Abstract
Plants are sessile organisms, and they can not move away under abiotic or biotic stresses. Thus plants have evolved a set of genes that response to adverse environment to modulate gene expression. In this study, we characterized and functionally studied an ERF transcription factor from Artemisia annua, AaERF1, which plays an important role in biotic stress responses. The AaERF1 promoter had been cloned and GUS staining results of AaERF1 promoter-GUS transgenic A. annua showed that AaERF1 is expressed ubiquitiously in all organs. Several putative cis-acting elements such as W-box, TGA-box and Py-rich element, which are involved in defense responsiveness, are present in the promoter. The expression of AaERF1 can be induced vigorously by methyl jasmonate as well as by ethephon and wounding, implying that AaERF1 may activate some of the defense genes via the jasmonic acid and ethylene signaling pathways of A. annua. The results of electrophoretic mobility shift assay (EMSA) and yeast one-hybrid experiments showed that AaERF1 was able to bind to the GCC box cis-acting element in vitro and in yeast. Ectopic expression of AaERF1 could enhance the expression levels of the defense marker genes PLANT DEFENSIN1.2 (PDF1.2) and BASIC CHITINASE (ChiB), and increase the resistance to Botrytis cinerea in the 35S::AaERF1 transgenic Arabidopsis. The down-regulated expression level of AaERF1 evidently reduced the resistance to B. cinerea in A. annua. The overall results showed that AaERF1 positively regulated the resistance to B. cinerea in A. annua.
Introduction
The necrotrophic fungus Botrytis cinerea causes significant economic losses throughout the world as a destructive pathogen of a broad spectrum of plant species [1]. Plants are sessile, thus they have evolved some gene families to cope with pathogen attack through complex adaptive responses. The AP2/ERF transcription factors are one of the most important families that are involved in plant response to biotic and abiotic stresses as well as in the development of various plant species [2]. The AP2/ERF transcription factors, which have the conserved AP2/ERF transcription factors binding domains of 57–66 amino acids, constitute a large multigene family divided into five subfamilies named AP2, CBF/DREB, ERF, RAV and the fifth (comprising members not assigned to other four groups) [3], [4]. The AP2 subfamily proteins contain two repeated AP2/ERF domains, while the RAV family proteins contain a B3 domain and a single AP2/ERF domain. In contrast to the AP2 and RAV subfamily members, the CBF/DREB and ERF subfamily proteins contain single AP2/ERF domain [5]. The genes in the CBF/DREB subfamily play a crucial role in the resistance of plants to abiotic stresses by recognizing the dehydration responsive or cold-repeat element (DRE/CRT) with a core motif of A/GCCGAC [6]. The ERF subfamily is often involved in the response to plant stresses like pathogenesis by modulating the expression of their target genes via binding to the cis-acting element AGCCGCC, known as the GCC box in their promoters [5].
Jasmonic acid (JA) and ethylene (ET) are two important hormones that act synergistically during plant resistance to necrotrophic pathogens such as B. cinerea by activating some ERF genes, which are responsive to both JA and ET treatment. For example, in Arabidopsis, two ERF genes, ERF1 and ORA59, are induced by JA and ET [7], [8]. Overexpression of either ERF1 or ORA59 resulted in constitutive expression of the defense marker genes, PLANT DEFENSIN1.2 (PDF1.2) and BASIC CHITINASE (B-CHI), thus enhancing the resistance to B. cinerea [7], [8]. A recent study also showed that another two ERF genes, ERF5 and ERF6, play redundant roles as positive regulators of JA/ET-mediated defense against B. cinerea in Arabidopsis [9]. Thus, at least four ERFs play key role in integrating the JA and ET signal in disease resistance. In previous studies, our group also showed that two ERFs from Gossypium barbadense, showed positive effects on disease resistance [10], [11]. ERF genes from other species, such as tomato (TSRF1 and Pti4), soybean (GmERF3) and Bupleurum kaoi (BkERF1, BkERF2.1 and BkERF2.2) also showed positive effects on disease resistance, suggesting the ERF genes have a conserved role in diverse species to counteract with plant pathogens [5], [12]–[16].
Artemisia annua L. is an important medicinal plant that produces artemisinin, which is widely used in malaria treatment. A recent study has shown that JA can increase artemisinin production by inducing two ERF genes in A. annua. AaERF1 and AaERF2, both of which directly bind to the CRTDREHVCBF2 (CBF2) and RAV1AAT (RAA) motifs present in both ADS and CYP71AV1 promoters to activate those key enzymes in artemisinin biosynthesis pathway [17]. Our previous study showed that wounding stress also significantly elevated the artemisinin content by increasing ADS and CYP71AV1 expression levels [18]. Compared to the great effort in artemisinin biosynthesis pathway, little is known about the disease resistance in A. annua. Since ERFs are key regulators that integrate JA and ET signals in disease resistance, it is attempted to establish whether AaERF1 has a role in disease resistance. Thus, our research focused on the function of AaERF1 in plant antifungal field and illustrated that AaERF1, which could bind to GCC box in in vitro and in yeast, positively regulated the resistance to B. cinerea in A. annua.
Results
AaERF1 is Ubiquitously Expressed in A. annua
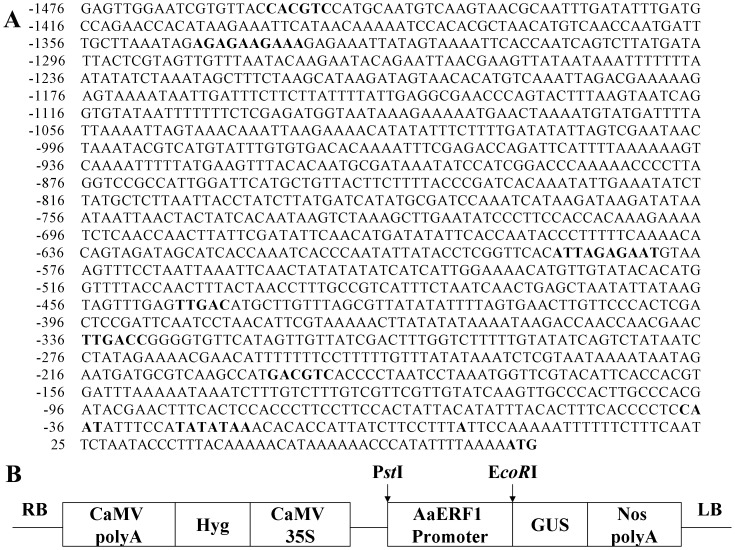
The promoter sequence of AaERF1(JQ513909)was cloned by genomic walking (Figure 1A). To observe the expression pattern of AaERF1 in details, the AaERF1 promoter was subcloned to the pCAMBIA1391Z vector (Figure 1B) and then AaERF1 promoter-GUS transgenic A. annua plants were generated. Six lines of the transgenic A. annua plants expressing the GUS and three lines for the wild-type background were prepared. All the lines showed similar fusion protein expression. GUS activity was detected in all tissues examined, including roots, stems, leaves and flowers (Figure 2A, 2B, 2C and 2D). In 1-month-old plants, GUS activity was high in root tips, stems and leaves (Figure 2A, 2B and 2C). During the flowering period, GUS activity was also detected in flower buds. So, AaERF1 is ubiquitously expressed in A. annua. From Figure 2B and 2C, GUS expression was also detected in the glandular trichomes and T-shaped trichomes. No signals were observed in the negative control plants transformed with pCAMBIA1391 empty vector (Figure S1).
