ABSTRACT
The processes and mechanisms of community assembly and its relationships to community functioning are central issues in ecology. Both deterministic and stochastic factors play important roles in shaping community composition and structure, but the connection between community assembly and ecosystem functioning remains elusive, especially in microbial communities. Here, we used microbial electrolysis cell reactors as a model system to examine the roles of stochastic assembly in determining microbial community structure and functions. Under identical environmental conditions with the same source community, ecological drift (i.e., initial stochastic colonization) and subsequent biotic interactions created dramatically different communities with little overlap among 14 identical reactors, indicating that stochastic assembly played dominant roles in determining microbial community structure. Neutral community modeling analysis revealed that deterministic factors also played significant roles in shaping microbial community structure in these reactors. Most importantly, the newly formed communities differed substantially in community functions (e.g., H2 production), which showed strong linkages to community structure. This study is the first to demonstrate that stochastic assembly plays a dominant role in determining not only community structure but also ecosystem functions. Elucidating the links among community assembly, biodiversity, and ecosystem functioning is critical to understanding ecosystem functioning, biodiversity preservation, and ecosystem management.
IMPORTANCE
Microorganisms are the most diverse group of life known on earth. Although it is well documented that microbial natural biodiversity is extremely high, it is not clear why such high diversity is generated and maintained. Numerous studies have established the roles of niche-based deterministic factors (e.g., pH, temperature, and salt) in shaping microbial biodiversity, the importance of stochastic processes in generating microbial biodiversity is rarely appreciated. Moreover, while microorganisms mediate many ecosystem processes, the relationship between microbial diversity and ecosystem functioning remains largely elusive. Using a well-controlled laboratory system, this study provides empirical support for the dominant role of stochastic assembly in creating variations of microbial diversity and the first explicit evidence for the critical role of community assembly in influencing ecosystem functioning. The results presented in this study represent important contributions to the understanding of the mechanisms, especially stochastic processes, involved in shaping microbial biodiversity.
Introduction
Biodiversity is the heart of community ecology, but the mechanisms and factors controlling biodiversity, especially β-diversity, are poorly understood (1). Various ecological/evolutionary processes and factors interact to shape the patterns of species composition and diversity on various scales (2). One important ecological/evolutionary process is community assembly, which is generally referred to as the construction and maintenance of local communities through sequential arrival of potential colonists from an external species pool (2–4). Knowledge about the processes and factors controlling community assembly is critical to our understanding of the patterns of species composition and diversity because it provides a conceptual framework for which and how many species live in a particular locality (5). However, the mechanisms of community assembly and many of the critical factors shaping species composition and structure remain controversial (1, 6). The traditional niche-based theory asserts that the deterministic factors, such as species traits, interspecies interactions (e.g., competition, predation, mutualism), and environmental conditions, govern species diversity and composition of a local community (7, 8), and hence community composition should converge toward a single pattern under similar environmental conditions, while communities under different environmental conditions will have divergent configurations (5). In contrast to niche-based theory, neutral theory (9) assumes that many natural community patterns can be generated by only considering ecological drift, i.e., stochastic processes of birth, death, colonization, extinction, and speciation (9–11). Ecological drift leads to dispersal-assembled communities whose members are present mainly or solely owing to dispersal (immigration) rather than adaptation to their habitats (12).
Stochasticity in community assembly could cause considerable site-to-site variability in species compositions, known as β-diversity, even under identical environmental conditions (1, 5, 6), which could also lead to alternative stable states in community structure (6, 13, 14). However, illustrating the existence of stochastic assembly and its relative roles in determining community composition is very challenging because it is extremely difficult, if not impossible, to manipulate or reconstruct assembly history in natural ecosystems (5) and to ensure that the initial conditions (e.g., initial density, initial environmental heterogeneity) and environmental conditions are identical (6, 13). In addition, the links between community assembly and functions remain elusive (5). It is not clear whether the dispersal-assembled communities with divergent structures are functionally different. However, discerning the relationships between community assembly and ecosystem functioning is even more difficult (1, 5, 15), especially in microbial communities.
It is well documented that microbial communities in nature are highly diverse (16, 17), but little is known about the factors shaping microbial community diversity and assembly. Although deterministic factors (18), such as carbon and nutrient resource heterogeneity (17), differences in environmental conditions (19, 20), spatial isolation (17), and species traits and/or interspecies interactions (18), play key roles in shaping soil microbial community structure, they alone are not sufficient to explain the extremely high diversity of microbial communities observed in nature (21). Several recent studies (22–24), showed that a substantial part of variations in microbial community compositions could not be explained by spatial distance and environmental variables, despite exhaustive measurements of all routinely measured environmental variables. A part of the microbial community variations observed must be a result of stochastic processes of community assembly through differential colonization history, ecological drift (e.g., stochastic birth, death, colonization, extinction, and speciation), and/or dispersal limitation (1, 9, 10, 25–27). However, explicit evidence for the existence of stochastic assembly and its relationships to community functioning is lacking.
The objectives of this study were to (i) illustrate the possible existence of stochastic assembly in microbial communities under identical environmental conditions from the same source community, (ii) understand the relative roles of deterministic versus stochastic factors in determining microbial community composition, and (iii) determine the possible linkages between community assembly and community functioning. In this study, we used a microbial electrolysis cell (MEC) as a model system to examine the importance of stochastic assembly in determining microbial community structure and functions. We hypothesize that stochastic process of colonization is critical to the assembly of the MEC microbial communities but the roles of deterministic processes such as priority effects are not negligible (28). Our results supported this hypothesis by demonstrating that stochastic assembly plays a dominant role in determining not only community structure but also ecosystem functions. These results have important implications on understanding ecosystem functioning, biodiversity preservation, and ecosystem management.
