Background: The gel-forming mucin genes at chr11p15.5 encode major components of the epithelial mucus layer.
Results: CTCF occupancy correlates with mucin gene expression and active loci form three-dimensional looped structures.
Conclusion: CTCF has a pivotal role in coordinating gene expression and response to lipopolysaccharide.
Significance: The study provides novel therapeutic prospects for epithelial diseases associated with mucin hypersecretion.
Keywords: Chromatin Structure, Gene Regulation, Glycobiology, Mucins, Mucus, CTCF, Chromatin Looping, Gel-forming Mucin Genes
Abstract
Four of the genes that encode gel-forming mucins, which are major components of the mucus layer protecting many epithelial surfaces, are clustered at chromosome 11p15.5 and show both cell- and tissue-specific expression patterns. We aimed to determine whether the individual genes were coordinately regulated by mechanisms involving higher order chromatin structure. CCCTC-binding factor (CTCF) sites were predicted in silico and CTCF occupancy then evaluated by chromatin immunoprecipitation. CTCF was found at many sites across the gene cluster, and its binding was correlated with mucin gene expression. Next, siRNA-mediated depletion of CTCF was shown to increase MUC2 expression in A549 lung carcinoma cells and both MUC6 and MUC5AC expression in LS180 colon carcinoma cells. These changes correlated with loss of CTCF binding at multiple sites, although others retained occupancy. In cells actively expressing the mucins, the gene cluster was shown by chromosome conformation capture to form looped three-dimensional structures with direct interactions between the MUC2 promoter region, regions 30 kb 5′ to it, close to the MUC6 promoter and others near the 3′ end of MUC5AC, >170 kb away. Finally, to demonstrate the importance of CTCF binding to mucin gene expression, Calu-3 lung carcinoma cells were exposed to lipopolysaccharide (LPS). LPS increased the expression of MUC2 and MUC5AC and reduced MUC5B. CTCF occupancy was concurrently depleted at specific binding sites close to these genes. These data suggest that CTCF binding and cell type-specific long-range interactions across the 11p15.5 gene cluster are critical mechanisms for coordinating gel-forming mucin gene expression.
Introduction
Recent advances in the methodology for analysis of transcriptional regulatory mechanisms genome-wide are revealing many novel functions for non-coding sequences (1). These elements are generally cis-acting but may be located at a considerable distance (>100 kb) from the genes they control. The new insights into the control of expression of individual genes are extensive; however, the opportunity to reveal regulatory mechanisms for gene clusters and gene networks across the genome is particularly exciting. Many functionally linked gene clusters such as the homeobox, β-globin, and gel-forming mucin loci are thought to have evolved by duplication of an ancestral gene followed by evolution of genes with both common and divergent properties (2, 3). The mechanisms that subsequently coordinate the whole gene cluster in addition to the individual loci within it are of considerable interest. We sought to determine the regulatory mechanisms controlling gene expression within the gel-forming mucin gene cluster located at chromosome 11p15.5 because synchronized expression of these genes is likely critical for normal function of epithelia lining many organs. Mucins are high molecular weight glycoproteins that contain tandem repeat regions enriched for serine, threonine, and proline, which are modified by O-glycosylation. These glycoproteins may be membrane-bound or secreted (gel-forming), and together, both types of mucin generate the characteristic mucus layer that protects epithelial surfaces in the intestine, pancreas, kidney, lungs, and many other organs (4, 5). Mucins also have additional critical functions in vivo and their misregulation is a characteristic feature of many disorders, including chronic obstructive pulmonary disease, cystic fibrosis, asthma, gastrointestinal inflammatory diseases, and lung, colon, and pancreatic carcinomas (6–10). The four gel-forming mucin genes at chromosome 11p15.5 include in order (telomeric to centromeric) MUC6, MUC2, MUC5AC, and MUC5B and encompass a ∼400-kb region (11). The expression of the four mucin genes is cell type- and tissue-specific: MUC6 is one of the two major gastric mucins (12) and is restricted to neck mucus cells (13). MUC2 is highly expressed in goblet cells within the intestinal epithelium and at lower levels in the epithelium lining conducting airways (8, 14); MUC5AC is expressed in surface goblet cells in the airway and the stomach (8, 15, 16) where its pattern of expression does not overlap with MUC6; and MUC5B expression is largely localized to submucosal gland cells within the respiratory tract (8, 16, 17),
Extensive analysis of the transcriptional regulation of individual genes encoding the gel-forming mucins has already been pursued and demonstrated that many of the gene promoters are activated by similar mechanisms. These include inflammatory mediators such as cytokines (interleukin 1β (IL-1β), IL-4, IL-6, IL-9, IL-12, IL-13 and tumor necrosis factor α (TNFα)); growth factors (epidermal growth factor (EGF), transforming growth factor α (TGF-α)) and bacterial LPS (reviewed in Ref. 10). Moreover, the four gene promoters contain functional binding sites for many general transcription factors, including Sp1, Sp3, CREB, AP-1, NF-κB, and c-Myc (reviewed in Refs. 10, 18) as would be expected from their mechanisms of activation. Recruitment of diverse transcription factors to these mucin genes results in their epigenetic modification by mechanisms, including DNA methylation, histone methylation, and histone acetylation/deacetylation (19, 20). Despite this wealth of information on the regulation of expression of MUC6, MUC2, MUC5AC, and MUC5B, the mechanisms that coordinate the expression of the whole gene cluster have not yet been examined. However, these more global regulatory mechanisms are critically important in ensuring the appropriate composition of the mucus gel in specific epithelial environments and that a well orchestrated response is generated to epithelial insults. Because individual mucus glycoproteins have quite different biochemical and biophysical properties, it is clear that misexpression will have functional consequences.
We tested the hypothesis that the 11p15.5 mucin gene cluster is coordinately regulated by modifications in higher order chromatin structure that enhance or restrict access to individual gene promoters within the cluster. Moreover, that the three-dimensional organization of this genomic region modulates the interaction of cis-acting enhancers or repressors with the individual gene promoters. Long range interactions are known to regulate other multigene clusters, including the T-helper type 2 cytokine, α-globin, major histocompatibility class II (MHC-II), and β-globin loci in a cell- and tissue-specific manner (21–26). At these and many other loci, CCCTC-binding factor (CTCF)3 interaction with insulator elements is important to prevent enhancers activating inappropriate promoters; it also isolates distinct chromatin domains by associating with boundary elements and prevents the spread of heterochromatin (27). CTCF is an 82-kDa protein that binds to variations of the consensus CCCTC motif using its 11 zinc finger domains (28–30). It contributes to the three-dimensional organization of chromosomes throughout the genome (31–33). CTCF sites are enriched in intergenic and intronic regions in comparison with proximal-promoter regions (33, 34), and although 40–60% of these sites are ubiquitous, the remainder are important in cell- and tissue-specific regulation (32, 35).
