Summary
How cell fate becomes restricted during somatic cell differentiation is a long-lasting question in biology. Epigenetic mechanisms not present in pluripotent cells and acquired during embryonic development are expected to stabilize the differentiated state of somatic cells and thereby restrict their ability to convert to another fate. The histone variant macroH2A acts as a component of an epigenetic multilayer that heritably maintains the silent X chromosome and has been shown to restrict tumor development. Here we show that macroH2A marks the differentiated cell state during mouse embryogenesis. MacroH2A.1 was found to be present at low levels upon the establishment of pluripotency in the inner cell mass and epiblast, but it was highly enriched in the trophectoderm and differentiated somatic cells later in mouse development. Chromatin immunoprecipitation revealed that macroH2A.1 is incorporated in the chromatin of regulatory regions of pluripotency genes in somatic cells such as mouse embryonic fibroblasts and adult neural stem cells, but not in embryonic stem cells. Removal of macroH2A.1, macroH2A.2 or both increased the efficiency of induced pluripotency up to 25-fold. The obtained induced pluripotent stem cells reactivated pluripotency genes, silenced retroviral transgenes and contributed to chimeras. In addition, overexpression of macroH2A isoforms prevented efficient reprogramming of epiblast stem cells to naïve pluripotency. In summary, our study identifies for the first time a link between an epigenetic mark and cell fate restriction during somatic cell differentiation, which helps to maintain cell identity and antagonizes induction of a pluripotent stem cell state.
Key words: Cell commitment, Epigenetic stability, Induced pluripotency, macroH2A, Nuclear reprogramming
Introduction
The epigenome of somatic cells is highly stable. This fundamental principle ensures tissue homeostasis, which prevents epigenetic reprogramming of somatic cells to diseased cell states. In spite of this stability, somatic cells can be reprogrammed to an embryonic stem cell (ESC) like state by a number of procedures, including nuclear transfer, cell fusion and overexpression of transcription factors (Gurdon et al., 1958; Tada et al., 1997; Tada et al., 2001; Takahashi and Yamanaka, 2006). These procedures are, however, very inefficient, reflecting a resistance toward reprogramming and the high stability of the differentiated state of somatic cells. The epigenetic factors that restrict somatic cell nuclear reprogramming to pluripotency are ill-defined. Resistance to reprogramming is acquired gradually during embryogenesis as pluripotent cells differentiate into somatic cell lineages (Foshay et al., 2012; Gurdon and Melton, 2008). Several layers of epigenetic modifications are likely to be involved and these may be acquired progressively as cells become more restricted during development. Transcription factors, DNA methylation, non-coding RNAs and histone modifications are known to play a role in the maintenance of epigenetic states. However, whether histone variants also restrict somatic cell reprogramming to induced pluripotency is not known.
Histone variants macroH2A are characterized by their large size, which is three times that of canonical histone H2A (Pehrson and Fried, 1992) and are generally associated with heterochromatin (Buschbeck and Di Croce, 2010). There are two macroH2A genes in the mouse (Costanzi and Pehrson, 2001; Pehrson and Fried, 1992), termed macroH2A.1 and macroH2A.2 in the new histone variant nomenclature, and previously referred to as macroH2A1 and macroH2A2 (Talbert et al., 2012). MacroH2A.1 can be alternatively spliced to give rise to macroH2A.1.1 or macroH2A.1.2 (Rasmussen et al., 1999; Timinszky et al., 2009), each of these possessing another variant (macroH2A.1.s3, macroH2A.1.s4 for macroH2A.1.1, and macroH2A.1.s1, macroH2A.1.s2 for macroH2A.1.2) (Bernstein et al., 2008). In mouse and in humans, in addition to alternative macroH2A.1 splicing, macroH2A.1.s3 and macroH2A.1.s1 also each differ from macroH2A.1.s4 and macroH2A.1.s2, respectively, by a single Lysine residue, the function of which, if any, is currently unknown (Bernstein et al., 2008).
MacroH2A histone variants incorporate into chromatin via their histone H2A-like region, which is attached via a linker region to a bulky non-histone (macro) domain that projects out of the nucleosome (Chakravarthy et al., 2005). All the three regions of macroH2A are independently associated with transcriptional repression (Buschbeck and Di Croce, 2010).
MacroH2As are developmentally regulated (Chang et al., 2010; Chang et al., 2005; Pehrson et al., 1997). In vivo, maternal macroH2A.1 is removed from pronuclei prior to the onset of somatic macroH2A expression at the eight-cell stage in the preimplantation embryo, and becomes enriched on the inactive X chromosome (Xi) by the morula stage (Chang et al., 2010; Costanzi et al., 2000). MacroH2A.1 is also enriched on the Xi in somatic cells, but not on the Xi of epiblast stem cells, derived from the post-implantation embryo (Costanzi and Pehrson, 1998; Pasque et al., 2011a). In the mouse embryo, macroH2A expression has not been investigated after E3.5. In general, macroH2A.1.1 has been found to be expressed in cells derived from differentiated tissues with low proliferation, whereas macroH2A.1.2 tends to be more widely expressed including in proliferative cells (Pehrson et al., 1997; Rasmussen et al., 1999).
Mice lacking macroH2A.1 or macroH2A.2 are fertile and viable, and display only mild phenotype, including metabolic defects, suggesting that macroH2A is not strictly required for embryonic development in the mouse (Boulard et al., 2010; Buschbeck and Di Croce, 2010; Changolkar et al., 2007). Removal of both macroH2A.1 and macroH2A.2 from female mouse ESCs was found to impair ESCs differentiation in one study (Creppe et al., 2012a; Creppe et al., 2012b), while it was not found to prevent X chromosome inactivation and differentiation by another group (Tanasijevic and Rasmussen, 2011). Using the heterologous system of mammalian nuclear transfer to Xenopus oocytes it was previously shown that the histone variant macroH2A is a component of the epigenetic mechanisms of gene silencing present in somatic cells. However, whether this bears significance for somatic cell epigenome resistance to reprogramming towards pluripotency has not been addressed (Pasque et al., 2011b).
Here, we report that during mouse embryogenesis, macroH2A.1 becomes highly expressed and incorporated into chromatin in somatic cell lineages. Importantly, macroH2A.1 is a structural component of the repressed chromatin of pluripotency genes in somatic cells and acts as an epigenetic barrier to the induction of naïve pluripotency. Our study demonstrates the importance of macroH2A histone variants in maintenance of cellular identity.
Results
MacroH2A is a hallmark of somatic cell differentiation in vivo
To investigate the expression pattern of macroH2A in vivo we carried out wholemount immunofluorescence of E3.5 and E4.5 mouse blastocysts against macroH2A.1 and pluripotency transcription factors Oct4 and Nanog to delineate the ICM. In early blastocysts (E3.5), prior to the establishment of naïve pluripotency, nuclear macroH2A.1 was detected throughout the embryo irrespective of cell lineage (Fig. 1A), which was consistent with a previous report (Costanzi et al., 2000). In contrast, in E4.5 blastocysts, we detected a dramatic decrease of nuclear macroH2A.1 in the ICM in comparison to the trophectoderm with only heterochromatic foci remaining enriched for this histone variant (Fig. 1B). This suggests that the establishment of naïve pluripotency in vivo is marked by a downregulation of macroH2A.1 expression.
Fig. 1.
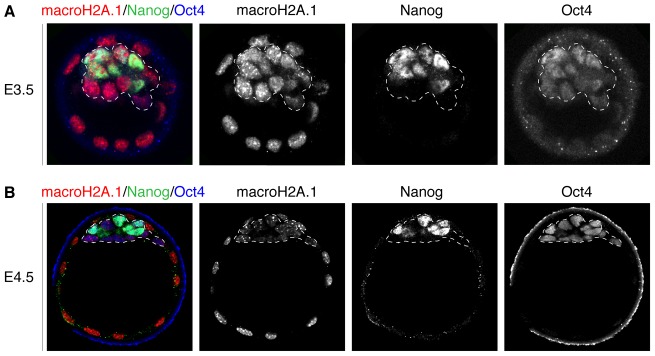
MacroH2A.1 is downregulated in the naïve pluripotent epiblast. (A) E3.5 female mouse blastocyst wholemount immunofluorescence against macroH2A.1 (red in merge panel), Oct4 (blue in merge panel) and Nanog (green in merge panel). Nuclear macroH2A.1 is detected at equivalent levels in cells of the trophectoderm and those of the ICM (dashed lines). Note that naïve pluripotency has not been established in the ICM at this stage. Images are projected confocal Z-sections. (B) E4.5 female mouse blastocyst wholemount immunofluorescence against macroH2A.1 (red in merge panel), Oct4 (blue in merge panel) and Nanog (green in merge panel). Nuclear macroH2A.1 is detected at higher levels in cells of the trophectoderm, and is downregulated in cells of the ICM (dashed line), and this is at a stage when naïve pluripotency has been established. Images are projected confocal Z-sections.
