved caspase-3 or PI uptake in the untreated control samples was subtracted. Asterisks indicate statistically significant differences between MARCH.GFP-transfected and GFP only-transfected control cells at the indicated concentration of TRAIL (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
At Steady-state TRAIL-R1 Is a Substrate for MARCH-8-mediated Ubiquitination
The fact that MARCH ligase activity could regulate TRAIL-R1 cell surface expression, suggested that TRAIL-R1 might be a target for ubiquitination. To examine this, a WT TRAIL-R1.mRFP chimera was expressed in MCF-7Casp-3 cells, together with FLAG-tagged ubiquitin. Upon immunoprecipitation (IP) with anti-mRFP antibody from Nonidet P-40 lysates, a distinct FLAG-reactive smear was revealed in association with WT TRAIL-R1.mRFP, but not with mRFP alone (−) (Fig. 4A, lanes 1 and 2). This suggests that ubiquitin is associated with TRAIL-R1 at steady-state.
Conceptually, the ubiquitin isolated with TRAIL-R1 could be appended to a TRAIL-R1 interacting protein rather than to TRAIL-R1 itself. To test whether ubiquitin was directly linked to the receptor, we expressed a TRAIL-R1 truncation mutant lacking the C-terminal death domain (TRAIL-R1 ΔWT). TRAIL-R1 WT and ΔWT were compared side by side for their migration pattern (Fig. 4B, lanes 2 and 4). TRAIL-R1 ΔWT migrated faster than TRAIL-R1 WT, as expected. Importantly, this was paralleled by a similar change in migration pattern of the co-isolated ubiquitin (Fig. 4B, lanes 6 and 8), indicating that ubiquitin was appended to TRAIL-R1 itself. Furthermore, ubiquitin was still present in TRAIL-R1 isolate obtained from cell lysate that had been denatured by boiling in SDS, prior to immunoprecipitation of TRAIL-R1 (Fig. 4C, lanes 1 and 2). Collectively, these data indicate that TRAIL-R1 itself is a substrate for steady-state ubiquitination by a endogenous machinery. Upon overexpression of MARCH-8, ubiquitination of TRAIL-R1 was increased (Fig. 4A, lanes 2 and 5), suggesting that MARCH-8 ubiquitinates TRAIL-R1.
Lysine 273 Is Important for Ubiquitination of TRAIL-R1
Next, we aimed to identify the potential ubiquitin acceptor site(s) in TRAIL-R1. The ligase domain of mammalian and viral MARCH proteins approximates its target close to the cytoplasmic face of the membrane (15, 20). TRAIL-R1 has a single membrane-proximal lysine residue (Lys-273) in the cytoplasmic region preceding the death domain (Fig. 4D and supplemental Fig. S5A). Notably, this lysine residue is highly conserved among primates, which like humans, and unlike other species have two TRAIL receptors (supplemental Fig. S5B). To study the possible involvement of Lys-273 in TRAIL-R1 ubiquitination, this amino acid was mutated to an alanine residue and steady-state ubiquitination of the TRAIL-R1 K273A mutant in MCF-7Casp-3 cells was examined. Comparable amounts of WT and K273A mutant TRAIL-R1 were isolated, but less ubiquitin was detected in association with the mutant receptor than with the WT receptor (Fig. 4A, lanes 2 and 3). The total cellular expression levels of TRAIL-R1 WT and K273A and ubiquitin are depicted in supplemental Fig. S4. Similar results were obtained when TRAIL-R1 WT and K273A were isolated from denatured cell lysates (Fig. 4C, lanes 2 and 3). These results indicate that Lys-273 is either the acceptor site of ubiquitin or important for interaction with the responsible ligase.
Upon mutation of Lys-273 in TRAIL-R1, ubiquitination of the receptor was reduced but not completely lost (Fig. 4, A and B). Moreover, upon overexpression of MARCH-8, ubiquitination of both TRAIL-R1 WT and K273A was increased (Fig. 4A). These data indicate that Lys-273 is a potential ubiquitin acceptor site in TRAIL-R1, but not the only one.
It has been shown that viral MARCH family members can target nonlysine residues for ubiquitination, specifically cysteine, threonine, and serine (33, 34). Ubiquitination on cysteine, threonine, and/or serine can also occur by an endogenous mechanism in mammalian cells (35), but the ligase(s) involved have not yet been identified. For these reasons, we examined whether TRAIL-R1 was ubiquitinated on the cysteines or the serine that are present in its membrane-proximal region (Fig. 4D). To simplify acceptor residue identification, we generated a C-terminal truncated receptor, leaving only the 14 most membrane-proximal cytoplasmic amino acids. In this truncated receptor, the lysine, cysteine, or serine residues were mutated, either alone or in combination (Fig. 4D). The TRAIL-R1 mutants lacking the membrane-proximal serine or cysteine residues were still ubiquitinated (Fig. 4E). In the absence of Lys-273, however, all ubiquitination of TRAIL-R1 was lost. Together, these data indicate that TRAIL-R1 is ubiquitinated at steady-state by an endogenous ligase. In TRAIL-R1, Lys-273 but also residues more than 14 amino acids distal from the plasma membrane are potential ubiquitin acceptor sites.