RESULTS AND DISCUSSION
Microbial communities in MEC reactors as model systems for studying community assembly.
MEC technology is a promising new technology for efficient and sustainable hydrogen production from biodegradable organic matter (29). MECs are bioreactors in which microorganisms oxidize organic matter by electrolysis and the liberated protons and electrons can then form H2 with a small energy input (31–33). In this process, exoelectrogenic microorganisms from wastewater generally attach to the anode of a MEC to form a biofilm and release protons and electrons through oxidation of organic matters. The released protons and electrons flow to the cathode to produce hydrogen at the cathodic side through the addition of a small voltage (0.7 V) to the circuit (29–31).
The establishment and dynamics of the biofilm communities in MEC reactors are ideal for examining the role of stochastic processes (i.e., colonization and extinction) in community assembly (34) (Fig. 1). Many replicate reactors can be set up with the same wastewater inoculum and operated under identical environmental conditions. The wastewater inoculum generally contains many different microbial populations (Fig. 1A) that can potentially colonize the anode by random chance. Due to such stochastic colonization, considerable site-to-site variation (unpredictability) in community composition could exist under identical environmental conditions (Fig. 1B). Such unpredictability could also be intensified by subsequent deterministic processes through species selection, priority effects, and population interactions (Fig. 1C). After initial colonization, some species could come off anode biofilm and be lost during medium exchanges, which is a proxy for the species extinction process. Thus, the final community structure could be dependent on stochastic ecological drift (i.e., initial stochastic colonization and subsequent extinction) as well as priority effects and species interactions.
FIG 1 .
Stochastic community assembly processes in the MECs. The diagram shows a schematic of neutral dynamics in the assembly of the MEC reactor biofilm community. It is assumed that the regional pool has 20 individuals of 5 species (A, B, C, D, and E), with each species having a different abundance (A). During the initial inoculation, different species colonize the anodes of the MECs to produce current. Due to the stochastic process of colonization, the established biofilm composition varies considerably among different MEC reactors to form 4 different community structure states (B). Following that, the reactor solutions were replaced with new sterile medium every 24 h. Due to competition for resources and space, some species could detach from the biofilm and subsequently be lost during medium exchange, whereas some species could recolonize the anodes, which further creates variation of the communities among different bioreactors (C). Thus, even though these reactors were operated under identical conditions with the same source community, the community structures were quite different due to ecological drift in colonization.
Using well-controlled laboratory systems such as the MEC bioreactors adopted here has several unique advantages for studying community assembly (18, 35–38). First, the small size and short generation times of microorganisms allow us to manipulate and monitor the ecologically meaningful influences of stochastic and deterministic factors on microbial communities in tractable experimental units and on short-time scales. Second, many replicate reactors can be set up with the same wastewater inoculum so that any differences of initial conditions (e.g., initial population densities and spatial heterogeneity) can be minimized. Third, the environmental conditions can be well controlled, and hence the reactor systems can be operated under identical environmental conditions, which is very difficult to achieve in natural settings (35). As a consequence, the effects of compounding factors on experimental results can be minimized. In addition, the laboratory systems are well-controlled closed systems, so that all important functional parameters of interest can be monitored at the whole-system level to allow establishment of reliable relationships of community structure to function, which is extremely difficult to achieve in nature.
Divergent communities obtained under identical environmental conditions from the same source community.
A total of 14 replicate MEC reactors were set up, and all were inoculated with the same wastewater from a wastewater treatment plant. To facilitate stochastic colonization, the reactors were initially inoculated with wastewater twice for 96 h to allow microbial cells to randomly colonize the anode for biofilm formation. After the initial colonization, the reactors were operated in fed-batch mode, in which the reactor solutions were replaced with fresh sterilized medium (~26 ml) every 24 h. These reactors were operated under identical conditions (e.g., temperature, substrate feeding, applied voltage, reactor structure, and materials) for 55 days. During this period, both stochastic processes (e.g., recolonization and extinction) and deterministic processes (e.g., species selection, priority effects, and species interactions) could shape the patterns of β-diversity among these reactor communities. As expected, the physical and chemical conditions in these reactors appeared to be very stable over the experimental periods, as indicated by effluent pH (see Fig. S1 in the supplemental material) and current generation (see Fig. S2). The functions of these reactors as measured by the yields of H2, CH4, and CO2 (see Fig. S3A, B, and C, respectively) were also relatively stable over time for most reactors after initial fluctuations.
The functional gene structures of these microbial communities were determined with GeoChip (19, 39), a high-throughput metagenomic technology for dissecting the functional gene structure of microbial communities important to biogeochemistry, ecology, and environmental sciences (19, 23, 40–45). GeoChip (version 3.0) contains about 28,000 probes covering approximately 57,000 genes from 292 functional gene families involved in carbon, nitrogen, phosphorus, and sulfur cycles, energy metabolism, antibiotic resistance, metal resistance, and organic contaminant degradation (46). GeoChip hybridization is able to provide species- and strain-level resolutions (47, 48). Many previous studies demonstrated that GeoChip is a powerful metagenomic technology for dissecting functional gene structure of microbial communities and for linking community structure to functions (19, 23, 33, 39–41, 43–46).