Figure 1. Sequence of AaERF1 promoter region and construction of AaERF1 promoter-GUS vector.
(A) The sequence of AaERF1 promoter region. The transcription initiation site and the translation start site are in bold, underlined letters. Numbers indicate the position relative to the transcription start site. Putative cis-acting regulatory elements involved in defense responsiveness in AaERF1 promoter are in bold. (B) Construction of AaERF1 promoter-GUS vector. PstI and EcoRI are the enzymes used in the construction.
Figure 2. Localization of AaERF1 expression using GUS staining of promoter:GUS transgenic plants.
GUS activity is revealed by histochemical staining. (A) Root. (B) Stem. (C) Leaf. (D) Flower buds.
Prediction of cis-acting Elements of Promoter Region of AaERF1
Putative cis-acting elements of the promoter were predicted using the PLANTCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Figure 1A; Table 1). A putative TATA box sequence was found at -27 bp, and the putative CAAT box sequence was located at -38 bp. The 5′-UTR pyrimidine-rich stretch site is a cis-acting element conferring high transcription levels. Such an element was found at position -1345 to -1336 as shown in Figure 1A. A TC-rich repeat, which is involved in defense and stress response, was localized to position -590 to -581. A TGA-box element (TGACGTCA), which is involved in plant defense responsiveness, was found at position -209 to -201. A G/C-box element (CACGTC), which is involved in light-induction or hormone control, was found at position -1458 to -1453. The W box is a fungal elicitor responsive element, which was present at positions -547 to -542 bp and -336 to -332 bp in AaERF1 promoter. A search for the regulatory elements in AaERF1 promoter also carried EIRE box. The above cis-acting elements are summarized in Table 1. Nearly all these cis-acting elements are related to defense responsiveness. Consequently, AaERF1 may be a defense responsiveness transcription factor in A. annua.
Table 1. Putative cis-acting regulatory elements involved in defense responsiveness in AaERF1 promoter.
| Cis-elements | Motif and position | Putative function |
| 5-UTR pyrimidine-rich stretch consensus:TTTCTTCTCT | −1345 AGAGAAGAAA -1336 | cis-acting element conferring high transcription |
| EIRE-box: TTGACC | −336 TTGACC -331 | elicitor responsive element |
| W-box consensus: TTGAC | −547 TTGAC -542; -336 TTGAC -332 | fungal elicitor responsive element |
| TGA-box: TGACGTCA | −209 TGACGTCA -201 | cis-acting element conferring plant defense responsiveness |
| G/C-box consensus: CACGTC | −1458 CACGTC -1453 | cis-acting element involved in light-induction or hormone control |
| TC-rich repeats: ATTTTCTTCA | −590 ATTAGAGAAT -581 | cis-acting element involved in defense and stress responsiveness |
Expression Profiling Analysis of AaERF1 after Hormone and Stress Treatments
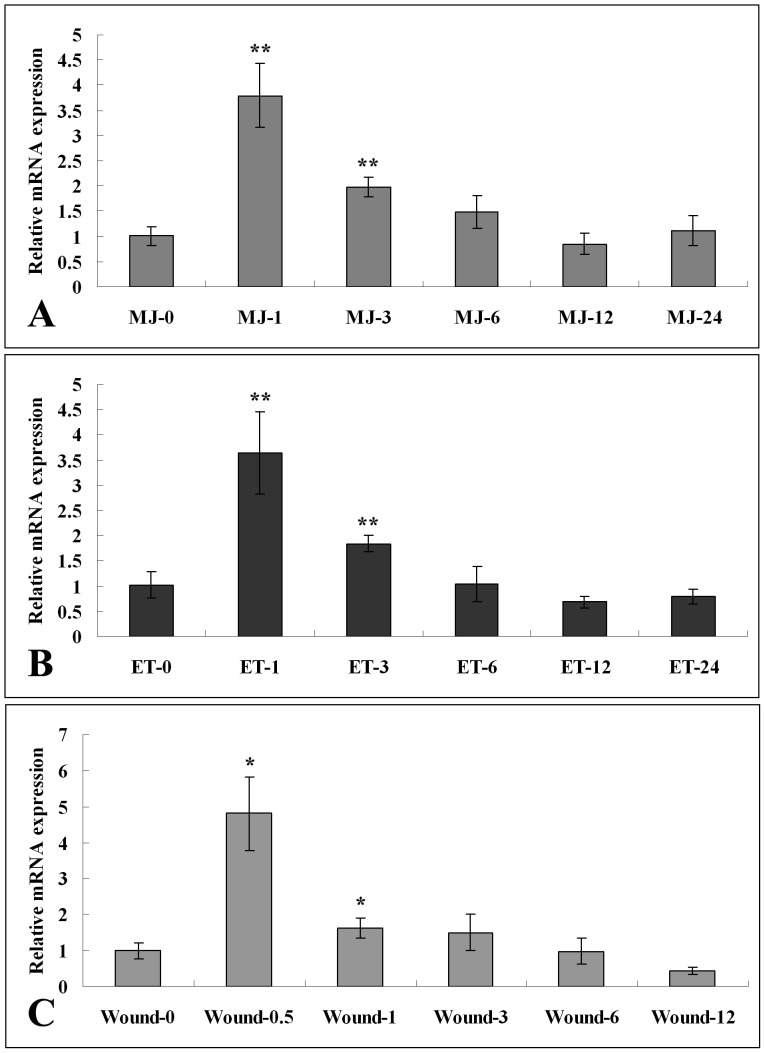
In this study, RT-Q-PCR analysis was used to obtain the expression pattern of AaERF1 after hormone and stress treatments including MeJA (100 µM), ethephon (500 µM) and wound treatments. The transcript level of AaERF1 was increased rapidly and peaked within 1 h after MeJA treatment, followed by a gradually decline (Figure 3A). The treatment with ethephon shows a similar expression pattern with the treatment of MeJA (Figure 3B). The transcript level of AaERF1 was also sensitive to stress treatments. Wounding could induce a significant accumulation of AaERF1 transcript in a short time period (0.5 h). Then the transcript level was quickly decreased (Figure 3C). The statistics analysis showed that the observed differences were statistically significant.
Figure 3. Expression patterns of AaERF1 in response to hormone and stress treatments by RT-Q-PCR.
