Summary
Tyrosine-kinase-based signal transduction mediated by modular protein domains is critical for cellular function. The Src homology (SH)2 domain is an important conductor of intracellular signaling that binds to phosphorylated tyrosines on acceptor proteins, producing molecular complexes responsible for signal relay. Cortactin is a cytoskeletal protein and tyrosine kinase substrate that regulates actin-based motility through interactions with SH2-domain-containing proteins. The Src kinase SH2 domain mediates cortactin binding and tyrosine phosphorylation, but how Src interacts with cortactin is unknown. Here we demonstrate that Src binds cortactin through cystine bonding between Src C185 in the SH2 domain within the phosphotyrosine binding pocket and cortactin C112/246 in the cortactin repeats domain, independent of tyrosine phosphorylation. Interaction studies show that the presence of reducing agents ablates Src-cortactin binding, eliminates cortactin phosphorylation by Src, and prevents Src SH2 domain binding to cortactin. Tandem MS/MS sequencing demonstrates cystine bond formation between Src C185 and cortactin C112/246. Mutational studies indicate that an intact cystine binding interface is required for Src-mediated cortactin phosphorylation, cell migration, and pre-invadopodia formation. Our results identify a novel phosphotyrosine-independent binding mode between the Src SH2 domain and cortactin. Besides Src, one quarter of all SH2 domains contain cysteines at or near the analogous Src C185 position. This provides a potential alternative mechanism to tyrosine phosphorylation for cysteine-containing SH2 domains to bind cognate ligands that may be widespread in propagating signals regulating diverse cellular functions.
Key words: Src, SH2 domain, Cortactin, Cystine
Introduction
Signal transduction through protein-protein interactions is essential for cellular function and is mediated by specialized protein domains. The SH2 domain is one of the initially discovered and best-characterized protein interaction motifs (Koch et al., 1991). SH2 domains are ∼100 amino acids in length, and genomic analysis indicates that 121 SH2 domains are found in 115 individual human proteins that participate in a wide range of signaling events (Liu et al., 2006). SH2 domains function by binding to phosphorylated tyrosine residues in target proteins typically specified by residues in positions −2 to +4 of the phosphotyrosine (Machida et al., 2007; Songyang et al., 1993). SH2 domains are structurally conserved, consisting of a β-sheet flanked by opposing α-helices (Pawson et al., 2001). A positively charged binding pocket within the β-sheet contains the canonical FLVRES sequence, where arginine βB5 forms the critical electrostatic bond with two oxygen atoms in the phosphotyrosine to generate domain-ligand binding (Kuriyan and Cowburn, 1997). While this contact is central for SH2-phosphotyrosine interactions, carboxyl-terminal residues within the β-sheet create variable binding interfaces (“specificity pockets”) that dictate ligand specificity based on the residues flanking the phosphotyrosine (Bae et al., 2009; Müller and Knapp, 2009; Yaffe, 2002). Though well characterized in terms of phosphotyrosine ligand binding, emerging reports have determined that select SH2 domains bind certain ligands independent of tyrosine phosphorylation (Bae et al., 2009; King et al., 2000; Liao et al., 2007). These interactions are mediated by ligand binding to regions on the SH2 domain that either include or exclude participation of the phosphotyrosine binding cleft, potentially increasing the variability and complexity of SH2 domain function in signal relay systems.