Our data demonstrate that CTCF binding across the gel-forming mucin gene cluster shows cell type specificity. Moreover, that depletion of CTCF by siRNA-mediated knockdown results in loss of occupancy only at certain sites, and these changes can alter the expression of individual mucin genes. Using quantitative chromosome conformation capture (q3C), we show long range interactions between the MUC2 promoter and sites in MUC6 and the adjacent adaptor-related protein complex 2 α 2 subunit (AP2A2) gene. Finally, we determine that LPS-induced up-regulation of MUC2 and MUC5AC and repression of MUC5B transcription in Calu-3 cells is accompanied by depletion of CTCF binding at specific sites. These data demonstrate the biological importance of the coordinate regulatory mechanisms across the gene cluster.
EXPERIMENTAL PROCEDURES
Cell Culture
LS180, LS174T (36), HT-29 (37) (colon carcinoma), A549 (38), Calu-3 (39) (lung carcinoma), BxPC-3 (40), (pancreatic adenocarcinoma), HEK293 (41) (human embryonic kidney) were grown in Dulbecco's modified Eagle's medium (DMEM), and Capan-1 (39) (pancreatic adenocarcinoma) was grown in Iscove's modified Dulbecco's medium; both media were supplemented with 10% fetal bovine serum (FBS). Primary human fibroblasts were cultured in Minimal essential medium (MEM) with 15% FBS.
Quantitative RT-PCR (qRT-PCR)
RNA was isolated from 2–3 days postconfluent cells using TRIzol® (Invitrogen), and a TaqMan® reverse transcription kit (Applied Biosystems) was used to make cDNA. Expression of the four mucin genes was measured using primers shown in supplemental Table 1. 18 S rRNA was used as an internal control.
Absolute Quantification of Expression
qRT-PCR was used to obtain expression values for the four mucin genes in all of the cells. Each mucin gene was set to 1.0 in LS180 cells, and the other cells were calculated relative to LS180. For absolute quantification, cloned cDNAs of the four mucin genes were used as follows MUC6 (42), MUC2 (43), MUC5AC (44), and MUC5B (45). Specific picomolar amounts (determined based on plasmid size) of each cloned plasmid were used in qRT-PCR to generate a standard curve for each mucin. Using the quantitative RT-PCR data from LS180 cDNA, the pmol amounts of each mucin were calculated for this line. These absolute numbers were then multiplied by the relative expression data for each cell line to give absolute values for each mucin gene.
ChIP
ChIP was done as described previously (46). 10 μg of CTCF antibody (Millipore 07-729) and rabbit IgG (Millipore 12-370) were used for each immunoprecipitation. Enrichment is shown relative to IgG and the primers used in SYBR® Green assays are listed in supplemental Table 1.
siRNA-mediated Depletion of CTCF
StealthTM CTCF siRNA (47) and non-targeting siRNA medium GC negative control duplex were used (Invitrogen). Knockdown experiments in A549 and LS180 were performed using LipofectamineTM RNAiMAX (Invitrogen). For A549, the cells were transfected with 30 pmol of CTCF or negative control siRNAs, 48 h after plating. For LS180, cells were reverse transfected with 50 pmol of CTCF and negative control siRNA. Cells were harvested 72 h after transfection to assay CTCF depletion by Western blot, changes in mRNA expression, and for ChIP.
Western Blots
A549 and LS180 were lysed with NET Buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA) with 1% (v/v) protease inhibitor mixture (Sigma) and 1% (w/v) Triton X-100. The antibodies used were CTCF (Millipore 07-729) and β-tubulin (Sigma-Aldrich, T 4026), and protein was detected using ECL. Films were scanned, and proteins were quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/).
q3C
q3C was performed as described previously (48, 49). The restriction enzyme used for digestion was BglII. Baits and interacting region primers used are listed in supplemental Table 1.
Lipopolysaccharide Treatment
Calu-3 cells were serum-starved from 24 h and treated either with PBS or with 200 ng/ml Pseudomonas aeruginosa LPS in PBS from Sigma (L9134).
RESULTS
Differential Mucin Gene Expression Patterns in Colon, Lung, and Pancreatic Adenocarcinoma Cell Lines
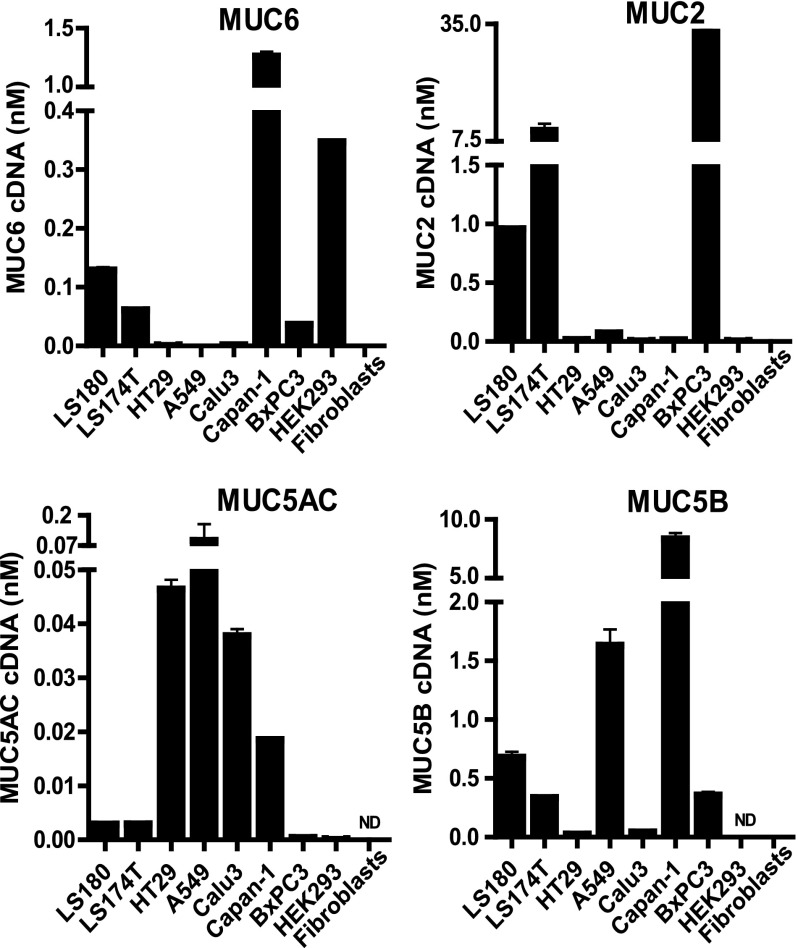
The chr.11p15.5 gel-forming mucins have distinct expression patterns that vary by cell and tissue type. In normal cells, only one gel-forming mucin is usually expressed (13); however, in cancer cell lines, multiple mucin genes may be expressed, thus providing a more informative model in which to examine coordinate gene regulation across the cluster. Although mucin gene expression data exist for many cell types, these are rarely quantitative. Hence, we first generated robust qRT-PCR assays for each of the four gel-forming mucin genes. To facilitate comparison of individual mucin genes within a cell type, we measured mucin gene expression using absolute quantification (Fig. 1). For each cell type, the individual expression values, normalized to 18 S rRNA, were set relative to LS180, a colon adenocarcinoma cell line that expresses all four genes. Next, picomolar amounts of the cDNA for each gel-forming mucin gene in LS180 were calculated. These absolute amounts were multiplied by the relative expression values of each mucin in the other cell lines. For example, if cell line A expressed 2-fold more MUC6 than LS180, using the picomolar amount for MUC6 in LS180 (∼0.1), line A would show an expression value of ∼0.2. The relative and absolute expression data sets, both show tissue- and cell type-specific differences in mucin gene expression. To illustrate, the lung carcinoma cell lines A549 and Calu-3 both express MUC5AC, but only A549 expresses MUC5B. Moreover, although LS180 and LS174T are derived from the same colon carcinoma cell line, LS174T expresses 10-fold more MUC2 (Fig. 1). These distinct gel-forming mucin gene expression profiles and the expression of several mucins by a single cancer cell suggest that there may be a mechanism to facilitate expression of certain genes and the repression of others.