MARCH-1 and -8 Interact With and Down-regulate WT TRAIL-R1, but Not TRAIL-R1 K273A
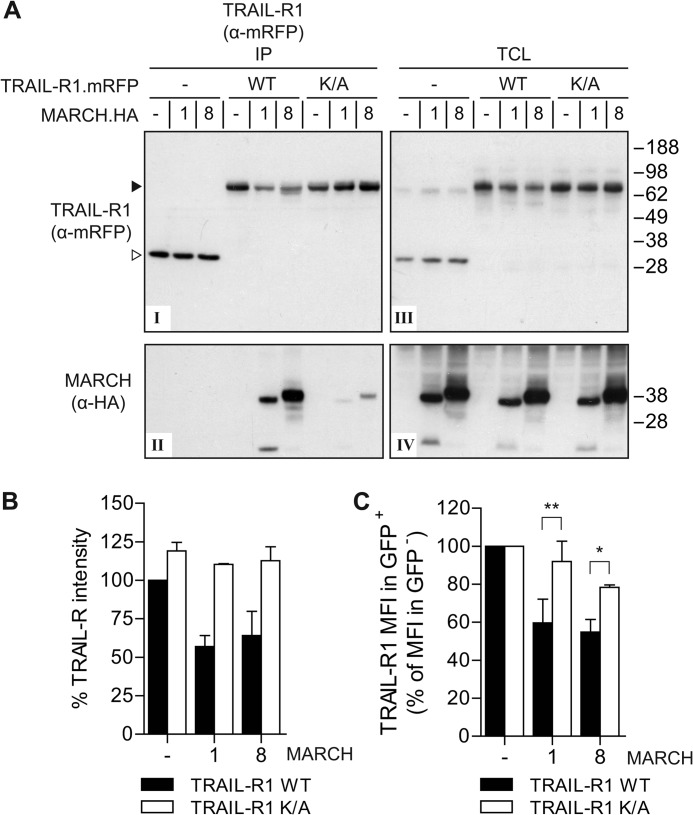
MARCH-8 is known to interact with its ubiquitination substrate CD86 (16). We therefore examined whether TRAIL-R1 could interact with MARCH-1 or -8. WT TRAIL-R1.mRFP or the K273A mutant were co-expressed with HA-tagged MARCH-1 or -8. Next, the TRAIL receptors were isolated by immunoprecipitation and isolates were immunoblotted with anti-HA antibody to detect associated MARCH proteins. Both MARCH proteins were detected at high stochiometry in the WT TRAIL-R1 immunoprecipitates (IP, Fig. 5A, panels I and II), indicating that TRAIL-R1 forms a complex with MARCH-1 or -8 at steady state. Strikingly, MARCH-1 and -8 interacted to a much lesser extent with the TRAIL-R1 K273A mutant, indicating that Lys-273 enables or greatly strengthens the interaction of TRAIL-R1 with MARCH-1 and MARCH-8 (Fig. 5A, panels I and II).
FIGURE 5.
MARCH-1 and -8 interact with and down-regulate wild-type TRAIL-R1, but not the TRAIL-R1 K273A mutant. A, MCF-7Casp-3 cells were transfected to express mRFP only (−), mRFP-tagged WT TRAIL-R1 or K273A (K/A) mutant, together with HA-tagged MARCH-1, MARCH-8, or empty vector (−), as indicated. Immunoprecipitation was performed with α-mRFP antibody and immunoprecipitates (IP) were analyzed by immunoblotting with α-mRFP and α-HA antibodies to detect TRAIL-R1 and MARCH-1/8, respectively. Panel I, mRFP detection in IP of TRAIL-R1.mRFP and control mRFP; panel II, MARCH-1 and -8 detection in IP of TRAIL-R1.mRFP and control mRFP; panel III, mRFP detection (TRAIL-R1.mRFP or RFP only) in total cell lysates (TCL); panel IV, MARCH-1 and -8 detection in TCL. B, quantification of TRAIL-R1 down-regulation in total cell lysates. Total protein levels of WT TRAIL-R1 and the K/A mutant in TCL of control cells (−), or those expressing HA-tagged MARCH-1 or -8 were quantified from Western blots as depicted in panel III of A and plotted as percentage of the WT TRAIL-R1.mRFP expression in control cells. Data represent mean ± S.D. of values from the experiment depicted in A and 2 additional experiments. C, impact of MARCH-1 or MARCH-8 on WT and K/A mutant TRAIL-R1 cell surface expression. Cells stably expressing WT or K273A TRAIL-R1.mRFP were transfected to express GFP (−), GFP-tagged MARCH-1 or MARCH-8, and stained with antibody to TRAIL-R1 as outlined for Fig. 2. Quantification of 2–4 independent experiments assessing TRAIL-R1 MFI in GFP+ cells as the percentage of TRAIL-R1 MFI in GFP− cells, whereby the values in control cells were set at 100%. Values represent mean ± S.D. Asterisks indicate statistically significant differences between TRAIL-R1 WT or K/A mutant (Student's t test; *, p < 0.05, **, p < 0.01).
Immunoblotting for TRAIL-R1 on total cell lysates revealed that total cellular protein levels of WT TRAIL-R1 were reduced by co-expression of MARCH-1 or -8 (Fig. 5A, panels III and IV). In contrast, the total cellular level of the TRAIL-R1 K273A mutant was unaffected by either MARCH protein, suggesting that TRAIL-R1 down-regulation occurred by virtue of its interaction with MARCH-1 or -8. These findings were substantiated by quantification of TRAIL-R1 levels in total cell lysates from multiple independent experiments (Fig. 5B).
Furthermore, we analyzed the impact of MARCH-1 and MARCH-8 on the cell surface expression of TRAIL-R1 WT and its K273A mutant. Flow cytometric analysis showed that the TRAIL-R1 K273A mutant was much less efficiently down-regulated from the cell surface by MARCH-1 or MARCH-8 than the WT TRAIL-R1 (Fig. 5C).
