ABSTRACT
Hydrogenotrophic methanogenic Archaea require reduced ferredoxin as an anaplerotic source of electrons for methanogenesis. H2 oxidation by the hydrogenase Eha provides these electrons, consistent with an H2 requirement for growth. Here we report the identification of alternative pathways of ferredoxin reduction in Methanococcus maripaludis that operate independently of Eha to stimulate methanogenesis. A suppressor mutation that increased expression of the glycolytic enzyme glyceraldehyde-3-phosphate:ferredoxin oxidoreductase resulted in a strain capable of H2-independent ferredoxin reduction and growth with formate as the sole electron donor. In this background, it was possible to eliminate all seven hydrogenases of M. maripaludis. Alternatively, carbon monoxide oxidation by carbon monoxide dehydrogenase could also generate reduced ferredoxin that feeds into methanogenesis. In either case, the reduced ferredoxin generated was inefficient at stimulating methanogenesis, resulting in a slow growth phenotype. As methanogenesis is limited by the availability of reduced ferredoxin under these conditions, other electron donors, such as reduced coenzyme F420, should be abundant. Indeed, when F420-reducing hydrogenase was reintroduced into the hydrogenase-free mutant, the equilibrium of H2 production via an F420-dependent formate:H2 lyase activity shifted markedly toward H2 compared to the wild type.
IMPORTANCE
Hydrogenotrophic methanogens are thought to require H2 as a substrate for growth and methanogenesis. Here we show alternative pathways in methanogenic metabolism that alleviate this H2 requirement and demonstrate, for the first time, a hydrogenotrophic methanogen that is capable of growth in the complete absence of H2. The demonstration of alternative pathways in methanogenic metabolism suggests that this important group of organisms is metabolically more versatile than previously thought.
Introduction
Methanogenic Archaea can be grouped into two physiologically distinct groups, the methylotrophs and the hydrogenotrophs (1). Methylotrophic methanogens are relatively versatile, as their substrate repertoire for methanogenesis includes H2 and CO2, acetate, methyl compounds, such as methanol and methylamines, and CO. Hydrogenotrophic methanogens are more restricted, using H2, formate, or for a few species, certain alcohols as electron donors for CO2 reduction to CH4.
Hydrogenotrophic methanogens are distinct from methylotrophic methanogens in their use of electron bifurcation as an energy-conserving step in methanogenesis from CO2 (1–4). Two pairs of electrons enter the methanogenic pathway at a protein complex that contains two key enzymes, heterodisulfide reductase (Hdr) and formylmethanofuran dehydrogenase (Fwd). One pair of electrons is used by Hdr in an exergonic reaction to reduce the heterodisulfide CoM-S-S–CoB to the sulfhydryl coenzymes HS–CoB and HS-CoM, which in turn serve as reductants for reduction of the methyl group on coenzyme M (CH3-S-CoM) to CH4. The other pair of electrons is used in an endergonic reaction by Fwd to reduce CO2 to the formyl group of formyl-methanofuran (formyl-MFR) (for a diagram of the methanogenic pathway, see reference 4).
By coupling the final, methane-producing step to the initial, CO2-reducing step, electron bifurcation renders the pathway cyclic. The cyclic model of methanogenesis was recently named the Wolfe cycle in honor of the contributions of Ralph S. Wolfe (5). The cyclic nature of the pathway can explain in part why hydrogenotrophs have not evolved the metabolic versatility of the methylotrophs. The latter organisms use acetate by oxidizing the carbonyl to provide electrons for reduction of the methyl to methane, and they use methyl compounds by disproportionation where some of the substrate is oxidized and some is reduced to methane. However, in hydrogenotrophs, the stoichiometric coupling of the methane-producing step to the recruitment of CO2 into the pathway prohibits the input of additional intermediates. Furthermore, in the case of acetate, only one pair of electrons is available from the oxidation of the carbonyl, yet electron bifurcation requires that two pairs of electrons must feed into the heterodisulfide reductase complex. Although many hydrogenotrophic methanogens contain acetyl coenzyme A (acetyl-CoA) synthase/CO dehydrogenase (ACS/CODH) (the essential enzyme for aceticlastic methanogenesis), they appear to use it only anabolically for CO2 fixation and not catabolically for acetate utilization.
Either H2 or formate donates electrons to all four reduction steps in methanogenesis from CO2. Both substrates supply electrons directly to the Hdr complex via an Hdr-associated hydrogenase or an Hdr-associated formate dehydrogenase (Fdh) (2), thus providing for the first and last reduction steps in the pathway. The two intermediate reduction steps use the coenzyme F420 as the direct electron donor, and H2 or formate directly reduces F420 via an F420-reducing hydrogenase or Fdh. Hence, H2 or formate alone can provide the stoichiometric reducing requirements for CO2 reduction to methane. However, as another consequence of the Wolfe cycle, the levels of intermediates in methanogenesis must be anaplerotically maintained in order to avoid decaying flux in the pathway (5). In the model species Methanococcus maripaludis we showed how this occurs by genetically eliminating six of the seven hydrogenases encoded in the genome (4). The resulting mutant, designated ∆6H2ase, was unable to use H2 as the stoichiometric electron donor for methanogenesis. The mutant also lacked a formate-H2 lyase activity found in the wild-type strain. As a result of these two deficiencies, the ∆6H2ase mutant required both formate and H2 for growth and methanogenesis. While formate was needed stoichiometrically for the four reduction steps of methanogenesis, H2 was required only in small amounts sufficient to support the anaplerotic needs of methanogenesis. In the ∆6H2ase mutant, only one hydrogenase remained: the membrane-bound energy-converting hydrogenase Eha, which harvests chemiosmotic energy to drive the reduction of a low-potential ferredoxin which in turn is thought to reduce CO2 to formyl-MFR. Hence, during growth on H2 or formate, Eha is essential in wild-type M. maripaludis to anaplerotically replenish methanogenesis at the first reduction step. Indeed, a knockout of the genes encoding Eha was not successful. Consistent with these results, H2 stimulated methanogenesis from formate in cell suspensions (4).