In this study, the β-diversity was represented by Sorensen’s incidence-based (DS) and Bray-Curtis’s abundance-based (DBC) indexes, which both range from 0 to 1. Substantial variations in community structure were observed among these reactor communities, as indicated by both Sorensen and Bray-Curtis dissimilarities (DS = 0.47 ± 0.11; DBC = 0.54 ± 0.11). Similarly, high variations were also observed at the level of individual functional gene groups based on DS and DBc (see Table S1 in the supplemental material). Also, four distinct groups can be visualized in the detrended correspondence analysis (DCA) ordination space (Fig. 2): (i) group A, which consists of only one reactor (number 9); (ii) group B, which is comprised of six reactors (numbers 2 to 7); (iii) group C, which contains five reactors (number 1, 8, 10, 11, 13, and 14); and (iv) group D, which encompasses one reactor (number 12). Interestingly, SIMPER (similarity percentage) analysis indicated that the functional populations involved in metal resistance and antibiotic resistance rather than typical electricigens (e.g., metal-reducing bacteria) were most important in driving the separation of the communities among different reactors, as observed in Fig. 2 (see Fig. S4 in the supplemental material), which implies that selection could be an important process after initial colonization of these reactors, because the functioning of the resistance genes is critical for successful competition and survival. In addition, as revealed by three nonparametric tests, multiple response permutation procedure (MRPP), permutational multivariate analysis of variance (Adonis), and analysis of similarity (ANOSIM), the overall community structures among these four groups were all significantly different (P < 0.05) (Table 1). Similar observations were obtained at the level of individual functional gene groups (see Table S2 in the supplemental material). Finally, as shown above, two groups (B and C) contained more than three samples, so that further statistical tests could be performed. Similarly, the community structures between these two groups were also significantly different with all three methods (Table 1). These results suggest that these MEC reactor communities were substantially different and most likely formed at least two distinct community states. Though the functions of these MEC communities were relatively stable (see Fig. S3A, B, and C) during the experimental period after initial 10 days, it is less clear whether the structures of these community states were stable over time. Further experiments with specific environmental perturbations and/or species invasions need to be designed to unequivocally test the stability of these multiple community states.
FIG 2 .

Detrended correspondence analysis (DCA) of GeoChip hybridization data showing the relationships of microbial community functional gene structures among different reactors. Four distinct groups can be defined based on the DCA ordination, which could represent different alternative community states. The overall community composition and structure among these groups were also all significantly different, as shown in Table 1.
TABLE 1 .
Significance tests of the differences of β-diversity among reactor communities
| Communities | Dissimilarity measurement | Adonisa |
ANOSIM b |
MRPPc |
|||
|---|---|---|---|---|---|---|---|
| F | P | R | P | δ | P | ||
| All four reactors groups | DS | 2.035 | 0.007 | 0.422 | 0.007 | 0.418 | 0.006 |
| DBC | 2.638 | 0.001 | 0.612 | 0.001 | 0.463 | 0.001 | |
| Reactor group B versus C | DS | 2.796 | 0.008 | 0.33 | 0.01 | 0.418 | 0.007 |
| DBC | 3.342 | 0.009 | 0.45 | 0.012 | 0.463 | 0.011 | |
Nonparametric multivariate analysis of variance with the Adonis function. The degree of freedom of Adonis for the case of “All four reactor groups” was 3, and that for the case of “Reactor group B versus C” was 1.
ANOSIM, analysis of similarities.
MRPP, multiresponse permutation procedure. MRPP is a nonparametric procedure that does not depend on assumptions such as normally distributed data or homogeneous variances but rather depends on the internal variability of the data.
Processes and mechanisms controlling community structure.
As shown above, high β-diversity was observed among these MEC reactors under identical environmental and operational conditions. Three distinct community assembly mechanisms can cause high β-diversity in ecological communities (1): (i) purely deterministic processes, in which community composition and diversity are determined by species interactions, habitat heterogeneity, and environmental conditions; (ii) purely stochastic processes, in which community compositions and diversity are generated by stochastic birth, death, colonization, and extinction; and (iii) interactions between stochastic and deterministic processes, in which stochastic variations in colonization history could lead to more intense deterministic priority effects that could intensify the unpredictability from ecological drift. Since these reactors were operated under identical environmental and operational conditions with the same inoculum, it is less likely that the purely deterministic processes play a significant role in the assembly of these communities. To test this hypothesis, a null model analysis was performed, and permutational analysis of multivariate dispersions (PERMDISP) was used to test the significance of the differences of β-diversity in these reactor communities from the null model (1, 6). If ecological drift (e.g., stochastic colonization and extinction) and possibly priority effects leading to multiple stable equilibria (5, 6) play dominant roles in community assembly, the β-diversity observed will be statistically indistinguishable from the random null expectation. On the other hand, if community assembly is shaped by purely deterministic processes, the β-diversity observed will be significantly lower than the random null expectation (6). Interestingly, PERMDISP test revealed that the observed β-diversity was indistinguishable from the null expectation (F1,26 = 0.003 and P = 0.962 for Sorensen dissimilarity; F1,26 = 0.015 and P = 0.897 for Bray-Curtis dissimilarity), suggesting that purely stochastic processes and/or the interactive effects of stochastic (colonization and extinction) and deterministic (e.g., priority effects and competition for space) processes could play prominent roles in shaping β-diversity in these MEC reactors.