A. Relative expression levels of AaERF1 after MeJA (100 µM). B. Relative expression levels of AaERF1 after ethephon (500 µM). C. Relative expression levels of AaERF1 after wound treatment. Total RNA was isolated respectively from A. annua leaves under different treatments for different periods of time (0 h, 0.5 h, 1 h, 3 h, 6 h, 12 h and 24 h) followed by RT-Q-PCR analysis with the gene-specific primers AaERF1-RT-F and AaERF1-RT-R. Values indicate the mean fold relative to sample 0 h. Actin is used as a control for normalization. Data are averages ± SE from three independent experiments.
Comparative and Bioinformatic Analyses of AaERF1
The results of the BLAST-Protein (BLASTP) online (http://www.ncbi.nlm.gov/blast) showed that the AaERF1 protein had a highly conserved AP2 domain with other ERF proteins, including Arabidopsis AtERF1, AtERF2, ORCA3, LeERF1, NtERF1, TaERF3 and ORA59 (Figure S2A). This domain is divided into two conserved segments of YRG and RAYG, in which a β-sheet and α-helix are predicted (β-α motif; see Figure S2A). A phylogenetic tree of ERF proteins was drawn using the CLUSTAL X program. The phylogenetic tree demonstrated that ERF proteins originated from a common ancestor and diverged into several groups (Figure S2B). According to the phylogenetic tree, the protein of AaERF1 had close evolutionary relationships to AtERF2, LeERF1, NtERF1 and TaERF3 which showed that they might share similar functions in disease resistance (Figure S2B).
AaERF1 Protein Interacts with the GCC Box in vitro
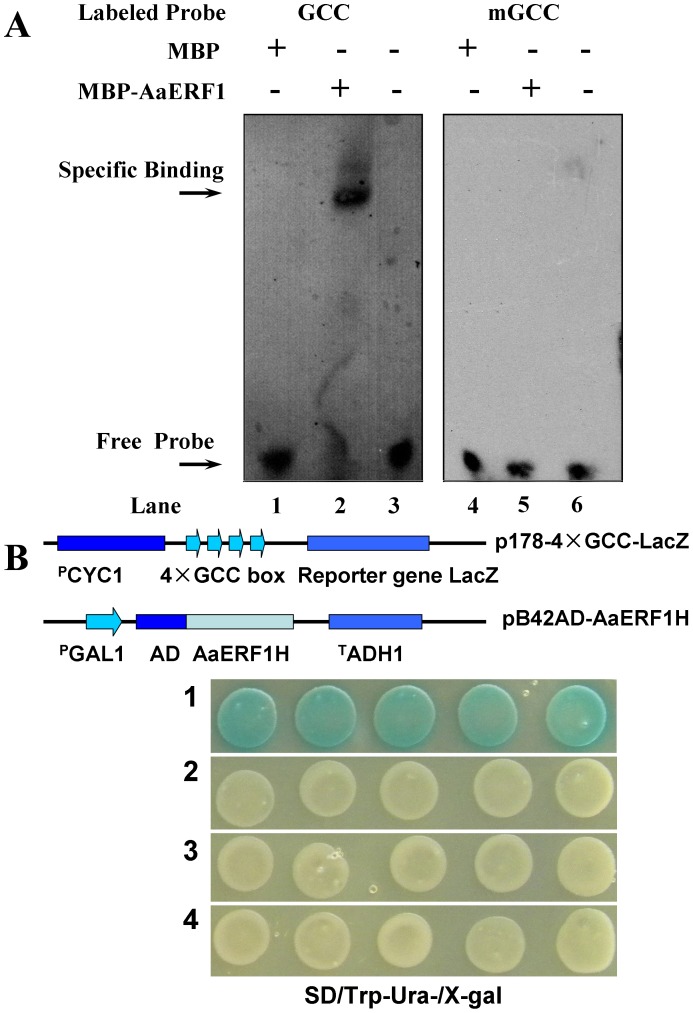
Since the AP2 domain of AaERF1 contained the key amino acids to bind the GCC box, the recombinant MBP-AaERF1 protein was constructed and overexpressed in E. coli BL21, purified, and used to examine the DNA binding ability in vitro. The purified MBP-AaERF1 protein was mixed, respectively, with the labeled wild-type GCC probe or a mutated GCC probe in the binding reaction. The results of EMSA showed that the gel mobility shift was specific to the MBP-AaERF1 protein with the labeled GCC probe (lane 2 in Figure 4A). As expected, there were no shifted bands in the combination of MBP-AaERF1 plus the mutated GCC (mGCC) probe (lane 5 in Figure 4A) and in the negative controls, including MBP with the labeled GCC probe (lane 1) or mGCC probe (lane 4), and only the labeled GCC probe (lane 3) or mGCC probe (lane 6) (Figure 4A). The results demonstrated that AaERF1 was able to bind to the GCC box cis-acting element, but not to the mutated GCC box in vitro.
Figure 4. The DNA binding ability of AaERF1 via GCC box.
A. Electrophoretic mobility shift assays on DNA binding of AaERF1. Lane 1: negative controls contain MBP plus labelled GCC probe; lane 2: the MBP–AaERF1 protein plus labelled GCC probe; lane 3: only labelled GCC probe; lane 4: negative controls contain MBP plus labelled mutated GCC probe; lane 5: MBP–AaERF1 plus labelled mutated GCC probe; lane 6: only labelled mutated GCC probe. The protein–GCC probe complex and free probes are indicated respectively by two arrows. B. GCC box binding analysis of AaERF1 using the yeast one-hybrid system. Sketch maps show the construction of vectors used in this experiment. Photographs show the growth behavior of transformants on SD/Trp−Ura−/X-gal medium. Sector 1: p178-4×GCC-LacZ+pB42AD-AaERF1; sector 2: p178+ pB42AD-AaERF1; sector 3: p178-4×GCC-LacZ+pB42AD; sector 4: p178+ pB42AD.
AaERF1 can Bind to the GCC Box in Yeast
The yeast one-hybrid system is a stable system to study the DNA binding ability of transcription factors [19]. The results of yeast one-hybrid and β-galactosidase activity assays indicated that only the hybrid cells containing the combination of pB42AD::AaERF1 and p178-4×GCC-LacZ showed β-galactosidase activity compared with other combinations, including pB42AD with p178-LacZ, pB42AD::AaERF1 with p178-4×GCC-LacZ, pB42AD::AaERF1 with p178-LacZ, and pB42AD with p178-4×GCC-LacZ. The results demonstrated that AaERF1 could bind to the GCC box cis-acting element in yeast cells (Figure 4B).