FIGURE 1.
Absolute quantification of mucin gene expression in airway, intestinal, and pancreatic cell lines. Gene expression for the four gel-forming mucin genes was calculated using standard curves generated on cloned cDNAs for each gene. Absolute expression, in picomolar amounts, was calculated in LS180 cDNA.
The Mucin Gene Cluster Is Demarcated by Multiple CTCF Binding Sites
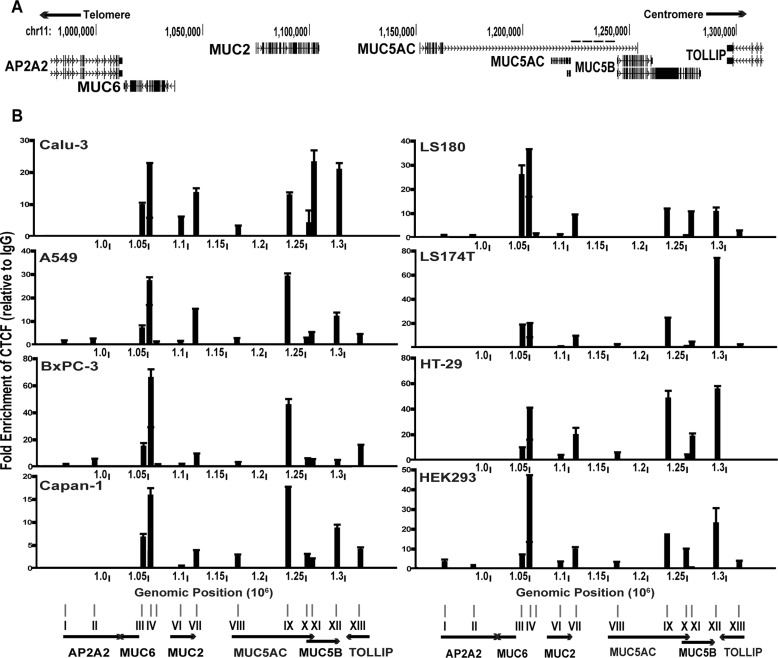
The mucin gene cluster at chromosome 11p15.5 encompasses ∼400 kb and contains four genes in the order MUC6, MUC2, MUC5AC, and MUC5B (Fig. 2A), with MUC6 telomeric and MUC5B centromeric. MUC6 is transcribed on the reverse strand, and MUC2, MUC5AC, and MUC5B are transcribed on the forward strand. There is a known gap in the HG19 assembly of the human genome sequence within the tandem repeat region of the MUC5AC gene (marked with dashed line on Fig. 2A). However, this does not impact interpretation of our data because the cDNA of MUC5AC has been cloned (44), enabling accurate expression studies and primer sets used in q3C lie outside this region.
FIGURE 2.
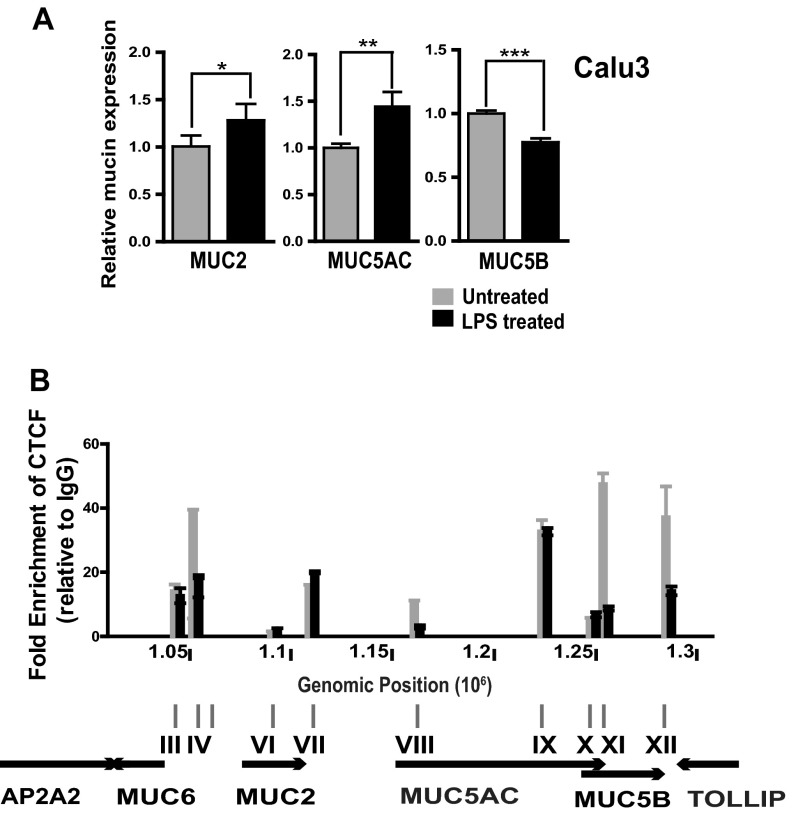
Differential occupancy of CTCF at predicted binding sites across the 11p15.5 mucin gene cluster. A, genomic organization of the gel-forming mucin gene cluster. Shown is the UCSC Genome Browser HG19 view of the four mucin genes on chr11p15.5. The dashed line above MUC5AC indicates a gap in the sequence. B, ChIP for CTCF in each cell line. Data are presented as fold enrichment over rabbit IgG isotype control. For each cell line, ChIP was performed at least twice, and representative data (consistent profiles in all replicates) are shown.