Here we show that the essentiality of H2 and Eha does not always hold, and that in M. maripaludis, there are at least two additional pathways by which the anaplerotic requirements of methanogenesis can be satisfied. We delete genes encoding Eha in a strain derived from the ∆6H2ase mutant that expresses one of these pathways, thus eliminating all H2 metabolism in a hydrogenotrophic methanogen. By reintroducing a single hydrogenase—the F420-reducing hydrogenase—into the hydrogenase-free strain and observing its effects in isolation from other H2-metabolizing pathways, we find that substantial electron flux is diverted to H2 production. These findings show that hydrogenotrophic methanogens are metabolically more versatile than previously thought.
RESULTS
CO stimulates growth of the ∆6H2ase mutant in the absence of H2.
Eha provides anaplerotic input to the methanogenic pathway by acting as a supplement to electron bifurcation for the reduction of a ferredoxin that in turn is used to reduce CO2 to formyl-MFR (4). However, other pathways of ferredoxin reduction/oxidation exist in methanogenic Archaea. The ACS/CODH enzyme complex converts CO2 to CO, which ultimately becomes the carbonyl carbon of acetyl-CoA (6, 7). This reduction is dependent on reduced ferredoxin as an electron donor.
To test whether the reverse reaction of ACS/CODH—CO oxidation to CO2 with the production of reduced ferredoxin—can stimulate methanogenesis in an H2-independent manner, the ∆6H2ase mutant was grown on formate medium with or without the addition of H2 or CO to the culture headspace. As expected, H2 promoted robust growth. In addition, CO promoted growth in the absence of H2 (Fig. 1). Growth rates were much lower with CO in place of H2, suggesting that ferredoxin reduction by Eha is preferred. Growth promoted by CO was directly attributable to ACS/CODH, since when the CO2-reducing subunits were genetically eliminated (∆6H2ase-∆cdh), growth no longer occurred with CO and formate as the only electron donors (Fig. 1).
FIG 1 .
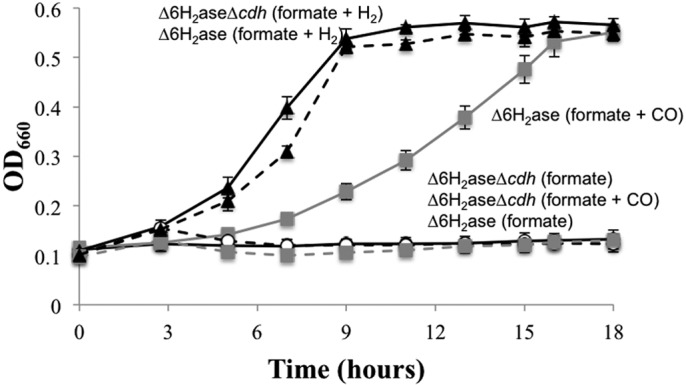
Growth of the ∆6H2ase mutant with CO and formate. The ∆6H2ase mutant (solid black lines) and the ∆6H2ase ∆cdh mutant (broken lines) were grown with formate plus H2 (black symbols), formate plus 5% CO (gray symbols), or formate alone (white symbols). Data points are averages of three cultures, and error bars represent 1 standard deviation around the mean.
Isolation of a suppressor mutation that allows growth of the ∆6H2ase mutant on formate alone.
The growth of the ∆6H2ase mutant with formate and CO suggests that ferredoxin reduction by Eha is not necessary for growth, provided alternative mechanisms to anaplerotically stimulate methanogenesis are present. CH3-S-CoM addition to cell extracts stimulates methanogenesis (8, 9), and methanogenic Archaea are capable of CH3-S-CoM uptake via a poorly characterized and very inefficient activity (10, 11). Therefore, we tried to grow the ∆6H2ase mutant on formate in the absence of H2 in the presence and absence of CH3-S-CoM. Surprisingly, regardless of the presence or absence of CH3-S-CoM, after prolonged incubation of nine independent cultures, all nine grew (Fig. 2). Upon transfer to new medium, each strain tested routinely grew to maximum optical density at 660 nm (OD660) within 24 h. These results suggested that independent suppressor mutations (∆6H2asesup) were generated that allowed for growth on formate alone. Suppressor strains that were generated in the presence of CH3-S-CoM grew well in its absence. Hence, although CH3-S-CoM failed to stimulate growth, mutants developed that had a novel mechanism to anaplerotically stimulate methanogenesis.
FIG 2 .

Generation of suppressor strains of the ∆6H2ase mutant capable of H2-independent growth. The ∆6H2ase mutant was grown in formate-containing medium without H2 or CO. The medium contained 1,000 µM (black symbols), 100 µM (gray symbols), or 0 µM (white symbols) CH3-S-CoM (three replicates each); however, CH3-S-CoM had no stimulatory effect on growth. Each curve represents growth in a single tube.