Next, we carried out wholemount immunofluorescence of E6.5 X-GFP female mouse conceptus against macroH2A.1 and GFP. The paternally derived X-GFP transgene was used as a marker to specifically delineate cells of the epiblast, whose mosaic expression reflects random X chromosome inactivation in female embryos (Hadjantonakis et al., 2001). Remarkably, macroH2A.1 expression was undetectable in the pluripotent epiblast, but it was detected strongly in the nuclei of visceral endoderm cells and to a lesser extent in the nuclei of trophectoderm cells (Fig. 2A; supplementary material Fig. S1A). This mirrors the expression level of macroH2A in cultured cell lines derived from these embryonic lineages, namely epiblast stem cells (EpiSCs) and trophoblast stem (TS) cells (Pasque et al., 2011a).
Fig. 2.
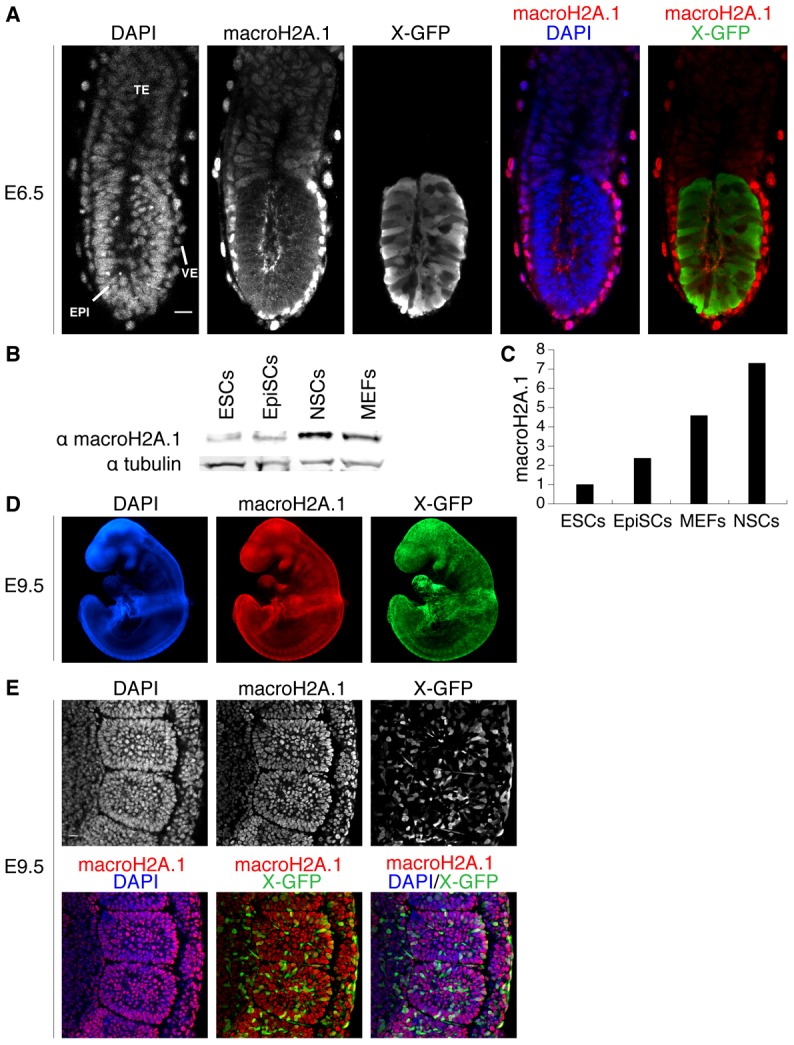
MacroH2A.1 becomes highly expressed during somatic lineage development. (A) E6.5 female X-GFP mouse conceptus wholemount immunofluorescence against macroH2A.1 (red in merge panel) and GFP (green in merge panel). MacroH2A.1 is highly expressed in the visceral endoderm (VE) and to some extent in the extra embryonic ectoderm (TE) but is not detected in the epiblast (EPI), precursor of all somatic lineages (mosaic X-GFP expression due to random X chromosome inactivation). DAPI is in blue. Images are projected confocal Z-sections. (B) Western blot analysis of macroH2A.1 in ESCs, EpiSCs, NSCs and MEFs. All cells are female. Tubulin was used as a loading control. (C) Quantification of western blot analysis shown in B. MacroH2A.1 signal was normalized to tubulin and ESCs levels set to 1. MacroH2A.1 expression is lowest in ESCs and increases with the differentiated state in EpiSCs, MEFs and adult NSCs. (D) E9.5 female X-GFP mouse embryo wholemount immunofluorescence against macroH2A.1 (red) and GFP (green). MacroH2A.1 is expressed throughout all tissues of the embryo at this stage. DAPI is in blue. Images acquired using epifluorescence microscopy. (E) E9.5 X-GFP mouse embryo immunofluorescence against macroH2A.1 (red) and GFP (green) showing a portion of the lateral plate mesoderm (including somites). Nuclear macroH2A.1 is detected in all somatic cells. Mosaic X-GFP expression is due to random X chromosome inactivation. DAPI is in blue. Images are projected confocal Z-sections. Scale bars: 20 µm.
Subsequently, we compared macroH2A.1 expression in in vitro cultures of pluripotent and somatic cells. We found that macroH2A.1 was quantitatively more abundant in mouse embryonic fibroblasts (MEFs) and adult neural stem cells (NSCs) compared to undifferentiated ESCs and EpiSCs (Fig. 2B,C). To assess whether macroH2A.1 expression is correlated with the differentiated state of cells in developing mouse embryos, we probed its expression later in development, in E9.5 embryos, in which somatic cell lineage determination has occurred. Nuclear macroH2A.1 was strongly detected throughout all somatic cells of E9.5 embryos, irrespective of their sex and X chromosome inactivation status (Fig. 2D,E; supplementary material Fig. S1B). The accumulation of nuclear macroH2A.1 in differentiated cells was in striking contrast to its low levels in mouse pluripotent ICM, post-implantation epiblast, germline (Hajkova et al., 2008) and zygote (Chang et al., 2010; Nashun et al., 2010). We conclude that macroH2A.1 marks the post-gastrulation embryo concomitantly with somatic cell lineage commitment and thereby can be considered a hallmark of somatic cell differentiation in vivo.
MacroH2A.1 is a chromatin component of repressed pluripotency genes in differentiated cells
Genome-wide analysis of macroH2A variants established that they generally localize to heterochromatic regions (Barzily-Rokni et al., 2011; Buschbeck et al., 2009; Changolkar et al., 2010; Creppe et al., 2012a; Gamble et al., 2010). At the same time, they can also be found in the coding regions of a small proportion of genes whose expression level inversely correlates with the amount of macroH2A incorporated in their chromatin (Gamble and Kraus, 2010; Gamble et al., 2010). Since the presence of macroH2A is mostly considered to have a repressive effect on genes and its abundance correlates with cell differentiation, we hypothesized that it may be incorporated in the chromatin of repressed pluripotency genes in somatic cells. We carried out ChIP analysis in undifferentiated ESCs, MEFs and adult NSCs. In ESCs, macroH2A.1 was detected at low levels in the regulatory sequences of pluripotency genes, Oct4 and Sox2, lineage-specific genes, B-Globin and Thy1, and the housekeeping genes GAPDH and c-Jun (Fig. 3). In somatic cells, both in mouse embryonic fibroblasts (MEFs) and in NSCs, macroH2A.1 became markedly enriched in the regulatory sequences of Oct4, B-Globin and Thy1, correlating with the repressed state of these genes in these cells. MacroH2A.1 did not become enriched in the GAPDH and c-Jun promoter, active in all cell types analyzed (Fig. 3). MacroH2A.1 was highly enriched in the chromatin of Sox2 regulatory regions in MEFs but not in NSCs in which this gene is highly expressed. In conclusion, the histone variant macroH2A.1 incorporates into the nucleosomes in the regulatory sequences of both repressed pluripotency and lineage-specific genes in somatic cells linking macroH2A not only to gene silencing but also to the stabilization of the differentiated cell state.
Fig. 3.