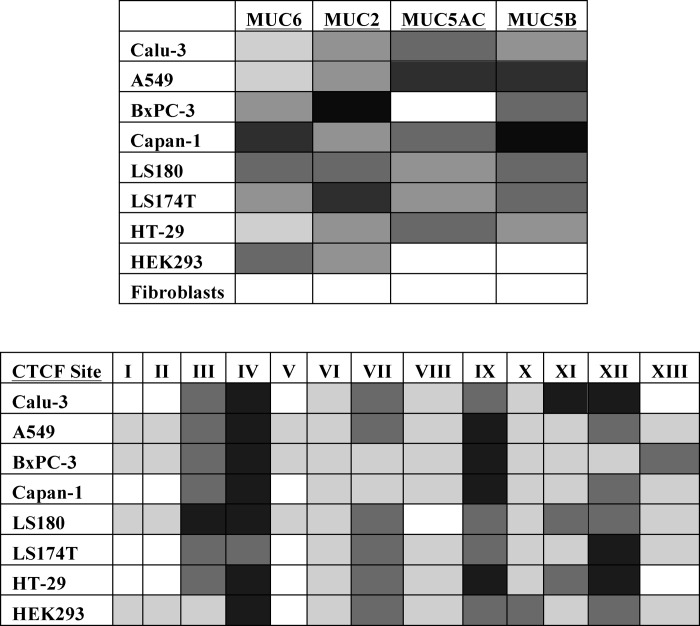
CTCF is known to regulate many multigene clusters to facilitate appropriate expression of each gene through modifying higher order chromatin structure and/or by binding to enhancer-blocking insulator elements. Thus, we used chromatin immunoprecipitation (ChIP) with an antibody specific for CTCF to examine its occupancy at predicted binding sites across the cluster in relevant cell types. Thirteen CTCF binding sites of 26 seen in ENCODE data (1) (derived from 47 cell lines) were analyzed by qRT-PCR (Fig. 2B, sites I–XIII). The choice of sites for analysis was made based on their location (all are intronic or intergenic), and CTCF occupancy was shown in relevant cell types. Among these sites several, for example sites IV and IX, show ubiquitous occupancy of CTCF in all cell types, although the levels of enrichment are variable. At other sites such as VII, XI, and XII, CTCF occupancy differs greatly between cell types, ranging from very low enrichment to the highest level of occupancy of all sites in a cell line (compare site XI in Calu-3 and A549). Interestingly, sites V, VI, and VIII, which are predicted to bind CTCF based on ENCODE data, show no enrichment despite adjacent sites showing high occupancy (e.g. site IV). The CTCF binding profile shows some correlation with mucin gene expression patterns, irrespective of the tissue origin of the cell line. For example, in Calu-3 (airway) and HT-29 (colon) cell lines, which express only MUC5AC (Fig. 1), the overall CTCF binding profile is very similar with the exception of site XI, at the 5′ end of MUC5B. In contrast, if both MUC5AC and MUC5B are expressed, as in A549 (airway) and Capan-1 (pancreatic) lines, CTCF occupancy at sites near the 5′ end (XI) and 3′ (XII) to MUC5B decreases (Fig. 2B). LS180 and LS174T, which are derivatives of the same colon carcinoma cell line, express all four gel-forming mucins. However, MUC2 expression levels are higher in LS174T (Fig. 1), and this apparently correlates with reduced CTCF occupancy at sites between MUC6 and MUC2 (sites III, IV) in comparison with LS180. The pancreatic cells express either very high levels of MUC2 (BxPC-3) or MUC6 (Capan-1) but not both. In these lines, there is overlap between sites of CTCF occupancy even though the gene expression profile is different. However, overall occupancy of CTCF across the gene cluster is higher in BxPC-3, which expresses very low levels of all the other genes, in contrast to Capan-1, which expresses several genes. Having established the CTCF occupancy profile of each cell line in relation to its expression of the gel-forming mucin genes, we next examined the effect of altering CTCF binding profiles on gene expression patterns.
Changes in CTCF Occupancy Alter Gel-forming Mucin Gene Expression
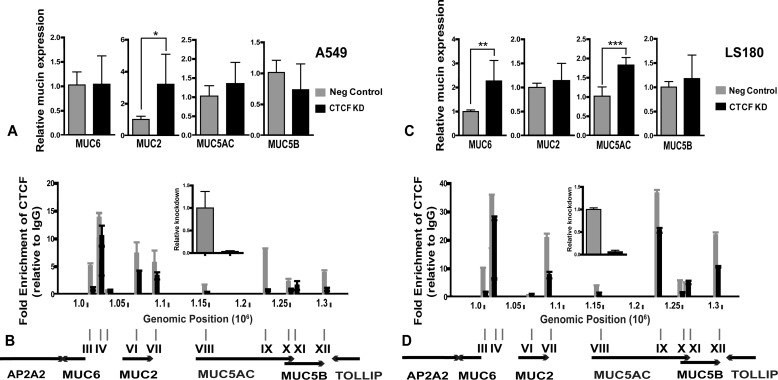
To determine whether CTCF occupancy at sites within the chr11p15.5 mucin gene cluster impacts gene expression, we used specific siRNAs to deplete CTCF. We first utilized A549 lung carcinoma cells, which express MUC5AC and MUC5B (Fig. 1). Efficient (∼90%) knockdown of CTCF resulted in an ∼3-fold increase in MUC2 expression (p < 0.05) in comparison with cells treated with a non-targeting siRNA control (Fig. 3A). Concurrent slight increases in MUC5AC and decreases in MUC5B expression after CTCF depletion were not statistically significant, and MUC6 levels did not change. To determine whether these alterations in mucin gene expression correlated with changes in CTCF occupancy at any sites, we performed ChIP for CTCF in negative control siRNA and CTCF siRNA-treated cells (Fig. 3B). Multiple CTCF-binding sites across the gene cluster showed selective depletion of CTCF occupancy after siRNA treatment. Of particular interest was loss of enrichment at sites VI and VII within the MUC2 gene, consistent with a dramatic increase in its expression, and at site IX, which is located within MUC5AC and close to the 5′ end of MUC5B because both genes are highly expressed in A549 cells. In contrast, CTCF knockdown had only a slight impact on occupancy at site IV, which lies between MUC6 and MUC2. This observation may underlie the apparent lack of transcriptional response of the MUC6 gene to CTCF depletion.
FIGURE 3.
siRNA-mediated depletion of CTCF augments specific mucin gene expression and causes differential loss of CTCF occupancy. A and C, changes in mucin gene expression after CTCF knockdown (KD; A, A549; C, LS180) were determined by RT-qPCR in negative control siRNA and CTCF siRNA treated cells. For each mucin gene data were normalized by setting negative (Neg) control siRNA expression to 1. B and D, selective loss of CTCF occupancy after knockdown (B, A549; D, LS180). ChIP for CTCF was carried out on cells transfected with a non-targeting (control) siRNA or an siRNA targeting CTCF. Data are presented as fold enrichment over rabbit IgG isotype control. ChIP was done at least twice, and representative data (consistent profiles in all replicates) are shown.
Next, we evaluated the effect of CTCF knockdown in LS180 colon carcinoma cells, which have a different mucin gene expression pattern from A549 and express all four gel-forming mucin genes (Fig. 1). siRNA-mediated CTCF depletion resulted in a significant increase in both MUC6 (p < 0.01) and MUC5AC (p < 0.001) expression relative to cells treated with a non-targeting siRNA (Fig. 3C). To identify the changes in CTCF occupancy after its depletion, we again performed ChIP for CTCF in negative control and CTCF siRNA-treated cells. Most sites showed loss of CTCF enrichment though the relative levels of depletion vary by site (Fig. 3D). None of the changes showed close correlation with expression of MUC6 and MUC5AC (with the exception of site VII), perhaps because both genes are already expressed in this line. Because CTCF knockdown in both A549 and LS180 cells increased mucin gene expression, this factor may be establishing a repressive chromatin environment at the gene cluster.