We conclude that TRAIL-R1 can form a complex with MARCH-1 and -8, supported by the Lys-273 residue in the cytoplasmic tail of TRAIL-R1. Moreover, MARCH-1 and -8 down-regulated cell surface expression of WT TRAIL-R1, but not the K273A mutant. Together, these data indicate that Lys-273 is involved in the interaction of TRAIL-R1 with MARCH-1/8, and its down-regulation by these ligases.
MARCH-8 Targets TRAIL-R1 for Lysosomal Degradation
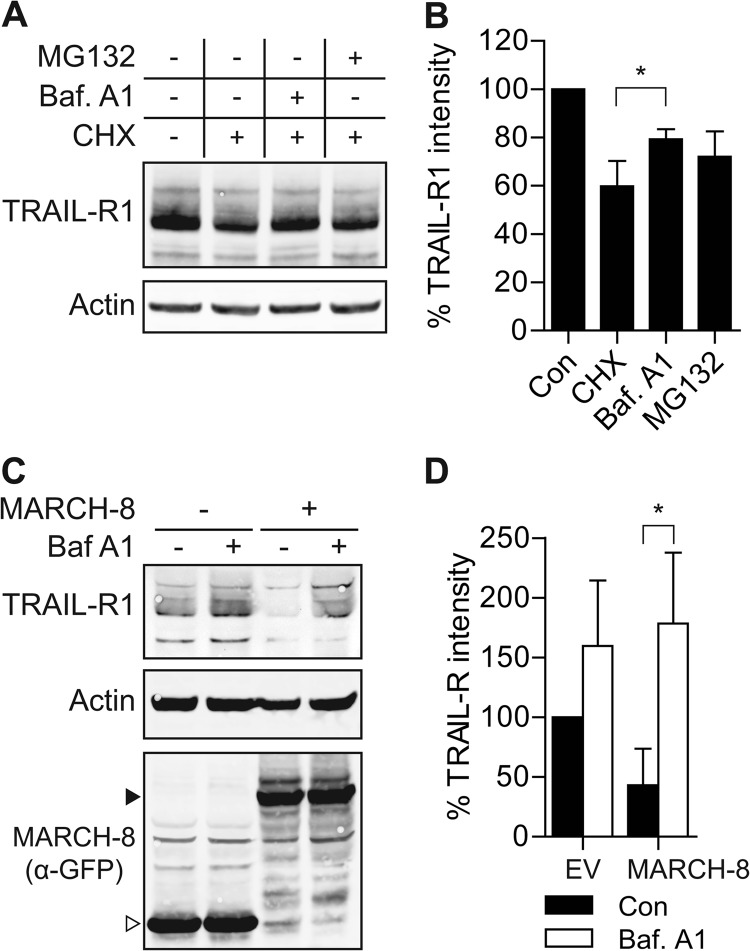
Because MARCH-1 and -8 down-regulated endogenous TRAIL-R1 from the cell surface (Fig. 1) and reduced total expression levels of exogenous TRAIL-R1 (Fig. 5), we hypothesized that MARCH-1 and -8 targeted TRAIL-R1 for degradation. To test this, we first studied the mechanism by which endogenous TRAIL-R1 was turned over at steady state. For this purpose, we used H358 lung cancer cells, as they express larger amounts of TRAIL-R1 than MCF-7Casp-3 cells (supplemental Fig. S6). After a 16-h treatment with the translation inhibitor cycloheximide (CHX), endogenous TRAIL-R1 levels were reduced to about 60% of untreated control levels (Fig. 6, A and B). Recoveries of TRAIL-R1 from Nonidet P-40 lysates and lysates prepared with more stringent RIPA buffer were similar, indicating that the receptor did not reside in compartments that were insoluble after Nonidet P-40 lysis (results not shown). Thus, CHX treatment visualized steady-state degradation of endogenous TRAIL-R1. This degradation could be blocked by bafilomycin A1 (BafA1), a lysosomal inhibitor (Fig. 6, A and B). Inhibiting the proteasome with MG132 also partially inhibited TRAIL-R1 degradation. Statistical evaluation of data from three independent experiments indicated that rescue of TRAIL-R1 by BafA1, but not rescue by MG132, was significant (Fig. 6B). This indicates that at steady-state, a large pool of endogenous TRAIL-R1 is targeted for degradation in lysosomes.
FIGURE 6.
MARCH-8 targets endogenous TRAIL-R1 for lysosomal degradation. A and B, H358 cells were treated with cycloheximide (CHX; 50 μg/ml) alone, or with CHX in combination with bafilomycin A1 (Baf A1; 200 nm) or MG132 (10 μm) for 16 h. Total cell lysates were analyzed by immunoblotting with α-TRAIL-R1 and α-actin antibodies. A, data of a representative experiment. B, mean ± S.D. of quantified values from 3 independent experiments. TRAIL-R1 intensity was corrected for actin expression, the control value was set to 100%. Asterisk indicates statistically significant difference (Student's t test; *, p < 0.05). C and D, MCF-7Casp-3 cells were transfected to express GFP only (−), or GFP-tagged MARCH-8. Cells were treated with BafA1 (100 nm) or left untreated for 16 h. Total cell lysates of GFP+ cells, obtained by flow cytometric sorting, were analyzed by immunoblotting with α-TRAIL-R1 and α-actin antibodies, and α-GFP antibody to detect MARCH-8. Solid and open arrowheads indicate, respectively, MARCH-8.GFP and GFP only. C, data of a representative experiment. D, mean ± S.D. of quantified values from 3 independent experiments. TRAIL-R1 intensity was corrected for actin expression, control value was set to 100%. Asterisk indicates a statistically significant difference (Student's t test; *, p < 0.05).