To further determine whether the deterministic factors such as priority effects are important in the assembly of these reactor communities, the experimental data were tested against the neutral community models (49, 50). Our null hypothesis is that these reactor communities are controlled solely by ecological drift (e.g., stochastic colonization, birth, death, and extinction), and thus the gene abundance distributions follow neutral dynamics, as predicted by neutral community models (51). By fitting Etienne’s neutral model (49) with the modeling parameters of θ = 367.578 and m = 0.0029, the null hypothesis was rejected for all MEC reactors based on both a Kolmogorov-Smirnov (KS) test and χ2 tests (P < 0.001) (see Table S3 in the supplemental material), suggesting that these local reactor communities might not be purely neutrally assembled. Therefore, it appears that both stochastic processes and deterministic priority effects could play roles in shaping β-diversity among these reactor communities.
The potential role of deterministic priority effects in the assembly of these reactor communities is also supported by the differences of local diversity in individual reactors (α-diversity). Since these reactors were inoculated with the same inoculum, the probabilities of all species or population colonizing the anodes of individual reactors should be more or less similar, and consequently the gene richness would be similar across all reactors if only stochastic processes alone determine community composition. Surprisingly, the functional gene richness and Shannon-Weaver diversity detected varied substantially among communities in these reactors, ranging from 584 to 2,485 genes. A total of 3,703 functional genes were detected, but only 3.4% of genes were shared among all reactors. None of the individual reactors contained more than 21.4% of the functional genes detected across all reactors. In addition, all major functional gene or phylogenetic groups on the GeoChip were detected in these reactors, but their relative abundance varied considerably (see Fig. S5A and B in the supplemental material), especially for various metal-reducing bacteria and several other dominant bacteria (see Fig. S5C). Such differences in gene richness and functional gene groups/populations are most likely due to strong selection after initial colonization.
Distinct differences in functions of the microbial communities formed under identical environmental conditions.
Although stochastic assembly leads to multiple community states, it is not clear whether such community assembly processes affect community functioning. In this study, the functions of the MEC reactor communities were assessed mainly based on the production of various gases: H2, CH4, and CO2. Based on the yields of H2 and CH4, these reactors can also be divided into four groups, which are consistent with those based on community structure by DCA. Gas production varied significantly among these four groups (Fig. 3), as indicated by analysis of variance (ANOVA) (F3,10 = 50.93 and P < 0.05 for H2; F3,10 = 16.83 and P < 0.05 for CH4; and F3,10 = 7.01 and P < 0.05 for CO2). For instance, group A produced about 76 times more H2 than group D did. In addition, Mantel tests showed that there were significant correlations between the changes of gas yields and community β-diversity measured by Sorensen dissimilarity (rM = 0.394, P = 0.019) or Bray-Curtis dissimilarity (rM = 0.384, P = 0.023), indicating that the functional differences among these reactors are most likely due to the differences in community structure. Therefore, our results suggested that stochastic assembly (stochastic colonization and extinction) leads to multiple community states with distinct community functions.
FIG 3 .
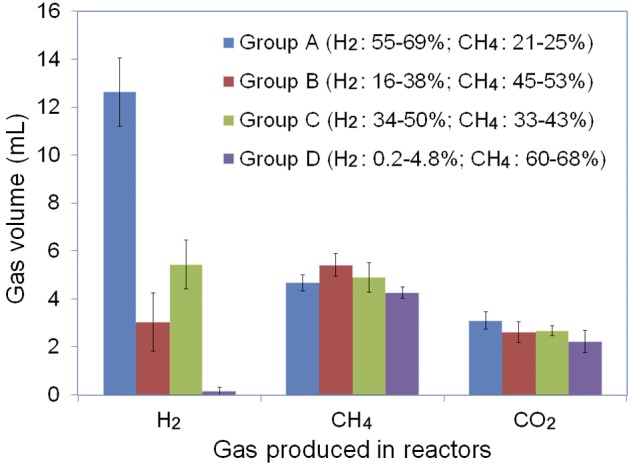
Average yields of H2, CH4, and CO2 for the four reactor groups as shown in Fig. 2. The average yields and standard deviations of each gas were obtained based on individual measurements across experimental time among reactors. H2 yields were dramatically different among these reactors, whereas lesser variations were observed for CH4 and CO2.
Relative roles of stochastic versus deterministic factors in determining community structure and functions.
To delineate the relative contributions of stochastic and deterministic factors to the assembly of these reactor communities, a canonical correspondence analysis (CCA)-based variation partitioning analysis (VPA) was performed (Fig. 4). CCA revealed 4 deterministic variables—effluent pH, H2, CH4, and CO2—that were significant predictors of community composition (Monte Carlo permutation: overall, F = 1.72 and P = 0.005; pH, F = 1.84 and P = 0.01; H2, F = 2.36 and P = 0.005, CH4, F = 1.48 and P = 0.088; CO2, F = 1.65 and P = 0.042) (see Fig. S6 in the supplemental material). Since these reactors were operated under identical conditions, the only measurable abiotic factor is the change in the effluent pH. Also, because the variations of gas production are most likely caused by the differences of community structure and their interactions (e.g., competition and mutualistic interactions), gas yields can, in turn, serve as a proxy for measuring the effects of biotic deterministic factors. Therefore, we can partition the variations in community composition into three main components: (i) biotic components (H2, CH4, and CO2), (ii) abiotic components (effluent pH), and (iii) an unexplained component (Fig. 4). VPA revealed that gases explained 30.1% (F = 1.59, P = 0.005) of variations in community structure, whereas pH explained 7.2% (F = 1.15, P = 0.35). Over half of the variations (56.6%) in community structure could be not explained by all the deterministic factors measured. Such unexplained components are most likely due to stochastic processes, because no other additional routine deterministic factors can be included. These results suggest that stochastic factors could have played major roles in determining the structure and functions of these MEC communities.
