Summary
The temporal and spatial regulation of protein synthesis plays an important role in the control of neural physiology. In axons and dendrites, translationally repressed mRNAs are actively transported to their destinations in a variety of ribonucleoprotein particles (RNPs). A subset of these neuronal RNPs has been shown to contain proteins associated with mRNA processing bodies (P bodies). P bodies are a class of highly conserved cytoplasmic granules that have been linked to both mRNA decay and translational repression via general and miRNA-mediated pathways. Here, we characterize functions for HPat/Pat1 (also known as Patr-1), a core component of P bodies, at the glutamatergic larval Drosophila neuromuscular junction (NMJ). We show that hpat mutants exhibit a strong synaptic hyperplasia at the NMJ. The synaptic defects observed in hpat mutants are associated with rearrangement of the axonal microtubule cytoskeleton suggesting that HPat negatively regulates presynaptic microtubule-based growth during NMJ development. Consistent with this, overexpression of HPat also blocks the rapid growth of presynaptic boutons induced by spaced depolarization. Finally, we demonstrate that HPat interacts genetically with the catalytic subunit of the deadenylase complex (twin/CCR4) and the miRNA pathway (Argonaute 1) to control bouton formation. We propose that HPat is required to target mRNAs involved in the control of microtubule architecture and synaptic terminal growth for repression, presumably in P bodies, via both general and miRNA-mediated mechanisms.
Key words: P bodies, Ribonucleoprotein particles, Pat1, Synaptogenesis, Neuromuscular junction
Introduction
In neurons, the local regulation of mRNA translation is required to control processes ranging from axon guidance and synaptogenesis to the persistent modifications in synaptic efficacy associated with long-term synaptic plasticity (Jung et al., 2012; Kindler and Kreienkamp, 2012). The spatial and temporal control of mRNA translation is regulated by cis-acting elements, most often found in their untranslated regions (UTRs), acting in concert with trans-acting factors such as RNA-binding proteins (RBPs) and microRNAs (miRNAs). In axons and dendrites, these translationally silent mRNAs are actively transported to their destination associated with a heterogeneous group of RNPs that include RNA transport granules, stress granules (SGs), and neuronal P bodies (Sossin and DesGroseillers, 2006).
P bodies are highly conserved cytoplasmic RNPs linked to both mRNA decay and translational repression pathways. In the former, mRNAs are targeted for deadenylation followed by decapping and then 5′-to-3′ exonucleolytic degradation (Cougot et al., 2004; Sheth and Parker, 2003). In the latter, mRNAs are deadenylated and targeted for translational repression and storage within larger P body aggregates (Teixeira et al., 2005). Orthologous RNPs can be found in Drosophila melanogaster that contain the fly decapping enzyme (Dcp2), enhancers of decapping (Dhh1, Dcp1, Edc3, and Pat1), the 5′-to-3′ exoribonuclease (Xrn1), and the miRNA RNA induced silencing complex (miRISC) proteins (Ago1, Ago2 and GW182; Eulalio et al., 2007a). Neurons in Drosophila and mammals contain populations of specialized P bodies with largely unknown functions (Barbee et al., 2006; Cougot et al., 2008; Zeitelhofer et al., 2008a). Interestingly, neuronal P bodies exhibit a significant amount of overlap with components of RNA transport granules including the Fragile X Mental Retardation Protein (FMRP). P bodies in dendrites of cultured hippocampal neurons exhibit motorized movements and re-localize towards distal sites in response to synaptic activation (Cougot et al., 2008). Additionally, acute synaptic stimulation results in a significant decrease in the number of dendritic P bodies suggesting that they can disassemble after neural activity (Zeitelhofer et al., 2008a). Most dendritic P bodies lack Xrn1, a catalytic component in the 5′-to-3′ decay pathway (Cougot et al., 2008). Together, these data support a model where neuronal P bodies accumulate translationally repressed mRNAs that are released from storage, and potentially repression, following synaptic activity.
The assembly of P bodies is influenced by a balance between translational activation and repression (Franks and Lykke-Andersen, 2008). P body assembly is a stepwise process where key P body components, notably Pat1 and a complex of Lsm proteins, are first recruited to an mRNA to form a P body monomer. Under certain cellular conditions these monomers can recruit additional P body components to form visible P body aggregates. The Pat1 protein has been proposed to act as a key scaffolding molecule during this assembly process (Braun et al., 2010; Pilkington and Parker, 2008). In support of this, it has recently been shown in Saccharomyces cerevisiae that P body assembly and disassembly can be regulated by the direct phosphorylation of Pat1 by the cAMP-dependent protein kinase, PKA (Ramachandran et al., 2011). Although Pat1 has no recognizable functional domains or motifs, it plays essential roles in both the translational repression and mRNA decay pathways (Marnef and Standart, 2010). As such, the single yeast and invertebrate Pat1 orthologs have dual functions in the control of deadenylation and decapping (Boag et al., 2008; Haas et al., 2010; Pilkington and Parker, 2008). In contrast, gene duplication in vertebrates has led to the evolution of two Pat1 paralogs (named Pat1a and Pat1b), with distinct functions in translational repression and decapping (Braun et al., 2010; Ozgur et al., 2010). Together, these data suggest that Pat1 proteins may be functioning at a pivotal point where the decision is made between targeting a specific mRNA for repression and storage in P bodies or for decapping followed by 5′-to-3′ exonucleolytic degradation.
Aside from their roles in mRNA metabolism, very little is known about the physiological functions of either P bodies or most P body components in neurons in vivo. In this study, we ask if HPat, the fly ortholog of Pat1, plays a role in the control of synaptic terminal (i.e. bouton) growth at a model glutamatergic synapse, the larval Drosophila NMJ. Surprisingly, we find that HPat is a strong negative regulator of bouton growth both during development and following acute chemically induced synaptic stimulation. Specifically, we show that HPat has both a pre- and postsynaptic function in the control of synaptogenesis during these processes. Synaptic hyperplasia observed in hpat mutants correlates strongly with a disruption in the organization of the axonal microtubule cytoskeleton suggesting that HPat negatively regulates presynaptic microtubule-based growth. Finally, we demonstrate that HPat interacts genetically with catalytic components of the deadenylase, but not the decapping, machinery to control bouton formation. Together, our findings suggest a model where HPat is directing specific neuronal mRNAs required the growth of synaptic boutons for repression, presumably within neuronal P bodies.
Results
hpat is an essential gene
Most alleles of hpat (also known as patr-1 or protein associated with topo II related-1) were recessive lethal and homozygous hpat mutant individuals died at the late third-instar or early pupal stage without obvious morphological defects (Fig. 1A; data not shown). However, to study the function of HPat, we first attempted to generate additional hpat alleles. By mobilizing a P element insertion located within the hpat 5′ UTR (hpatEY10289), we isolated two partial deletions of the EY10289 insertion (hpatd17 and hpatd3; supplementary material Fig. S1A). The hpatd17 allele was found to be homozygous viable. Interestingly, in hpatd17, a ∼10.2 kb deletion occurred within the central region of the P element leaving ∼600 bp of flanking insert sequence. In contrast, hpatd3 was recessive lethal. In hpatd3, a ∼3.9 kb deletion occurred within the central region of the P element leaving ∼6.9 kb of flanking P element sequence. The lethality of hpatd3 was not complemented by either hpatEY10289 or a second lethal P element insertion, hpatl(3)06442 (Fig. 1A; data not shown). Moreover, hpatd3 was lethal in trans to a genetic deficiency, Df(3R)Exel8165, which deletes the entire hpat locus (Fig. 1B). The lethality of hpatd3 was rescued by the introduction of a genomic DNA fragment (P21M20) that contained the entire hpat locus plus flanking genomic sequence that includes two uncharacterized upstream genes (CG3995 and CG5220; Fig. 1B). One copy of the P21M20 fragment was not sufficient to rescue lethality. hpatd3 was not lethal in trans to a recessive lethal allele of the nearest gene, CG3995 (CG3995LL03178; no alleles are available for CG5220; data not shown). Based on these results, we conclude that hpatd3 specifically affects hpat function, and that hpat is an essential gene.
Fig. 1.