AaERF1-overexpression in Arabidopsis Causes the Increase of Disease Resistance to B. cinerea
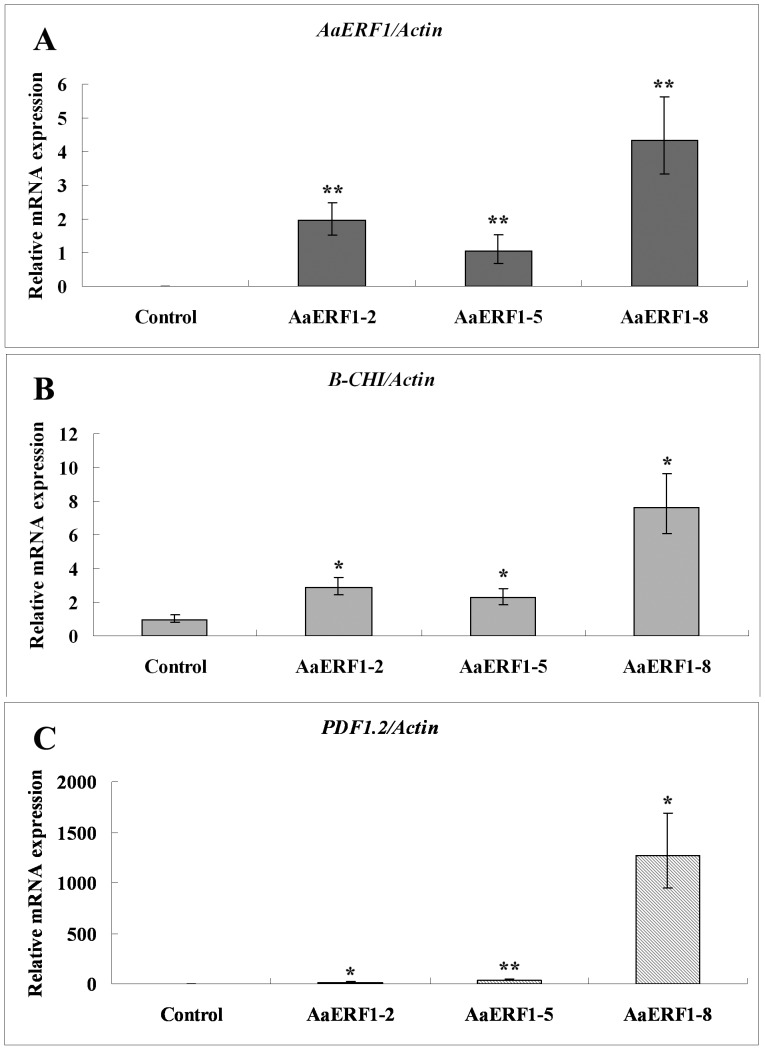
The transgenic Arabidopsis plants were first confirmed by kanamycin-resistant screening and genomic DNA-based PCR, and then three transgenic lines were chosen for further analysis. The control experiment involving the transfer of empty plasmid p2300+ to Arabidopsis was also conducted. The results showed that the transcript levels of AaERF1 had a significant increase in AaERF1-overexpression lines (Figure 5A). Correspondingly, Chi-B was shown to be elevated between 2.3- and 7.7-fold in AaERF1-overexpression lines (Figure 5B). The transcript levels of PDF1.2 were elevated between 15- and 1269-fold than that of the control (Figure 5C). The statistics analysis showed that the observed differences were statistically significant.
Figure 5. The expression levels of AaERF1, Chi-B and PDF1.2 in 35S::AaERF1 transgenic Arabidopsis analyzed by RT-Q-PCR.
Vertical bars represent standard deviation. A. The expression of AaERF1 in the control and transgenic Arabidopsis plants. Values indicate the mean fold relative to sample the AaERF1-5 transgenic plants. B. The expression of Chi-B in the control and transgenic Arabidopsis plants. Values indicate the mean fold relative to sample the pCAMBIA2300+ empty vector transgenic plants C. The expression of PDF1.2 in the control and transgenic Arabidopsis plants. Values indicate the mean fold relative to sample the pCAMBIA2300+ empty vector transgenic plants. Actin is used as a control for normalization. Data are averages ± SE from three independent experiments.
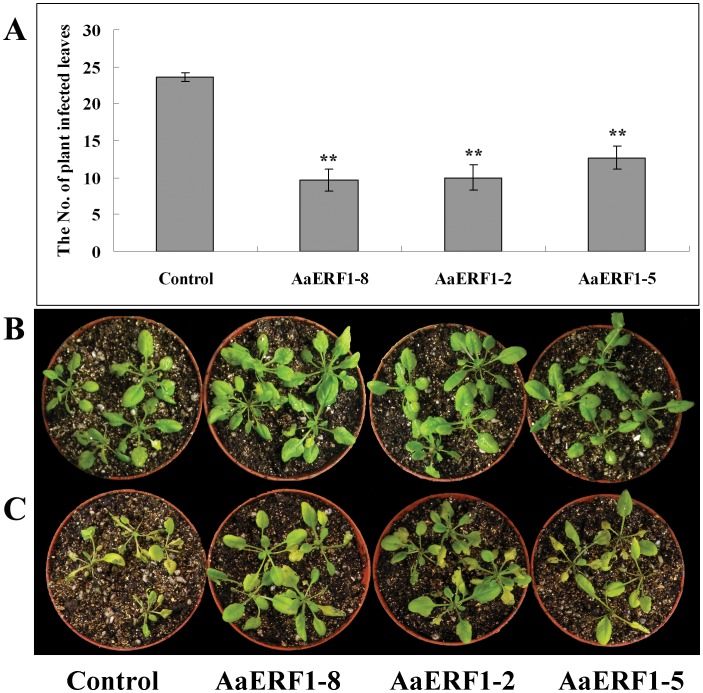
The AaERF1-overexpression lines were observed following inoculation with B. cinerea. For each of the AaERF1-overexpression lines, we observed a significant reduction in the development of disease symptoms in independent inoculation experiments. Four days following inoculation with B. cinerea, 79% of the control plants showed symptoms of infection, whereas only between 32% and 42% of the leaves from AaERF1-overexpression lines were symptomatic (Figure 6A, 6C). The statistics analysis showed that the observed differences were statistically significant. The control plants turned dry and died, while most of the AaERF1-overexpression plants were growing well (Figure 6B, 6C). The results showed that the overexpression of AaERF1 could increase the disease resistance to B. cinerea in Arabidopsis.
Figure 6. The 35S: AaERF1 lines show increased disease resistance.
A. The numbers of control and the three independent 35S: AaERF1 transgenic Arabidopsis lines showing disease symptoms 4 d after inoculation with Botrytis cinerea. Average data with standard errors from three biological replicates are shown. B. The control and 35S: AaERF1 lines, without inoculation with Botrytis cinerea. C. The control and 35S: AaERF1, 4 d after inoculation with Botrytis cinerea, with 35S: AaERF1 plants showing reduced disease symptoms (see “Materials and Methods” for description).