Deletion of eha is possible due to a suppressor mutation.
A suppressor mutation that allows growth of ∆6H2asesup on formate alone could have produced a novel H2 production activity, or it could have generated a new ferredoxin reducing activity that is independent of H2. If a novel H2 production pathway were responsible, eha would still be essential (4). We attempted deletion of the genes encoding the active site subunits of Eha (ehaNO) in one of the ∆6H2asesup strains (12, 13). Deletion of ehaNO was indeed possible, suggesting that another ferredoxin reduction activity is present in the suppressor background. The new strain (∆7H2asesup) lacks the genes encoding the active sites for all genomically encoded hydrogenases: ∆vhuAU ∆vhcA ∆fruA ∆frcA ∆hmd ∆ehbN ∆ehaNO. The ∆7H2asesup mutant grew in the absence of H2, and H2 did not stimulate growth (Fig. 3A).
FIG 3 .
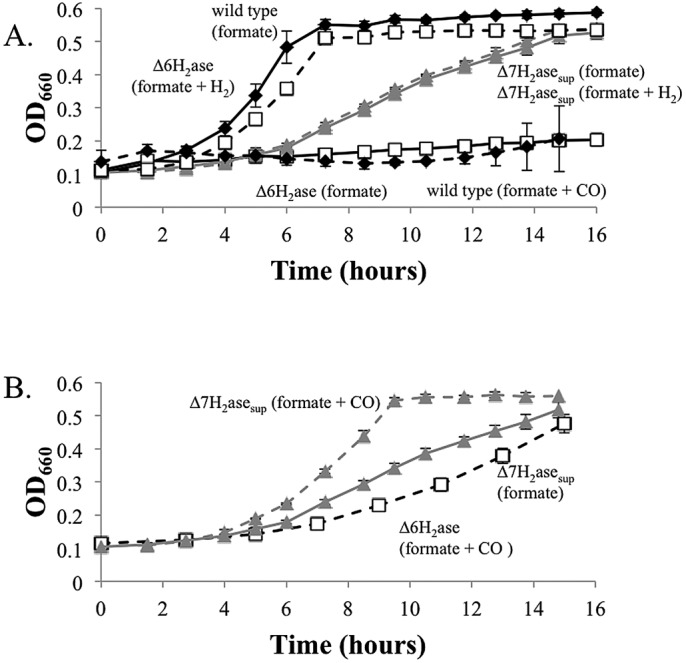
Growth of the ∆7H2asesup mutant. (A) Growth of the ∆7H2asesup mutant with formate alone or formate plus H2. (B) Growth of the ∆7H2asesup mutant with formate + CO. The ∆7H2asesup mutant grown on formate (from Fig. 3A) and the ∆6H2ase mutant grown on formate plus CO (from Fig. 1) are shown for comparison. Wild-type strain MM901 (black symbols) and the ∆7H2asesup (gray symbols) and ∆6H2ase (white symbols) mutants were studied. Broken lines indicate that the cultures were grown with H2 or 5% CO in the culture headspace. Data points are averages of three cultures, and error bars represent 1 standard deviation around the mean.
CO is a potent inhibitor of nickel-containing hydrogenases (14, 15). Five percent CO inhibited wild-type M. maripaludis grown on formate but had no inhibitory effect on the ∆7H2asesup mutant. In fact, the ∆7H2asesup mutant grew better on formate in the presence of 5% CO than it did on formate alone (Fig. 3B), suggesting that in the ∆7H2asesup mutant, like the ∆6H2ase mutant, CO was oxidized to CO2, leading to the production of reduced ferredoxin. The growth of the ∆7H2asesup mutant is apparently limited by the availability of reduced ferredoxin, and CO oxidation partially relieves its slow growth phenotype. The reduced ferredoxins generated by the suppressor activity and ACS/CODH appear additive in stimulating methanogenesis. The ∆7H2asesup mutant grew faster with formate and CO than either the ∆7H2asesup mutant grown with formate alone or the ∆6H2ase mutant grown with formate and CO.
Genome sequencing reveals a suppressor mutation allowing for growth of the ∆6H2asesup and ∆7H2asesup mutants on formate alone.
To determine the genetic background that allowed growth of the ∆6H2asesup and ∆7H2asesup mutants on formate in the absence of H2 or CO, we performed Illumina sequencing on the ∆7H2asesup mutant and six of the isolated suppressors and compared the sequence to the ∆6H2ase parent (see Table S1 in the supplemental material). Four of six of the ∆6H2asesup mutants, as well as the ∆7H2asesup mutant, shared a common insertion (AT at position 931341) in an intergenic region directly upstream of the gene for glyceraldehyde-3-phosphate (G3P):ferredoxin oxidoreductase (GAPOR) (see Fig. S1 in the supplemental material). The insertion generated the sequence AATATATA upstream of GAPOR which is very similar to the consensus methanogen promoter TTTA(T/A)ATA (16). Therefore, it appears that generation of a promoter upstream of GAPOR increases expression of this ferredoxin-reducing enzyme to allow for H2-independent growth. Sanger sequencing confirmed the presence of this mutation in the ∆7H2asesup mutant and its absence in the ∆6H2ase mutant. The nature of the suppressor mutations allowing growth in the other two ∆6H2asesup strains was not readily apparent from the genome sequencing data.
Overexpression of GAPOR in the ∆6H2ase mutant allows growth without H2.