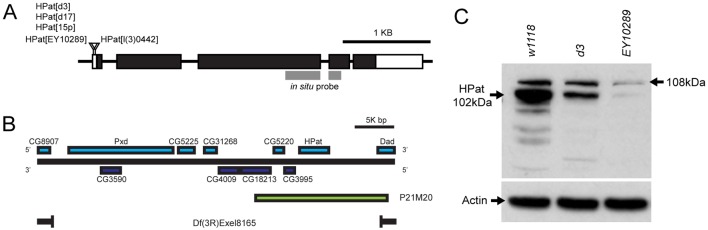
Drosophila HPat alleles and antibodies. (A) Diagram of the hpat gene showing intron and exon boundaries. Also shown are the insertion sites for two P elements (hpatEY10289 and hpatl(3)0442) in the hpat 5′ UTR and the region targeted by the hpat antisense in situ construct. In hpatd3 and hpatd17, deletions occurred within the hpatEY10289 P element such that the hpat coding sequence was unaffected. Instead, the size of the P element inserted into the hpat 5′ UTR was reduced. hpat15p is a precise excision of hpatEY10289. (B) Diagram of the hpat gene region. Shown are sequences included in the P21M20 rescue transgene and missing from the Df(3R)Exel8165 deficiency. (C) Western blot showing HPat expression levels in extract from whole w1118, hpatd3 and hpatEY10289 homozygous larvae. Rabbit HPat antibodies recognize 108 kDa and 102 kDa bands corresponding to two predicted isoforms. Both bands are significantly reduced in hpatd3 mutants and almost eliminated in hpatEY10289 homozygotes. Smaller bands in the w1118 control and hpat mutants are likely proteolytic fragments of the HPat protein.
To further characterize lethal hpat alleles and examine HPat expression, we produced two polyclonal antibodies against the HPat protein. These antibodies recognized major bands of ∼102 kDa and 108 kDa on immunoblots of extracts from whole Drosophila third-instar larvae (Fig. 1C). Evidence from gene prediction algorithms and cDNA analysis suggest that these bands likely represent two isoforms resulting from alternative transcriptional start sites (supplementary material Fig. S1B; data not shown). The expression level of both isoforms is significantly reduced, but not completely eliminated, in extract from hpatd3 and hpatEY10289 homozygous larvae (the 102 kDa band is reduced to 38% and 2% of controls respectively; the 108 kDa band is reduced to 66% and 15%; Fig. 1C; supplementary material Fig. S1C). In contrast to whole larval immunoblots, HPat antibody staining appears to be equally disrupted in hpatd3 and hpatEY10289 mutant ventral ganglia and muscle (supplementary material Fig. S2). This suggests that the hpatd3 allele may have a strong effect on the tissue-specific expression of HPat. Together, these data suggest that our HPat antibodies are specific for the HPat protein and indicate that hpatd3 and hpatEY10289 are hypomorphic, and not null, alleles of hpat.
HPat is expressed in the larval central nervous system (CNS)
We next asked if HPat was expressed in the larval nervous system in vivo. First, we verified hpat expression in larval brains by RNA in situ hybridization (Fig. 2A). When probed with an antisense riboprobe against mRNA sequence spanning exons 3 and 4 (see probe location in Fig. 1A), the presence of hpat mRNA was detected throughout the larval CNS when compared to sense controls. Consistent with these results, hpat mRNA was also detected in total RNA isolated from the larval CNS by quantitative real-time PCR (RT-qPCR; data not shown).
Fig. 2.
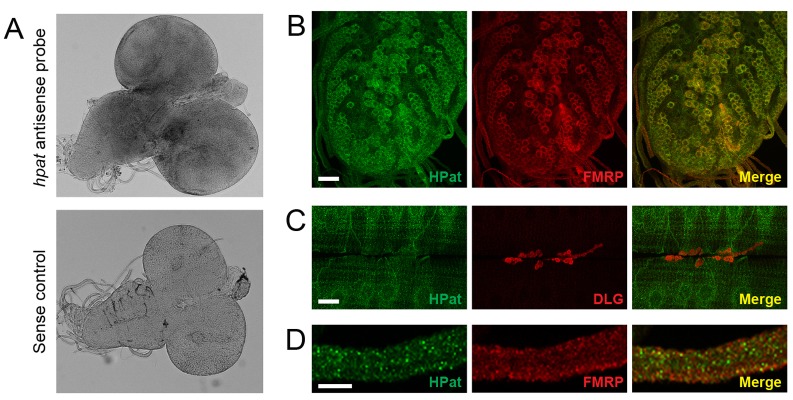
HPat is expressed in the larval nervous system. (A) Whole-mount RNA in situ hybridization of w1118 larval brains stained with either antisense or sense riboprobes directed against the hpat mRNA. (B) Larval ventral ganglion stained with rat antibodies against HPat (green) and/or FMRP (red) showing punctate cytoplasmic staining in the soma of most neurons. There is a significant amount of overlap between HPat and FMRP in the merged image. Scale bar: 20 µm. (C) NMJs double stained with antibodies against HPat (green) and postsynaptic Dlg (red). HPat exhibits punctate perinuclear localization in muscle but is not enriched at the NMJ. Scale bar: 20 µm. (D) Peripheral nerves exiting the ventral ganglion showing localization of both HPat (green) and FMRP (red). In the merged images in B and D, yellow spots indicate co-localization between HPat- and FMRP-containing punctae. Scale bar: 10 µm.
We then stained larval brains with polyclonal antibodies against HPat. We found that the HPat protein exhibited a specific and punctate staining pattern in the cytoplasm and neuropile of most neurons within the third-instar larval ventral ganglion (Fig. 2B; supplementary material Fig. S2A). We also observed high levels of HPat expression in the optic lobes and larval eye imaginal discs (data not shown). To examine synaptic enrichment at the NMJ, we performed double labeling with antibodies against HPat and a postsynaptic marker (Discs Large or “Dlg”; the fly ortholog of postsynaptic density protein 95 or “PSD-95”). We observed that HPat was not enriched at peripheral NMJ synapses (Fig. 2C). HPat was moderately expressed in muscle and exhibited a specific punctate staining pattern with distinct perinuclear enrichment (Fig. 2C; supplementary material Fig. S2B). Thus, we conclude that HPat is a cytoplasmic protein that is expressed in neurons and muscle but is not detectably enriched at the neuromuscular synapse.
We further examined subcellular localization of the HPat protein in neurons in vivo by determining if HPat co-localized with FMRP, a component of neuronal transport granules. As expected, HPat strongly co-localized to FMRP-expressing neurons in the larval ventral ganglion (merged image in Fig. 2B). Both HPat and FMRP also partially co-localize within punctae in peripheral nerves exiting the larval CNS (merged image in Fig. 2D; data not shown). We found that 48% (±11%; n = 203) of HPat-positive foci also contained FMRP. Conversely, 33% (±10%; n = 182) of FMRP-positive foci also contained HPat. Both HPat and FMRP-containing granules disappear from peripheral nerves prior to reaching the NMJ (data not shown). HPat-containing punctae are significantly reduced in peripheral nerves and soma of most cells in the ventral ganglion of hpatd3 and hpatEY10289 mutants (supplementary material Fig. S2). This suggests that HPat may specifically localize to a distinct population of P bodies in the larval CNS.
HPat regulates synaptic terminal growth during development of the larval NMJ
Having demonstrated that HPat localized to neurons in the larval CNS and that it formed cytoplasmic foci reminiscent of neuronal P bodies, we next asked if HPat had a function in the control of synaptic development. To investigate this, we took advantage of the larval Drosophila NMJ, a genetic model system for the study of glutamatergic synapses in the mammalian brain (Schuster, 2006). Foremost among its useful properties, its terminal synaptic boutons exhibit a highly plastic behavior during development (Collins and DiAntonio, 2007). For this analysis, we examined two third-instar larval NMJs in abdominal segment 3. First, we examined the synapse at muscle 6/7. This NMJ contains two types of glutamatergic boutons – 1b (big) and 1s (small) that are derived from distinct neurons (Rohrbough et al., 2000). Type 1b boutons are highly plastic can be easily distinguished by their larger size and higher levels of the postsynaptic density marker, Dlg (Menon et al., 2004). Second, we examined the NMJ at muscle 4 (Zhang et al., 2001). This NMJ is innervated by a single neuron, contains only type 1b boutons, and allows for the quantification of synaptic branching, an additional measure of synaptic complexity.