Down-regulated Expression Level of AaERF1 in A. annua Causes the Reduction of Disease Resistance to B. cinerea
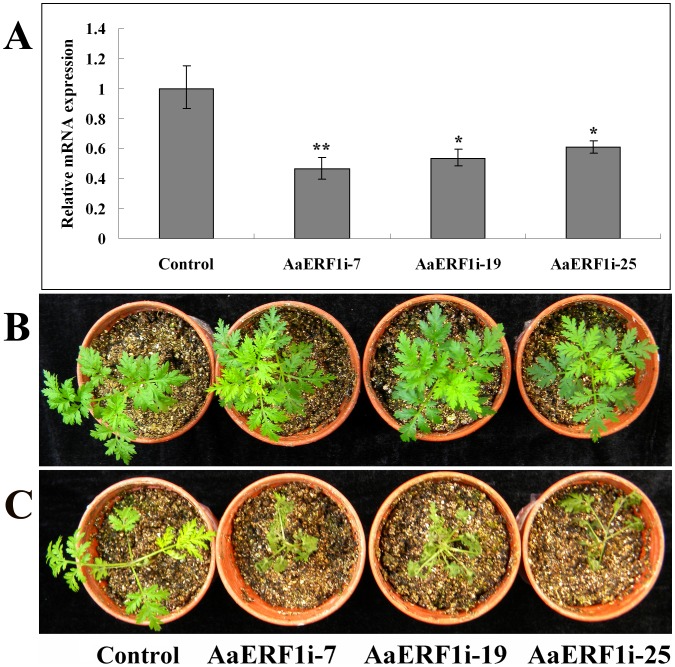
Here, we constructed the RNAi vector of AaERF1 and transformed it into A. annua. The control experiment involving the transfer of empty plasmid pCAMBIA2300+ to A. annua was also conducted. The transgenic plants were first confirmed by genomic DNA-based PCR using the 35S forward primer, AaERF1 reverse primer and the reverse primer of kanamycin-resistant gene (Figure S3), and then three independent transgenic lines were chosen for further analysis. In the RNAi transgenic lines, the transcript levels of AaERF1 were suppressed to 46–61% of the control level (Figure 7A). The statistics analysis showed that the observed differences were statistically significant.
Figure 7. The RNAi lines of AaERF1 show decreased disease resistance.
A. The expression of AaERF1 in the empty vector and AaERF1i transgenic A. annua plants. Error bars are SE (n = 3). B. The empty vector and AaERF1i lines, without inoculation with Botrytis cinerea. C. The empty vector and AaERF1i lines, 6 d after inoculation with Botrytis cinerea, with AaERF1i lines showing increased disease symptoms. The experiment was performed three times with similar results.
The three independent AaERF1i lines were inoculated with B. cinerea. The results showed that each of the AaERF1i lines had a significant reduction in the disease symptoms in three independent inoculations. Six days following inoculation with B. cinerea, most of the leaves in AaERF1i lines were dry and dead, while most of the the control plants were growing well (Figure 7B). The results showed that AaERF1 was a positive regulator to the disease resistance to B. cinerea in A. annua.
Discussion
The putative cis-acting elements of AaERF1 promoter were predicted as shown in Figure1A and summarized in Table 1. The W box (TTGAC) is the binding site for members of the WRKY family of transcription factors [20]. The importance of W boxes was illustrated by studies on Arabidopsis transcription during systemic-acquired resistance [21]. Previous reports indicated that the G-box–related hexamers(CACNTG,CACATG and (T/C)ACGTG)are the binding sites of MYC2 [22]–[24]. MYC2 is a negative regulator of the JA-responsive pathogen defense genes PDF1.2 and B-CHI [25]. At -209bp of AaERF1 promoter, there is a TGA motif, which is a perfect binding site for TGA transcription factors. TGA transcription factors are essential for the activation of JA and ET dependent defense mechanisms in Arabidopsis [26]. In addition, we identified the TC-rich repeats (ATTTTCTCCA) in the promoter of AaERF1, which was previously described in tobacco (Nicotiana tabacum) as cis-acting elements involved in defence and stress responsiveness [27]. All above elements are related to the disease resistance, implying that AaERF1 may have a similar function.
The results of RT-Q-PCR showed that the transcript level of AaERF1 was increased rapidly and peaked within 1 h after MeJA, ethephon and wound treatments, followed by a gradual decline. Jasmonates and ethylene are considered the major signal compounds for wound-induced gene expression in plants [28]. Methyl jasmonate was reported as a volatile compound emitted from the leaves of A. tridentata subspecies tridentata resulting in the induction of defense-related genes in nearby tomato plants [29]. So, the transcript of AaERF1 can be induced vigorously by MeJA, ethephon or wound treatments, implying that AaERF1 may play an important role in the JA and ET signaling pathways and have some function in disease resistance of A. annua.
The bioinformatic analysis showed that the AP2/ERF domain of AaERF1 contained a β-sheet and α-helix (β-α motif; see Figure S2A), all of which are important for DNA binding with the GCC Box [30], [31]. The phylogenetic tree analysis of AaERF1 showed that AaERF1 had close relationship with AtERF2 and TaERF3. AtERF2 and TaERF3 have been well characterized and their functions were mainly related to disease resistance, at least in part, via binding to the GCC box in the promoter region of downstream genes [19], [32]–[34]. So, all above analysis implied that the protein of AaERF1 has a function in disease resistance and may have the GCC Box binding ability.
From the results of EMSA and yeast one-hybrid experiment, we know that AaERF1 was able to bind to the GCC box cis-acting element in vitro and in yeast cells. The ERF subfamily of proteins recognizes the cis-acting element GCC box, which is mainly involved in the response to biotic stresses like pathogenesis [5]. Enhancement of disease resistance in plants has been achieved by overexpressing ERF proteins, such as Arabidopsis AtERF1 [8], [35], AtERF2 [31] and rice OsBIERF3 [36]. So, we infer that the overexpression of AaERF1 could enhance the disease resistance in plants.
PDF1.2 and Chi-B in Arabidopsis were marker genes of the resistance to several fungi, including B. cinerea [35], [37]. The results of RT-Q-PCR showed that the transcripts of AaERF1, Chi-B and PDF1.2 showed an obvious correlated increase in AaERF1-overexpression lines, which were similar with the overexpression of ORA59 in Arabadopsis [8] (Figure 5A, 5B and 5C). After the inoculation with B. cinerea, the control lines dried and died, while most of the AaERF1-overexpression lines were growing well (Figure 6). The results showed that overexpression of AaERF1 could increase the resistance to B. cinerea in Arabidopsis.
Six days after inoculated with B. cinerea, nearly all the AaERF1i transgenic A. annua showed symptoms of infection, while the control plant were growing well (Figure 7B). Yu et al. showed that AaERF1 could directly bind to the CBF2 and RAA motifs present in both ADS and CYP71AV1 promoters [17]. In the AaERF1i transgenic lines, as a result of reduced ADS and CYP71AV1 gene expression, the contents of artemisinin and artemisinic acid were decreased to 76–58% and 55–30% of the wild-type level, respectively [17]. For large amounts of specialized metabolites are considered briefly and related to demonstrated or presumed roles in plant defense [38], [39], the reduction of artemisinin and artemisinic acid may result in reduction of the resistance to B. cinerea in A. annua. From the above results, we conclude that AaERF1 is a positive regulator of the resistance to B. cinerea in A. annua.