Long Range Interactions at the Gel-forming Mucin Gene Cluster
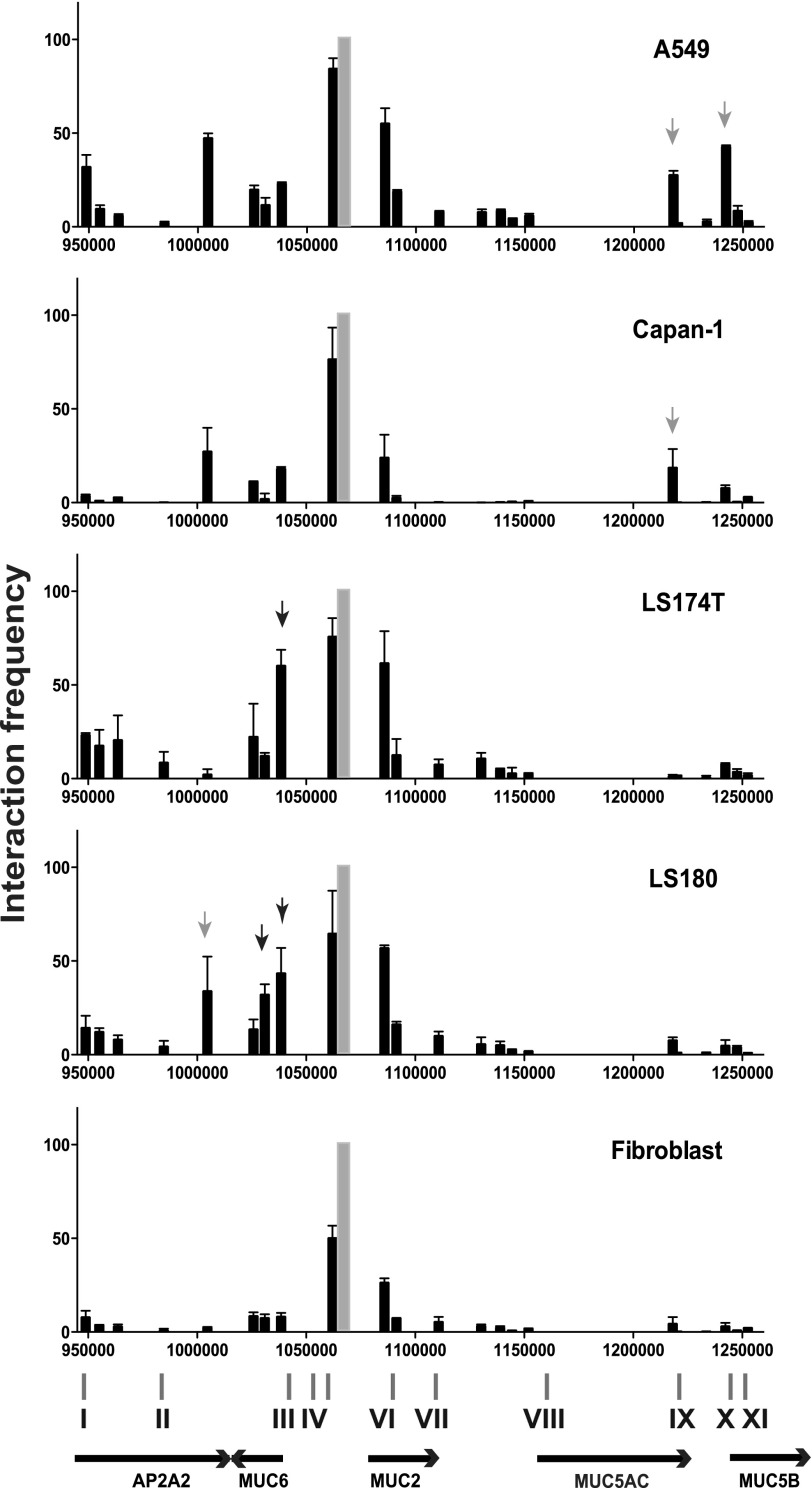
We next used q3C to test the hypothesis that some aspects of coordinate regulation of the mucin genes at chr11p15.5 are mediated by the long range interactions of cis-acting elements across the cluster. We first measured interactions between the MUC2 promoter and other regions in the gene cluster. In skin fibroblasts, which do not express any gel forming mucins, there are no interactions across the gene cluster, except for the expected interaction of regions <10 kb away from the MUC2 promoter bait (Fig. 4). In contrast, in LS174T and LS180 cells, which express high levels of all four genes, higher interactions were observed between the MUC2 promoter and elements close to the MUC6 5′ end (marked by black arrows in Fig. 4 at chr11:1,031,000–1,038,000). In LS174T cells, which express substantially higher levels of MUC2 (10-fold) than LS180 cells, the interaction frequencies across the region appeared greater. In addition, in LS180, a higher interaction frequency was seen between the MUC2 promoter and the 3′ end of the AP2A2 gene (at chr 11:1,005,000, gray arrow). In Capan-1 cells, which express abundant MUC6 but no MUC2, the interaction frequency between the MUC2 promoter and the MUC6 5′ end (chr11:1,038,000) was low, though slightly higher with the 3′ end of the AP2A2 gene (chr11:1,005,000). The interactions between the MUC6 and MUC2 promoters in cell lines that express both genes were confirmed with a 3C bait at the MUC6 gene promoter (supplemental Fig. 2). Interactions between the MUC2 promoter and the 3′ end of the mucin gene cluster were generally very low, with the exception of one region close to the 3′ end of the MUC5AC gene (chr11:1,218,000, light gray arrow) in Capan-1 cells. Capan-1 express much higher levels of MUC5AC and particularly MUC5B than LS180 or LS174T, and this may be relevant to the three-dimensional structure of the mucin gene cluster in these cells. Consistent with this hypothesis, A549 airway epithelial cells, which express high levels of MUC5AC and MUC5B show interactions between the MUC2 promoter and regions within the MUC5AC and MUC5B genes (Fig. 4, chr11:1,218,000 and 1,242,000, light gray arrows).
FIGURE 4.
Long range interactions are evident across the 11p15.5 mucin gene cluster. 3C (q3C) interactions are shown in A549, Capan-1, LS180, and LS174T cell lines and fibroblasts. A fixed forward primer and a Taqman probe were designed within a BglII fragment at the MUC2 promoter (bait, gray bar), and multiple reverse primers were generated within distal BglII fragments (black bars) across the 11p15.5 mucin gene cluster. The x axis represents the genomic location on chromosome 11; the y axis represents the interaction frequency relative to a control fragment adjacent to the bait. Experiments were performed at least twice for each cell type, and data shown are from a single representative 3C experiment. Error bars represent the S.E. of at least two qPCR reactions for each fragment. Arrows denote BglII fragments interacting with the bait as described in the text.