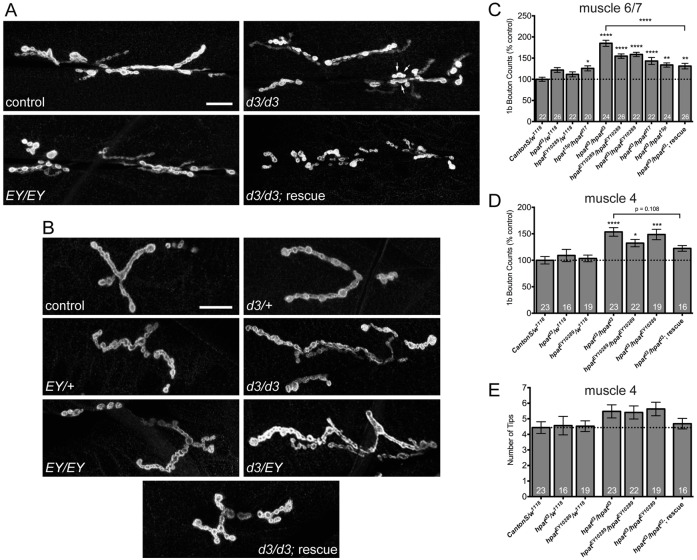
Are there structural defects at the NMJ associated with hpat loss-of-function? To address this question, we analyzed NMJ phenotypes in hpat mutants (hpatd3, hpatd17 and hpatEY10289). Surprisingly, we found a robust and highly significant hyperplasia of muscle 6/7 synaptic terminals in hpatd3 homozygous mutants compared to controls (Fig. 3A,C). This overgrowth was measured by counting the number of type 1b synaptic boutons per NMJ (85% increase over controls; P<0.0001). Importantly, the hpatd3 mutant phenotype was significantly suppressed by the introduction of two copies of an hpat genomic rescue construct (P21M20; described in Fig. 1B; Fig. 3A,C; P<0.0001). The overgrowth phenotype observed in hpatd3 homozygous mutants was confirmed by examining 1b bouton numbers in hpatEY10289 homozygous (54% increase; P<0.0001), hpatd3/hpatEY10289 (59% increase; P<0.0001) and hpatd3/hpatd17 mutant backgrounds (43% increase; P<0.0001). Similar increases were observed when quantifying type 1s synaptic boutons (supplementary material Fig. S3A). As a control for genetic background effects, we also generated a revertant line by precise excision of the EY10289 insertion (called hpat15p). When crossed to the homozygous viable hpatd17 allele (hpat15p/hpatd17), there is a small, but significant, increase in bouton numbers (Fig. 3C; supplementary material Fig. S3A; 1b boutons = 26% increase over controls; P<0.05). When crossed to the hpatd3 allele (hpat15p/hpatd3), there is a slightly greater increase (Fig. 3C; 34% increase; P<0.001). Together, these data suggest that genetic background may have a weak effect on bouton formation. However, this effect on 1b bouton numbers was not significantly greater than that observed in hpatd3 or hpatEY10289 heterozygotes (Fig. 3C; hpat15p/hpatd17 = 3% increase over hpatd3/+; and hpat15p/hpatd3 = 10% increase over hpatd3/+). Moreover, both 1b and 1s bouton numbers in both lethal hpat mutants (hpatd3 and hpatEY10289) remain significantly higher than hpat15p/hpatd17 mutant larvae (data not shown).
Fig. 3.
HPat negatively regulates synaptic terminal growth during larval development. (A) Representative images of muscle 6/7 and (B) muscle 4 NMJs from abdominal segment A3 labeled with an antibody against Dlg. Scale bar: 20 µm. Arrows in the hpatd3/hpatd3 panel in A indicate a cluster of terminal synaptic boutons. (C) Quantification of the number of type 1b synaptic boutons at muscle 6/7 and (D) muscle 4 synaptic clefts. (E) Quantification of the number of total bouton branch tips at muscle 4 for each indicated genotype. Unless otherwise indicated, stars denote statistical significance compared to Canton-S/w1118 controls (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; one-way ANOVA; Tukey’s post-hoc test).
Confirming our results with the two strongest alleles of hpat (hpatd3 and hpatEY10289), we also observed a significant increase in the number of boutons at the muscle 4 NMJ (Fig. 3B,D). As expected, hpatd3 homozygotes exhibited a 53% increase over controls (P<0.0001). Again, we validated this result by examining bouton numbers in hpatEY10289 homozygotes (32% increase; P<0.05) and hpatd3/hpatEY1089 mutants (49% increase; P<0.001). While introduction of two copies of the P21M20 rescue construct did substantially reduce type 1b bouton numbers compared to hpatd3 homozygotes, this decrease was not statistically significant (P = 0.108). This might be due to the generally weaker effect hpat appears to have on the muscle 4 NMJ compared to muscle 6/7. Interestingly, quantification of muscle 4 synaptic branches revealed that hpat had no significant impact on that particular aspect of synaptic complexity (Fig. 3E). This observation suggests that hpat may specifically be involved in the control of new bouton growth.
We next asked if hpat had a specific pre- or postsynaptic function in the control of synaptic terminal development. To address this question, we analyzed bouton numbers in transgenic lines where the HPat protein was overexpressed using either a motor neuron- or muscle-specific Gal4 driver (C380-Gal4 and 24B-Gal4, respectively). As expected, presynaptic expression of transgenic HPat revealed the opposite phenotype as hpat mutants (Fig. 4A,B). hpat neuronal overexpression NMJs displayed a modest, but statistically significant, decrease in type 1b boutons (13% decrease; P<0.01). Comparable results were observed using a second motor neuron driver (D42-Gal4) – these NMJs exhibited a similar decrease in bouton number (12%; P<0.05). In contrast, postsynaptic overexpression of HPat resulted in no significant change in type 1b bouton numbers when compared to controls (Fig. 4A,B). Unlike in hpat mutants, quantification of type 1s boutons did not follow a similar trend (supplementary material Fig. S4A). Surprisingly, postsynaptic overexpression of HPat using 24B-Gal4 resulted in a significant increase in 1s boutons compared to controls (37% increase; P<0.05). Unfortunately, HPat levels could not be sufficiently depleted by RNAi so these results could not be confirmed by cell-autonomous hpat reduction-in-function (data not shown). Taken together, the lack of a strong and consistent phenotype following HPat overexpression suggests that hpat does not have a specific role on either side of the synaptic cleft. Thus, we conclude that both muscle and neuronal hpat function is likely required for normal synaptic terminal development.
Fig. 4.
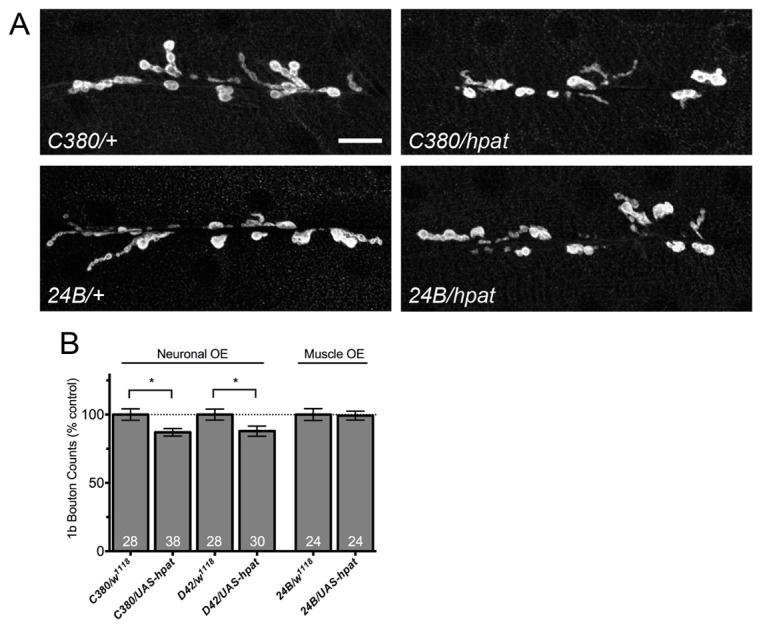
HPat overexpression does not have a strong effect on synaptic terminal growth during larval development. (A) Representative images of muscle 6/7 NMJs from abdominal segment A3 labeled with antibodies against Dlg. Scale bar: 20 µm. (B) Quantification of type 1b boutons at muscle 6/7 clefts. Asterisks indicate statistical significance compared to controls (*P<0.05; Student’s t-test). There is a modest decrease in type 1b bouton numbers when UAS-hpat is driven by presynaptic (C380 and D42) but not postsynaptic (24B) Gal4 drivers. OE, overexpression.
HPat regulates organization of the presynaptic microtubule cytoskeleton
One thing we noticed in our analysis of hpatd3 mutants was the presence of clusters of terminal synaptic boutons, a phenotype that is suggestive of abnormal presynaptic overgrowth (Fig. 3A, arrows). In Drosophila, terminal synaptic boutons can also be identified by the presence of distinct “loops” in the microtubule cytoskeleton (Roos et al., 2000). These loops are believed to identify sites of active bouton division. Based on the synaptic terminal overgrowth phenotype we observed in hpat mutants (Fig. 3A–D), we predicted that we would see a similar increase in the number of microtubule loops per NMJ (muscle 6/7; abdominal segment 3) when compared to controls. To address this hypothesis, we asked whether presynaptic microtubule architecture was disrupted in the two strongest hpat mutant backgrounds.