Although the generation of a putative promoter sequence upstream of GAPOR suggests that overexpression of this gene leads to growth, the nature of the mutation could also result in a promoter reading in the opposite direction (see Fig. S1 in the supplemental material). Therefore, instead of engineering the mutation on the chromosome of the ∆6H2ase mutant to test its efficacy at stimulating growth, we chose to overexpress GAPOR on a replicative vector to recapitulate the effect and avoid possible overexpression of a second operon. GAPOR was placed under control of the Methanococcus vanneilii histone promoter on the replicative vector pLW40neo (17) and introduced into the ∆6H2ase background. The resulting strain displayed moderate growth in the absence of H2 and robust growth in the presence of H2, verifying that either GAPOR or Eha could be used to generate the reduced ferredoxin required for growth (Fig. 4).
FIG 4 .

Growth of the ∆6H2ase mutant overexpressing GAPOR in the presence and absence of H2. The ∆6H2ase mutant overexpressing GAPOR grown on formate plus H2 (black symbols), the ∆6H2ase mutant overexpressing GAPOR grown on formate alone (gray symbols), and the ∆6H2ase mutant grown on formate alone (white symbols) were examined.
The ∆7H2asesup mutant expressing F420-reducing hydrogenase can produce substantial amounts of H2.
The ∆7H2asesup mutant grows more slowly than the wild type on formate, suggesting that reduced ferredoxin is limiting. Reduced ferredoxin limitation of methanogenesis implies that other reduced cofactors that feed into the pathway, such as F420H2, are present in excess. Wild-type M. maripaludis possesses an F420-dependent formate:H2 lyase activity that is catalyzed by Fdh and F420-reducing hydrogenase. The wild-type strain grown on formate can accumulate H2 in the culture headspace to a concentration of 0.16% ± 0.02% of the gas phase at 2 atm pressure (mean ± standard deviation [SD] for three biological replicates) (18, 19). An excess of F420H2 should drive the equilibrium of this activity toward increased H2 production. frc encoding F420-reducing hydrogenase was placed on the replicative vector pLW40 (17) and reintroduced into the ∆7H2asesup mutant to restore formate:H2 lyase activity. When grown with formate as the only electron donor for methanogenesis, the ∆7H2asesup-frc mutant was capable of producing H2 up to a concentration of 2.32% ± 0.79% (Fig. 5A; see Fig. S2 in the supplemental material). When cultures entered stationary phase, H2 reuptake occurred, presumably due to depletion of formate and an equilibrium shift back toward F420H2 production from H2. The ∆7H2asesup mutant, which lacks formate:H2 lyase activity, was incapable of H2 production.
FIG 5 .

Growth and H2 production by the ∆7H2asesup mutant expressing F420-reducing hydrogenase (∆7H 2asesup-frc). (A) Growth on formate (black symbols) and H2 production (gray symbols) by the wild-type strain MM901 (circles), the ∆7H2asesup mutant (squares), and the ∆7H2asesup-frc mutant (triangles) in batch culture. Data are from a single representative experiment, but two replicate experiments gave similar results (see Fig. S2 in the supplemental material). (B) CH4 (black curve) and H2 production (bars) of the ∆7H2asesup-frc mutant in continuous culture. Actively growing cultures (gray bars) and cultures after metronidazole (50 µg ml−1) was added to completely oxidize ferredoxin (white bars) are shown. The medium dilution rate, gas flow rate, and culture optical density are shown on the x axis.
We also attempted continuous culture of the ∆7H2asesup-frc mutant to assess how additional factors affect the equilibrium of formate:H2 lyase activity. The ∆7H2asesup-frc mutant was maintained at a low OD660 under conditions where the medium dilution rate (0.125 liters h−1) slightly exceeded the growth rate to ensure that the culture was continuously growing. Under these conditions, the ∆7H2asesup-frc mutant produced between 100 and 500 µmol H2 gdw−1 (gdw stands for grams [dry weight]) (Fig. 5B). The higher production rates were observed when the gas flow rate was increased from 25 to 230 ml min−1, demonstrating that increased removal of H2 results in an equilibrium shift toward even greater H2 production. When the growth rate was allowed to exceed the medium dilution rate (0.031 liters h−1), H2 production was not greatly affected.
As reduced ferredoxin limitation of growth appears to lead to increased H2 production, we sought to take this to the extreme case of the absence of reduced ferredoxin. Metronidazole, an antibiotic capable of oxidizing ferredoxin (20), was added to the chemostat, and after 1 h, H2 production was found to have increased 5-fold to ~2.5 mmol gdw−1 h−1. With increased gas flow, this rate approached 5 mmol gdw−1 h−1. Robust H2 production upon metronidazole addition verifies that limiting reduced ferredoxin leads to an excess of F420H2.
DISCUSSION
Hydrogenotrophic methanogens have unappreciated metabolic versatility.
Although they do not share pathways of methylotrophic methanogens, hydrogenotrophic methanogens have a different kind of metabolic versatility. We have shown previously that this is due in part to the ability of hydrogenotrophs to use either formate or H2 as the electron donor to all four reduction steps of methanogenesis (2, 4). In addition, we have shown here that hydrogenotrophs also have versatility in their pathways of ferredoxin reduction for the anaplerotic electron input to methanogenesis. First, our results show clearly that H2 is not the only possible reductant for this purpose. The ∆6H2ase mutant grows in the complete absence of H2 as long as CO is present along with formate. In the case of the ∆7H2asesup strain, even CO was not needed, and formate was the sole electron donor. H2 addition to cultures of the ∆7H2asesup mutant grown on formate had no stimulatory effect, confirming that H2 uptake did not occur (Fig. 3B). CO, a hydrogenase inhibitor, also had no inhibitory effect in the ∆7H2asesup background but did in the wild type (14, 15). Additionally, the ∆7H2asesup mutant lacked all formate:H2 lyase activity (Fig. 5A). Taken together, these data confirm that all hydrogenase activity had been eliminated. The ∆6H2ase, ∆6H2asesup and ∆7H2asesup strains represent, to our knowledge, the first examples of hydrogenotrophic methanogens capable of growth in the complete absence of H2.