Lipopolysaccharide Exposure Decreases CTCF Occupancy across the Cluster in Association with Activation of Mucin Gene Expression
Among the many known in vivo activators of gel-forming mucin gene expression are bacterial exoproducts such as LPS, a component of the outer membrane of Gram-negative bacteria. To elucidate the biological relevance of CTCF binding at the gel-forming mucin cluster, we exposed Calu-3 cells to 200 ng/ml LPS from P. aeruginosa. This led to a significant increase in MUC2 and MUC5AC expression as expected (50–52), a significant (∼25%) decrease in MUC5B expression (Fig. 5A), and no change in MUC6 expression (data not shown). Next, we measured CTCF occupancy by ChIP across the mucin gene cluster in untreated and LPS-treated cells. The results showed that LPS activation was accompanied by a partial loss of CTCF binding at several sites across the cluster, including site IV, which is located between MUC6 and MUC2 and sites XI and XII, which are located at the 5′ and 3′ end of MUC5B, respectively (Fig. 5B). These data suggest that loss of CTCF binding at the gel-forming mucin gene cluster is an integral part of the activation of these genes in response to LPS treatment.
FIGURE 5.
LPS alters CTCF occupancy across the mucin gene cluster and changes gene expression levels. A, changes in mucin gene expression after exposure to 200 ng/ml LPS were determined by qRT-PCR. For each mucin gene, data were normalized by setting expression in PBS-treated control cells to 1. B, LPS treatment alters CTCF occupancy across the mucin cluster. ChIP for CTCF was carried out after exposure of cells to 200 ng/ml LPS or to PBS. Data are shown as fold enrichment over a rabbit IgG isotype control. ChIP was performed twice, and representative data (consistent profiles in all replicates) are shown.
DISCUSSION
Coordinate regulation of multigene clusters such as the gel-forming mucin gene complex on chromosome 11p15.5 often involves organization of the chromatin into higher order structures, which may be mediated by CTCF. Here, we examined the mechanistic role of CTCF in coordinating the complex pattern of regulation of the gel-forming mucin genes and mediating the long range interactions across the cluster. First, we profiled the expression of the four genes, MUC6, MUC2, MUC5AC, and MUC5B in colon, lung, and pancreatic cancer cells, for the first time using absolute quantification of transcripts. Next, we determined CTCF binding in these cell types by ChIP and found that in cells where the mucin gene cluster was active, CTCF bound at multiple sites. In contrast in fibroblasts, where the mucin genes are not expressed, CTCF binding is only seen at ubiquitous sites, and the cluster is marked by repressive histone modifications such as H3K9Me3 and H3K27Me3 (ENCODE data in supplemental Fig. 1). Interestingly, the predicted CTCF sites in the genes adjacent to the mucins, AP2A2 5′ and Toll interacting protein (TOLLIP) 3′, show relatively low occupancy in the cancer cell lines that we examined (Fig. 2B). Two CTCF binding sites (IV and IX) showed occupancy in all mucin-expressing cell lines, irrespective of gene expression patterns, and these likely correspond to ubiquitous sites observed across the genome. However, several sites showed selective CTCF occupancy that correlated, at least in part, with gene expression profiles (Table 1). For example, Calu-3 and HT-29 express only MUC5AC at high levels (Fig. 1), and CTCF occupancy in both cell types is similar (Fig. 2B) with the exception of site XI. In comparison in A549 and Capan-1 cells, which both express MUC5B and MUC5AC, lower CTCF occupancy is seen at sites XI and XII, which are located near the 5′ (XI) and 3′ (XII) end of MUC5B (Fig. 2B). LS180 and LS174T express all four mucin genes, although at different abundance, and our absolute expression data showed that LS174T has 10-fold more MUC2 transcript compared with LS180 (Fig. 1). Concurrently, CTCF occupancy shown by ChIP at sites III and IV, which are located between MUC6 and MUC2, is decreased in LS174T in comparison with LS180 (Fig. 2B).
TABLE 1.
Absolute quantification of mucin gene expression and CTCF occupancy across the 11p15.5 region
Absolute quantification of mucin gene expression and CTCF occupancy across the 11p15.5 region. Gene expression data from Fig. 1 for the four gel-forming mucin genes are classified into none, very low, low, medium, high, and very high expression for each mucin in each cell line. White, none; pale gray, very low; light gray, low; medium gray, mid; dark gray, high; black, very high. CTCF ChIP data from Fig. 2B are shown with low, medium, or high occupancy for each of the 13 sites in each cell line. White, N/A, pale gray, low; medium gray, mid; dark gray, high.
siRNA-mediated depletion of CTCF in A549 cells increased MUC2 expression ∼3-fold (Fig. 3A) and in LS180 increased both MUC6 and MUC5AC expression (Fig. 3C). ChIP for CTCF subsequent to its knockdown showed loss of enrichment at most sites in both cell lines, although the effect was generally less at one ubiquitous CTCF site (IV) than at the cell-selective CTCF binding sites (Fig. 3, B and D). These data show that CTCF is a negative regulator of gel-forming mucin gene expression. It is likely based on data from other loci (29, 53, 54) that CTCF suppresses gene expression by recruiting a repressive complex such as the polycomb group repressor complex 2 or SIN3A and associated histone deacetylases. Consistent with these observations, previous reports showed an increase in MUC2 expression following histone deacetylase 2 depletion (19) and that expression of MUC2 was apparently dependent on H3K9 and H3K27 acetylation in the 5′-flanking region (55). However, our data showing that CTCF depletion increases expression of MUC2, MUC5AC, and MUC6 in different cell lines are in contrast with observations that only MUC2 and MUC5B are subject to epigenetic regulation, with MUC5AC being rarely influenced and MUC6 not at all (19).
To elucidate the biological relevance of CTCF binding at the 11p15.5 mucin gene cluster, LPS from P. aeruginosa was used to modulate mucin gene expression. LPS treatment significantly increased MUC2 and MUC5AC expression (Fig. 5A), and this correlated with and likely resulted from decreased CTCF binding at several sites across the cluster (Fig. 5B). Moreover, LPS treatment significantly reduced MUC5B expression (∼25%), suggesting a coordinated regulatory mechanism for the gene cluster, whereby up-regulation of one or more gel-forming mucins is accompanied by down-regulation of others. The mechanism whereby MUC5B expression is decreased may involve the establishment of repressive histone modifications, consistent with previous observations on the regulation of MUC5B (19). These results are consistent with our data on siRNA-mediated depletion of CTCF, which show that CTCF is a repressor of mucin gene expression (Fig. 3). They also suggest that one mechanism of LPS-induction of gel-forming mucin gene expression is the inhibition of CTCF binding at certain sites across the cluster. This inhibition may also involve direct or indirect interactions with NF-κB at the mucin gene cluster. The impact of LPS activation on individual mucin gene promoters was studied previously (56, 57). P. aeruginosa LPS interacts with Toll-like receptor 4, which activates the Src/Ras/MAPK/pp90rsk pathway resulting in nuclear localization of NF-κB and its binding to elements close to the MUC2 and MUC5AC promoters (reviewed in Ref. 10). Our results suggest that LPS may have a more global role across the gene cluster. These data also suggest that CTCF may play an important role in other inflammatory pathways that up-regulate mucin gene expression but are activated by agents other than LPS.