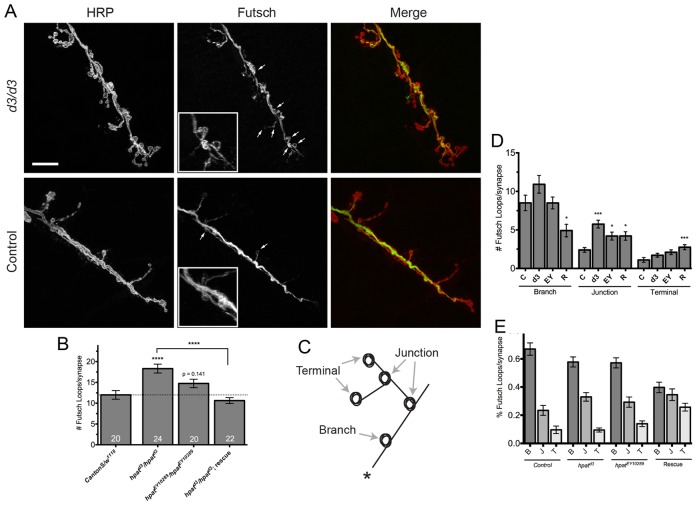
To examine the microtubule cytoskeleton, we first double stained hpatd3 and hpatEY10289 mutant larvae with antibodies against a presynaptic membrane marker (horseradish peroxidase or “HRP”) and Futsch, the Drosophila ortholog of microtubule-associated protein 1B or “MAP1B”. Futsch staining tightly co-localizes with microtubule markers and can be used to identify synaptic microtubule loops (Roos et al., 2000). As predicted, we found that hpatd3 homozygous larvae exhibited a robust and highly significant increase in the number of Futsch-positive loops compared to controls (Fig. 5A,B; mean number of Futsch loops per NMJ: control = 12±1.1; hpatd3 mutants = 18.3±1.1; 53% increase; P<0.0001). These loops were often found in clusters off the main synaptic arbor (Fig. 5A, inset). The hpatd3 mutant phenotype was completely suppressed by the introduction of two copies of an hpat genomic rescue construct (Fig. 5B; P21M20 rescue = 10.6±0.7; P<0.0001). Interestingly, while we did see an increase in the total number of Futsch loops in hpatEY10289 homozygous mutant larvae compared to controls, this increase was not statistically significant (hpatEY10289 mutants = 14.8±1.0; 23% increase; P = 0.141). This is consistent with the generally weaker phenotype observed in hpatEY10289 mutants when examining bouton development (Fig. 3A–D). Next, we identified significant changes in the distribution of Futsch loops in three regions of the larval NMJ (Fig. 5C; main synaptic arbors, synaptic terminal junctions and terminal boutons). Interestingly, we found that there was a significant increase in the number of Futsch loops at branch junctions in both hpatd3 mutants (5.8±0.5; 140% increase; P<0.001) and hpatEY10289 mutants (4.2±0.5; 75% increase; P<0.05) compared to controls (2.4±0.3; Fig. 5D). Despite this increase, the relative frequency of Futsch loops occurring in each region remained consistent (Fig. 5E). In contrast, hpatd3/hpatd3 rescue larvae did show a significant redistribution in the number of Futsch loops in each synaptic region compared to control and hpat mutant genotypes (Fig. 5D,E). This may be because two copies of the P21M20 transgene only partially rescue specific synaptic phenotypes (Fig. 3A–D). Finally, we observed that Futsch staining in hpatd3 mutants was generally weaker and more punctate than in controls (compare Futsch staining in Fig. 5A). These data suggest that, directly or indirectly, the futsch mRNA expression may be a target for regulation by the HPat protein.
Fig. 5.
HPat regulates structure of the presynaptic microtubule cytoskeleton. (A) Representative images of muscle 6/7 NMJs from abdominal segment A3 labeled with antibodies against presynaptic HRP (red) and Futsch (green) to show microtubule loops. Arrows indicate examples of Futsch loops. The inset in the hpatd3/hpatd3 panel is a magnified region showing an abnormal cluster of Futsch-positive loops. Scale bar: 20 µm. (B) Quantification of the number of Futsch-positive loops per NMJ at muscle 6/7 clefts. Unless otherwise indicated, asterisks indicate statistical significance compared to control NMJs (***P<0.001; one-way ANOVA; Tukey’s post-hoc test). (D–E) Genotypes here are as follows: “C” (CantonS/w1118; n = 20) control; “d3” (hpatd3/hpatd3; n = 24); “EY” (hpatEY10289/hpatEY10289; n = 20); and “R” (hpatd3/hpatd3; P21M20/P21M20; n = 24) rescue. (C) Cartoon diagram showing three regions of the NMJ where Futsch loops were typically observed. The regions quantified were the main synaptic arbors (“Branch” or “B”), terminal synaptic boutons (“Terminal” or “T”), and junctions where two or more branches meet (“Junction” or “J”). In this figure, the synaptic arbor marked by a “*” would be the site of innervation. (D) Quantification of the location of Futsch-positive loops in hpat mutants in each of the regions shown in C. The most significant increase is observed at the point where two synaptic arbors meet (*P<0.05; ***P<0.001; Kruskal–Wallis; Dunn’s post-hoc test). (E) The relative distribution of Futsch-positive loops in each indicated synaptic region.
In summary, we have used multiple approaches to show that HPat is involved in the control of synaptic terminal growth during larval NMJ development. More specifically, HPat appears to have a function on both the presynaptic and postsynaptic side of the synaptic cleft, at least in part, by controlling organization of the presynaptic microtubule cytoskeleton.
HPat regulates activity-dependent bouton formation
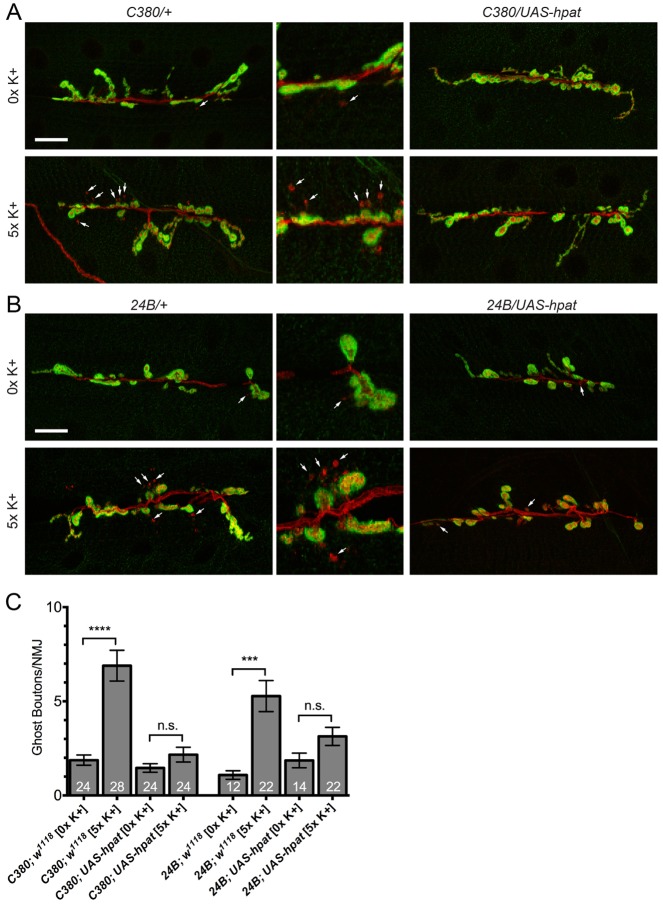
Because hpat regulates synaptic terminal growth during larval development, we speculated that it might also be involved in the control of activity-dependent bouton formation. To test this hypothesis, we used a model system that allowed us to assay activity-dependent plasticity (Ataman et al., 2008) at the larval NMJ. Treatment of partially dissected larval preparations with a spaced high K+ depolarization paradigm induces the formation of immature synaptic boutons known as “ghost boutons” (Ataman et al., 2008). These presynaptic varicosities have been shown to contain synaptic vesicles but lack active zones and postsynaptic structures (Ataman et al., 2006). We analyzed transgenic lines that expressed HPat under the control of either a motor neuron-specific (C380-Gal4) or muscle-specific driver (24B-Gal4) and stained larval preparations with antibodies against HRP and Dlg (Fig. 6A,B). In our hands, stimulated control larval preparations exhibited a significant increase in the number of ghost boutons compared to pseudo-stimulated controls (Fig. 6A–C; mean number of ghost boutons per NMJ: C380/+ unstimulated control = 1.9±0.3 or stimulated control = 6.9±0.8; P<0.0001; 24B/+ unstimulated control = 1.1±0.2 or stimulated control = 5.3±0.8; P<0.001). As we predicted, both pre- and postsynaptic overexpression of HPat suppressed this phenotype. Following presynaptic expression, the number of ghost boutons observed per NMJ was reduced to baseline levels (Fig. 6A,C; unstimulated C380; UAS-hpat = 1.5±0.2; stimulated C380; UAS-hpat = 2.2±0.4). Interestingly, the postsynaptic overexpression of HPat also significantly suppressed ghost bouton formation (Fig. 6B,C; unstimulated 24B; UAS-hpat = 1.9±0.4; stimulated 24B; UAS-hpat = 3.1±0.5). While there is some genotypic variation, the ratio of unstimulated to stimulated ghost boutons drops to similar levels using both drivers (C380-Gal4 = 3.6 to 1.4; 24B-Gal4 = 4.8 to 1.6). Based on these data, we conclude that HPat is a very strong negative regulator of activity-dependent synaptic terminal growth at the larval NMJ. Furthermore, these results support our model that hpat has a function on both sides of the synaptic cleft.