H2-independent growth occurs via novel electron flow pathways.
Here we have demonstrated two pathways by which H2 may be replaced as the anaplerotic electron donor for methanogenesis. Like Eha with H2, both pathways reduce ferredoxin which then presumably reduces CO2 to formyl-MFR. First, CO served this purpose in the ∆6H2ase and ∆7H2asesup strains. To our knowledge, only one other study reported the use of CO for methanogenesis in a hydrogenotrophic methanogen. In that report, CO as the sole substrate supported slow growth of Methanothermobacter thermautotrophicus by disproportionation to CO2 and CH4, and an F420-reducing carbon monoxide dehydrogenase activity was found (21). This differs from our results in which formate as well as CO was present. In addition, our ∆6H2ase and ∆7H2asesup mutants are capable of producing F420H2 directly from formate, so an F420-reducing carbon monoxide dehydrogenase activity is likely of little importance. Instead, the oxidation of CO leads to reduced ferredoxin. CO formation from H2 and CO2 with reduced ferredoxin as an intermediate has been observed in cell suspensions of hydrogenotrophic methanogens possessing ACS/CODH (7, 22), and the process we observed here, which depended on ACS/CODH, appears to be the reverse.
The CO utilization demonstrated here for M. maripaludis contrasts with methylotrophic methanogens. First, in methylotrophic methanogens, CO can be converted to methane and CO2 or to formate and acetate with the concomitant generation of ATP (23, 24). In contrast, 5% CO, as was used in our growth experiments, is insufficient as a stoichiometric electron donor for the amount of growth observed and functioned only anaplerotically. In both kinds of methanogens, CO oxidation results in reduced ferredoxin, but only the methylotrophs are known to use reduced ferredoxin as a stoichiometric source of electrons for methanogenesis. Second, methylotrophic methanogens can carry out methanogenesis solely from acetate by using ACS/CODH to cleave acetyl-CoA. M. maripaludis can utilize acetate anabolically (25) and can also use ACS/CODH anabolically for CO2 fixation to acetyl-CoA (6). Nevertheless, although CO utilization as an anaplerotic stimulant of methanogenesis in M. maripaludis occurred via ACS/CODH, M. maripaludis is apparently unable to use acetate for methanogenesis, even anaplerotically. Thus, our ∆6H2ase mutant, which was always grown in the presence of acetate and Casamino Acids, would not grow on formate in the absence of H2 without a suppressor mutation occurring.
As an additional novel pathway, overexpression of GAPOR could substitute for Eha and H2. GAPOR is found throughout the Archaea (26–29) and functions in glycolysis, catalyzing the oxidation of G3P to 3-phosphoglycerate (3-PG) with the concomitant reduction of ferredoxin (26, 29). The corresponding gluconeogenic reactions are catalyzed by G3P dehydrogenase (GAPDH), a NADPH-dependent enzyme, and phosphoglycerate kinase (PGK), an ATP-dependent enzyme (Fig. 6). Running both pathways simultaneously would result in:
FIG 6 .

Glyceraldehyde-3-phosphate:ferredoxin oxidoreductase cycle for ATP-dependent ferredoxin reduction. The GAPOR cycle of M. maripaludis as originally described by Park et al. (29) is shown with potential input from F420H2. Fdh, formate dehydrogenase; Fno, F420H2:NADP+ oxidoreductase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GAPOR, glyceraldehyde-3-phosphate: ferredoxin oxidoreductase; PGK, phosphoglycerate kinase. G3P, glyceraldehyde-3-phosphate; 1,3-DPG, 1,3-diphosphoglycerate; 3-PG, 3-phosphoglycerate.
NADPH + ATP + Fdox → NADP+ + ADP + Pi + Fdred where Fd is ferredoxin, Fdox is oxidized Fd, and Fdred is reduced Fd.
M. maripaludis also encodes an F420H2:NADP+ oxidoreductase (Fno) and the F420-dependent Fdh (12, 30). When these activities are taken into account, an ATP-dependent F420H2:ferredoxin oxidoreductase activity is possible (Fig. 6):
F420H2 + ATP + Fdox → F420 + ADP + Pi + Fdred
This pathway evidently operates in the ∆6H2asesup strains and the ∆7H2asesup strain, as well as in the ∆6H2ase strain with GAPOR overexpressed on a plasmid.
Reduced ferredoxin abundance versus inefficient electron transfer.