FIG 4 .

CCA-based variation partitioning analysis showing the importance of abiotic (pH) and biotic (gases) deterministic factors in explaining the variations of microbial community functional structures. More than half of the variations in community structure could be not explained by all the deterministic factors measured and are most likely due to stochastic processes.
Conclusions and implications.
Understanding the factors causing variations in β-diversity is an important but poorly understood issue in ecology because they are the key mechanisms influencing global variation in biodiversity (1, 52). Using a well-controlled laboratory system, this study provides an explicit evidence of the dominant roles of stochastic assembly (i.e., stochastic colonization) in creating variations of microbial community composition and structure, which is also supported by several previous observations (25, 53). Under identical environmental and operational conditions, the initial stochastic colonization generates substantially different microbial communities with very little overlap from the same source community, indicating that ecological drift could play a crucial role in controlling microbial community composition and should be taken into account in developing a mechanistic understanding of microbial community structure (25, 51). Second, this study also highlights the importance of the interactions of both stochastic and deterministic processes in shaping microbial community composition. After initial stochastic colonization, deterministic processes (species selection, priority effects) could play a critical role in maintaining or enhancing the community variations created by stochastic assembly. These results are consistent with several other studies from macrobial communities under various conditions (1, 6) as well as microbial communities (54). Therefore, the phenomenon of the interactive effects of both stochastic and deterministic processes on community composition could be general to many ecosystems, but their relative importance will be dependent on particular ecosystems, the organisms’ traits, and environmental conditions. Third, more importantly, our findings provide the first empirical support for the critical role of community assembly in influencing ecosystem functioning, a central issue in ecological and environmental sciences (15). Under identical environmental conditions, the same source community can evolve into several communities not only with different structures but also with distinct functions. Mantel tests showed that the differences in community functions are primarily due to the variations in community structure, indicating the critical role of biodiversity in determining ecosystem functions. In addition, our findings have important practical implications in biodiversity conservation, ecosystem management, and engineering applications. For example, many environmental engineering studies currently have no replicates or do not have enough replicates. Our results indicate that unpredictability in replicate reactors is a consequence of stochastic processes in community assembly and that the study of replicates improves the chances of obtaining desirable microbial biofilm communities for environmental engineering purposes. However, it should be noticed that it is not clear whether the phenomenon found in this artificial bioreactor community is applicable to microbial communities from other habitats in natural settings such as soils, marine sediments, and groundwater. Thus, more rigorous tests on the generality of this phenomenon in various ecosystems are needed before it is considered a fundamental principle of microbial ecology.
MATERIALS AND METHODS
The details for all materials and methods used in this study are provided in the supplemental material. Briefly, fourteen single chamber reactors were set up and inoculated with the same mixture of wastewater (50%) from the wastewater treatment plant in Norman, OK, and growth medium (50%). The collected wastewater was allowed to settle to remove big particles, with no obvious flocs observed, and hence the wastewater should be homogeneous. The single-chamber reactors were made of polycarbonate, as previously described (31). All reactors were started up as replicates in direct MEC mode (55) at a fixed applied voltage of 0.7 V (model 3645A; Circuit Specialists, Inc.). All reactors were incubated at room temperature (~22°C) for 48 h to allow microorganisms in the wastewater to randomly colonize the anode brushes. To enhance biofilm establishment on the anode, the original reactor solution was replaced with the same fresh mixture of the wastewater and growth medium and incubated under the same conditions for another 48 h. After biofilm establishment, all reactors were operated in a fed-batch mode with a 24-h cycle for about two months by replacing the reactor solution with fresh sterile growth medium every 24 h. After each medium change, the chambers were purged using extremely pure N2 (99.998%) for 10 min to remove oxygen.
The gas samples were collected daily for gas chromatography analysis of hydrogen, carbon dioxide, and methane. Microbial samples were archived at –80°C. Community DNA from representative samples was extracted using the mechanical grinding and SDS-based chemical lysis method (56). The functional gene compositions and structures of these communities were analyzed with GeoChip as described previously (19, 41). Many of our previous studies indicated that GeoChip hybridization-based detection is quantitative (19, 44, 47, 57, 58). Various types of statistical analyses, including DCA, CCA, VPA, ANOVA, nonparametric analyses (e.g., MRPP, ANOSIM, and Adonis), and randomization tests of β-diversity (e.g., EcoSim, PERMDISP), are described elsewhere (1, 6, 24, 55). In addition, the neutral community modeling analyses were performed as previously described (49, 50). Both the χ2 test and Kolmogorov-Smirnov (KS) test were used to assess the difference between the observed distribution and expected frequency distribution generated from neutral models.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods.