Next, we addressed MARCH-8-induced degradation of endogenous TRAIL-R1 in our main model system, MCF-7Casp-3 cells. Cells were transfected to express MARCH-8.GFP or GFP alone, treated with BafA1 for 16 h or left untreated. Endogenous TRAIL-R1 expression was determined in the transfected populations that were isolated by flow cytometric sorting. In cells that did not express MARCH-8, BafA1 treatment increased TRAIL-R1 expression (Fig. 6C). MARCH-8 expression considerably reduced endogenous TRAIL-R1 levels, which was reverted by blocking lysosomal degradation with BafA1 (Fig. 6C). Statistical evaluation of data from three independent experiments indicated that BafA1 treatment rescued MARCH-8-induced degradation of TRAIL-R1 (Fig. 6D). These experiments indicate that MARCH-8 targets endogenous TRAIL-R1 in MCF-7Casp-3 cells for lysosomal degradation at steady state.
TRAIL-R1 Is Ubiquitinated and Down-regulated by Endogenous MARCH-8
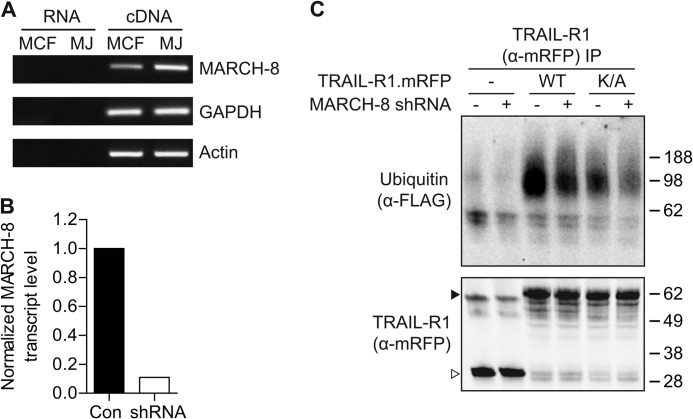
The findings outlined above strongly suggested that endogenous MARCH proteins were responsible for TRAIL-R1 ubiquitination and its down-regulation. To test the involvement of endogenous MARCH proteins, we used RNA interference. RT-PCR revealed that both MCF-7Casp-3 and Mel JuSo cells express MARCH-8 (Fig. 7A). A short hairpin (sh)RNA construct was made that efficiently silenced MARCH-8 expression, as tested on endogenous MARCH-8 mRNA (Fig. 7B).
FIGURE 7.
TRAIL-R1 is a substrate of endogenous MARCH-8. A, endogenous MARCH-8 expression in MCF-7Casp-3 (MCF-7) and Mel Juso (MJ) cells, as determined by RT-PCR on cDNA. Non-reverse transcribed RNA (RNA) was used as a control template to exclude amplification of genomic DNA. B, down-regulation of endogenous MARCH-8 by RNAi. MCF-7Casp-3 cells were transfected with MARCH-8 shRNA, together with GFP. MARCH-8 shRNA expressing cells (GFP+) and nonexpressing cells (GFP−) cells were separated by flow cytometric sorting and analyzed for endogenous MARCH-8 transcript levels by quantitative RT-PCR. Signals that were corrected for GAPDH and MARCH-8 transcript levels in the GFP+ population were normalized to the levels in the GFP− population. C, MCF-7Casp-3 cells were transfected to express mRFP alone, WT TRAIL-R1.mRFP, or K273A TRAIL-R1.mRFP, together with FLAG-ubiquitin and either an empty vector, or the MARCH-8 targeting shRNA. TRAIL-R1 was isolated by immunoprecipitation with α-mRFP antibody and immunoprecipitates (IP) were analyzed by immunoblotting for TRAIL-R1 (α-mRFP) or ubiquitin (α-FLAG). Solid and open arrowheads indicate, respectively, TRAIL-R1.mRFP and mRFP only. Data shown are representative of 3 independent experiments.
The validated MARCH-8 shRNA was subsequently used to test whether endogenous MARCH-8 was responsible for the ubiquitination of TRAIL-R1. MCF-7Casp-3 cells were transfected to express WT or K273A TRAIL-R1 and FLAG-ubiquitin, together with either a control vector or a MARCH-8 shRNA. Silencing of MARCH-8 reduced the level of WT TRAIL-R1 ubiquitination (Fig. 7C). Interestingly, ubiquitination of TRAIL-R1 K273A was also reduced to some extent upon MARCH-8 silencing (Fig. 7C). Therefore, we conclude that TRAIL-R1 is a substrate for ubiquitination by endogenous MARCH-8 that potentially targets lysine 273, but also one or more additional residues.