The CO-dependent and GAPOR-dependent pathways of ferredoxin reduction appear less efficient than Eha in supplying anaplerotic electrons to methanogenesis, since the ∆6H2ase strain with formate and CO and the ∆7H2asesup strain with formate alone grew more slowly than Eha+ strains with formate and H2. Indeed, combining the two pathways by including CO with formate for growth of the ∆7H2asesup strain increased growth in an additive manner. The low efficiency of the alternative pathways may be due to low concentrations of reduced ferredoxin produced compared to what can be produced by Eha, or the reduced ferredoxin could be abundant but inefficient at transferring electrons to Fwd. The latter interpretation is consistent with the proposal that the ferredoxin pools for anabolism and catabolism are normally separated in M. maripaludis (4), since the anabolic ferredoxin-reducing hydrogenase Ehb and the anaplerotic ferredoxin-reducing Eha reduce ferredoxins that substitute inefficiently for each other (4, 25).
Robust H2 production by the ∆7H2asesup-frc mutant suggests abundant F420H2.
A low availability of reduced ferredoxin, limiting methanogenesis and growth, should result in a buildup of F420H2. The equilibrium between F420H2 and H2 should then result in significant H2 production upon reintroducing F420-reducing hydrogenase into the ∆7H2asesup strain. This proved to be the case. Furthermore, the enhancement of H2 production by the addition of metronidazole supported the notion that an abundance of F420H2 resulted from a depletion of reduced ferredoxin.
MATERIALS AND METHODS
Growth conditions.
All strains were grown as described previously in McCas medium containing 200 mM sodium formate and 200 mM morpholinepropanesulfonic acid (MOPS) (pH 7) buffer (19, 31). H2-CO2 (20:80; 40 lb/in2) or N2-CO2 (20:80; 15 lb/in2) was added to the culture headspace unless otherwise indicated. Antibiotics (neomycin sulfate [5 mg ml−1], puromycin [2.5 µg ml−1], or metronidazole [50 µg ml−1]) were used where appropriate. For growth with CO in the culture headspace, 50% CO was injected to a final concentration of 5% (vol/vol). For growth curves, triplicate cultures were inoculated with a 10% inoculum, and the optical density at 660 nm (OD660) was monitored.
For growth under continuous culture, a modified version of a previously established chemostat system was employed (32–34). The ∆7H2asesup-frc mutant was grown under steady-state conditions with 380 mM sodium formate. NaCl was removed to maintain osmotic balance. Casamino Acids (0.2% [wt/vol]) and ampicillin (100 µg ml−1) were included in the growth medium. A gas mixture containing Ar−CO2-1% H2S (29:8:3) was used. Medium and gas flow rates are indicated in the text. The pH of steady-state samples was monitored and maintained at 6.95 by the automated addition of 10% H2SO4. Agitation in the vessel was maintained at 50 rpm throughout steady-state growth.
For continuous culture, the ∆7H2ase-frc mutant was grown to a maximum OD660 of 0.7 with a medium dilution rate of 0.083 liters h−1 and a gas flow rate of 26 ml ⋅ min−1. Medium dilution was changed to 0.125 liters h−1 until the culture OD660 dropped to 0.28 at which point the first sample was taken. The low OD660 ensured the culture was constantly growing with excess formate. When the medium dilution rate was changed throughout the course of sampling, culture was given at least 24 h to equilibrate before samples were collected. For changes in the gas flow rate, the gas phase was allowed at least 1 h to equilibrate before sample collection.
Generation of mutants and plasmids.
Strains used in this study are listed in Table 1. Plasmids and primers can be found in Table S2 in the supplemental material. Strain MM901, a derivative of M. maripaludis S2 that lacks uracil phosphoribosyl transferase (upt), was used as the wild-type strain (2, 35). Mutants were constructed using methods described in reference 31 and modified in reference 2. Briefly, to generate deletion mutants, a PCR product containing an in-frame deletion of the gene of interest was ligated into the vector pCRuptneo (neo stands for neomycin) (2). This was then transformed into strain MM901. Neomycin sulfate (5 mg ml−1) was used to select for a merodiploid, and 6-azauracil (250 µg ml−1) was used as negative selection to isolate the mutant of interest. To place genes onto a replicative vector, PCR products were cloned directly into pLW40 or pLW40neo and transformed into the strain of interest. Either neomycin sulfate (5 mg ml−1) or puromycin (2.5 µg ml−1) was included as appropriate. Suppressor strains of the ∆6H2ase mutant capable of growth in the absence of H2 were grown in formate medium either with or without the addition of 100 µM or 1 mM ammonium 2-(methylthio)ethanesulfonate (CH3-S-CoM); however, CH3-S-CoM was found to have no stimulatory effect on growth and was excluded from all subsequent experiments. All mutants were confirmed by PCR screen, and deletion of ehaNO was also confirmed by Southern blotting and Illumina sequencing (see below).
TABLE 1 .
Methanococcus maripaludis strains used in this study
| Strain | Description | Relevant locus | Reference |
|---|---|---|---|
| MM901 | Wild-type M. maripaludis with an in-frame deletion of upt | See reference | 2 |
| MM1284 | Δ6H2ase, MM901 ΔvhuAU ΔvhcA ΔfruA ΔfrcA Δhmd ΔehbN | See reference | 4 |
| MM1310 | Δ7H2asesup, MM1316 ΔehaNO | Mmp1461, Mmp1462 | This study |
| MM1315 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1316 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1317 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1318 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1319 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1320 | Δ6H2ase mutant with a suppressor allowing for growth without H2 | This study | |
| MM1327 | MM1284 with a deletion of CO dehydrogenase (cdh) | Mmp0983-0985 | This study |
| MM1338 | MM1284 with GAPOR overexpressed on pLW40neo | Mmp0945 | This study |
| MM1339 | MM1310 with F420-reducing hydrogenase (frc) on pLW40 | Mmp0817-0820 | This study |
Genome sequencing.