pH changes during experimental periods for all 14 MEC reactors, and inflow solution. The effluent pH varied slightly among different reactors but was very stable over time for individual reactors. Download
Current production during the experimental period for individual reactors. All 14 reactors produced current, although some degrees of variations were observed among reactors. Current production was also fairly stable over the experimental periods for each reactor. Download
Stability of gas production over experimental time. (A) H2; (B) CH4; (C) CO2. The variation of the yield of gases (H2, CH4, or CO2) over time was measured by coefficients of variation (CVs) of the production of each gas for each reactor. The CV at each time point was calculated based on the average and standard deviations of gas yield over a 15-day window. By shifting the window to the next time points, the dynamic changes of CVs in gas production over time can be visualized as shown here. Little change in the CVs of gas yields were observed for most (H2) or all (CH4 and CO2) of the reactors after initial 10 days, suggesting that the functional performance for each reactor in terms of H2 production was relatively stable for these reactors after the initial 10 days. However, the H2 yields varied substantially among different reactors, whereas the CO2 yields were very consistent among different reactors. The CH4 yields also varied considerably among different reactors. Download
Distribution of the functional groups which were most important in separating different bioreactor communities as shown in Fig. 2. The most important genes/populations were identified by SIMPER (similarity percentage) analysis (in PAST software). Only the functional genes belonging to the top 10% contributors (i.e., 90% cumulative cutoff value) are shown in this figure. The number of genes/populations of each functional group is listed in parentheses, and all 59 genes/populations were included in the pie diagram. Download
Abundance changes of functional gene groups (A), phylogenetic groups (B), and key metal-reducing bacteria and several other dominant groups based on cytochrome genes (C). The overall functional and phylogenetic diversity patterns of these MEC reactor communities were substantially different at the levels of major functional categories (A) or phyla/classes (B), although the reactors were inoculated with the same inoculum and operated under identical conditions. Geobacter, Shewanella, Desulfovibrio, and Anaeromyxobacter are typical metal-reducing bacteria. Pseudomonas and Rhodobacter are commonly found in MEC reactors. They could play important roles in substrate oxidation, electron transfer, and hydrogen production. As shown in panel C, their abundance varied substantially among reactors. Download
Canonical correspondence analysis (CCA) showing the relationships between community functional structure and environmental variables. Significant relationships (P < 0.1) were observed between community structure and all environmental variables as whole or individual gases. Download
Average β-diversity of the MEC reactor communities based on individual functional gene categories.
Significance tests of the differences of β-diversity among the four reactor groups based on individual functional gene categories.
Estimated P values of the goodness of fit to a neutral community model for different microbial communities in these reactors. The relative abundance of signal intensity × 105 was used to fit the neutral community model. The metacommunity parameters are θ = 367.578 and m = 0.0029. All communities are not fitted with the neutral community model as indicated by both the Kolmogorov-Smirnov (KS) test and χ2 test.
ACKNOWLEDGMENTS
We thank James H. Brown and James M. Tiedje for editing this paper. This work was supported by the State Key Joint Laboratory of Environment Simulation and Pollution Control (grant 11Z03ESPCT) at Tsinghua University, the National Creative Research Groups Project (grant 50821002), and the State Key Laboratory of Urban Water Resource and Environment (grant no. 2010DX11 and no. 2011TS09). The development of the GeoChips and associated computational pipelines used in this study were supported by ENIGMA (Ecosystems and Networks Integrated with Genes and Molecular Assemblies) through the Office of Science, Office of Biological and Environmental Research, the U. S. Department of Energy under Contract No.DE-AC02-05CH11231.
Footnotes
Citation Zhou J, Liu W, Deng Y, Jiang Y, Xue K, He, Van Nostrand JD, Wu L, Yang Y, and Wang A. 2013. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4(2):e00584-12. doi:10.1128/mBio.00584-12.