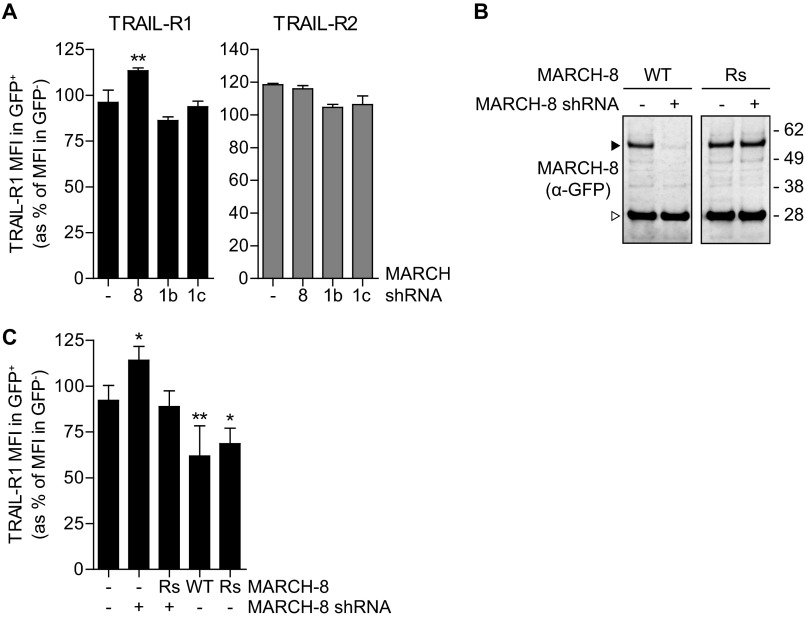
We next used MARCH-8 shRNA to test whether endogenous MARCH-8 regulated endogenous TRAIL-R1 cell surface expression. MCF-7Casp-3 cells were transfected with either a control vector or a MARCH-8 targeting shRNA. Subsequently, cell surface expression of TRAIL-R1 and TRAIL-R2 was examined by flow cytometry. Expression of a MARCH-8 shRNA significantly up-regulated TRAIL-R1 cell surface expression, whereas TRAIL-R2 surface expression was unaffected (Fig. 8A). In contrast, expression of two different validated MARCH-1 targeting shRNAs had no effect on either TRAIL-R1 or TRAIL-R2 cell surface expression (supplemental Fig. S7 and Fig. 8A).
FIGURE 8.
Endogenous MARCH-8 regulates cell surface expression of endogenous TRAIL-R1. A, MCF-7Casp-3 cells were transfected to express GFP (−), together with either a control vector (−), with MARCH-8 targeting shRNA (8), or with two different MARCH-1-targeting shRNAs (1b and 1c). Endogenous TRAIL-R1 cell surface expression was determined by flow cytometric analysis and data were evaluated as described in the legend to Fig. 2. Data represent mean values ± S.D. from 3 independent experiments. Asterisks indicate statistically significant differences compared with GFP-transfected control cells (one-way analysis of variance, Bonferroni correction; ***, p < 0.001). B, validation of the MARCH-8 rescue construct. MCF-7Casp-3 cells were transfected to express GFP-tagged MARCH-8 WT, or a MARCH-8 rescue (Rs) variant carrying silent mutations to allow escape from RNAi. Cells were cotransfected with empty vector (−) or MARCH-8 shRNA construct. MARCH-8.GFP expression was analyzed by immunoblotting for GFP in total cell lysates. C, MCF-7Casp-3 cells were transfected to express GFP (−), GFP-tagged MARCH-8 WT or Rs alone (−), or in conjunction MARCH-8 targeting shRNA (+). Endogenous TRAIL-R1 cell surface expression was determined by flow cytometric analysis and data were evaluated as described in the legend to Fig. 2. Data represent mean ± S.D. from at least 4 independent experiments. Asterisks indicate statistically significant differences compared with GFP-transfected control cells (one-way analysis of variance, Bonferroni correction; ***, p < 0.001).
The specificity of the RNAi effect was confirmed by using a MARCH-8 rescue (Rs) construct that could escape from shRNA-mediated down-regulation (Fig. 8B). MCF-7Casp-3 cells were transfected to express GFP-tagged MARCH-8 WT or MARCH-8 Rs, in the presence or absence of MARCH-8 shRNA. Both WT and Rs MARCH-8 down-regulated cell surface levels of TRAIL-R1, whereas silencing of MARCH-8 expression with shRNA significantly increased TRAIL-R1 cell surface levels (Fig. 8C). Up-regulation of TRAIL-R1 by MARCH-8 shRNA was overruled by co-expression of the untargeted MARCH-8 Rs (Fig. 8C). These data show that steady-state cell surface levels of endogenous TRAIL-R1 are regulated by endogenous MARCH-8. The collective data indicate that endogenous MARCH-8 ubiquitinates TRAIL-R1 to attenuate its cell surface expression.
DISCUSSION
We here present TRAIL-R1 as the first physiological substrate of the endogenous MARCH-8 ubiquitin ligase. Hereby, TRAIL-R1 joins MHC class II and CD86 (22, 24), as the only endogenous substrates that are thus far identified for the entire mammalian MARCH family. Moreover, we identify Lys-273 in the cytoplasmic tail of TRAIL-R1 as one of the potential ubiquitin acceptor sites for MARCH-8. For the viral MARCH-related ligases K3 and K5, it has been demonstrated very elegantly that ubiquitin transfer does not rely on the amino acid context of the ubiquitination site, but its distance from the plasma membrane, which was optimal at 15 amino acids for K3 and 10 amino acids for K5 (36). This can easily be understood from the topology of the ligases. In K3 and K5, the amino-terminal RING domain is spaced by 28 amino acids from the first transmembrane segment. This constrains the RING domain in its ability to reach an acceptor site in its target that likewise is fixed in the membrane (37). Interestingly, this spacing is highly conserved in all nine mammalian transmembrane MARCH proteins (analysis by SMART) and strongly suggests that all MARCH proteins ubiquitinate their targets on residues close to the plasma membrane. The ubiquitination sites identified for MARCH-1, -4, -8, and/or -9 in MHC class I, -II and CD86 are indeed membrane-proximal (15, 23, 24).