Genome sequencing was carried out for the ∆6H2ase mutant (MM1284), the ∆7H2asesup mutant (MM1310), and six of the ∆6H2ase strains that had developed suppressor mutations (MM1315, MM1316, MM1317, MM1318, MM1319, and MM1320). ∆6H2ase strains were isolated by dilution to extinction before processing. High-molecular-weight DNA for sequencing was extracted using the Qiagen PureGene kit according to the manufacturer’s instructions. Genomes were sequenced as described in reference 36, using unpaired 36-bp reads with the Illumina Genome Analyzer GA IIX according to the manufacturer’s instructions (Illumina, San Diego, CA). For each genome, a random-fragment library was constructed using a custom protocol. Briefly, genomic DNA (gDNA) samples were sheared using a Bioruptor UCD-200 (Diagenode Inc., Denville, NJ), and end repaired using an End-It DNA end repair kit (Epicentre). Repaired fragments were subjected to A tailing using Taq DNA polymerase (Roche Inc., USA, Chicago, IL), and custom adaptors ligated to A-tailed fragments using T4 DNA ligase (New England Biolabs, Beverly, MA). Libraries were size selected using automated electrophoresis on a Pippen Prep (Sage Science, Beverly, MA) and assessed for size range and concentration using a Qubit (Invitrogen Inc., Carlsbad, CA) and a Bioanalyzer (Agilent Inc., San Diego, CA).
Raw sequence data were compared directly to the M. maripaludis strain S2 reference genome (NCBI reference NC_005791.1) to identify possible suppressor mutations allowing for H2-independent growth (see Table S1 in the supplemental material). Putative mutations upstream of the gene encoding glyceraldehyde-3-phosphate:ferredoxin oxidoreductase (GAPOR) were identified and verified in ∆6H2ase and ∆7H2asesup mutants with Sanger sequencing (GeneWiz, South Plainfield, NJ).
CH4 and H2 measurements.
To measure H2 from batch culture, 2.5 ml of culture headspace was collected and stored for no longer than 24 h in a 5-ml serum vial preflushed with 100% N2. To measure gas concentration from chemostat-grown cells, gas was collected directly into 5-ml serum vials after flushing with at least 350 ml of chemostat gas outflow. For batch culture measurements, the headspace contained a pressure of 2 atm. H2 and CH4 concentrations were measured by gas chromatography as described previously (4). To completely oxidize ferredoxin, metronidazole (50 µg ml−1) was used. Upon the addition of metronidazole, gas flow through the chemostat vessel was allowed to equilibrate for 1 h prior to collection and analysis. Rates of H2 production were calculated assuming a liter of cell material at an OD660 of 1.0 yields 0.34 gram (dry weight) (18).
SUPPLEMENTAL MATERIAL
Genomic context of a suppressor mutation allowing for H2-independent growth of the Δ7H2asesup mutant Download
Growth and H2 production by the Δ7H2asesup-frc mutant. Growth on formate (black symbols) and H2 production (gray symbols) by the wild-type strains(circles), the Δ7H2asesup mutant (squares), and the Δ7H2asesup-frc mutant (triangles) in batch culture. Download
Genome sequencing results for Δ6H2ase, Δ7H2asesup, and Δ6H2asesup mutants. Mutations due to the upt and hydrogenase deletions are not shown.
Primers and plasmids used in this study
ACKNOWLEDGMENTS
We thank David Stahl for use of the gas chromatograph for H2 measurements.
This work was supported by grant DE-FG02-05ER15709 from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy. K.C.C. was supported in part by the Public Health Service National Research Service award T32 GM07270 from the National Institute of General Medical Sciences.
Footnotes
Citation Costa KC, Lie TJ, Jacobs MA, Leigh JA. 2013. H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis. mBio 4(2):e00062-13. doi:10.1128/mBio.00062-13.