In conclusion, the promoter of AaERF1 was cloned by genomic walking and the GUS staining results of AaERF1 promoter-GUS transgenic A. annua showed that AaERF1 is ubiquitously expressed in A. annua. The expression of AaERF1 can be induced vigorously by MeJA, ethephon and wound treatments, implying that AaERF1 may activate some of the defense genes via the JA and ET signaling pathways of A. annua. Electrophoretic mobility shift assay (EMSA) and yeast one-hybrid results showed that AaERF1 was able to bind to the GCC box cis-acting element in vitro and in yeast. The overexpression of AaERF1 could enhance the expression levels of Chi-B and PDF1.2 and increase the resistance to B. cinerea in the 35S::AaERF1 transgenic Arabidopsis. The down-regulated expression level of AaERF1 evidently reduced the resistance to B. cinerea in A. annua. These data suggested that AaERF1 could not only regulate the artemisinin biosynthetic pathway, but also play important roles as a positive regulator of the resistance to B. cinerea in A. annua.
Materials and Methods
Plant Materials
The seeds of A. annua were obtained from the School of Life Sciences, Southwest University in Chongqing, P.R. China. The plants of A. annua were grown in a greenhouse. Arabidopsis thaliana ecotype Columbia-0 was used in this study and grown under 16 h light (70 mmol m-2s-1) and 8 h dark cycle at 22°C. Different tissues of A. annua and Arabidopsis plants were collected for RNA extraction using plant RNA isolation reagent (Tiangen Biotech, Beijing) following the manufacturer’s instructions. The concentration of the purified RNA was quantified with a nucleic acid analyser (Nanodrop-1000, Nano).
Isolation and Analysis the AaERF1 Promoter
The upstream region of AaERF1 was amplified from the genomic DNA using the Genome Walker Kit (Clontech, Canada). The AaERF1-specific primers (AaERF1-sp1, AaERF1-sp2, Adaptor Prime1 and Adaptor Prime2) were used following the manufacturer's recommended procedures. The products amplified from the final reaction products were electrophoresed in 1% agarose gel, and a 1543 bp fragment was eluted from the gel and cloned into the pMD18-T-simple vector. The insert DNA was sequenced by Shenzhen Genomics Institute. The sequence obtained was searched for putative cis-acting elements previously characterized using the PlantCare software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
β-galactosidase (GUS) Expression in Transgenic A. annua
To generate the AaERF1 promoter-GUS construct, the 5′-flanking DNA of the AaERF1 coding region was amplified with AaERF1-PF and AaERF1-PR. The 1.5 kb PCR fragment was cloned into the pCAMBIA1391Z vector for sequence confirmation. The construct was transformed into A. annua plants as described previously [40]. Histochemical staining for GUS activity in transgenic plants was performed as the protocol described previously [41]. Plants transformed with pCAMBIA1391Z were used as a parallel negative control.
Hormonal and Stress Treatments
A. annua plants grown in MS medium for 2 weeks were treated with solutions of 100 µM MeJA (Sigma Aldrich, USA) and 500 µM Ethephon. Ethephon, an ethylene releaser, was used as ethylene replacement [42]. Since ethephon on hydrolysis releases ethylene and phosphorus, therefore the disodium hydrogen phosphate buffer (Na2HPO3, 5 mM) was prepared for the decomposition of ethephon. 100 mL ethephon solution was mixed with 100 mL disodium hydrogen phosphate (5 mM). Ten plants were transfered to new petri dishes and pooled for each treatment. For wound induction, the same mixed plants of A. annua were cut some 2–3 mm nicks and kept at 22°C under humidified conditions. For the hormonal treatments, all the plants were collected before treatment (0 h) and at different time point after treatment (1 h, 3 h, 6 h, 12 h, 24 h) for the gene expression analysis. For the wound treatments, a time point of 0.5 h was added.
Expression Pattern Analysis of AaERF1 by RT-Q-PCR
Expression patterns of AaERF1 in response to hormonal and stress treatments of A. annua were analyzed using the RT-Q-PCR method. The expression levels of AaERF1, Chi-B and PDF1.2 in AaERF1-overexpressing Arabidopsis were also analysed by this method. All RNA samples were digested with DNase I (RNase-free) prior to use. Aliquots of 0.4 µg total RNA were employed in the reverse transcriptase reaction using random hexamer primers for the synthesis of first strand cDNA. The amplification reactions of qRT-PCR were performed on an iCycler iQTM Real-Time PCR Machines (Bio-Rad, Watford, UK) with gene-specific primers (Table S1), and the SYBR ExScript RT-PCR kit (Takara, Shiga, Japan) protocol to confirm changes of gene expression. The RT-Q-PCR cycling was performed at 37°C (3 min), 95°C (4 min), 40 cycles at 95°C (15 s), 56°C (40 s), 72°C (40 s). The expression of all genes was normalized against the expression of the endogenous control gene (Actin). Values displayed are mean fold change as calculated by the 2-ΔΔCT method. All experiments were repeated three times.
Comparative and Bioinformatic Analysis
Based on the sequence of AaERF1 (JN162091), one pair of primers (AaERF1-F and AaERF1-R) were designed, synthesized and used to amplify the full-length sequence of AaERF1 from A. annua. Comparative and bioinformatic analysis of AaERF1 were carried out online at the websites, http://www.ncbi.nlm.nih.gov and http://cn.expasy.org. Sequence analysis was performed using DNAMAN software (Lynnon Biosoft, USA) and Vector NTI software (Invitrogen). The phylogenetic analysis of AaERF1 protein and ERF proteins from other species was carried out by alignment with CLUSTAL X (1.81) using default parameters. A phylogenetic tree was constructed by neighbor-joining method using software MEGA version 3.1 [43], [44].
Electrophoretic Mobility Shift Assay
The AaERF1 cDNA sequence was cloned into the EcoRI and PstI sites of the pMAL-C2 vector to produce a MBP-AaERF1 fusion construct (New England BioLabs). The sequenced pMAL-C2-AaERF1 construct was introduced into E. coli BL21 for expression. Fusion proteins were expressed in BL21 cells by adding 0.5 mM IPTG to culture medium for 7 h at 28°C and purified using amylose resin (New England BioLabs). The purified recombinant protein was quantified using the Bradford assay (2-D Quant Kit, Amersham Biosciences Corp., San Francisco, CA, USA). The 3′ end biotin-labelled oligonucleotides for the GCC box and the mutated mGCC box were synthesized (Sangon) and equimolar pairs were annealed using the protocol provided by Sigma (Table 1). The binding reaction, gel preparation and electrophoretic mobility-shift assay (EMSA) were performed following the protocol of Zhang et al. [45].
Yeast one-hybrid
To analyze the GCC-binding activity of AaERF1 protein, the entire encoding regions of AaERF1 was fused into the BamHI-XhoI sites of the activation domain of the pB42AD vector (pAD). Reporter vectors containing the 4 × GCC lacZ was prepared and integrated into the yeast strain EGY48, inserted upstream of the minimal promoter element (MP) and the reporter gene lacZ existed in the vector p178. Vectors were introduced into yeast strain through LiAc mediated transformation method (Clontech, Shanghai, China). The cells were grown on tryptophan- and uracil- deficient SD medium for 2–3 days at 30°C, and then transferred to 5-bromo-4-chloro-3-indolyl-beta-Dgalactopyranoside (X-gal) containing plates for color change observation [46].