Lysine 273 of TRAIL-R1 also conforms to this rule, as it is located 11 amino acids from the transmembrane segment. However, MARCH-8 overexpression still increased ubiquitination of TRAIL-R1 K273A (Fig. 4A) and MARCH-8 knockdown reduced ubiquitination of this mutant (Fig. 7C). Therefore, Lys-273 is not the only possible target residue in TRAIL-R1 for MARCH-8. We have ruled out unconventional ubiquitination of membrane-proximal cysteine and serine residues. This leaves more membrane-distal residues as potential ubiquitin acceptor sites. Surprisingly, the first lysine residue after Lys-273 is located 108 residues from the membrane (supplemental Fig. S5A). Interestingly, it was recently described that exogenous MARCH-8 ubiquitinates a IL-1 receptor accessory protein on a lysine located 124 amino acids from the membrane (38).
We found that MARCH-1 and -8 interact with TRAIL-R1 at high stoichiometry, in a detergent-resistant manner. The current literature suggests that interaction between MARCH-type ligases and their substrates primarily depends on the transmembrane regions of the partners. This was reported for the viral ligase K5 (39–40), but also for MARCH-1 (41–43) and MARCH-8 (16, 44). Possibly, Lys-273 directly interacts with MARCH-1 and MARCH-8. Alternatively, Lys-273 may be the ubiquitination site and interaction with the MARCH ligases is stabilized by the ubiquitination event itself.
We found that upon deliberate expression, several MARCH family ligases could attenuate cell surface expression of TRAIL-R1. The ligases preferentially down-regulated TRAIL-R1 and had little impact on TRAIL-R2, as observed in breast carcinoma and melanoma cell lines. MARCH-1 and -8 also down-regulated CD95 in the study by Bartee et al. (15) and we confirmed that out of MARCH-1, -2, -4, -8, and -9, these two ligases had the most profound effect on cell surface expression of CD95 (supplemental Fig. S2). The selectivity of MARCH ligases to down-regulate TRAIL-R1 and CD95 with preference over TRAIL-R2 may reflect availability of ubiquitination sites. However, all three receptors have lysine residues at membrane-proximal locations (supplemental Fig. S5C). We therefore consider that the inefficient targeting of TRAIL-R2 may rather be due to a difference in membrane microdomain localization and/or endosomal trafficking of TRAIL-R2 compared with the other two receptors. We observed that the cell surface levels of TRAIL-R1, but not TRAIL-R2 increased upon transient overexpression of K44A dynamin-1. This may imply that TRAIL-R2 is turned over at the steady-state by a dynamin-independent mechanism, but it may also indicate a difference between the two receptors with regards to the dynamics of this process.
Current knowledge on the mechanism of action of the MARCH ligases indicates that they may down-regulate cell surface expression of TRAIL-R1 by various mechanisms. For the closely related MARCH-1 and MARCH-8, available data argue that they down-regulate cell surface expression of their, partially shared, target proteins by promoting lysosomal degradation. MARCH-1 and -8 can accomplish this in different ways that may depend on the target and cell type. In B cells and dendritic cells, MARCH-1 attenuated cell surface expression of its endogenous substrate MHC class II by promoting its transport toward lysosomes, but not its endocytosis (22, 25). MARCH-8 on the other hand down-regulated cell surface expression of its targets CD86 and MHC class II by promoting their endocytosis from the plasma membrane (16, 23, 45). In the case of recently identified targets CD44 and CD98 (46, 47), MARCH-8 diverted them away from recycling endosomes and into the lysosomal pathway. This process required the TSG101 component of the ESCRT-1 complex, suggesting an impact of MARCH-8 on cargo sorting in multivesicular bodies (47). This is reminiscent of the action of viral K3 (48) and underlines that MARCH-8 can promote lysosomal sorting of membrane proteins at different locations in the endosomal route. In case of TRAIL-R1, MARCH-8 may likewise impact before and/or after endocytosis, but our data indicate that it targets TRAIL-R1 for lysosomal degradation, thereby decreasing cell surface and total protein expression levels.
Proteasomal inhibition also inhibited TRAIL-R1 degradation to some extent (Fig. 6A). Possibly, there is a pool of TRAIL-R1 that is directly degraded by the proteasome. Alternatively, proteasome inhibition might have impacted on TRAIL-R1 degradation in an indirect manner, e.g. by affecting endosomal routing (49, 50) or gene expression (51).
The closely related MARCH-1 and MARCH-8 both interacted with TRAIL-R1 and down-regulated it from the cell surface. However, in MCF-7 cells, only silencing of MARCH-8 and not MARCH-1 had an impact on the cell surface expression of endogenous TRAIL-R1. This may reflect differential expression, because MARCH-1 is primarily found in lymphoid tissues, whereas MARCH-8 is more ubiquitously expressed (15, 20).
In the breast cancer cells we have studied, TRAIL receptors signaled for apoptosis from the cell surface rather than from endosomes (data not shown), in agreement with previous findings in B-lymphoma and cervix carcinoma cells (11, 12). Mechanisms that attenuate TRAIL receptor cell surface expression can therefore be expected to affect TRAIL receptor signaling. In normal physiology, the TRAIL receptors are targeted by membrane bound TRAIL that is expressed by natural killer cells. In experimental cancer therapy, TRAIL receptors are targeted by soluble recombinant TRAIL, but also by receptor-selective agonistic antibodies, to induce tumor-specific cell death (4). This novel function of MARCH-8 may therefore have implications both in a physiological setting, as well as in future cancer therapy.