We investigated long range interactions across the gel-forming mucin gene cluster by q3C and found higher order structures across the region. Using the MUC2 promoter as the bait region, we observed interactions with the 5′ end of MUC6 and the 3′ end of AP2A2 (Fig. 4). The MUC2 promoter interaction with the 5′ end of MUC6 may depend on both genes being expressed, as seen in LS180 and LS174T. If MUC2 is silent (Capan-1 and A549), its promoter does not interact with the 5′ end of MUC6 but still interacts with the 3′ end of AP2A2 (Fig. 4). Another long range interaction that seems dependent on gene activity is between the MUC2 promoter and regions close to the 3′ end of MUC5AC and the 5′ end of MUC5B in Capan-1 and A549. These long range interactions may be critical for facilitating expression of the gel-forming mucin genes, creating chromatin loops to prevent inappropriate cross-regulation between adjacent genes such as MUC6 and MUC2 and at the same time providing feedback to communicate gene expression patterns to other loci in the cluster. The characterization of an extra level of transcriptional regulation across the gel-forming mucin gene cluster, in addition to the control of individual genes by their respective promoters, may be relevant to human disease. Many diseases of the airway epithelium such as chronic obstructive pulmonary disease, asthma, and cystic fibrosis, gastrointestinal inflammatory diseases, and carcinomas are associated with gel-forming mucin hypersecretion. The identification of CTCF as a critical mediator in controlling gene expression at this cluster may provide novel therapeutic opportunities because the protein is known to recruit repressive complexes that are suitable for pharmacological manipulation.
Supplementary Material
Acknowledgments
We thank Drs. Gunnar Hansson (MUC2), Thécla Lesuffleur (MUC5AC), and Marie-Pierre Buisine and Isabelle Van Seuningen (MUC5B) for the mucin cDNAs clones.
This work was supported in part by the National Institutes of Health Grants R01 HL094585 and R01 HD068901 and the Cystic Fibrosis Foundation.

This article contains supplemental Table 1 and Figs. 1 and 2.
- CTCF
- CCCTC-binding factor
- q3C
- quantitative chromosome conformation capture
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Rosenbloom K. R., Dreszer T. R., Pheasant M., Barber G. P., Meyer L. R., Pohl A., Raney B. J., Wang T., Hinrichs A. S., Zweig A. S., Fujita P. A., Learned K., Rhead B., Smith K. E., Kuhn R. M., Karolchik D., Haussler D., Kent W. J. (2010) ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38, D620–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sproul D., Gilbert N., Bickmore W. A. (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6, 775–781 [DOI] [PubMed] [Google Scholar]
- 3. Desseyn J. L., Aubert J. P., Porchet N., Laine A. (2000) Evolution of the Large Secreted Gel-Forming Mucins. Mol. Biol. Evol. 17, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 4. Thornton D. J., Rousseau K., McGuckin M. A. (2008) Structure and Function of the Polymeric Mucins in Airways Mucus. Annu. Rev. Physiol. 70, 459–486 [DOI] [PubMed] [Google Scholar]
- 5. Hollingsworth M. A., Swanson B. J. (2004) Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4, 45–60 [DOI] [PubMed] [Google Scholar]
- 6. Tam A., Wadsworth S., Dorscheid D., Man S. F., Sin D. D. (2011) The airway epithelium: more than just a structural barrier. Ther. Adv. Respir. Dis. 5, 255–273 [DOI] [PubMed] [Google Scholar]
- 7. Rose M. C., Voynow J. A. (2006) Respiratory Tract Mucin Genes and Mucin Glycoproteins in Health and Disease. Physiol. Rev. 86, 245–278 [DOI] [PubMed] [Google Scholar]
- 8. Copin M. C., Devisme L., Buisine M. P., Marquette C. H., Wurtz A., Aubert J. P., Gosselin B., Porchet N. (2000) From normal respiratory mucosa to epidermoid carcinoma: Expression of human mucin genes. Int. J. Cancer 86, 162–168 [DOI] [PubMed] [Google Scholar]
- 9. Balagué C., Audié J. P., Porchet N., Real F. X. (1995) In situ hybridization shows distinct patterns of mucin gene expression in normal, benign, and malignant pancreas tissues. Gastroenterology 109, 953–964 [DOI] [PubMed] [Google Scholar]
- 10. Van Seuningen I., Pigny P., Perrais M., Porchet N., Aubert J. P. (2001) Transcriptional regulation of the 11p15 mucin genes. Towards new biological tools in human therapy, in inflammatory diseases and cancer? Front. Biosci. 6, D1216–1234 [DOI] [PubMed] [Google Scholar]
- 11. Pigny P., Guyonnet-Duperat V., Hill A. S., Pratt W. S., Galiegue-Zouitina S., d'Hooge M. C., Laine A., Van-Seuningen I., Degand P., Gum J. R., Kim Y. S., Swallow D. M., Aubert J. P., Porchet N. (1996) Human mucin genes assigned to 11p15.5: identification and organization of a cluster of genes. Genomics 38, 340–352 [DOI] [PubMed] [Google Scholar]
- 12. Toribara N. W., Roberton A. M., Ho S. B., Kuo W. L., Gum E., Hicks J. W., Gum J. R., Jr., Byrd J. C., Siddiki B., Kim Y. S. (1993) Human gastric mucin. Identification of a unique species by expression cloning. J. Biol. Chem. 268, 5879–5885 [PubMed] [Google Scholar]
- 13. Reid C. J., Harris A. (1998) Developmental expression of mucin genes in the human gastrointestinal system. Gut 42, 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gum J. R., Byrd J. C., Hicks J. W., Toribara N. W., Lamport D. T., Kim Y. S. (1989) Molecular cloning of human intestinal mucin cDNAs. Sequence analysis and evidence for genetic polymorphism. J. Biol. Chem. 264, 6480–6487 [PubMed] [Google Scholar]
- 15. Guyonnet Duperat V., Audie J. P., Debailleul V., Laine A., Buisine M. P., Galiegue-Zouitina S., Pigny P., Degand P., Aubert J. P., Porchet N. (1995) Characterization of the human mucin gene MUC5AC: a consensus cysteine-rich domain for 11p15 mucin genes? Biochem. J. 305, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy M. G., Rahmani M., Hernandez J. R., Alexander S. N., Ehre C., Ho S. B., Evans C. M. (2011) Mucin production during prenatal and postnatal murine lung development. Am. J. Respir. Cell Mol. Biol. 44, 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reid C. J., Gould S., Harris A. (1997) Developmental Expression of Mucin Genes in the Human Respiratory Tract. Am. J. Respir. Cell Mol. Biol. 17, 592–598 [DOI] [PubMed] [Google Scholar]
- 18. Thai P., Loukoianov A., Wachi S., Wu R. (2008) Regulation of Airway Mucin Gene Expression. Annu. Rev. Physiol. 70, 405–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vincent A., Perrais M., Desseyn J. L., Aubert J. P., Pigny P., Van Seuningen I. (2007) Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene 26, 6566–6576 [DOI] [PubMed] [Google Scholar]