Overexpression in A. thaliana
The full-length AaERF1 coding sequence was amplified with primers AaERF1-F and AaERF1-R by Platinum PrimeSTAR HS DNA polymerase (Takara, Shiga, Japan) and subcloned into pMD18-T simple vector. The pMD18-T-AaERF1 vector was digested by SacI and BamHI. The full-length ORF of AaERF1 was cloned into the BamHI and SacI sites of the pCAMBIA2300+ vector under the 35S promoter to generate pCAMBIA2300-35S::AaERF1::NOS. The construct was transferred into Agrobacterium tumefaciens GV3101, and then introduced into A. thaliana (ecotype Columbia) plants using the floral dip method [47]. Transgenic plants were selected on MS plates containing 50 µg/mL kanamycin. PCR was performed to verify the transgenic status of the screened plants.
RNAi in A. annua
The 201 bp fragment of AaERF1, corresponding to AaERF1 cDNA from nucleotides 179–378, was cloned from A. annua by RT–PCR. In RT–PCR, AaERF1i-F (with XbaI and XhoI sites) and AaERF1i-R (with BamHI and HindIII sites) were used as the forward and reverse primers, respectively. The amplified fragment was cloned into pMD18-T simple vector and sequenced. After confirmation by sequencing, the fragments were forwardly and reversely placed on the two end sides of the GUS intron in pBluescript SK+ to construct the hp structure. Then the expression cassette was excised with SacI and KpnI from pBluescript SK+ containing the AaERF1 hp structure and ligated into the expression vector pCAMBIA2300+ to get the final hp AaERF1i-containing vector pCAMBIA2300:: p35S-hairpin AaERF1-nos. pCAMBIA2300 vector containing only nptII (neomycin phosphotransferase gene conferring resistance to kanamycin) was used as the control vector in transformation. The pCAMBIA2300:: p35S-hairpin AaERF1-nos and pCAMBIA2300+ vectors were then transferred into A. tumefaciens strain EHA105 by a conventional freezing and thawing method, and the resulting strains were used in the transformation of A. annua. The transformation of A. annua was performed following the protocol of Zhang et al. [40]. All the primers used in this study are in the Table S1.
Pathogen Infections
B. cinerea was grown on potato dextrose agar plates for about 2 weeks at 26°C. Spores were collected from the cultures and washed twice with sterile water. Washed spores were suspended by 10 mL of sterile water, and the suspension was filtered through miracloth to remove mycelia. Four-week-old plants were spray with spore suspensions (2 × 105 spores mL-1) and maintained under high humidity. Disease development was observed over the following 6 d. Inoculated plants were scored based on the presence of any disease symptoms after 4 d inoculation with B. cinerea, including chlorosis of the leaves, curling and necrosis of the leaves. For each of the control and transgenic plant lines, three biological replicates were performed in parallel.
Supporting Information
Gus-staining of transgenic A. annua using the pCAMBIA1391Z empty vector plasmid.
(TIF)
Comparison of AP2/ERF domain sequences and dendrogram of ERF proteins. A. Amino acid alignment of the AP2/ERF domains between AaERF1 and ERF proteins. Highly conserved residues in all the sequences are indicated in white with black background and only partially conserved residues in ERF proteins are showed in black with grey background. One α-helix and three β-sheets are marked above the corresponding sequences. The YRG and RAYD elements are indicated with solid lines below the consensus sequence. B. A phylogenetic tree of the ERF proteins was constructed. Alignments were made in Clustal X using the default parameters. Accession numbers for the AP2/ERF proteins used are as follows: AtERF1, AF076277; AtERF2, NM124093; AtERF3, XP002894264; AtERF4, NM112384; AtERF5, NM124094; AtERF6, Q8VZ91; AtERF7, NM112922; AtERF8, Q9MAI5; AtERF9, Q9FE67; AtERF10, Q9ZWA2; ERF1, AAD03545; ORA59, NM100497; LeERF1, Q84XB3; TaERF3, EF570122; NtERF1, Q40476;ORCA3, EU072424; GmERF3, EU681278; AaERF1, JN162091).
(TIF)
Analysis of transgenic A.annua plants by PCR. A. PCR analysis of 35S forward primer and AaERF1 reverse primer in AaERF1-RNAi transgenic plants. M: DNA size marker DL2000, V: empty-vector transgenic A. annua, C: water control, P: positive control. B. PCR analysis of 35S-forward primer and the reserse prmer of kanamycin-resistant gene in AaERF1-RNAi transgenic plants.
(TIF)
Primers used in this study.
(DOC)
Acknowledgments
We thank Dr. Qiong Wu (Shanghai Jiao Tong University) for providing the bacterial strain of Botrytis cinerea. Note: The novel nucleotide sequence data published here has been deposited in the EMBL/DDBJ/GenBank databases under accession number JQ513909.