Supplementary Material
Acknowledgments
We thank Drs. Henning Walczak, Klaus Früh, and Eric Bartee for kindly providing reagents, Lennert Janssen and the employees of the flow cytometry and digital microscopy facilities of our institute for expert technical assistance and Dr. Marcel Verheij for support.
This work was supported by Grants NKI 2004-3079 and NKI 2008-4110 from the Dutch Cancer Society and Grant ECHO 700.57.008 from the Netherlands Organization for Scientific Research (to J. B.).

This article contains supplemental ”Methods“ and Figs. S1–S7.
- TRAIL
- TNF-related apoptosis inducing ligand
- BafA1
- bafilomycin A1
- IZ
- isoleucine-zippered
- MARCH
- membrane-associated RING-CH
- mRFP
- monomeric red fluorescent protein
- R
- receptor
- RING
- really interesting new gene
- Rs
- rescue
- IP
- immunoprecipitation
- CHX
- cycloheximide
- MFI
- mean fluorescence intensity.
REFERENCES
- 1. Smyth M. J., Cretney E., Takeda K., Wiltrout R. H., Sedger L. M., Kayagaki N., Yagita H., Okumura K. (2001) Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon γ-dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 193, 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takeda K., Hayakawa Y., Smyth M. J., Kayagaki N., Yamaguchi N., Kakuta S., Iwakura Y., Yagita H., Okumura K. (2001) Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 7, 94–100 [DOI] [PubMed] [Google Scholar]
- 3. Grosse-Wilde A., Voloshanenko O., Bailey S. L., Longton G. M., Schaefer U., Csernok A. I., Schütz G., Greiner E. F., Kemp C. J., Walczak H. (2008) TRAIL-R deficiency in mice enhances lymph node metastasis without affecting primary tumor development. J. Clin. Invest. 118, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashkenazi A., Herbst R. S. (2008) To kill a tumor cell. The potential of proapoptotic receptor agonists. J. Clin. Invest. 118, 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimberley F. C., Screaton G. R. (2004) Following a TRAIL. Update on a ligand and its five receptors. Cell Res. 14, 359–372 [DOI] [PubMed] [Google Scholar]
- 6. Peter M. E., Krammer P. H. (2003) The CD95(APO-1//Fas) DISC and beyond. Cell Death Differ. 10, 26–35 [DOI] [PubMed] [Google Scholar]
- 7. Sprick M. R., Rieser E., Stahl H., Grosse-Wilde A., Weigand M. A., Walczak H. (2002) Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but cannot functionally substitute caspase-8. EMBO J. 21, 4520–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaudhary P. M., Eby M., Jasmin A., Bookwalter A., Murray J., Hood L. (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity 7, 821–830 [DOI] [PubMed] [Google Scholar]
- 9. Lee K. H., Feig C., Tchikov V., Schickel R., Hallas C., Schütze S., Peter M. E., Chan A. C. (2006) The role of receptor internalization in CD95 signaling. EMBO J. 25, 1009–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schneider-Brachert W., Tchikov V., Neumeyer J., Jakob M., Winoto-Morbach S., Held-Feindt J., Heinrich M., Merkel O., Ehrenschwender M., Adam D., Mentlein R., Kabelitz D., Schütze S. (2004) Compartmentalization of TNF receptor 1 signaling. Internalized TNF receptosomes as death signaling vesicles. Immunity 21, 415–428 [DOI] [PubMed] [Google Scholar]
- 11. Austin C. D., Lawrence D. A., Peden A. A., Varfolomeev E. E., Totpal K., De Mazière A. M., Klumperman J., Arnott D., Pham V., Scheller R. H., Ashkenazi A. (2006) Death-receptor activation halts clathrin-dependent endocytosis. Proc. Natl. Acad. Sci. U.S.A. 103, 10283–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohlhaas S. L., Craxton A., Sun X. M., Pinkoski M. J., Cohen G. M. (2007) Receptor-mediated endocytosis is not required for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. J. Biol. Chem. 282, 12831–12841 [DOI] [PubMed] [Google Scholar]
- 13. Doherty G. J., McMahon H. T. (2009) Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 [DOI] [PubMed] [Google Scholar]
- 14. Hicke L., Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 15. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., Früh K. (2004) Down-regulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78, 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goto E., Ishido S., Sato Y., Ohgimoto S., Ohgimoto K., Nagano-Fujii M., Hotta H. (2003) c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 278, 14657–14668 [DOI] [PubMed] [Google Scholar]
- 17. Morokuma Y., Nakamura N., Kato A., Notoya M., Yamamoto Y., Sakai Y., Fukuda H., Yamashina S., Hirata Y., Hirose S. (2007) MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J. Biol. Chem. 282, 24806–24815 [DOI] [PubMed] [Google Scholar]
- 18. Lehner P. J., Hoer S., Dodd R., Duncan L. M. (2005) Down-regulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol. Rev. 207, 112–125 [DOI] [PubMed] [Google Scholar]
- 19. Nathan J. A., Lehner P. J. (2009) The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp. Cell Res. 315, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 20. Wang X., Herr R. A., Hansen T. (2008) Viral and cellular MARCH ubiquitin ligases and cancer. Semin. Cancer Biol. 18, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., Gatti E. (2008) MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. U.S.A. 105, 3491–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., Toyooka K., Matsuoka K., Hotta H., Yamamoto A., Ishido S. (2007) Novel regulation of MHC class II function in B cells. EMBO J. 26, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohmura-Hoshino M., Matsuki Y., Aoki M., Goto E., Mito M., Uematsu M., Kakiuchi T., Hotta H., Ishido S. (2006) Inhibition of MHC class II expression and immune responses by c-MIR. J. Immunol. 177, 341–354 [DOI] [PubMed] [Google Scholar]