Fig. 6.
Neuronal HPat negatively regulates activity-dependent synaptic terminal growth. (A,B) Representative images of muscle 6/7 NMJs from abdominal segment A3 labeled with antibodies against presynaptic HRP (red) and postsynaptic Dlg (green). HPat expression was driven pre- or postsynaptically using the C380-Gal4 and 24B-Gal4 drivers. Arrows indicate the presence of immature ghost boutons (HRP+ Dlg−). Middle panels are magnified images of regions indicated by arrows in 0×K+ and 5×K+ treatment groups. Stimulated (5×K+) larvae of each genotype were taken through a spaced high-K+ paradigm. Unstimulated (0×K+) larvae were taken through a low K+ pseudo-stimulation paradigm. Scale bars: 20 µm. (C) Quantification of the number of ghost boutons per NMJ at muscle 6/7 clefts. Asterisks indicate statistical significance between the indicated groups (***P<0.001; ****P<0.0001; Kruskal–Wallis; Dunn’s post-hoc test). Note the a 3- to 4-fold increase in the number of ghost boutons is observed at control NMJs following high-K+ depolarization. This increase is almost completely suppressed by both pre- and postsynaptic overexpression of HPat (down to a 1.4- and 1.5-fold increase, respectively). n.s., not significant.
Neuronal HPat interacts genetically with components of the deadenylation machinery
It has previously been shown that HPat and its orthologs have important cellular functions in the control of mRNA storage and decay (Marnef and Standart, 2010). Based on these observations, we postulated that HPat might be regulating synaptic terminal growth at the NMJ through one (or both) of these pathways. To test this hypothesis, we asked if hpat interacted genetically with twin (fly CCR4) or dcp2, key catalytic components of the deadenylase and decapping complexes respectively, to control synaptic terminal growth during development (Fig. 7A; Rehwinkel et al., 2005; Temme et al., 2004). We have found that hpatd3 heterozygotes did not show an increase in 1b bouton numbers compared to controls (Fig. 3C; Fig. 7B). Similarly, twin and dcp2 heterozygotes did not exhibit a significant increase over baseline numbers of boutons (Fig. 7B). However, if either twin or dcp2 interact with hpat to control bouton growth, we predicted that we would see enhanced synaptic hyperplasia in trans-heterozygous larvae.
Fig. 7.
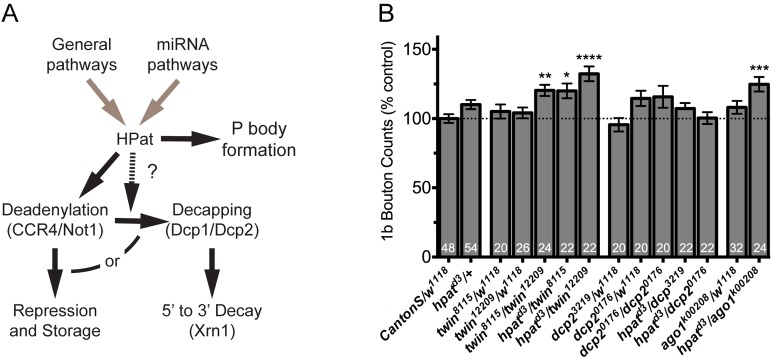
HPat interacts with components of the deadenylase complex and miRNA pathway to control synaptic terminal growth during larval development. (A) Diagram outlining major events associated with mRNA storage and decay in P bodies. mRNAs can be targeted to P bodies via general and miRNA-mediated pathways. HPat may interact with these pathways to direct mRNAs for: (1) deadenylation (by CCR4/Not1) followed by translational repression and storage; or (2) deadenylation followed by decapping (by Dcp1/Dcp2) and then 5′ to 3′ decay (by Xrn1). (B) Quantification of the number of type 1b boutons at muscle 6/7 clefts in genetic interaction experiments. Trans-heterozygous larvae were generated by crossing the hpatd3 allele to indicated alleles of each genes (e.g. hpatd3/+; dcp2BG01766/+ or hpatd3/+; twinGS12209/+). Asterisks indicate statistical significance compared to the control (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; one-way ANOVA; Tukey’s post-hoc test). There is a significant genetic interaction between HPat and both CCR4 and Ago1 in the control of synaptic terminal growth during development. In contrast, there is no significant genetic interaction between Dcp2 and HPat.
As predicted, in hpat and twin trans-heterozygotes, we did observe a highly significant overgrowth compared to controls (Fig. 7B; twinGS8115/+; hpatd3/+ = 20% increase; twinGS12209/+; hpatd3/+ = 39% increase; P<0.05 and P<0.0001, respectively; both twinGS8115 and twinGS12209 are lethal hypomorphic alleles of twin; Zaessinger et al., 2006). Nearly identical results were observed in twinGS8115/twinGS12209 mutants (20% increase; P<0.01). The synaptic terminal overgrowth phenotype observed in trans-heterozygote and twinGS8115/twinGS12209 mutant larvae was not as robust as observed in hpat mutants alone. This suggests that, while hpat can interact genetically with twin/CCR4 to control synaptic terminal growth, twin is not a limiting factor for hpat function. In contrast to twin, no significant increase was observed in hpat and dcp2 trans-heterozygotes compared to controls (Fig. 7B). Two lethal alleles of dcp2 were examined: dcp2BG01766 and dcp2GS3219. Only dcp2BG01766 homozygous mutants resulted in viable third-instar larvae and they did not exhibit a significant synaptic hyperplasia at the NMJ (Fig. 7B; dcp2GS3219 homozygotes and dcp2BG01766/dcp2GS3219 mutants did not survive to the third-instar stage; data not shown). In both twin and dcp2 mutants, type 1s boutons showed a substantial amount of variability (supplementary material Fig. S5A). Together, these data indicate that HPat does not require the decapping machinery to regulate synaptic terminal growth during development. More specifically, this suggests that HPat may be involved in targeting some key mRNAs involved in the control of new synaptic bouton growth for repression and not for 5′-to-3′ mRNA decay.
HPat is involved in miRNA-mediated control of synaptic terminal growth
It has previously been proposed that P bodies represent sites of mRNA regulation by the miRNA pathway (Eulalio et al., 2007b; Liu et al., 2005b). Furthermore, is has been demonstrated that the Drosophila Argonaute 1 (Ago1) protein, a key effector of miRNA activity in flies, is required for the regulation of synaptic terminal growth at the NMJ (Jin et al., 2004). Two lines of evidence led us to ask whether hpat interacted genetically with ago1 to control synaptic structure at the NMJ. First, in Drosophila S2 cells, HPat is required for miRNA-mediated post-transcriptional gene silencing of some Ago1-regulated mRNAs (Fig. 7A; Eulalio et al., 2007c). Second, HPat copurifies with GW182, a core P body component and key factor in translation repression and mRNA degradation by miRNAs (Jäger and Dorner, 2010). Compared to controls, hpatd3/+; ago1l(2)k00208/+ trans-heterozygotes exhibited a moderate (25%) but highly significant (P<0.001) increase in the number of type 1b boutons per NMJ (Fig. 7B). Again, the phenotype observed in hpatd3/+; ago1l(2)k00208/+ trans-heterozygotes is not as strong as that observed in hpatd3 homozygous mutants alone. This suggests that key mRNAs involved in the control of microtubule organization and/or synaptic terminal growth may associate with HPat, and presumably P bodies, through both general- and miRNA-mediated mechanisms (Fig. 7A).
Discussion
The immediate translation of synapse-localized mRNAs is thought to play an essential role in the control of long-term synaptic plasticity (Jung et al., 2012; Kindler and Kreienkamp, 2012). A growing amount of evidence suggests that this process is mediated by factors that are associated with a variety of neuronal RNPs, including P bodies (Zeitelhofer et al., 2008b). Until recently, few experiments have been carried out that directly examined functional roles for P body proteins in the control of neural physiology (Barbee et al., 2006; Hillebrand et al., 2010). In this report, we provide novel data indicating that HPat/Pat1, a core component of P bodies, is required to control synaptic terminal growth at the Drosophila NMJ during development (Figs. 3 and 4). Significantly, we also provide the first functional evidence that a component of P bodies is required to regulate rapid activity-dependent growth of presynaptic boutons at this synapse (Fig. 6). In support of these observations, we demonstrate that the synaptic hyperplasia observed during NMJ development in hpat mutants correlates with reorganization of the microtubule cytoskeleton in axon terminals (Fig. 5). Mechanistically, HPat interacts genetically with the deadenylase, but not the decapping, pathway to control bouton formation (Fig. 7). Together, these data suggest that HPat may target key synaptic mRNAs for deadenylation and translational repression, presumably in P bodies, and not for degradation. This supports the hypothesis that one major function of neuronal P bodies is to sequester and actively transport translationally quiescent mRNAs from the soma to distant sites of translational activation, either in axon terminals or dendritic spines (Barbee et al., 2006; Cougot et al., 2008; Zeitelhofer et al., 2008a).
