Abstract
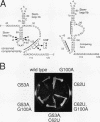
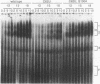
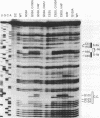
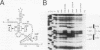
Mutations in stem-loop IIa of yeast U2 RNA cause cold-sensitive growth and cold-sensitive U2 small nuclear ribonucleoprotein function in vitro. Cold-sensitive U2 small nuclear RNA adopts an alternative conformation that occludes the loop and disrupts the stem but does so at both restrictive and permissive temperatures. To determine whether alternative U2 RNA structure causes the defects, we tested second-site mutations in U2 predicted to disrupt the alternative conformation. We find that such mutations efficiently suppress the cold-sensitive phenotypes and partially restore correct U2 RNA folding. A genetic search for additional suppressors of cold sensitivity revealed two unexpected mutations in the base of an adjacent stem-loop. Direct probing of RNA structure in vivo indicates that the suppressors of cold sensitivity act to improve the stability of the essential stem relative to competing alternative structures by disrupting the alternative structures. We suggest that many of the numerous cold-sensitive mutations in a variety of RNAs and RNA-binding proteins could be a result of changes in the stability of a functional RNA conformation relative to a competing structure. The presence of an evolutionarily conserved U2 sequence positioned to form an alternative structure argues that this region of U2 is dynamic during the assembly or function of the U2 small nuclear ribonucleoprotein.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Igel A. H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990 Dec;4(12A):2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Sproat B. S., Lamond A. I. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992 Sep 11;20(17):4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S. E., Tyc K., Kastner B., Reichelt J., Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993 Jan;13(1):307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoo B. B., Sherman F., Azubalis D. A., Fjellstedt T. A., Mehnert D., Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979 Sep;93(1):51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Cui Z., Mason T. L. A single nucleotide substitution at the rib2 locus of the yeast mitochondrial gene for 21S rRNA confers resistance to erythromycin and cold-sensitive ribosome assembly. Curr Genet. 1989 Oct;16(4):273–279. doi: 10.1007/BF00422114. [DOI] [PubMed] [Google Scholar]
- Dammel C. S., Noller H. F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993 Apr;7(4):660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Cross-linking of U2 snRNA using nitrogen mustard. Evidence for higher order structure. J Biol Chem. 1992 Mar 5;267(7):4497–4502. [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991 Aug 29;352(6338):821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- Fearon K., Mason T. L. Structure and function of MRP20 and MRP49, the nuclear genes for two proteins of the 54 S subunit of the yeast mitochondrial ribosome. J Biol Chem. 1992 Mar 15;267(8):5162–5170. [PubMed] [Google Scholar]
- Fortner D. M., Troy R. G., Brow D. A. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev. 1994 Jan;8(2):221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci U S A. 1969 Jun;63(2):384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Igel A. H., Ares M., Jr Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature. 1988 Aug 4;334(6181):450–453. doi: 10.1038/334450a0. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson D. J., Rahe B., Pringle J., Beggs J. D. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991 Feb 21;349(6311):715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of Drosophila pre-mRNAs. Nucleic Acids Res. 1985 Jul 11;13(13):4971–4981. doi: 10.1093/nar/13.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Madhani H. D., Bordonné R., Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990 Dec;4(12B):2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992 Nov 27;71(5):819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Miraglia L., Seiwert S., Igel A. H., Ares M., Jr Limited functional equivalence of phylogenetic variation in small nuclear RNA: yeast U2 RNA with altered branchpoint complementarity inhibits splicing and produces a dominant lethal phenotype. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7061–7065. doi: 10.1073/pnas.88.16.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986 Jun 20;45(6):869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H. F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Shannon K. W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991 May;5(5):773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Strauss E. J., Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991 Apr;5(4):629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- Sweeney R., Yao C. H., Yao M. C. A mutation in the large subunit ribosomal RNA gene of Tetrahymena confers anisomycin resistance and cold sensitivity. Genetics. 1991 Feb;127(2):327–334. doi: 10.1093/genetics/127.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991 May;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai P. C., Kessler D. P., Ingraham J. Cold-sensitive mutations in Salmonella typhimurium which affect ribosome synthesis. J Bacteriol. 1969 Mar;97(3):1298–1304. doi: 10.1128/jb.97.3.1298-1304.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Guthrie C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985 Dec 30;4(13B):3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wu J. A., Manley J. L. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991 Aug 29;352(6338):818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- Yano R., Yura T. Suppression of the Escherichia coli rpoH opal mutation by ribosomes lacking S15 protein. J Bacteriol. 1989 Mar;171(3):1712–1717. doi: 10.1128/jb.171.3.1712-1717.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavanelli M. I., Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991 Dec;5(12B):2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]