- 20. Yamada N., Kitamoto S., Yokoyama S., Hamada T., Goto M., Tsutsumida H., Higashi M., Yonezawa S. (2011) Epigenetic regulation of mucin genes in human cancers. Clin. Epigenetics 2, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spilianakis C. G., Flavell R. A. (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 22. Vernimmen D., De Gobbi M., Sloane-Stanley J. A., Wood W. G., Higgs D. R. (2007) Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26, 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Majumder P., Gomez J. A., Chadwick B. P., Boss J. M. (2008) The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 205, 785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Majumder P., Boss J. M. (2010) CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell Biol. 30, 4211–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 26. Hou C., Dale R., Dean A. (2010) Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. U.S.A. 107, 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krivega I., Dean A. (2012) Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev. 22, 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lobanenkov V. V., Nicolas R. H., Adler V. V., Paterson H., Klenova E. M., Polotskaja A. V., Goodwin G. H. (1990) A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 [PubMed] [Google Scholar]
- 29. Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., Neiman P. E., Collins S. J., Lobanenkov V. V. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell Biol. 16, 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohlsson R., Renkawitz R., Lobanenkov V. (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends in Genetics 17, 520–527 [DOI] [PubMed] [Google Scholar]
- 31. Bell A. C., West A. G., Felsenfeld G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- 32. Phillips J. E., Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyle A. P., Song L., Lee B. K., London D., Keefe D., Birney E., Iyer V. R., Crawford G. E., Furey T. S. (2011) High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 21, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., Ren B. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuddapah S., Jothi R., Schones D. E., Roh T. Y., Cui K., Zhao K. (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tom B. H., Rutzky L. P., Jakstys M. M., Oyasu R., Kaye C. I., Kahan B. D. (1976) Human colonic adenocarcinoma cells. I. Establishment and description of a new line. In Vitro. 12, 180–191 [DOI] [PubMed] [Google Scholar]
- 37. Fogh J., Trempe G. (1975) New human tumor cell lines in Human Tumor Cells in Vitro. (Fogh J., Ed.) pp. 115–141, Plenum Publishing Corp., New York [Google Scholar]
- 38. Lieber M., Smith B., Szakal A., Nelson-Rees W., Todaro G. (1976) A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int. J. Cancer 17, 62–70 [DOI] [PubMed] [Google Scholar]
- 39. Shen B. Q., Finkbeiner W. E., Wine J. J., Mrsny R. J., Widdicombe J. H. (1994) Calu-3: a human airway epithelial cell line that shows cAMP-dependent Cl− secretion. Am. J. Physiol. 266, L439–501 [DOI] [PubMed] [Google Scholar]
- 40. Tan M. H., Nowak N. J., Loor R., Ochi H., Sandberg A. A., Lopez C., Pickren J. W., Berjian R., Douglass H. O., Jr., Chu T. M. (1986) Characterization of a new primary human pancreatic tumor line. Cancer Invest. 4, 15–23 [DOI] [PubMed] [Google Scholar]
- 41. Graham F. L., Smiley J., Russell W. C., Nairn R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74 [DOI] [PubMed] [Google Scholar]
- 42. Leir S. H., Harris A. (2011) MUC6 mucin expression inhibits tumor cell invasion. Exp. Cell Res. 317, 2408–2419 [DOI] [PubMed] [Google Scholar]
- 43. Lidell M. E., Johansson M. E., Mörgelin M., Asker N., Gum J. R., Jr., Kim Y. S., Hansson G. C. (2003) The recombinant C-terminus of the human MUC2 mucin forms dimers in Chinese-hamster ovary cells and heterodimers with full-length MUC2 in LS 174T cells. Biochem. J. 372, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lesuffleur T., Roche F., Hill A. S., Lacasa M., Fox M., Swallow D. M., Zweibaum A., Real F. X. (1995) Characterization of a mucin cDNA clone isolated from HT-29 mucus-secreting cells. The 3′ end of MUC5AC? J. Biol. Chem. 270, 13665–13673 [DOI] [PubMed] [Google Scholar]
- 45. Porchet N., Dufosse J., Audie J. P., Duperat V. G., Perini J. M., Nguyen V. C., Degand P., Aubert J. P. (1991) Structural Features of the Core Proteins of Human Airway Mucins Ascertained by cDNA Cloning. Am. Rev. Resp. Dis. 144, S15–18 [DOI] [PubMed] [Google Scholar]
- 46. Ott C. J., Blackledge N. P., Kerschner J. L., Leir S. H., Crawford G. E., Cotton C. U., Harris A. (2009) Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. U.S.A. 106, 19934–19939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 48. Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 49. Blackledge N. P., Ott C. J., Gillen A. E., Harris A. (2009) An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 37, 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dohrman A., Miyata S., Gallup M., Li J. D., Chapelin C., Coste A., Escudier E., Nadel J., Basbaum C. (1998) Mucin gene (MUC 2 and MUC 5AC) upregulation by Gram-positive and Gram-negative bacteria. Biochim. Biophys. Acta 1406, 251–259 [DOI] [PubMed] [Google Scholar]
- 51. Smirnova M. G., Guo L., Birchall J. P., Pearson J. P. (2003) LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell. Immunol. 221, 42–49 [DOI] [PubMed] [Google Scholar]
- 52. Hauber H. P., Goldmann T., Vollmer E., Wollenberg B., Zabel P. (2007) Effect of Dexamethasone and ACC on Bacteria-Induced Mucin Expression in Human Airway Mucosa. Am. J. Respir. Cell Mol. Biol. 37, 606–616 [DOI] [PubMed] [Google Scholar]
- 53. Li T., Hu J. F., Qiu X., Ling J., Chen H., Wang S., Hou A., Vu T. H., Hoffman A. R. (2008) CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell Biol. 28, 6473–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lutz M., Burke L. J., Barreto G., Goeman F., Greb H., Arnold R., Schultheiss H., Brehm A., Kouzarides T., Lobanenkov V., Renkawitz R. (2000) Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 28, 1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamada N., Hamada T., Goto M., Tsutsumida H., Higashi M., Nomoto M., Yonezawa S. (2006) MUC2 expression is regulated by histone H3 modification and DNA methylation in pancreatic cancer. Int. J. Cancer 119, 1850–1857 [DOI] [PubMed] [Google Scholar]
- 56. Li D., Gallup M., Fan N., Szymkowski D. E., Basbaum C. B. (1998) Cloning of the Amino-terminal and 5′-Flanking Region of the Human MUC5AC Mucin Gene and Transcriptional Up-regulation by Bacterial Exoproducts. J. Biol. Chem. 273, 6812–6820 [DOI] [PubMed] [Google Scholar]
- 57. Li J. D., Dohrman A. F., Gallup M., Miyata S., Gum J. R., Kim Y. S., Nadel J. A., Prince A., Basbaum C. B. (1997) Transcriptional activation of mucin by Pseudomonas aeruginosa lipopolysaccharide in the pathogenesis of cystic fibrosis lung disease. Proc. Natl. Acad. Sci. U.S.A. 94, 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.