Synaptic functions for HPat
The Pat1 proteins have been directly implicated in two important cellular processes: (1) the assembly of P bodies; and (2) the coupling of deadenylation to decapping (Marnef and Standart, 2010). In higher eukaryotes, functions for Pat1 appear to depend heavily on both cellular context and RNP composition. For example, vertebrates have two Pat1 proteins, Pat1a and Pat1b, each having tissue-specific functions in translational repression or mRNA degradation (Marnef et al., 2010; Nakamura et al., 2010; Ozgur et al., 2010). In somatic cells, vertebrate Pat1b interacts strongly with Rck (vertebrate Dhh1)-positive P bodies, is required for P body assembly, and provides a physical link between deadenylation and decapping. In oocytes, Pat1a interacts weakly with Rck and instead associates with deadenylated mRNAs in cytoplasmic polyadenylation element binding protein (CPEB)-containing translational repression complexes (Marnef et al., 2010; Ozgur et al., 2010). Similarly, in the nematode Caenorhabditis elegans, a single HPat ortholog (PATR-1) exhibits distinct tissue-specific functions. In somatic cells, PATR-1 localizes to particles containing CGH-1 (worm Dhh1) that are involved in mRNA decapping and degradation (Boag et al., 2008). These RNPs are thought to be analogous to P bodies that also contain DCAP-2 (Dcp2), ALG-1 (Argonaute), and AIN-1 (GW182; Ding et al., 2005). Unlike in vertebrates, it is currently unclear if worm PATR-1 can also localize to distinct RNPs in ovaries and oocytes that are specifically involved in translational repression.
The Drosophila HPat protein has a well-defined role in the regulation of mRNA degradation. First, as a core component of P bodies, epitope tagged HPat can co-immunoprecipitate with the decapping enzyme (Dcp2), the decapping activators (Dhh1, Dcp1, Edc3, Edc4 and Lsm1-7), components of the CCR4-NOT deadenylase complex, and the 5′-to-3′ exoribonuclease (Xrn1) (Braun et al., 2010; Haas et al., 2010). Second, when HPat is artificially tethered to a reporter mRNA in vitro, its binding is sufficient to trigger deadenylation followed by decapping (Haas et al., 2010). Finally, co-depletion of both Drosophila HPat and Me31B (fly Dhh1) inhibits decapping caused by either miRNAs or tethering of GW182 to reporter mRNAs (Eulalio et al., 2007c). Based on these observations, we were surprised to find that hpat did not interact genetically with dcp2 to control synaptic terminal growth (Fig. 7B). However, similar results were seen in homozygous dcp2 mutant larvae. Taken together, these observations suggest that HPat is not involved in targeting key mRNAs involved in bouton formation for decay.
In contrast, two independent lines of evidence indicate that regulation of deadenylation is important for synaptic terminal growth during development. First, core components of the deadenylase complex were identified in a genetic screen for factors that control dendrite morphogenesis in Drosophila sensory neurons (Parrish et al., 2006). siRNA knockdowns of twin/CCR4 and NOT1 were found to significantly increase dendritic arborization and dendrite branch length respectively. Second, we show here that two hypomorphic twin/CCR4 mutants cause synaptic overgrowth at the NMJ (Fig. 7B). Interestingly, in vitro data indicates that in the absence of HPat protein, only decapping but not deadenylation is inhibited (Eulalio et al., 2007c; Haas et al., 2010). However, our in vivo data suggests that, at least at the Drosophila NMJ, hpat exhibits a significant genetic interaction only with twin/CCR4 (Fig. 7B). Our interpretation of these observations is that HPat is contributing to the deadenylation of specific neuronal mRNAs involved in the control of synaptic terminal growth. This HPat-mediated deadenylation immediately targets these mRNAs for decapping and decay. This hypothesis is consistent with a model for Pat1 function in yeast, where Pat1 first inhibits translation and then, in an independent step, acts as a scaffold to recruit components of the decapping complex to repressed mRNAs (Nissan et al., 2010). The decay process could conceivably be arrested if components of the decapping complex (e.g. Dcp2 and enhancers of decapping) were inaccessible. Furthermore, under certain cellular conditions, repressed mRNAs that are stored within P bodies have been shown to be capable of re-entering the translating pool (Brengues et al., 2005). An interesting but unresolved question is whether specific HPat-containing granules involved in the repression of mRNAs associated with synaptic terminal growth contain proteins required for mRNA decay.
Targeting of synaptic mRNAs to P bodies by the miRNA pathway
It has previously been shown that Ago1 is involved in the control of synaptic terminal growth at the Drosophila NMJ (Jin et al., 2004). Since this time, a number of studies have demonstrated that the miRNA pathway is intimately involved in the control of synaptic development and plasticity (Siegel et al., 2011). Three lines of evidence suggest that P bodies are closely associated with miRNA-mediated regulation of these processes. First, Me31B, a core component of fly P bodies, is required for both miRNA-mediated repression and the control of dendrite morphogenesis in Drosophila sensory neurons (Barbee et al., 2006; Hillebrand et al., 2010). Second, P bodies in cultured hippocampal neurons contain the GW182 protein, miRNAs, and miRNA-repressed mRNAs (Cougot et al., 2008; Cougot et al., 2012). GW182 has been shown to be an important functional component of P bodies and the miRISC. Inhibition of GW182 expression leads to the disruption of P body assembly and miRNA-mediated repression (Ding et al., 2005; Jakymiw et al., 2005; Liu et al., 2005a; Rehwinkel et al., 2005). GW182 also interacts directly with the Ago proteins to facilitate binding of miRNAs to target mRNAs in order to direct their deadenylation and translational silencing or their deadenylation followed by exonucleolytic degradation (Carthew and Sontheimer, 2009; Eulalio et al., 2007c). Finally, a genetic interaction linking hpat to the miRISC (i.e. ago1) indicates that the miRNA pathway is targeting key neuronal mRNAs involved in the control of synaptic terminal growth for HPat-mediated deadenylation and repression in P bodies (Fig. 7B). The generally weaker phenotype observed in hpatd3/+; ago1l(2)k00208/+ trans-heterozygotes compared to hpatd3 homozygotes (Fig. 3A–D; Fig. 7B; 25% and 85% increase in bouton numbers, respectively) suggests that this interaction is incomplete. Other key mRNAs are likely being directed to P bodies through general translational repression pathways (Fig. 7A). This role for the HPat protein in the control of some miRNA-mediated mRNA regulation is supported in published work. Genetic evidence indicates that HPat is required for the control of ∼15% of all mRNAs targeted for repression or decay by Ago1 (Eulalio et al., 2007c). HPat has also been shown to directly co-immunoprecipitate with GW182 by interacting with its C-terminal silencing domain (Jäger and Dorner, 2010). In turn, GW182 has recently been shown to co-immunoprecipitate with NOT1 in both flies and worms (Kuzuoglu-Ozturk et al., 2012; Temme et al., 2010). Based on published data, miRNA-mediated repression in neuronal P bodies is likely a dynamic process. For example, de-repression of the CAT-1 mRNA by miR-122 in human hepatocarcinoma cells is associated with its exit from P bodies and re-entry into the translating pool (Bhattacharyya et al., 2006).
Implications for translational regulation by P bodies
The data presented in this study provide support for two models of P body function in neurons. First, P bodies may assemble in both pre- and postsynaptic cells where they are required to deadenylate and package specific translationally repressed mRNAs destined for the synapse. Some of these mRNAs may have essential functions in the control of synaptic terminal growth. This model is supported by the following observations. Here, we show that HPat localizes to particles in the both muscle and neurons in the larval ventral ganglion (Fig. 2B,C; supplementary Fig. S2) and can form HPat-containing granules in peripheral nerves projecting towards the NMJ (Fig. 2D). Interestingly, we find that endogenous HPat does not localize to the synapse (Fig. 2C; supplementary material Fig. S2). This suggests that these particles are dissociating completely (or into sub-resolution P body monomers) prior to reaching the synapse, perhaps in response to synaptic activity at the NMJ (Zeitelhofer et al., 2008a). HPat overexpression may drive mRNAs into P bodies where they are sequestered from the translating pool resulting in an inhibition of synaptic terminal growth (Figs 4,6). Interestingly, our data indicate that activity-dependent growth is more sensitive to HPat overexpression than synaptogenesis during larval development. This may be because activity-dependent processes require immediate new protein synthesis. HPat overexpression may prevent key mRNAs from rapidly entering the translating pool. In contrast, synaptic development is significantly slower process that allows for alternative mechanisms to deliver key mRNAs and/or proteins required for synaptogenesis. In hpat mutants, some key mRNAs may remain in the translating pool throughout development where they can be inappropriately regulated and translated. As a result, the failure to repress translation results in synaptic hyperplasia (Fig. 3A,C). Second, an intriguing possibility is that some synapse-localized mRNAs are locally translated and then immediately deadenylated, repressed, and stored in P bodies. This is supported by recent work where BDNF stimulation has been shown to lead to the accumulation of Dcp1-positive P bodies both in the soma and in hippocampal neuron dendrites (Huang et al., 2012). This correlates strongly with BDNF-stimulated activation of specific miRNA transcription and miRNA-mediated translational regulation. These models are not mutually exclusive and will require further study to resolve.