REFERENCES
- 1. Chase JM. 2010. Stochastic community assembly causes higher biodiversity in more productive environments. Science 328:1388–1391 [DOI] [PubMed] [Google Scholar]
- 2. Fukami T. 2004. Community assembly along a species pool gradient: implications for multiple-scale patterns of species diversity. Popul. Ecol. 46:137–147 [Google Scholar]
- 3. Drake JA. 1990. The mechanics of community assembly and succession. J. Theor. Biol. 147:213–233 [Google Scholar]
- 4. Warren PH, Law R, Weatherby AJ. 2003. Mapping the assembly of protist communities in microcosms. Ecology 84:1001–1011 [Google Scholar]
- 5. Chase JM. 2003. Community assembly: when should history matter? Oecologia 136:489–498 [DOI] [PubMed] [Google Scholar]
- 6. Chase JM. 2007. Drought mediates the importance of stochastic community assembly. Proc. Natl. Acad. Sci. U. S. A. 104:17430–17434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fargione J, Brown CS, Tilman D. 2003. Community assembly and invasion: an experimental test of neutral versus niche processes. Proc. Natl. Acad. Sci. U. S. A. 100:8916–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31:34 [Google Scholar]
- 9. Hubbell SP. 2001. The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton, NJ. [DOI] [PubMed] [Google Scholar]
- 10. Bell G. 2001. Neutral macroecology. Science 293:2413–2418 [DOI] [PubMed] [Google Scholar]
- 11. Chave J. 2004. Neutral theory and community ecology. Ecol. Lett. 7:241–253 [Google Scholar]
- 12. Alonso D, Etienne RS, McKane AJ. 2006. The merits of neutral theory. Trends Ecol. Evol. 21:451–457 [DOI] [PubMed] [Google Scholar]
- 13. Jiang L, Patel SN. 2008. Community assembly in the presence of disturbance: A microcosm experiment. Ecology 89:1931–1940 [DOI] [PubMed] [Google Scholar]
- 14. Price JE, Morin PJ. 2009. Community convergence in a simple microbial food web. Ecol. Res. 24:587–595 [Google Scholar]
- 15. Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, Hooper DU, Huston MA, Raffaelli D, Schmid B, Tilman D, Wardle DA. 2001. Ecology—biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804–808 [DOI] [PubMed] [Google Scholar]
- 16. Daniel R. 2005. The metagenomics of soil. Nat. Rev. Microbiol. 3:470–478 [DOI] [PubMed] [Google Scholar]
- 17. Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316:1746–1748 [DOI] [PubMed] [Google Scholar]
- 19. He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, Van Nostrand JD, Hobbie SE, Reich PB, Zhou J. 2010. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol. Lett. 13:564–575 [DOI] [PubMed] [Google Scholar]
- 20. Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John R, Dalling JW, Harms KE, Yavitt JB, Stallard RF, Mirabello M, Hubbell SP, Valencia R, Navarrete H, Vallejo M, Foster RB. 2007. Soil nutrients influence spatial distributions of tropical tree species. Proc. Natl. Acad. Sci. U. S. A. 104:864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramette A, Tiedje JM. 2007. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. U. S. A. 104:2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Kang S, Schadt CW, Garten CT. 2008. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U. S. A. 105:7768–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 5:1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, Sloan WT. 2010. Combined niche and neutral effects in a microbial wastewater treatment community. Proc. Natl. Acad. Sci. U. S. A. 107:15345–15350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caruso T, Chan Y, Lacap DC, Lau MC, Mckay CP, Pointing SB. 2011. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 5:1406–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stegen JC, Lin X, Konopka AE, Fredrickson JK. 2012. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6:1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erable B, Vandecandelaere I, Faimali M, Delia ML, Etcheverry L, Vandamme P, Bergel A. 2010. Marine aerobic biofilm as biocathode catalyst. Bioelectrochemistry 78:51–56 [DOI] [PubMed] [Google Scholar]
- 29. Cheng S, Logan BE. 2007. Sustainable and efficient biohydrogen production via electrohydrogenesis. Proc. Natl. Acad. Sci. U. S. A. 104:18871–18873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Logan BE. 2009. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 7:375–381 [DOI] [PubMed] [Google Scholar]
- 31. Call D, Logan BE. 2008. Hydrogen production in a single chamber microbial electrolysis cell lacking a membrane. Environ. Sci. Technol. 42:3401–3406 [DOI] [PubMed] [Google Scholar]
- 32. Lee HS, Rittmann BE. 2010. Characterization of energy losses in an upflow single-chamber microbial electrolysis cell. Int. J. Hydrogen Energ. 35:920–927 [Google Scholar]
- 33. Lu L, Ren N, Zhao X, Wang H, Wu D, Xing D. 2011. Hydrogen production, methanogen inhibition and microbial community structures in psychrophilic single-chamber microbial electrolysis cells. Energy Environ. Sci. 4:1329–1336 [Google Scholar]
- 34. Battin TJ, Sloan WT, Kjelleberg S, Daims H, Head IM, Curtis TP, Eberl L. 2007. Microbial landscapes: new paths to biofilm research. Nat. Rev. Microbiol. 5:76–81 [DOI] [PubMed] [Google Scholar]
- 35. Zhou J. 2009. Predictive microbial ecology. Microb. Biotechnol. 2:154–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bell T. 2010. Experimental tests of the bacterial distance-decay relationship. ISME J. 4:1357–1365 [DOI] [PubMed] [Google Scholar]
- 37. Pfisterer AB, Schmid B. 2002. Diversity-dependent production can decrease the stability of ecosystem functioning. Nature 416:84–86 [DOI] [PubMed] [Google Scholar]
- 38. Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N. 2009. Initial community evenness favours functionality under selective stress. Nature 458:623–626 [DOI] [PubMed] [Google Scholar]
- 39. He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67–77 [DOI] [PubMed] [Google Scholar]
- 40. Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, Fei S, Deng S, He Z, Van Nostrand JD, Luo Y. 2012. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Change 2:106–110 [Google Scholar]
- 41. Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, Lee YJ, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D’haeseleer P, Hazen TC, Zhou J. 2012. Microbial gene functions enriched in the deepwater Horizon deep-sea oil plume. ISME J. 6:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A, He Z, Van Nostrand JD, Hazen TC, Stahl DA, Wall JD, Arkin AP. 2011. How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9:452–466 [DOI] [PubMed] [Google Scholar]