Funding Statement
This work was funded by China “863” Program (grant no. 2011AA100605), China Transgenic Research Program (grant no. 2011ZX08002–001), Ministry of Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jarvis WR (1980) Epidemiology. In: Coley-Smith JR, Verhoeff K and Jarvis WR, editors. The Biology of Botrytis. London: Academic Press, 181–217. [Google Scholar]
- 2. Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140: 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, et al. (2002) DNA-binding specificity of the ERF/AP2 domain of transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng JX, Liu D, Pan Y, Gong W, Ma LG, et al. (2005) An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol 59: 853–868. [DOI] [PubMed] [Google Scholar]
- 5. Zhang GY, Chen M, Li LC, Xu ZS, Chen XP, et al. (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60: 3781–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanism. Annu Rev Plant Biol 50: 571–599. [DOI] [PubMed] [Google Scholar]
- 7. Solano R, Stepanova A, Chao QM, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, et al. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H (2012) ERF5 and ERF6 Play Redundant Roles as Positive Regulators of JA/Et-Mediated Defense against Botrytis cinerea in Arabidopsis. PLOS One 7, doi:10.1371/journal.pone.0035995. [DOI] [PMC free article] [PubMed]
- 10. Qin J, Zuo K, Zhao J, Ling H, Cao Y, et al. (2006) Overexpression of GbERF confers alteration of ethylene-responsive gene expression and enhanced resistance to Pseudomonas syringae in transgenic tobacco. J Biosci 31: 255–263. [DOI] [PubMed] [Google Scholar]
- 11. Zuo KJ, Qin J, Zhao JY, Ling H, Zhang LD, et al. (2007) Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene 391: 80–90. [DOI] [PubMed] [Google Scholar]
- 12. Fischer U, Dröge-Laser W (2004) Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol Plant Microbe Interact 17: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 13. Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, et al. (2003) New members of the tomato ERF family show specific expression pattern and diverse DNA binding capacity to the GCC box element. FEBS Lett 550: 149–154. [DOI] [PubMed] [Google Scholar]
- 14. Zhang HB, Zhang DB, Chan J, Yang YH, Huang ZJ, et al. (2004) Tomato stressresponsive actor TSRF1 interacts with ethylene responsive lement GCC box and regulates pathogen resistance to Ralstonia olanacearum . Plant Mol Biol 55: 825–834. [DOI] [PubMed] [Google Scholar]
- 15. Zhang HB, Li W, Chen J, Yang YH, Zhang Z, et al. (2007) Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol Biol 63: 63–71. [DOI] [PubMed] [Google Scholar]
- 16. Liu WY, Chiou SJ, Ko CY, Lin TY (2011) Functional characterization of three ethylene response factor genes from Bupleurum kaoi indicates that BkERFs mediate resistance to Botrytis cinerea . J Plant Physiol 168: 375–381. [DOI] [PubMed] [Google Scholar]
- 17. Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, et al. (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant. 5: 353–365. [DOI] [PubMed] [Google Scholar]
- 18. Liu DH, Zhang LD, Li CX, Yang K, Wang YY, et al. (2010) Effect of wounding on gene expression involved in artemisinin biosynthesis and artemisinin production in Artemisia annua . Russ J Plant Physiol 57: 882–886. [Google Scholar]
- 19. Liang HX, Lu Y, Liu HX, Wang FD, Xin ZY, et al. (2008) A novel activator-type ERF of Thinopyrum intermedium, TiERF1, positively regulates defence responses. J Exp Bot 11: 3111–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rushton PJ, Torres JT, Parniske M, Wermert P, Hahlbrock K, et al. (1996) Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parseley PR1 genes. EMBO J 15: 5690–700. [PMC free article] [PubMed] [Google Scholar]
- 21. Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, et al. (2000) Arabidopsis MAP kinase 4 negatively regulated systemic acquired resistance. Cell 103: 1111–20. [DOI] [PubMed] [Google Scholar]
- 22. De Pater S, Pham K, Memelink J, Kijne J (1997) RAP-1 is an Arabidopsis MYC-like R protein homologue that binds to G-box sequence motifs. Plant Mol. Biol 34: 169–174. [DOI] [PubMed] [Google Scholar]
- 23. Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, et al. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dombrech B, Xue GP, Sprague SJ, Kirkegaar JA, Ross JJ, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zander M, Camera SL, Lamotte O, Métraux JP, Gatz C (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant Journal 61: 200–210. [DOI] [PubMed] [Google Scholar]
- 27. Klotz KL, Lagrimini LM (1996) Phytohormone control of the tobacco anionic peroxidase promoter. Plant Mol Biol 31: 565–573. [DOI] [PubMed] [Google Scholar]
- 28. Hiraga S, Ito H, Sasaki K, Yamakawa H, Mitsuhara I, et al. (2000) Wound-induced expression of a tobacco peroxidase is not enhanced by ethephon and suppressed by methyl jasmonate and coronatine. Plant Cell Physiol 41: 165–170. [DOI] [PubMed] [Google Scholar]
- 29. Farmer EE, Ryan CA (1990) Interplant communication: airborne methyljasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Nat. Acad Sci USA 87: 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (1998) A novel model of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J 17: 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mazarel M, Puthoff DP, Hart JK, Rodermel SR, Baum T (2002) Identification and characterization of a soybean ethylene responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol Plant Microbe Interact 15: 577–586. [DOI] [PubMed] [Google Scholar]
- 32. McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, et al. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and diseaseres istance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang ZY, Yao WL, Dong N, Liang HX, Liu HX, et al. (2007) A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments. J Exp Bot 58: 2993–3003. [DOI] [PubMed] [Google Scholar]
- 34. Jung J, Won SY, Suh SC, Kim H, Wing R, et al. (2007) The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 225: 575–588. [DOI] [PubMed] [Google Scholar]
- 35. Berrocal LM, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32. [DOI] [PubMed] [Google Scholar]
- 36. Cao YF, Wu YF, Zheng Z, Song FM (2006) Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol Mol Plant P 67: 202–211. [Google Scholar]
- 37. Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Currt Opin Plant Biol 1: 404–411. [DOI] [PubMed] [Google Scholar]
- 38. Levin DA (1973) The role of trichomes in plant defense. Q Rev Biol 48: 3–15. [Google Scholar]
- 39. Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711. [DOI] [PubMed] [Google Scholar]
- 40. Zhang L, Jing FY, Li FP, Li MY, Wang YL, et al. (2009) Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol Appl Bioc 52: 199–207. [DOI] [PubMed] [Google Scholar]
- 41. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang W, Wen CK (2010) Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiology and Biochemistry 48: 45–53. [DOI] [PubMed] [Google Scholar]
- 43. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 44. Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- 45. Zhang F, Zuo KJ, Zhang JQ, Liu X, Zhang LD, et al. (2010) An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J Exp Bot 13: 3599–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye R, Yao QH, Xu ZH, Xue HW (2004) Development of an efficient method for the isolation of factors involved in gene transcription during rice embryo development. Plant J 38: 348–357. [DOI] [PubMed] [Google Scholar]
- 47. Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gus-staining of transgenic A. annua using the pCAMBIA1391Z empty vector plasmid.
(TIF)
Comparison of AP2/ERF domain sequences and dendrogram of ERF proteins. A. Amino acid alignment of the AP2/ERF domains between AaERF1 and ERF proteins. Highly conserved residues in all the sequences are indicated in white with black background and only partially conserved residues in ERF proteins are showed in black with grey background. One α-helix and three β-sheets are marked above the corresponding sequences. The YRG and RAYD elements are indicated with solid lines below the consensus sequence. B. A phylogenetic tree of the ERF proteins was constructed. Alignments were made in Clustal X using the default parameters. Accession numbers for the AP2/ERF proteins used are as follows: AtERF1, AF076277; AtERF2, NM124093; AtERF3, XP002894264; AtERF4, NM112384; AtERF5, NM124094; AtERF6, Q8VZ91; AtERF7, NM112922; AtERF8, Q9MAI5; AtERF9, Q9FE67; AtERF10, Q9ZWA2; ERF1, AAD03545; ORA59, NM100497; LeERF1, Q84XB3; TaERF3, EF570122; NtERF1, Q40476;ORCA3, EU072424; GmERF3, EU681278; AaERF1, JN162091).
(TIF)
Analysis of transgenic A.annua plants by PCR. A. PCR analysis of 35S forward primer and AaERF1 reverse primer in AaERF1-RNAi transgenic plants. M: DNA size marker DL2000, V: empty-vector transgenic A. annua, C: water control, P: positive control. B. PCR analysis of 35S-forward primer and the reserse prmer of kanamycin-resistant gene in AaERF1-RNAi transgenic plants.
(TIF)
Primers used in this study.
(DOC)