Materials and Methods
Drosophila stocks
All fly stocks were raised at 25°C on standard Bloomington media. Strains were obtained from: w1118, Canton-S, hpatEY10289, hpatl(3)06442, C380-Gal4, D42-Gal4, 24B-Gal4, Df(3R)Exel8165, Δ2-3 transposase, dcp2BG01766 and ago1l(2)k00208 (Bloomington Drosophila Stock Center); CG3995LL03178, hpatLL03060, dcp2GS3219, twinGS12209 and twinGS8115 (Kyoto Drosophila Stock Center); and C380-Gal4, cha-Gal80 (S. Sanyal).
Generation of hpatd3 allele
Two new alleles for hpat were generated by imprecise mobilization of transposon P{EPgy2}Patr-1EY10289. Patr-1EY10289 is a homozygous lethal P element insertion line, in which the P{EPgy2} element is inserted in the 5′ UTR of HPat at a position 66 bp distal to the first initiation codon. We generated excision lines by crossing virgin patr-1EY10289 flies to males possessing Δ2–3 transposase. From 14 independent excision events, one allele (hpatd3) was found to not carry P element markers (w+ and y+) and retain homozygous lethality. A second allele (hpatd17) was found not to carry P element markers but was homozygous viable. Finally, a precise excision of EY10289 (hpatd15) was generated for use as a control for genetic background. The molecular lesions in all hpat alleles were characterized by PCR and DNA sequencing.
Construction of hpat transgenic lines
Gal4-responsive transgenic HPat lines were constructed as follows. The 2907 bp open reading frame of hpat was PCR amplified from EST clone RE36948 and cloned into pENTR and then into pTW (which contains the pUAST promoter) by Gateway cloning (Invitrogen). RE36948 and pTW were obtained through the Drosophila Genomics Resource Center. Then pTW-HPat was used as a substrate for P element-mediated germline transformation (Genetic Services, Inc.). Transformed lines were crossed to the indicated GAL4 driver stocks. A transgenic construct for rescue of hpat mutants was constructed as follows. A ∼21 kb genomic fragment containing the entire hpat locus plus about 5.4 kb of upstream and 10.4 kb of downstream DNA (P[acman] BAC CH322-21M20) was obtained through the BACPAC Resource Center. The entire BAC clone was used for PhiC31-mediated germline transformation (BestGene, Inc.) into an attP landing site on chromosome 2 (Bloomington Drosophila Stock Center stock no. 9736). One line (P21M20) was recovered and used to rescue hpatd3 phenotypes.
Generation of HPat antibodies
The full-length HPat coding region was cloned into pProExHTa (Invitrogen) to produce a 6×His-tagged HPat protein. The protein was expressed in E. coli BL21, and purified using Ni-NTA Agarose (Qiagen) under the denaturing condition. Purified protein was further purified by SDS-PAGE, dialyzed against PBS containing 2 M urea, and used to elicit polyclonal antibody production in rabbits and rats (MBL, Japan). Rabbit polyclonal antisera were affinity-purified with the same antigens immobilized on a HiTrap NHS-activated HP column (GE Healthcare).
Western analysis
Protein extracts were made as follows. Briefly, w1118, homozygous hpatd3, and homozygous hpatEY10289 wandering third-instar larvae were homogenized on ice using a motorized disposable microtube pestle in PBS containing EDTA-free protease inhibitor tablets (Roche). Immediately after homogenization, an equal volume of 2×Laemmli sample buffer (BioRad) was added and the sample was passed through a QIAshredder (Qiagen) and boiled for 10 min. The sample was clarified by centrifugation and the supernatant separated by SDS-PAGE. Protein was transferred to a PVDF membrane (Amersham), blocked with 5% milk in PBS, blotted with rabbit anti-HPat (this study) and mouse anti-beta Actin ab8224 (Abcam) antibodies, and detected using ECL Plus reagents (Amersham). Relative HPat and Actin expression levels were quantified using ImageJ v1.45 (NIH) image analysis software. Both hpatd3 and hpatEy10289 mutants were normalized to controls.
Immunohistochemistry and RNA in situ hybridization
To study NMJ morphology, wandering third-instar larvae were dissected in calcium-free HL3 (Stewart et al., 1994). Larval body wall preparations were processed essentially as previously described (Sanyal et al., 2003). Unless otherwise indicated, control genotypes used for analysis were F1 larvae from w1118 crossed to Canton-S. Heterozygous larvae were F1 larvae from the indicated genotype crossed to w1118. Primary antibodies used were mouse anti-Dlg or anti-Futsch 22C10 (Developmental Studies Hybridoma Bank) and goat anti-HRP-Dylight-594 (Jackson Labs). Secondary antibodies used were Alexa 488-conjugated anti-mouse IgG (Molecular Probes). To study HPat localization or co-localization with FMRP, CNS and larval body wall preparations from Canton-S flies were dissected and processed as described above. Primary antibodies used were rat anti-HPat (this study) and/or mouse anti-FMRP 6A15 (Abcam), and goat anti-HRP-Dylight-633 (Jackson Labs). Secondary antibodies used were Alexa 488- or 568-conjugated anti-rat, rabbit or mouse IgG (Molecular Probes). Quantification of HPat and FMRP punctae was done from 10 randomly selected ROIs using the JACoP plugin for ImageJ v1.45 (NIH). RNA in situ hybridization was carried out essentially as described previously (Park et al., 2003). Indicated RNA probes were labeled with DIG and visualized with NBT/BCIP.
NMJ morphological analysis
Laser scanning confocal microscopy was performed on an Olympus FluoView FV1000 microscope. Projections from confocal z-series stacks were obtained using a 60× (N.A. 1.35) or 100× (N.A. 1.4) objective and generated from stacks collected at intervals of 0.8 µm. Images were combined using FV1000 imaging software. Analysis of boutons was done essentially as previously described (Rohrbough et al., 2000). For each genotype, 1b and 1s synaptic boutons were counted in both hemisegments of abdominal segment A3 for a minimum of 10 larvae (unless otherwise indicated; 20 paired NMJs). For each experiment, images were randomized blindly and then scored blindly using the Cell Counting plugin for ImageJ v1.45 (NIH). There was no obvious difference in muscle size between mutant or transgenic groups and controls.
High potassium stimulation
High potassium stimulation for activity assays was essentially done as previously described (Ataman et al., 2008). Briefly, non-wandering third instar larvae were partially dissected leaving CNS intact. The larvae then received 2, 2, 2, 4 and 6-minute stimulation pulses with osmolarity-adjusted 90 mM KCl HL-3 (Roche et al., 2002). Each stimulation paradigm was separated by 15-minute rest steps in 5 mM KCl HL-3 (Stewart et al., 1994) and a final resting step of 74 minutes. Upon completion of the stimulation paradigm, larval preparations were processed as described above for NMJ morphology. Psuedo-stimulated control experiments were done in the same manner, except that the 5 mM KCl HL-3 replaced the 90 mM KCl HL-3 during mock stimulation steps. Ghost boutons were identified by the presence of HRP and absence of Dlg.
Statistical analysis
Statistical analysis (specific tests are indicated in the corresponding figure legends) including graphing was performed using Prism v6.0 (GraphPad software) and statistical significance was determined to be at P<0.05. Where indicated, data are normalized to controls and are presented as mean±s.e.m. The numbers indicated in the columns of all graphs are the number of individual NMJs from which measurements were taken for that genotype.
Supplementary Material
Acknowledgments
We thank R. Dhatt for help characterizing the hpatd3 allele; R. Sand, L. Rozeboom and B. Symmes for assistance analyzing NMJ phenotypes; B. Symmes for editorial assistance; J. Blankenship for useful discussions; Genetic Services, Inc. and BestGene, Inc. for germline transformation services; The Bloomington Drosophila Stock Center and Kyoto Drosophila Stock Center for fly lines; the Drosophila Genomics Resource Center for fly transformation vectors and cDNAs; and the BACPAC Resource Center for BAC clones. Flybase also provided essential information. Author contributions are as follows: conceived and designed the experiments: S.J.P., S.A.B.; analyzed data, S.J.P., K.R.N., S.F.R., S.A.B.; contributed reagents/materials, Y.K., A.N., M.R., S.A.B.; wrote the paper: S.J.P., S.A.B.