REFERENCES
- 1. Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579–591 [DOI] [PubMed] [Google Scholar]
- 2. Costa KC, Wong PM, Wang T, Lie TJ, Dodsworth JA, Swanson I, Burn JA, Hackett M, Leigh JA. 2010. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaster AK, Moll J, Parey K, Thauer RK. 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U. S. A. 108:2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA. 2012. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 109:15473–15478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thauer RK. 2012. The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. U. S. A. 109:15084–15085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. 2010. Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 8:447–460 [DOI] [PubMed] [Google Scholar]
- 7. Eikmanns B, Fuchs G, Thauer RK. 1985. Formation of carbon monoxide from CO2 and H2 by Methanobacterium thermoautotrophicum. Eur. J. Biochem. 146:149–154 [DOI] [PubMed] [Google Scholar]
- 8. Bobik TA, Wolfe RS. 1989. Activation of formylmethanofuran synthesis in cell extracts of Methanobacterium thermoautotrophicum. J. Bacteriol. 171:1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gunsalus RP, Wolfe RS. 1977. Stimulation of CO2 reduction to methane by methylcoenzyme M in extracts Methanobacterium. Biochem. Biophys. Res. Commun. 76:790–795 [DOI] [PubMed] [Google Scholar]
- 10. Dybas M, Konisky J. 1989. Transport of coenzyme M (2-mercaptoethanesulfonic acid) and methylcoenzyme M [(2-methylthio)ethanesulfonic acid] in Methanococcus voltae: identification of specific and general uptake systems. J. Bacteriol. 171:5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balch WE, Wolfe RS. 1979. Transport of coenzyme M (2-mercaptoethanesulfonic acid) in Methanobacterium ruminantium. J. Bacteriol. 137:264–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, Hackett M, Haydock AK, Kang A, Land ML, Levy R, Lie TJ, Major TA, Moore BC, Porat I, Palmeiri A, Rouse G, Saenphimmachak C, Söll D, Van Dien S, Wang T, Whitman WB, Xia Q, Zhang Y, Larimer FW, Olson MV, Leigh JA. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tersteegen A, Hedderich R. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Transcription of the operons and sequence analysis of the deduced proteins. Eur. J. Biochem. 264:930–943 [DOI] [PubMed] [Google Scholar]
- 14. Purec L, Krasna AI, Rittenberg D. 1962. The inhibition of hydrogenase by carbon monoxide and the reversal of this inhibition by light. Biochemistry 1:270–275 [DOI] [PubMed] [Google Scholar]
- 15. Pandelia ME, Ogata H, Currell LJ, Flores M, Lubitz W. 2010. Inhibition of the [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F by carbon monoxide: an FTIR and EPR spectroscopic study. Biochim. Biophys. Acta 1797:304–313 [DOI] [PubMed] [Google Scholar]
- 16. Reeve JN. 1993. Structure and organization of genes. In Ferry JG, Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman and Hall, New York, NY. [Google Scholar]
- 17. Dodsworth JA, Leigh JA. 2006. Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc. Natl. Acad. Sci. U. S. A. 103:9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lupa B, Hendrickson EL, Leigh JA, Whitman WB. 2008. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl. Environ. Microbiol. 74:6584–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hendrickson EL, Leigh JA. 2008. Roles of coenzyme F420-reducing hydrogenases and hydrogen- and F420-dependent methylenetetrahydromethanopterin dehydrogenases in reduction of F420 and production of hydrogen during methanogenesis. J. Bacteriol. 190:4818–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lockerby DL, Rabin HR, Bryan LE, Laishley EJ. 1984. Ferredoxin-linked reduction of metronidazole in Clostridium pasteurianum. Antimicrob. Agents Chemother. 26:665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Daniels L, Fuchs G, Thauer RK, Zeikus JG. 1977. Carbon monoxide oxidation by methanogenic bacteria. J. Bacteriol. 132:118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bott M, Thauer RK. 1987. Proton-motive-force-driven formation of CO from CO2 and H2 in methanogenic bacteria. Eur. J. Biochem. 168:407–412 [DOI] [PubMed] [Google Scholar]
- 23. Rother M, Metcalf WW. 2004. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. U. S. A. 101:16929–16934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lessner DJ, Li L, Li Q, Rejtar T, Andreev VP, Reichlen M, Hill K, Moran JJ, Karger BL, Ferry JG. 2006. An unconventional pathway for reduction of CO2 to methane in co-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U. S. A. 103:17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Porat I, Kim W, Hendrickson EL, Xia Q, Zhang Y, Wang T, Taub F, Moore BC, Anderson IJ, Hackett M, Leigh JA, Whitman WB. 2006. Disruption of the operon encoding Ehb hydrogenase limits anabolic CO2 assimilation in the archaeon Methanococcus maripaludis. J. Bacteriol. 188:1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukund S, Adams MW. 1995. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:8389–8392 [DOI] [PubMed] [Google Scholar]
- 27. Selig M, Xavier KB, Santos H, Schönheit P. 1997. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167:217–232 [DOI] [PubMed] [Google Scholar]
- 28. van der Oost J, Schut G, Kengen SW, Hagen WR, Thomm M, de Vos WM. 1998. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J. Biol. Chem. 273:28149–28154 [DOI] [PubMed] [Google Scholar]
- 29. Park MO, Mizutani T, Jones PR. 2007. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase from Methanococcus maripaludis. J. Bacteriol. 189:7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berk H, Thauer RK. 1998. F420H2: NADP oxidoreductase from Methanobacterium thermoautotrophicum: identification of the encoding gene via functional overexpression in Escherichia coli. FEBS Lett. 438:124–126 [DOI] [PubMed] [Google Scholar]
- 31. Moore BC, Leigh JA. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187:972–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haydock AK, Porat I, Whitman WB, Leigh JA. 2004. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. FEMS Microbiol. Lett. 238:85–91 [DOI] [PubMed] [Google Scholar]
- 33. Hendrickson EL, Haydock AK, Moore BC, Whitman WB, Leigh JA. 2007. Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc. Natl. Acad. Sci. U. S. A. 104:8930–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leigh JA. 2011. Growth of methanogens under defined hydrogen conditions. Methods Enzymol. 494:111–118 [DOI] [PubMed] [Google Scholar]
- 35. Whitman WB, Shieh J, Sohn S, Caras DS, Premachandran U. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235–240 [Google Scholar]
- 36. Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507 http://dx.doi.org/10.1371/journal.pone.0036507 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic context of a suppressor mutation allowing for H2-independent growth of the Δ7H2asesup mutant Download
Growth and H2 production by the Δ7H2asesup-frc mutant. Growth on formate (black symbols) and H2 production (gray symbols) by the wild-type strains(circles), the Δ7H2asesup mutant (squares), and the Δ7H2asesup-frc mutant (triangles) in batch culture. Download
Genome sequencing results for Δ6H2ase, Δ7H2asesup, and Δ6H2asesup mutants. Mutations due to the upt and hydrogenase deletions are not shown.
Primers and plasmids used in this study