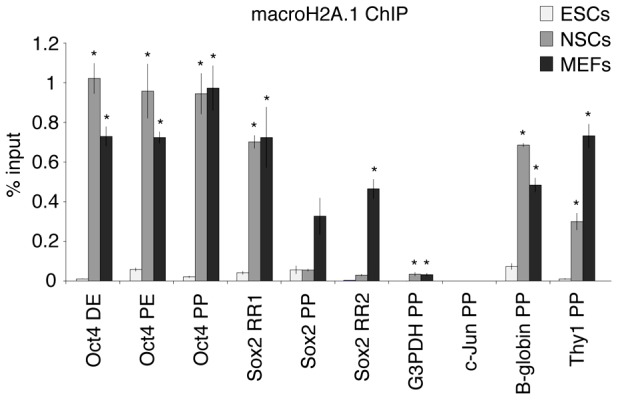
MacroH2A.1 marks chromatin of regulatory sequences of repressed pluripotency genes in differentiated cells. MacroH2A.1 ChIP analysis of pluripotent and lineage-specific gene regulatory regions in pluripotent (ESCs, light gray), and somatic cells (NSCs, gray; MEFs, dark gray). DE, distal enhancer; PE, proximal enhancer; PP, proximal promoter; RR1, regulatory region 1; RR2, regulatory region 2. Error bars depict the s.e.m. (n = 3). There were significant differences between ESCs and NSCs or MEFs, as indicated; *P<0.05; t-test one tail, type 3.
MacroH2A acts as an epigenetic barrier to induced pluripotency
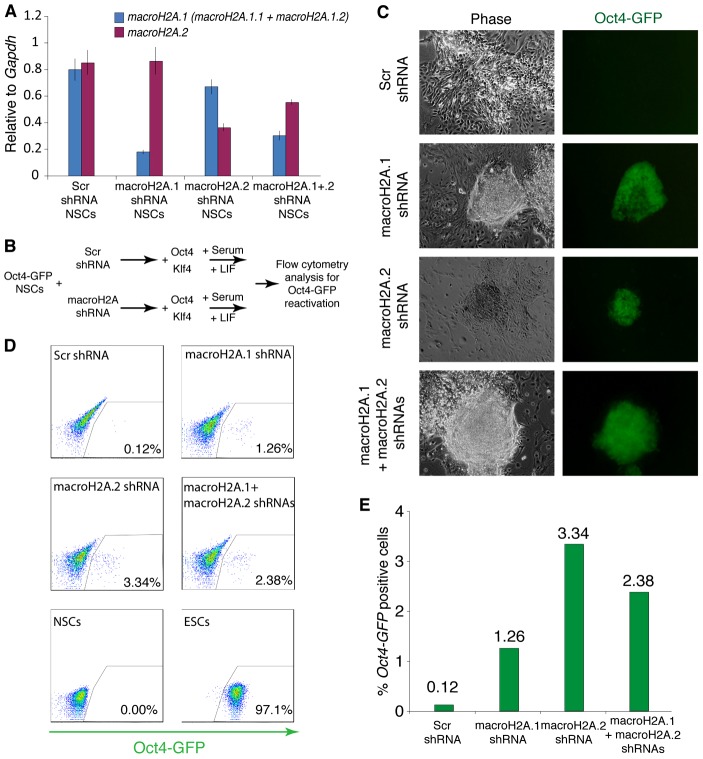
The correlation of macroH2A abundance with increased cell differentiation in vivo and its incorporation into the chromatin of repressed genes in somatic cells may indicate that histone variants macroH2A help stabilize the differentiated cell state. If this is the case, then macroH2A should represent an epigenetic barrier to somatic cell reprogramming toward pluripotency. To test that, we established adult female Oct4-GFP NSC lines stably expressing shRNAs targeting macroH2A.1 (macroH2A.1.1 and macroH2A.1.2), macroH2A.2, macroH2A.1 and macroH2A.2, or scrambled shRNA. qRT-PCR confirmed efficient macroH2A depletion in the established cell lines (Fig. 4A). NSCs can be reprogrammed to naïve pluripotency and form induced pluripotent stem cells (iPSCs) through retroviral expression of Oct4 and Klf4 transcription factors (Kim et al., 2008; Silva et al., 2008). The infected cells give rise to reprogramming intermediates, pre-iPSCs, in serum/leukemia inhibitory factor (LIF) culture conditions, and can be induced to undergo complete reprogramming, characterized by Oct4-GFP reporter transgene reactivation, upon the switch to culture conditions consisting of a combination of MEK/ERK-signaling inhibitor and GSK3β inhibitor (2i) with LIF (Silva et al., 2008). We infected stable macroH2A- or scramble-shRNA expressing NSC lines with retroviruses encoding Oct4 and Klf4 and cultured the cells in serum/LIF culture conditions (Fig. 4B). Remarkably, without switching to 2i/LIF culture conditions, we started to observe the emergence of Oct4-GFP positive colonies in macroH2A-depleted, but not in control infections 19-20 days post-infection (Fig. 4C). We performed flow cytometry analysis 24 days post-infection to assess the efficiency of reprogramming, as judged by Oct4-GFP reactivation (Fig. 4D). Infections of the macroH2A.1, macroH2A.2 and macroH2A.1 plus macroH2A.2 depleted cells contained 1.26%, 3.34% and 2.38% GFP-positive cells, respectively, indicating a 10- to 25-fold increase over control macroH2A containing cells (0.12% GFP positive cells) (Fig. 4E). We did not observe GFP positive colonies in the control infections within the experimental timeframe (Fig. 4C). We conclude that the removal of macroH2A.1 from somatic cells increases the efficiency of iPSCs generation.
Fig. 4.
MacroH2A acts as a barrier to somatic cell reprogramming to pluripotency. (A) qRT-PCR analysis of macroH2A.1 (blue) and macroH2A.2 (purple) expression in Oct4-GFP NSCs stably expressing Scr shRNA, macroH2A.1 shRNA, macroH2A.2 shRNA or both macroH2A.1 and macroH2A.2 shRNAs. Error bars indicate s.d. (B) Diagram showing the experimental set up. Stable Oct4-GFP NSCs lines expressing shRNAs against macroH2A.1, macroH2A.2 or both macroH2A.1 and macroH2A.2, or control scrambled sequence (Scr), were generated and induced to reprogram following retroviral-mediated Oct4 and Klf4 expression under serum and LIF culture conditions from 3 days after factor expression. (C) MacroH2A depletion improves the efficiency of reprogramming to pluripotency. Oct4-GFP-positive colonies obtained from expression of Oct4 and Klf4 in macroH2A shRNA Oct4-GFP NSCs cultured with serum and LIF in the absence of 2i. Note that no Oct4-GFP colonies were obtained from shRNA-expressing cells induced to reprogram within the experimental time frame. Images were taken 24 days following retroviral pluripotency gene expression. (D) Flow cytometry analysis of reprogramming cultures. The proportion of Oct4-GFP-positive cells 24 days after the induction of nuclear reprogramming is indicated (% total cells). Oct4-GFP NSCs and Oct4-GFP ESCs were used as controls. Results are representative of two experiments. (E) Reprogramming efficiency as judged by the proportion of Oct4-GFP-positive cells.
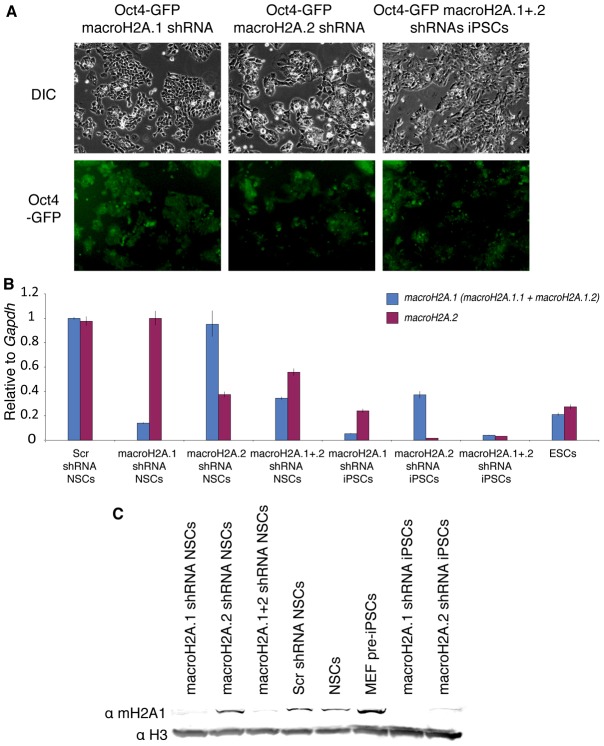
To characterize the obtained iPSCs, several colonies were picked and expanded. The established iPSC lines had characteristic ESC morphology and maintained Oct4-GFP expression (Fig. 5A). Since shRNAs against macroH2A were expressed from polymerase-III H1-RNA promoter, their expression was not silenced upon the establishment of pluripotency. Surprisingly, the obtained iPSCs exhibited almost complete loss of expression of the histone variant being knocked down (Fig. 5B,C), which potentially indicates strong selection for cells with the lowest macroH2A levels during the reprogramming process. qRT-PCR analysis of these shRNA iPSCs demonstrated complete silencing of the retroviral transgenes used to overexpress Oct4 and Klf4 in NSCs (Fig. 6A) and the upregulation of endogenous pluripotency markers Oct4, Nanog, Rex1 and Klf4 to levels comparable to ESCs (Fig. 6B). The obtained iPSCs also reactivated the silent X-chromosome as indicated by the loss of the trimethyl(me3)H3K27 nuclear focus (supplementary material Fig. S2). When injected into a kidney capsule, macroH2A shRNA iPSCs generated teratomas with tissues of three embryonic lineages present (Fig. 6C). Moreover, when injected into blastocysts, macroH2A shRNA iPSCs contributed to the embryo and generated coat color chimeras, demonstrating their wide developmental potential (Fig. 6D). Interestingly, some of the double macroH2A.1 and macroH2A.2 shRNA chimeras, but not macroH2A.1 or macroH2A.2 shRNA only, exhibited a range of developmental and morphological defects at different times after birth (supplementary material Fig. S3), which may be a result of improper maintenance of gene silencing in the absence of all macroH2A variants.