Footnotes
Funding
This work was funded by a grant from the National Institutes of Health [grant number R01DA26048 to S.A.B.]; and an Investigator Grant award to M.R. from the Science Foundation of Ireland. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.113043/-/DC1
References
- Ataman B., Ashley J., Gorczyca D., Gorczyca M., Mathew D., Wichmann C., Sigrist S. J., Budnik V. (2006). Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc. Natl. Acad. Sci. USA 103, 7841–7846 10.1073/pnas.0600387103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataman B., Ashley J., Gorczyca M., Ramachandran P., Fouquet W., Sigrist S. J., Budnik V. (2008). Rapid activity-dependent modifications in synaptic structure and function require bidirectional Wnt signaling. Neuron 57, 705–718 10.1016/j.neuron.2008.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. A., Estes P. S., Cziko A. M., Hillebrand J., Luedeman R. A., Coller J. M., Johnson N., Howlett I. C., Geng C., Ueda R.et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009 10.1016/j.neuron.2006.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 10.1016/j.cell.2006.04.031 [DOI] [PubMed] [Google Scholar]
- Boag P. R., Atalay A., Robida S., Reinke V., Blackwell T. K. (2008). Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 182, 543–557 10.1083/jcb.200801183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J. E., Tritschler F., Haas G., Igreja C., Truffault V., Weichenrieder O., Izaurralde E. (2010). The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 29, 2368–2380 10.1038/emboj.2010.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. A., DiAntonio A. (2007). Synaptic development: insights from Drosophila. Curr. Opin. Neurobiol. 17, 35–42 10.1016/j.conb.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Séraphin B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31–40 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Bhattacharyya S. N., Tapia–Arancibia L., Bordonné R., Filipowicz W., Bertrand E., Rage F. (2008). Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 28, 13793–13804 10.1523/JNEUROSCI.4155-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Cavalier A., Thomas D., Gillet R. (2012). The dual organization of P-bodies revealed by immunoelectron microscopy and electron tomography. J. Mol. Biol. 420, 17–28 10.1016/j.jmb.2012.03.027 [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. (2005). The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19, 437–447 10.1016/j.molcel.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm–Ansmant I., Izaurralde E. (2007a). P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8, 9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm–Ansmant I., Schweizer D., Izaurralde E. (2007b). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970–3981 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S. F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. (2007c). Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 21, 2558–2570 10.1101/gad.443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T. M., Lykke–Andersen J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G., Braun J. E., Igreja C., Tritschler F., Nishihara T., Izaurralde E. (2010). HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 189, 289–302 10.1083/jcb.200910141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillebrand J., Pan K., Kokaram A., Barbee S., Parker R., Ramaswami M. (2010). The Me31B dead-box helicase localizes to postsynaptic foci and regulates expression of a CaMKII Reporter mRNA in dendrites of Drosophila olfactory projection neurons. Front Neural Circuits 4, 121 10.3389/fncir.2010.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Ruiz C. R., Eyler E. C., Lin K., Meffert M. K. (2012). Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell 148, 933–946 10.1016/j.cell.2012.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger E., Dorner S. (2010). The decapping activator HPat a novel factor co-purifying with GW182 from Drosophila cells. RNA Biol. 7, 381–385 10.4161/rna.7.3.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J. C., Fritzler M. J., Chan E. K. (2005). Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7, 1167–1174 10.1038/ncb1334 [DOI] [PubMed] [Google Scholar]
- Jin P., Zarnescu D. C., Ceman S., Nakamoto M., Mowrey J., Jongens T. A., Nelson D. L., Moses K., Warren S. T. (2004). Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117 10.1038/nn1174 [DOI] [PubMed] [Google Scholar]
- Jung H., Yoon B. C., Holt C. E. (2012). Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci. 13, 597 10.1038/nrn3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S., Kreienkamp H. J. (2012). Dendritic mRNA targeting and translation. Adv. Exp. Med. Biol. 970, 285–305 10.1007/978-3-7091-0932-8_13 [DOI] [PubMed] [Google Scholar]
- Kuzuoglu–Ozturk D., Huntzinger E., Schmidt S., Izaurralde E. (2012). The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res 40, 5654–5665 10.1093/nar/gks218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. (2005a). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7, 1161–1166 10.1038/ncb1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia–Sanchez M. A., Hannon G. J., Parker R. (2005b). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719–723 10.1038/ncb1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A., Standart N. (2010). Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem. Soc. Trans. 38, 1602–1607 10.1042/BST0381602 [DOI] [PubMed] [Google Scholar]
- Marnef A., Maldonado M., Bugaut A., Balasubramanian S., Kress M., Weil D., Standart N. (2010). Distinct functions of maternal and somatic Pat1 protein paralogs. RNA 16, 2094–2107 10.1261/rna.2295410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A., Weil D., Standart N. (2011). RNA-related nuclear functions of human Pat1b,17 the P-body mRNA decay factor. Mol. Biol. Cell 23, 213–224 10.1091/mbc.E11-05-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Sanyal S., Habara Y., Sanchez R., Wharton R. P., Ramaswami M., Zinn K. (2004). The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44, 663–676 10.1016/j.neuron.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Tanaka K. J., Miyauchi M., Huang L., Tsujimoto M., Matsumoto K. (2010). Translational repression by the oocyte-specific protein P100 in Xenopus. Dev. Biol. 344, 272–283 10.1016/j.ydbio.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Nissan T., Rajyaguru P., She M., Song H., Parker R. (2010). Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39, 773–783 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S., Chekulaeva M., Stoecklin G. (2010). Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 30, 4308–4323 10.1128/MCB.00429-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Schroeder A. J., Helfrich–Förster C., Jackson F. R., Ewer J. (2003). Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 130, 2645–2656 10.1242/dev.00503 [DOI] [PubMed] [Google Scholar]
- Parrish J. Z., Kim M. D., Jan L. Y., Jan Y. N. (2006). Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 20, 820–835 10.1101/gad.1391006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington G. R., Parker R. (2008). Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 28, 1298–1312 10.1128/MCB.00936-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K. (2011). The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43, 973–981 10.1016/j.molcel.2011.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Behm–Ansmant I., Gatfield D., Izaurralde E. (2005). A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647 10.1261/rna.2191905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche J. P., Packard M. C., Moeckel–Cole S., Budnik V. (2002). Regulation of synaptic plasticity and synaptic vesicle dynamics by the PDZ protein Scribble. J. Neurosci. 22, 6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J., Grotewiel M. S., Davis R. L., Broadie K. (2000). Integrin-mediated regulation of synaptic morphology, transmission, and plasticity. J. Neurosci. 20, 6868–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J., Hummel T., Ng N., Klämbt C., Davis G. W. (2000). Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron 26, 371–382 10.1016/S0896-6273(00)81170-8 [DOI] [PubMed] [Google Scholar]
- Sanyal S., Narayanan R., Consoulas C., Ramaswami M. (2003). Evidence for cell autonomous AP1 function in regulation of Drosophila motor-neuron plasticity. BMC Neurosci. 4, 20 10.1186/1471-2202-4-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster C. M. (2006). Glutamatergic synapses of Drosophila neuromuscular junctions: a high-resolution model for the analysis of experience-dependent potentiation. Cell Tissue Res. 326, 287–299 10.1007/s00441-006-0290-5 [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Saba R., Schratt G. (2011). microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497 10.1016/j.gde.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Sossin W. S., DesGroseillers L. (2006). Intracellular trafficking of RNA in neurons. Traffic 7, 1581–1589 10.1111/j.1600-0854.2006.00500.x [DOI] [PubMed] [Google Scholar]
- Stewart B. A., Atwood H. L., Renger J. J., Wang J., Wu C. F. (1994). Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A 175, 179–191 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia–Sanchez M. A., Brengues M., Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. (2004). A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23, 2862–2871 10.1038/sj.emboj.7600273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C., Zhang L., Kremmer E., Ihling C., Chartier A., Sinz A., Simonelig M., Wahle E. (2010). Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16, 1356–1370 10.1261/rna.2145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaessinger S., Busseau I., Simonelig M. (2006). Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133, 4573–4583 10.1242/dev.02649 [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M., Karra D., Macchi P., Tolino M., Thomas S., Schwarz M., Kiebler M., Dahm R. (2008a). Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J. Neurosci. 28, 7555–7562 10.1523/JNEUROSCI.0104-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M., Macchi P., Dahm R. (2008b). Perplexing bodies: The putative roles of P-bodies in neurons. RNA Biol. 5, 244–248 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Bailey A. M., Matthies H. J., Renden R. B., Smith M. A., Speese S. D., Rubin G. M., Broadie K. (2001). Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 107, 591–603 10.1016/S0092-8674(01)00589-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.