- 24. Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., Shin J.-S. (2011) Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 187, 2966–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walseng E., Furuta K., Bosch B., Weih K. A., Matsuki Y., Bakke O., Ishido S., Roche P. A. (2010) Ubiquitination regulates MHC class II-peptide complex retention and degradation in dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20465–20470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maas C., Verbrugge I., de Vries E., Savich G., van de Kooij L. W., Tait S. W., Borst J. (2010) Smac/DIABLO release from mitochondria and XIAP inhibition are essential to limit clonogenicity of Type I tumor cells after TRAIL receptor stimulation. Cell Death Differ. 17, 1613–1623 [DOI] [PubMed] [Google Scholar]
- 27. Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. (2009) Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J. Cell Biol. 185, 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V. M., Neefjes J. (2007) Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kranenburg O., Verlaan I., Moolenaar W. H. (1999) Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J. Biol. Chem. 274, 35301–35304 [DOI] [PubMed] [Google Scholar]
- 30. Verbrugge I., de Vries E., Tait S. W., Wissink E. H., Walczak H., Verheij M., Borst J. (2008) Ionizing radiation modulates the TRAIL death-inducing signaling complex, allowing bypass of the mitochondrial apoptosis pathway. Oncogene 27, 574–584 [DOI] [PubMed] [Google Scholar]
- 31. Damke H., Baba T., Warnock D. E., Schmid S. L. (1994) Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol. 127, 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ganten T. M., Haas T. L., Sykora J., Stahl H., Sprick M. R., Fas S. C., Krueger A., Weigand M. A., Grosse-Wilde A., Stremmel W., Krammer P. H., Walczak H. (2004) Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitization of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ. 11, S86–S96 [DOI] [PubMed] [Google Scholar]
- 33. Cadwell K., Coscoy L. (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309, 127–130 [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007) Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tait S. W., de Vries E., Maas C., Keller A. M., D'Santos C. S., Borst J. (2007) Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J. Cell Biol. 179, 1453–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cadwell K., Coscoy L. (2008) The specificities of Kaposi's sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J. Virol. 82, 4184–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herr R. A., Harris J., Fang S., Wang X., Hansen T. H. (2009) Role of the RING-CH domain of viral ligase mK3 in ubiquitination of nonlysine and lysine MHC I residues. Traffic 10, 1301–1317 [DOI] [PubMed] [Google Scholar]
- 38. Chen R., Li M., Zhang Y., Zhou Q., Shu H. B. (2012) The E3 ubiquitin ligase MARCH8 negatively regulates IL-1β-induced NF-κB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 109, 14128–14133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coscoy L., Sanchez D. J., Ganem D. (2001) A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanchez D. J., Coscoy L., Ganem D. (2002) Functional organization of MIR2, a novel viral regulator of selective endocytosis. J. Biol. Chem. 277, 6124–6130 [DOI] [PubMed] [Google Scholar]
- 41. Corcoran K., Jabbour M., Bhagwandin C., Deymier M. J., Theisen D. L., Lybarger L. (2011) Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1). J. Biol. Chem. 286, 37168–37180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thibodeau J., Bourgeois-Daigneault M. C., Huppé G., Tremblay J., Aumont A., Houde M., Bartee E., Brunet A., Gauvreau M. E., de Gassart A., Gatti E., Baril M., Cloutier M., Bontron S., Früh K., Lamarre D., Steimle V. (2008) Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 38, 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tze L. E., Horikawa K., Domaschenz H., Howard D. R., Roots C. M., Rigby R. J., Way D. A., Ohmura-Hoshino M., Ishido S., Andoniou C. E., Degli-Esposti M. A., Goodnow C. C. (2011) CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J. Exp. Med. 208, 149–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jahnke M., Trowsdale J., Kelly A. P. (2012) Structural requirements for recognition of major histocompatibility complex class II by membrane-associated RING-CH (MARCH) protein E3 ligases. J. Biol. Chem. 287, 28779–28789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goto E., Mito-Yoshida M., Uematsu M., Aoki M., Matsuki Y., Ohmura-Hoshino M., Hotta H., Miyagishi M., Ishido S. (2008) An excellent monitoring system for surface ubiquitination-induced internalization in mammals. PLoS One 3, e1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bartee E., Eyster C. A., Viswanathan K., Mansouri M., Donaldson J. G., Früh K. (2010) Membrane-associated RING-CH proteins associate with Bap31 and target CD81 and CD44 to lysosomes. PLoS One 5, e15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eyster C. A., Cole N. B., Petersen S., Viswanathan K., Früh K., Donaldson J. G. (2011) MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol. Biol. Cell 22, 3218–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hewitt E. W., Duncan L., Mufti D., Baker J., Stevenson P. G., Lehner P. J. (2002) Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21, 2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Longva K. E., Blystad F. D., Stang E., Larsen A. M., Johannessen L. E., Madshus I. H. (2002) Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Kerkhof P., Alves dos Santos C. M., Sachse M., Klumperman J., Bu G., Strous G. J. (2001) Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell 12, 2556–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.