Fig. 5.
MacroH2A knockdown is maintained in the obtained iPSCs. (A) Morphology of the iPSCs obtained from reprogrammed macroH2A-depleted NSCs. (B) qRT-PCR analysis of macroH2A.1 and macroH2A.2 expression in NSCs, ESCs and iPSCs derived from macroH2A.1- or macroH2A.2-depleted NSCs. MacroH2A.1 (macroH2A.1.1 + macroH2A.1.2) is in blue, macroH2A.2 is in purple. Error bars indicate s.d. (C) Western blot analysis of macroH2A.1 in shRNA NSCs and macroH2A shRNA iPSCs. Histone H3 was used as a loading control.
Fig. 6.
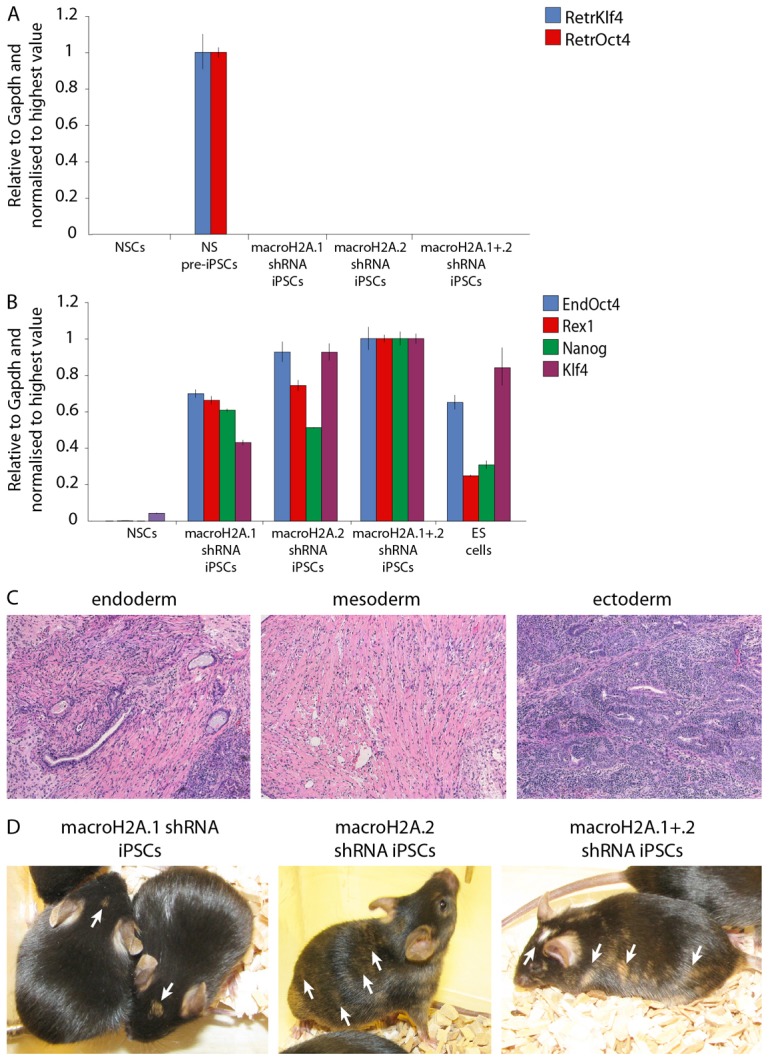
Characterization of macroH2A-depleted iPSCs. (A) qRT-PCR analysis of retrovirus expression in NSCs, NS Pre-iPSCs and in iPSCs derived from macroH2A.1 or macroH2A.2 NSCs. Klf4 retrovirus (RetrKlf4) is in blue and Oct4 retrovirus (RetrOct4) in red. (B) Gene expression analysis of the iPSCs depleted of macroH2A. qRT-PCR analysis of pluripotency gene expression in NSCs, ESCs and iPSCs derived from macroH2A.1 or macroH2A.2 NSCs. Endogenous Oct4 (EndOct4) is in blue, Rex1 in red, Klf4 in purple and Nanog in green. (C) Tissues of the three embryonic germ layers are present in the teratomas derived from macroH2A.2 shRNA expressing iPSCs. (D) Contribution of macroH2A-depleted iPSCs to chimeric mice. These were obtained after blastocyst injection of macroH2A shRNA iPSCs into C57/BL6 blastocysts. Agouti coat color indicates chimerism (arrows). Error bars indicate s.d.
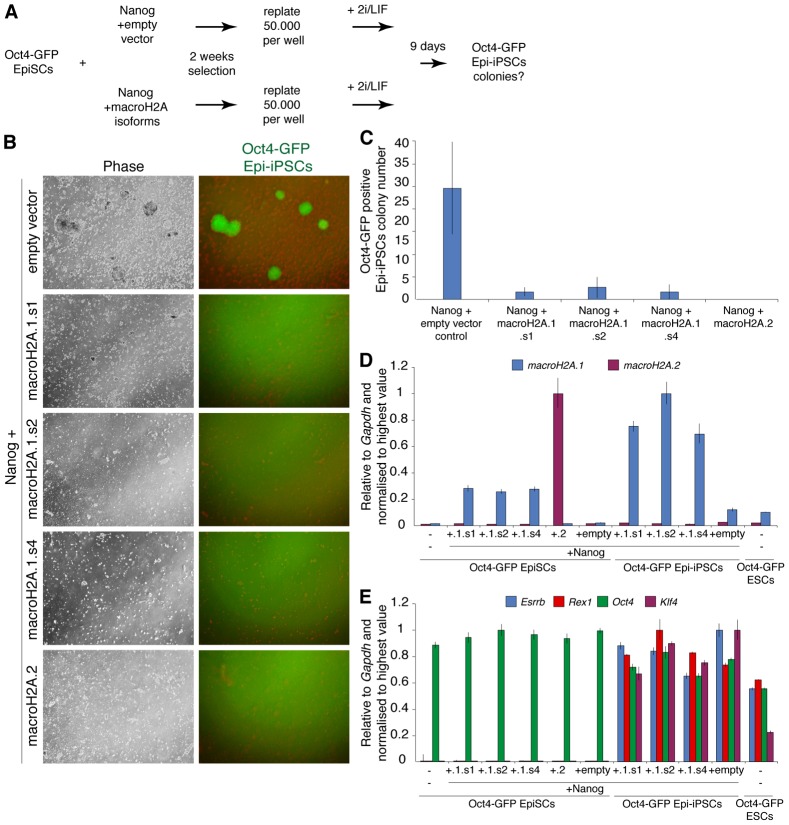
We hypothesized that if indeed macroH2A serves as a barrier to induced pluripotency, then its overexpression in EpiSCs, which naturally express lower levels of this histone variant compared to somatic cells (Fig. 2B,C), would decrease the efficiency of their conversion to naïve pluripotency. EpiSCs can be reprogrammed to naïve pluripotency by overexpression of a single factor such as Nanog or Klf4 together with a switch to 2i plus LIF culture conditions (Guo et al., 2009; Silva et al., 2009; Yang et al., 2010). We transfected EpiSCs carrying the Oct4-GFP reporter (distal enhancer) with transgenes encoding Nanog in combination with macroH2A isoforms or an empty vector control and kept them under dual selection for 2 weeks (Fig. 7A). MacroH2A isoforms macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4 or macroH2A.2 were used, and overexpression confirmed by qRT-PCR (Fig. 7D). The expression of Oct4 was not changed in the selected EpiSCs, and these maintained normal EpiSCs morphology (Fig. 7E). To induce reprogramming to naïve pluripotency, Oct4-GFP EpiSCs expressing combinations of transgenes were replated at equal densities and subsequently switched to 2i plus LIF conditions for 9 days (Fig. 7A). The appearance of EpiSCs derived iPSCs (Epi-iPSCs) colonies, characterized by response to 2i plus LIF culture conditions and Oct4-GFP reporter expression, was monitored. Remarkably, macroH2A overexpression in EpiSCs dramatically reduced the number of emerging 2i plus LIF Oct4-GFP positive Epi-iPSC colonies, indicating that macroH2A negatively regulates the efficiency of EpiSCs reprogramming to naïve pluripotency (Fig. 7B,C). Overexpression of individual macroH2A.1 isoforms reduced reprogramming efficiency by 9- to 15-fold and in the case of macroH2A.2 no colonies were obtained. To assay the expression of macroH2A and pluripotency genes in the obtained Epi-iPSCs, several of these were picked, expanded, and subjected to qRT-PCR analysis (Fig. 7D,E). The Epi-iPSCs obtained showed pluripotency gene expression levels comparable to control ESCs (Fig. 7E). We conclude that overexpression of macroH2A impairs the reprogramming of EpiSCs to naïve pluripotency.