- 43. Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D’haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 44. Wang F, Zhou H, Meng J, Peng X, Jiang L, Sun P, Zhang C, Van Nostrand JD, Deng Y, He Z, Wu L, Zhou J, Xiao X. 2009. GeoChip-based analysis of metabolic diversity of microbial communities at the Juan de Fuca ridge hydrothermal vent. Proc. Natl. Acad. Sci. U. S. A. 106:4840–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. 2010. Functional molecular ecological networks. mBio 1:e00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, Li X, Wu L, Gentry TJ, Yin Y, Liebich J, Hazen TC, Zhou J. 2010. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 4:1167–1179 [DOI] [PubMed] [Google Scholar]
- 47. Tiquia SM, Wu L, Chong SC, Passovets S, Xu D, Xu Y, Zhou J. 2004. Evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in environmental samples. BioTechniques 36:664–670 [DOI] [PubMed] [Google Scholar]
- 48. Liebich J, Schadt CW, Chong SC, He Z, Rhee SK, Zhou J. 2006. Improvement of oligonucleotide probe design criteria for functional gene microarrays in environmental applications. Appl. Environ. Microbiol. 72:1688–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Etienne RS. 2007. A neutral sampling formula for multiple samples and an “eXact” test of neutrality. Ecol. Lett. 10:608–618 [DOI] [PubMed] [Google Scholar]
- 50. Volkov I, Banavar JR, Hubbell SP, Maritan A. 2003. Neutral theory and relative species abundance in ecology. Nature 424:1035–1037 [DOI] [PubMed] [Google Scholar]
- 51. Woodcock S, van der Gast CJ, Bell T, Lunn M, Curtis TP, Head IM, Sloan WT. 2007. Neutral assembly of bacterial communities. FEMS Microbiol. Ecol. 62:171–180 [DOI] [PubMed] [Google Scholar]
- 52. Dornelas M, Connolly SR, Hughes TP. 2006. Coral reef diversity refutes the neutral theory of biodiversity. Nature 440:80–82 [DOI] [PubMed] [Google Scholar]
- 53. Hashsham SA, Fernandez AS, Dollhopf SL, Dazzo FB, Hickey RF, Tiedje JM, Criddle CS. 2000. Parallel processing of substrate correlates with greater functional stability in methanogenic bioreactor communities perturbed by glucose. Appl. Environ. Microbiol. 66:4050–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 4:337–345 [DOI] [PubMed] [Google Scholar]
- 55. Liu W, Wang A, Cheng S, Logan BE, Yu H, Deng Y, Nostrand JD, Wu L, He Z, Zhou J. 2010. Geochip-Based functional gene analysis of anodophilic communities in microbial electrolysis cells under different operational modes. Environ. Sci. Technol. 44:7729–7735 [DOI] [PubMed] [Google Scholar]
- 56. Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rhee SK, Liu X, Wu L, Chong SC, Wan X, Zhou J. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu L, Liu X, Schadt CW, Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods.
pH changes during experimental periods for all 14 MEC reactors, and inflow solution. The effluent pH varied slightly among different reactors but was very stable over time for individual reactors. Download
Current production during the experimental period for individual reactors. All 14 reactors produced current, although some degrees of variations were observed among reactors. Current production was also fairly stable over the experimental periods for each reactor. Download
Stability of gas production over experimental time. (A) H2; (B) CH4; (C) CO2. The variation of the yield of gases (H2, CH4, or CO2) over time was measured by coefficients of variation (CVs) of the production of each gas for each reactor. The CV at each time point was calculated based on the average and standard deviations of gas yield over a 15-day window. By shifting the window to the next time points, the dynamic changes of CVs in gas production over time can be visualized as shown here. Little change in the CVs of gas yields were observed for most (H2) or all (CH4 and CO2) of the reactors after initial 10 days, suggesting that the functional performance for each reactor in terms of H2 production was relatively stable for these reactors after the initial 10 days. However, the H2 yields varied substantially among different reactors, whereas the CO2 yields were very consistent among different reactors. The CH4 yields also varied considerably among different reactors. Download
Distribution of the functional groups which were most important in separating different bioreactor communities as shown in Fig. 2. The most important genes/populations were identified by SIMPER (similarity percentage) analysis (in PAST software). Only the functional genes belonging to the top 10% contributors (i.e., 90% cumulative cutoff value) are shown in this figure. The number of genes/populations of each functional group is listed in parentheses, and all 59 genes/populations were included in the pie diagram. Download
Abundance changes of functional gene groups (A), phylogenetic groups (B), and key metal-reducing bacteria and several other dominant groups based on cytochrome genes (C). The overall functional and phylogenetic diversity patterns of these MEC reactor communities were substantially different at the levels of major functional categories (A) or phyla/classes (B), although the reactors were inoculated with the same inoculum and operated under identical conditions. Geobacter, Shewanella, Desulfovibrio, and Anaeromyxobacter are typical metal-reducing bacteria. Pseudomonas and Rhodobacter are commonly found in MEC reactors. They could play important roles in substrate oxidation, electron transfer, and hydrogen production. As shown in panel C, their abundance varied substantially among reactors. Download
Canonical correspondence analysis (CCA) showing the relationships between community functional structure and environmental variables. Significant relationships (P < 0.1) were observed between community structure and all environmental variables as whole or individual gases. Download
Average β-diversity of the MEC reactor communities based on individual functional gene categories.
Significance tests of the differences of β-diversity among the four reactor groups based on individual functional gene categories.
Estimated P values of the goodness of fit to a neutral community model for different microbial communities in these reactors. The relative abundance of signal intensity × 105 was used to fit the neutral community model. The metacommunity parameters are θ = 367.578 and m = 0.0029. All communities are not fitted with the neutral community model as indicated by both the Kolmogorov-Smirnov (KS) test and χ2 test.