Fig. 7.
MacroH2A overexpression prevents efficient reprogramming of EpiSC to naïve pluripotency. (A) Diagram showing the experimental set up. Oct4-GFP EpiSCs were transfected with vectors encoding Nanog in combination with macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4, macroH2A.2 or empty vector control and grown for 2 weeks under dual selection. Selected cells were plated at a density of 50,000 cells per well of a six-well plate, switched to 2i plus LIF the next day and cultured for another 9 days before monitoring the presence of reprogrammed Oct4-GFP-positive Epi-iPSCs. (B) Representative phase and GFP images taken 9 days after the 2i plus LIF culture of Oct4-GFP EpiSC with Nanog and macroH2A variants or empty vector overexpression. (C) Oct4-GFP-positive Epi-iPSCs colony counts. The average number of Oct4-GFP-positive Epi-iPSCs per well 9 days after the switch of macroH2A overexpressing or control cells to 2i plus LIF conditions is indicated. Values represent the average colony number from three independent wells. (D) qRT-PCR analysis of macroH2A.1 (light blue) and macroH2A.2 (purple) in Oct4-GFP EpiSCs transfected with Nanog (+Nanog) and macroH2A.1.s1 (+.1.s1), macroH2A.1.s2 (+.1.s2), macroH2A.1.s4 (+.1.s4), macroH2A.2 (+.2) or empty vector control (+empty); and in the resulting Oct4-GFP Epi-iPSCs, as well as control Oct4-GFP ESCs. (E) qRT-PCR analysis of Esrrb (light blue), Rex1 (red), Oct4 (green) and Klf4 (purple) in Oct4-GFP EpiSCs transfected with Nanog (+Nanog) and macroH2A.1.s1 (+.1.s1), macroH2A.1.s2 (+.1.s2), macroH2A.1.s4 (+.1.s4), macroH2A.2 (+.2) or empty vector control (+empty); and in the resulting Oct4-GFP Epi-iPSCs and control Oct4-GFP ESCs. Please note control Oct4-GFP ESCs express Nanog, but do not overexpress Nanog. Error bars indicate s.d.
Given that macroH2A incorporates in silenced pluripotency genes in somatic cells and that it prevents efficient reprogramming to induced pluripotency, we sought to determine whether macroH2A overexpression in ESCs induces differentiation and/or pluripotency gene repression. Overexpression of macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4 and macroH2A.2 in ESCs did not induce cell death or differentiation and the obtained ESCs maintained ESC morphology, indicating that ectopic macroH2A expression is compatible with maintenance of pluripotency (Fig. 8A). ESCs overexpressing macroH2A isoforms showed a modest decrease in pluripotency gene expression and an increase in basal expression levels of the differentiation mesoderm marker T-Brachyury, but little changes in the expression of Gata4 (Fig. 8B,C). We conclude that ectopic macroH2A expression in ESCs, though leading to some downregulation of pluripotency markers and misexpression of a mesoderm marker, is not an inducer of ESCs differentiation.
Fig. 8.
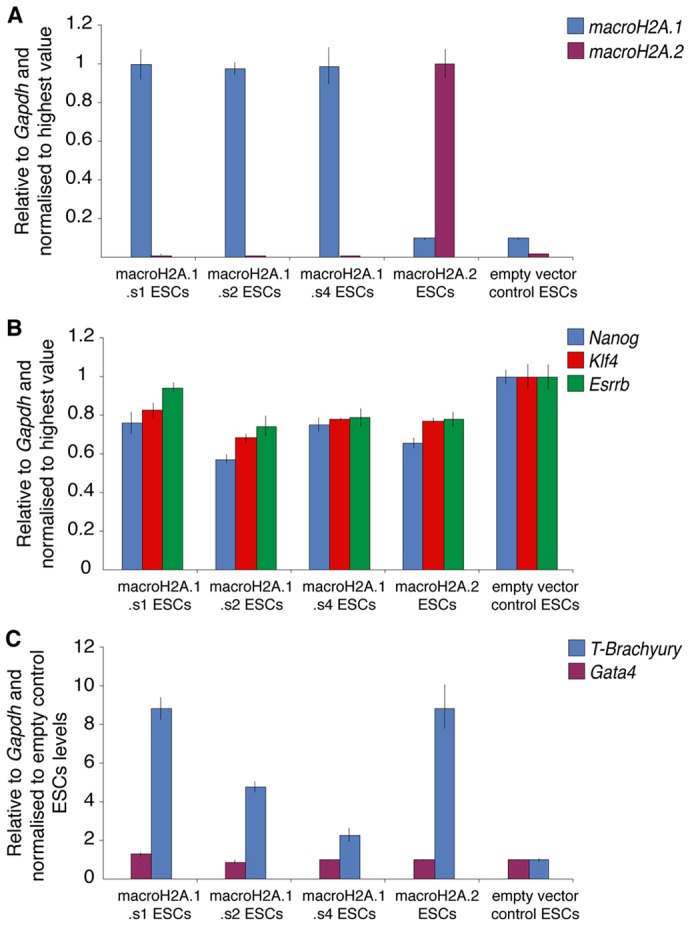
MacroH2A overexpression in ESCs. (A) qRT-PCR analysis of macroH2A.1 and macroH2A.2 expression in ESCs overexpressing macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4 and macroH2A.2, or empty vector control. MacroH2A.1 is in blue and macroH2A.2 in purple. (B) qRT-PCR analysis of Nanog, Klf4 and Esrrb expression in ESCs overexpressing macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4 and macroH2A.2, or empty vector control. Nanog is in blue, Klf4 in red and Esrrb in green. (C) qRT-PCR analysis of T-Brachyury and Gata4 expression in ESCs overexpressing macroH2A.1.s1, macroH2A.1.s2, macroH2A.1.s4 and macroH2A.2, or empty vector control. T-Brachyury is in blue and Gata4 in purple. Error bars indicate s.d.
The increased efficiency of somatic cell reprogramming upon the removal of macroH2A and the decreased efficiency of EpiSC reprogramming upon macroH2A overexpression, together with the fact that macroH2A is a chromatin component of repressed pluripotency genes in somatic cells indicates that macroH2A histone variants represent an epigenetic barrier to cell reprogramming to naïve pluripotency.
Discussion
In this study, we report that the histone variant macroH2A is present at low levels in the pluripotent cell compartment of pre- and post-implantation mouse embryos while it accumulates to high levels in somatic cells in vivo as well as in vitro. This observed correlation of macroH2A abundance with the degree of cell differentiation together with the known gene repression functions of these histone variants suggests that macroH2A is involved in gene repression during cell commitment. These findings are consistent with previous reports showing high macroH2A expression in differentiated cells both in mouse and human (Creppe et al., 2012a; Dai and Rasmussen, 2007; Pehrson et al., 1997).
In somatic cells, macroH2A depletion, though causing decreased stability of silencing of the inactive X chromosome and pluripotency genes (Csankovszki et al., 2001; Pasque et al., 2011a), is not sufficient to induce full reactivation of repressed genes, or a change in cell fate. At the same time, when exposed to reprogramming conditions, as we show in this study, cells depleted of macroH2A are induced to reprogram with higher efficiencies. Moreover, ectopic macroH2A expression strongly impairs EpiSCs reprogramming to naïve pluripotency. These results suggest that late incorporation of macroH2A into chromatin of repressed genes during development is important to stabilize their silenced state and thereby contribute to the maintenance of the differentiated state of somatic cells. Whether macroH2A also restricts direct reprogramming of one somatic cell type to another warrants further investigations.
A key mechanistic step during nuclear reprogramming may be chromatin decompaction, which gives reprogramming factors access to pluripotency gene regulatory regions. MacroH2A variants in a nucleosome have been shown to promote chromatin compaction by multiple mechanisms. We believe that the removal of macroH2A makes chromatin more accessible to respond to transcription factors Oct4 and Klf4, thereby facilitating transcriptional activation of target genes and cell state transitions during somatic cell reprogramming. MacroH2A has been reported to promote chromatin compaction (Abbott et al., 2005; Chakravarthy et al., 2012), recruitment of histone deacetylases (Chakravarthy et al., 2005), lower the affinity for SWI/SNF chromatin remodeling complexes (Chang et al., 2008), hinder transcription factor binding (Angelov et al., 2003) and associate with Polycomb proteins Ezh2 and Suz12 (Buschbeck et al., 2009). These repressive functions attributed to macroH2A variants, acting together with other epigenetic mechanisms such as DNA methylation and Polycomb, are likely to be mechanistically linked to the low efficiency of nuclear reprogramming. Indeed, genetic ablation of HDAC2 or its chemical inhibition improves the efficiency with which somatic cells can be induced to pluripotency (Huangfu et al., 2008).
Importantly, not only does macroH2A restrict induced pluripotency and transcriptional reactivation, but it also prevents progression of melanomas to the metastatic state (Kapoor et al., 2010). Indeed, loss of macroH2A.1.1 is seen in several cancer types and correlates with negative outcome for patients with lung or colorectal cancer (Kapoor et al., 2010; Novikov et al., 2011; Sporn and Jung, 2012; Sporn et al., 2009). This reinforces the view that loss of macroH2A is associated with decreased long-term somatic cell stability. Interestingly, macroH2A.1 shRNA iPSCs were devoid of detectable macroH2A.1, unlike macroH2A.1 shRNA NSCs, indicating a possible selection for cells with low macroH2A.1 expression during reprogramming to induced pluripotency. This seems particularly relevant in light of the increased metastatic and teratogenic state of macroH2A depleted cells (Creppe et al., 2012a; Kapoor et al., 2010). Thus, it would be of interest to explore the process of induced pluripotency as a route to identify novel developmentally regulated epigenetic barriers not only to nuclear reprogramming but also to cancer progression.
Overexpression of macroH2As in ESCs, EpiSCs or Epi-iPSCs did not lead to cell differentiation or detectable morphological changes, but for ESCs it did cause a small reduction in pluripotency gene expression and an increase in the expression of mesoderm inducer T-Brachyury. This suggests that high levels of multiple macroH2A isoforms are compatible with pluripotency. This is in agreement with the high expression of pluripotency genes in Epi-iPSCs in which high levels of macroH2A isoforms in combination with Nanog are expressed. At the same time, macroH2A overexpression could prone ESCs for differentiation. Interestingly, T-Brachyury expression was found to be impaired in ESCs depleted of macroH2A.1 and induced to differentiate using retinoic acid (Creppe et al., 2012a).
Genetic ablation of macroH2A.1 or macroH2A.2 is not lethal and results in fertile mice, strongly suggesting that individually these histone variants are dispensable for normal embryonic development (Changolkar et al., 2007; Changolkar et al., 2010). Double knockout mice were not reported to date, which leaves open the question whether there are compensatory mechanisms between the two variants. In this study, we observed developmental and morphological abnormalities in chimeric mice obtained from double macroH2A.1 and macroH2A.2 shRNA iPSCs, but not from iPSCs in which individual macroH2A variants were depleted (supplementary material Fig. S3). This may suggest that the knockout phenotype in previous reports could have been masked by potential compensatory mechanisms, which requires further confirmation by combined genetic ablation. Given that macroH2A is required to stabilize the silenced state of repressed genes, the interpretation of the knockout phenotype should include the analysis of susceptibility to cancer, the efficiency of regeneration and germ cell formation and the analysis of the effect of various stress conditions on an organism. It is also possible that macroH2As are not strictly required for embryogenesis, but play a role in maintenance of somatic cell identity, consistent with its known functions in maintenance of silenced states as well as acting as a barrier to reprogramming to pluripotency.
In summary, this study provides evidence that histone variants macroH2A are developmentally regulated epigenetic marks being produced and incorporated into the chromatin of repressed genes concomitantly with cell lineage commitment. Thus, for the first time we identified a link between an epigenetic mark and stabilization of the differentiated cell state that in turn antagonizes induction of pluripotency.
Materials and Methods
Cell culture
Pre-iPS, iPS and PLAT-E cells were cultured in GMEM containing 10% FCS, 1× NEAA, 1× Pen/Strep, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, supplemented with LIF. NSCs were maintained in homemade NS medium [DMEM/F-12, 1× NEAA, 0.1 mM 2-mercaptoethanol, 1× Pen/Strep, 1∶100 B27 supplement (Invitrogen), 1∶200 N2 supplement (PAA), 4.5 µM HEPES (PAA), 0.03 M glucose (Sigma-Aldrich), 120 µg/ml BSA (Invitrogen)] supplemented with 10 ng/ml of EGF and 20 ng/ml of FGF2. EpiSCs were grown and induced to reprogram as described previously (Guo et al., 2009; Silva et al., 2009).
Plasmids
pMXs-Oct3/4, pMXs-Klf4 (from Addgene). Retroviral infection and plasmids for macroH2A RNAi, as well as primers used to amplify macroH2A isoforms are described elsewhere (Pasque et al., 2011a). For EpiSC reprogramming experiments, plasmids pPB-CAG-macroH2A.1.s1-pA-PGK-hph, pPB-CAG-macroH2A.1.s2-pA-PGK-hph, pPB-CAG-macroH2A.1.s4-pA-PGK-hph, pPB-CAG-macroH2A.2-pA-PGK-hph and pPB-CAG-Nanog-IRES-bsd were used.
Retroviral infection
PLAT-E cells were transfected using FuGENE 6 transfection reagent (Roche) according to manufacturer's instructions. One day after the transfection complete medium change was performed and 8×105 NSCs were seeded per well of a six-well plate. The following day virus-containing supernatants from PLAT-E cultures were filtered through 0.45 µm cellulose acetate filters and mixed with polybrene (Sigma-Aldrich) to the final concentration of 4 µg/ml. The polybrene/virus mixture was applied to NSCs and these were incubated in virus-containing medium for 24 hours after which the medium was replaced with NS medium. For reprogramming experiments, the infected cells were switched to serum/LIF medium 3 days after infection.
Flow cytometry
Flow cytometry was performed using BD LSRFortessa cell analyzer with subsequent data analysis using FlowJo software. Cell sorting was performed using a MoFlo high-speed cell sorter.
ChIP analysis
Starting material: 2–10×106 cells, trypsinized off culture plates, washed in complete DMEM, and washed twice in PBS to remove excess serum.
Fixation: Cells are suspended in 0.3 ml PBS to which 0.1 ml 4% paraformaldehyde in 1× PBS is added for 7 minutes at room temperature and quenched with 0.4 ml 250 mM cold glycine (125 mM final concentration). Cells are then washed twice in 2× PBS (and stored at −80°C as pellets).
Lysis: Cell pellets are resuspended in Lysis buffer (made from 2× stock with protease inhibitor, SDS and DTT added on the day) to a final volume of not more than 270 ml in Protein Lobind 1.5 ml microcentrifuge tubes (Eppendorf). Cells kept on ice with occasional tapping of the tubes for 10 minutes.
Chromatin fragmentation: The lysate is sonicated sufficiently to yield chromatin fragments of 250–700 bps using a Bioruptor sonicator (twice for 7-10 minutes with 30 seconds on/off time, on ‘high’ power, Diagenode). Chromatin is cleared by centrifugation (10 minutes at 10,000 r.p.m. at 4°C). Fragmented chromatin was stored at −80°C.
Chromatin fragment size determination was established by purifying a known amount of the chromatin lysate by the purification procedure described below. Purified DNA was analyzed by gel electrophoresis, post stained with ethidium bromide for fragment sizes. Purified DNA was quantified by UV spectrophotometry (Nanodrop) in order to establish the approximate chromatin concentration of the lysates (approximately twice the DNA concentration). Chromatin lysate were aliquoted into units of ∼25 mg of chromatin (about 27 µl each from 107 cells).
Pre-adsorption of beads: An equal volume of Protein A and Protein G magnetic beads (DynaBead, Invitrogen) were mixed together (10 µl each per pull-down tube) and washed three times in RIPA Buffer. Beads were resuspended in SDS free RIPA with BSA (0.1 µg/µl, Sigma) and ssDNA (75 ng/µl, Salmon Sperm derived, Sigma) at a volume of 20 µl/IP and rotated for 30 minutes to 4 hours at 4°C. Beads were then washed once in RIPA (no SDS) and resuspended in RIPA (with BSA and ssDNA and without SDS, such that there were 20 µl/IP.
Immunocomplex and chromatin pull-down: A 1∶10 or 1∶20 dilution of chromatin was made in elution buffer and stored at −80°C as ‘input sample’. For each pull down series. 25 µg of chromatin was diluted 1∶10 in SDS-free RIPA buffer containing (0.1 µg/µl BSA (Sigma) and 75 ng/µl ssDNA (Salmon Sperm, Sigma-Aldrich). Antibody (20–40 µl total) diluted in SDS-free RIPA (with DNA and BSA) to give 1–10 mg (exact amount as per manufacturer's suggestion) were added to this and rotated at 4°C for 20–60 minutes. 20 µl of pre-adsorbed protein A/G beads were then added to each pull down and rotate for 4 hours or overnight at 4°C.
Capture of immunocomplexes and stringency wash: Beads were captured on magnetic rack, washed three times in 1 ml low wash buffer, washed in 1 ml high wash buffer. Complexes were eluted from protein A/G by adding 120 ml of elution buffer and incubated at 30°C for 15–20 minutes (avoid foaming). Chromatin was dissociated from beads magnetically and the solution transferred to new DNA LoBind tubes (Eppendorf).
DNA purification: RNase A was added (1 µl of DNase free 1 mg/ml to each tube) to ‘Input’ and chromatin samples pulled down and heated at 65°C for 6 hours or overnight. Samples were tapped up occasionally to move around. DNA was purified with PCR spin columns (Qiagen) and eluted with 2× 60 µl.
Buffers: ChIP lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 5 mM EDTA pH 8, 1% Triton X-100, 0.5% NP-40. 0.5 mM DTT, Protease inhibitor [Roche, Complete EDTA free] and 1% SDS added on the day of use). ChIP RIPA buffer, SDS free [50 mM Tris-HCl pH 8, 150 mM NaCl, 2 mM EDTA pH 8, 1% NP-40, 0.5% sodium deoxycholate, protease inhibitors (add freshly)]. ChIP low wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 150 mM NaCl, 20 mM Tris-HCl pH 8). ChIP high wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 500 mM NaCl, 20 mM Tris-HCl pH 8). ChIP elution buffer (1% SDS, 100 mM NaHCO3).
Antibodies used: macroH2A.1: Upstate 07-219 and Abcam ab37264. H3K9ac: Cell Signaling no. 9649. The primers used are listed in supplementary material Table S1.
Embryo collection and culture
F1 females from C57B16×CBA crosses were superovulated by intra-peritoneal injection of 10 IU of PMSG (Intervet) and 10 IU of hCG 48 hours later. Matings with F1 males\males expressing EGFP-H2B (Hadjantonakis and Papaioannou, 2004) were set up overnight, and the mice were separated the following morning. Mice were sacrificed by cervical dislocation and their oviducts were microdissected in M2 medium to collect the embryonic stage required. Zygotes (embryonic day 0.5; E0.5) were released by oviduct incision, cumulus cells were removed using short hyaluronidase treatment (1 mg/ml). Embryo culture was carried out at 37.5°C in pre-equilibrated KSOM medium under paraffin oil in an incubator with 5% CO2.
Wholemount immunofluorescence
Embryos at E3.5 and E4.5 stages were fixed for 10 minutes in 4% paraformaldehyde in 1× PBS at 37°C. Embryos at E6.5 and E9.5 were fixed for 2 hours in 4% paraformaldehyde in 1× PBS at 37°C. Fixed embryos were rinsed twice in PBS 0.2% Tween (PBT). Permeabilization was carried out during 20 minutes in 0.5% Triton X, followed by the three washes in 1× PBT. The embryos were blocked in 5% fetal bovine serum (FBS) in 1× PBT (blocking) overnight. Primary antibodies overnight incubations were performed at 4°C. Embryos were washed three times for 10–15 minutes in PBT, before detection using secondary antibodies, 2 hours at room temperature followed by three washes in PBT and counterstaining with DAPI. Embryos were imaged using confocal and epifluoresence microscopy. The primary antibodies used were: rabbit IgG anti-mouse macroH2A.1 (1∶100, kind gift from Dr Theodore Rasmussen), rat IgG2 anti-GFP (1∶500, Nacalai Tesque, Cat. No. 04404-26). The secondary antibodies used were: Alexa Fluor 594 anti-rabbit IgG (1∶200, Invitrogen), Alexa Fluor 488 anti-rat (1∶200, Invitrogen), Alexa Fluor 647 anti-mouse (1∶200, Invitrogen).
Immunofluorescence of cultured cells
Cells grown on glass slides, rinsed in 1× PBS, washed 30 seconds in CSK buffer (100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 10 mM PIPES pH 6.8), 30 seconds in 0.5% Triton X CSK buffer and fixed in 4% PFA 10 minutes before rinsing in 1× PBS 0.2% Tween 20 (PBT). Permeabilized cells were incubated in blocking buffer (5% FBS in 1× PBT) for 30 minutes, then incubated in blocking buffer with primary antibodies overnight at 4°C. The next day, slides were rinsed three times for 10 minutes in PBT and incubated for 30 minutes in blocking buffer with secondary antibodies at room temperature, followed by three washes in PBT and counterstained with DAPI. Slides were mounted with vectashield and imaged using confocal microscopy. The primary antibodies used were: rabbit IgG anti-macroH2A.1 (1∶100, Upstate, Cat. no. 07-219, Lot 32188), mouse IgG3 anti-H3K27me3 (1∶100, AbCam ab6002), mouse IgG2b anti-Oct4 (1∶200 Santa-Cruz C10, SC5279), rat IgG2a anti-Nanog (1∶200, eBioscience, clone eBioMLC-51, 14-5761-80). The secondary antibodies used were: Alexa Fluor 488 anti-mouse IgG (1∶200, Invitrogen), Alexa Fluor 488 goat anti-rat (1∶200, Invitrogen), Alexa Fluor 594 goat anti-rabbit IgG (1∶200, Invitrogen), Alexa Fluor 647 donkey anti-mouse (1∶200, Invitrogen).
Quantitative RT-PCR
Total RNA was isolated using RNeasy kit (QIAGEN) in accordance with manufacturer's protocol. Reverse transcription was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). qRT-PCR reactions were performed in triplicate using SYBR Green PCR Master Mix or TaqMan Universal PCR Master Mix (both Applied Biosystems). qRT-PCR primers and gene specific Taqman expression assays (Applied Biosystems) are listed in supplementary material Table S2. ΔCts with GAPDH were calculated and brought to power −2. The values were normalized to the highest value where indicated.
Western previously
As described previously (Pasque et al., 2011a) with minor modifications. 1×106 cell pellets were extracted in 200 µl of combined lysis/loading buffer (300 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 4.1% SDS, 20% glycerol, 0.0004% bromoblue, 175 mM Tris-Cl, 10% betamercaptoethanol), denatured 10 minutes at 98°C and incubated on ice for 5 minutes. 15 µl of extract were loaded onto a 12% Tris-HCl acrylamide gel (BioRad 161-1120), electrophoresed, and transferred to nitrocellulose membranes. Membranes were blocked in TBS containing 3% BSA. Primary antibodies were incubated overnight at 4°C. Primary antibodies used were: rabbit anti-macroH2A.1 (1 µg/ml, Abcam ab37264), rabbit anti-Histone H3 (1:4000, Abcam ab1791). After extensive TBS 0.05% Tween20 (TBS-T) washes, secondary antibody 680CW-conjugated goat anti-rabbit (LI-COR, 1:25,000) was incubated 30 minutes at room temperature. After extensive TBS-T washes, fluorescence was detected using the Odyssey® Infrared imaging system (LI-COR).
Teratoma assay
iPSCs were injected under the kidney capsule of anesthetized MF1 mice. Four weeks after the injection tumors were surgically dissected from the mice. Samples were fixed in PBS containing 4% formaldehyde and embedded in paraffin. Sections were stained with hematoxylin and eosin.
Blastocyst injection
Standard microinjection methodology using host blastocysts of C57BL/6 strain was employed.
Statistical analysis
Statistical differences were assessed with the unpaired Student's t-test. Data are presented as mean ± s.e.m. or mean ± s.d.; P-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The authors are indebt to Dr John Gurdon for his support as well as comments on the manuscript and to Dr Jennifer Nichols for advice on antibody staining on early embryos. We are very grateful to Dr Theodore Rasmussen for his kind gift of macroH2A.1 antibody. We thank William Mansfield and Charles-Etienne Dumeau for performing blastocyst injections and teratoma assays and Margaret McLeish for histological processing of the teratomas. Astrid Gillich, Richard P. Halley-Stott and Maryna Panamarova made equal contribution as second authors to this paper. The authors declare that they have no conflict of interest.
Footnotes
Funding
This work was supported by The Wellcome Trust [grant numbers RG54943, 081277 and RG44593]. J.S. is a Wellcome Trust Career Development Fellow [grant number WT086692]. M.P. and M.Z.-G. are supported by a Wellcome Trust Senior Fellowship to M.Z.-G. V.P. was also supported by a Wallonia-Brussels International Excellence Grant; A.R. and M.P. by the Darwin Trust of Edinburgh; and R.P.H.-S. by the Medical Research Council. Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.113019/-/DC1
References
- Abbott D. W., Chadwick B. P., Thambirajah A. A., Ausió J. (2005). Beyond the Xi: macroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 280, 16437–16445 10.1074/jbc.M500170200 [DOI] [PubMed] [Google Scholar]
- Angelov D., Molla A., Perche P-Y., Hans F., Côté J., Khochbin S., Bouvet P., Dimitrov S. (2003). The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11, 1033–1041 10.1016/S1097-2765(03)00100-X [DOI] [PubMed] [Google Scholar]
- Barzily–Rokni M., Friedman N., Ron–Bigger S., Isaac S., Michlin D., Eden A. (2011). Synergism between DNA methylation and macroH2A1 occupancy in epigenetic silencing of the tumor suppressor gene p16(CDKN2A). Nucleic Acids Res. 39, 1326–1335 10.1093/nar/gkq994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Muratore–Schroeder T. L., Diaz R. L., Chow J. C., Changolkar L. N., Shabanowitz J., Heard E., Pehrson J. R., Hunt D. F., Allis C. D. (2008). A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc. Natl. Acad. Sci. USA 105, 1533–1538 10.1073/pnas.0711632105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard M., Storck S., Cong R., Pinto R., Delage H., Bouvet P. (2010). Histone variant macroH2A1 deletion in mice causes female-specific steatosis. Epigenetics & Chromatin 3, 8 10.1186/1756-8935-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M., Di Croce L. (2010). Approaching the molecular and physiological function of macroH2A variants. Epigenetics 5, 118–123 10.4161/epi.5.2.11076 [DOI] [PubMed] [Google Scholar]
- Buschbeck M., Uribesalgo I., Wibowo I., Rué P., Martin D., Gutierrez A., Morey L., Guigó R., López–Schier H., Di Croce L. (2009). The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16, 1074–1079 10.1038/nsmb.1665 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S., Gundimella S. K. Y., Caron C., Perche P-Y., Pehrson J. R., Khochbin S., Luger K. (2005). Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25, 7616–7624 10.1128/MCB.25.17.7616-7624.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S., Patel A., Bowman G. D. (2012). The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Res. 40, 8285–8295 10.1093/nar/gks645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C., Ma Y., Jacobs S., Tian X. C., Yang X., Rasmussen T. P. (2005). A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev. Biol. 278, 367–380 10.1016/j.ydbio.2004.11.032 [DOI] [PubMed] [Google Scholar]
- Chang E. Y., Ferreira H., Somers J., Nusinow D. A., Owen–Hughes T., Narlikar G. J. (2008). MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry 47, 13726–13732 10.1021/bi8016944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C., Gao S., Sung L-Y., Corry G. N., Ma Y., Nagy Z. P., Tian X. C., Rasmussen T. P. (2010). Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell Reprogram 12, 43–53 10.1089/cell.2009.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar L. N., Costanzi C., Leu N. A., Chen D., McLaughlin K. J., Pehrson J. R. (2007). Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27, 2758–2764 10.1128/MCB.02334-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar L. N., Singh G., Cui K., Berletch J. B., Zhao K., Disteche C. M., Pehrson J. R. (2010). Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol. Cell. Biol. 30, 5473–5483 10.1128/MCB.00518-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C., Pehrson J. R. (1998). Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 10.1038/31275 [DOI] [PubMed] [Google Scholar]
- Costanzi C., Pehrson J. R. (2001). MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 276, 21776–21784 10.1074/jbc.M010919200 [DOI] [PubMed] [Google Scholar]
- Costanzi C., Stein P., Worrad D. M., Schultz R. M., Pehrson J. R. (2000). Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 127, 2283–2289 [DOI] [PubMed] [Google Scholar]
- Creppe C., Janich P., Cantariño N., Noguera M., Valero V., Musulén E., Douet J., Posavec M., Martín–Caballero J., Sumoy L.et al. (2012a). MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol. Cell. Biol. 32, 1442–1452 10.1128/MCB.06323-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C., Posavec M., Douet J., Buschbeck M. (2012b). MacroH2A in stem cells: a story beyond gene repression. Epigenomics 4, 221–227 10.2217/epi.12.8 [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Nagy A., Jaenisch R. (2001). Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153, 773–784 10.1083/jcb.153.4.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B., Rasmussen T. P. (2007). Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells 25, 2567–2574 10.1634/stemcells.2007-0131 [DOI] [PubMed] [Google Scholar]
- Foshay K. M., Looney T. J., Chari S., Mao F. F., Lee J. H., Zhang L., Fernandes C. J., Baker S. W., Clift K. L., Gaetz J.et al. (2012). Embryonic stem cells induce pluripotency in somatic cell fusion through biphasic reprogramming. Mol. Cell 46, 159–170 10.1016/j.molcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble M. J., Kraus W. L. (2010). Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle 9, 2566–2572 10.4161/cc.9.13.12144 [DOI] [PubMed] [Google Scholar]
- Gamble M. J., Frizzell K. M., Yang C., Krishnakumar R., Kraus W. L. (2010). The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 24, 21–32 10.1101/gad.1876110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J. B., Melton D. A. (2008). Nuclear reprogramming in cells. Science 322, 1811–1815 10.1126/science.1160810 [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Elsdale T. R., Fischberg M. (1958). Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182, 64–65 10.1038/182064a0 [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A-K., Papaioannou V. E. (2004). Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 4, 33 10.1186/1472-6750-4-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis A. K., Cox L. L., Tam P. P., Nagy A. (2001). An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis 29, 133–140 10.1002/gene.1016 [DOI] [PubMed] [Google Scholar]
- Hajkova P., Ancelin K., Waldmann T., Lacoste N., Lange U. C., Cesari F., Lee C., Almouzni G., Schneider R., Surani M. A. (2008). Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881 10.1038/nature06714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 10.1038/nbt1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Goldberg M. S., Cumberland L. K., Ratnakumar K., Segura M. F., Emanuel P. O., Menendez S., Vardabasso C., Leroy G., Vidal C. I.et al. (2010). The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 10.1038/nature09590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo–Bravo M. J., Ruau D., Han D. W., Zenke M.et al. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454, 646–650 10.1038/nature07061 [DOI] [PubMed] [Google Scholar]
- Nashun B., Yukawa M., Liu H., Akiyama T., Aoki F. (2010). Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137, 3785–3794 10.1242/dev.051805 [DOI] [PubMed] [Google Scholar]
- Novikov L., Park J. W., Chen H., Klerman H., Jalloh A. S., Gamble M. J. (2011). QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 31, 4244–4255 10.1128/MCB.05244-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Gillich A., Garrett N., Gurdon J. B. (2011a). Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 30, 2373–2387 10.1038/emboj.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Halley–Stott R. P., Gillich A., Garrett N., Gurdon J. B. (2011b). Epigenetic stability of repressed states involving the histone variant macroH2A revealed by nuclear transfer to Xenopus oocytes. Nucleus 2, 533–539 10.4161/nucl.2.6.17799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J. R., Fried V. A.1992). MacroH2A, a core histone containing a large nonhistone region. Science 2571398–1400 10.1126/science.1529340 [DOI] [PubMed] [Google Scholar]
- Pehrson J. R., Costanzi C., Dharia C. (1997). Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 65, 107–113 [DOI] [PubMed] [Google Scholar]
- Rasmussen T. P., Huang T., Mastrangelo M. A., Loring J., Panning B., Jaenisch R. (1999). Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 27, 3685–3689 10.1093/nar/27.18.3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253 10.1371/journal.pbio.0060253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn J. C., Jung B. (2012). Differential regulation and predictive potential of MacroH2A1 isoforms in colon cancer. Am. J. Pathol. 180, 2516–2526 10.1016/j.ajpath.2012.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn J. C., Kustatscher G., Hothorn T., Collado M., Serrano M., Muley T., Schnabel P., Ladurner A. G. (2009). Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 10.1038/onc.2009.26 [DOI] [PubMed] [Google Scholar]
- Tada M., Tada T., Lefebvre L., Barton S. C., Surani M. A. (1997). Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520 10.1093/emboj/16.21.6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. (2001). Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 10.1016/S0960-9822(01)00459-6 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Ahmad K., Almouzni G., Ausió J., Berger F., Bhalla P. L., Bonner W. M., Cande W. Z., Chadwick B. P., Chan S. W. L.et al. (2012). A unified phylogeny-based nomenclature for histone variants. Epigenetics & Chromatin 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanasijevic B., Rasmussen T. P. (2011). X chromosome inactivation and differentiation occur readily in ES cells doubly-deficient for macroH2A1 and macroH2A2. PLoS ONE 6, e21512 10.1371/journal.pone.0021512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G., Till S., Hassa P. O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E. H. K., Scheffzek K.et al. (2009). A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 16, 923–929 10.1038/nsmb.1664 [DOI] [PubMed] [Google Scholar]
- Yang J., van Oosten A. L., Theunissen T. W., Guo G., Silva J. C. R., Smith A. (2010). Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 7, 319–328 10.1016/j.stem.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.