Src and related tyrosine kinases are highly regulated enzymes where the SH2 domain plays a pivotal role in controlling kinase function. Binding of the Src SH2 domain to the phosphorylated carboxyl-terminal tyrosine 527 is key in maintaining Src in a closed autoinhibitory state (Okada and Nakagawa, 1989; Xu et al., 1999). Tyrosine 527 dephosphorylation results in an open conformation, allowing the kinase (SH1) domain to phosphorylate substrates. Elimination of tyrosine 527 results in constitutive kinase activation and neoplastic transformation (Yeatman, 2004). The Src SH2 domain also potentiates kinase activity through stable binding to several tyrosine phosphorylated substrates, notably focal adhesion kinase (FAK) and p130CAS (Burnham et al., 2000; Schaller et al., 1994). Src regulates cellular growth, division, adhesion, and motility through SH2-mediated interactions and subsequent cis and/or trans substrate tyrosine phosphorylation (Brown and Cooper, 1996). Elevated growth factor signaling in human cancer leads to Src hyperactivation, promoting tumor progression through increased growth and invasive potential. Increased Src activity accomplishes this by promoting tumor cell migration, invadopodia formation, and matrix metalloproteinase (MMP) activity (Guarino, 2010). These attributes have resulted in the development of kinase-targeted Src inhibitory compounds that are currently being evaluated for efficacy as anti-tumor and -metastatic therapeutics. (Elsberger et al., 2010).
Several actin-binding proteins serve as Src targets for SH2 domain binding and phosphorylation that modulate actin dynamics essential for whole and intracellular motility. Cortactin is a filamentous (F)-actin binding protein that regulates actin related protein (Arp)2/3-based actin network formation responsible for cortical actin-based membrane protrusion (Kirkbride et al., 2011). Src phosphorylates cortactin at three positions (Y421/466/482 in the murine form) within a proline (P)-rich domain near the carboxyl terminus (Huang et al., 1998). Cortactin tyrosine phosphorylation coincides with cellular membrane deforming events involving cortical actin remodeling, including cell migration, pathogen uptake, endocytosis, osmotic shock, synaptic remodeling, cell junction regulation, and invadopodia formation (Cosen-Binker and Kapus, 2006). Mechanistic insight to date indicates that cortactin tyrosine phosphorylation creates binding sites for SH2 domain-containing adaptor proteins. These include Crk during Shigella internalization (Bougnères et al., 2004) and Nck1 in invadopodia maturation (Oser et al., 2009). For Nck1, additional Nck1 domains mediate the assembly of N-WASp-containing macromolecular complexes that further enhance Arp2/3 actin network formation (Tehrani et al., 2007). Src-mediated cortactin phosphorylation also enhances binding of the cortactin carboxyl-terminal SH3 domain to proline-rich domains in target proteins (Ammer and Weed, 2008), although the molecular details of this process are currently unclear.
While the SH2 domain has been previously shown to be solely responsible for mediating Src association with cortactin (Okamura and Resh, 1995), the precise SH2 Src interaction site on cortactin is unknown. Here we demonstrate that Src associates with cortactin through a phosphotyrosine-independent SH2 domain interaction involving the formation of a cystine linkage. Deletion and mutational mapping indicates that cysteine residues 112 in the 1st and 246 in the 5th cortactin repeat represent two separate docking sites for the Src SH2 domain. Src and cortactin form a stable redox-sensitive linkage in cells that is required for cortactin phosphorylation. Molecular modeling of the Src SH2 domain shows peptides containing cortactin C112 and C246 dock within the Src phosphotyrosine-binding cleft, with cortactin cysteine residues in close proximity to Src C185 at position βC3. Tandem mass spectroscopy of the Src SH2 domain mixed with cortactin peptides demonstrates formation of cystine bonds between Src C185 and cortactin C112 and C246. Cells containing cortactin mutants lacking C112 and C246 display reduced cortactin tyrosine phosphorylation, motility, and adhesion. Cortactin C112/246 is required for the formation of initial (pre-) invadopodia complexes and extracellular matrix degradation. Our results indicate that Src interacts with cortactin independent of tyrosine phosphorylation through novel cystine bonding within the Src SH2 domain phosphotyrosine-binding region. Sequence inspection indicates that 25% of all SH2 domains contain a cysteine at, or in close proximity to βC3, pointing to potential widespread usage of cystine-based SH2 domain interactions in numerous signaling pathways.
Results
The Src SH2 domain binds cortactin independent of tyrosine phosphorylation
Src and other tyrosine kinases phosphorylate cortactin on tyrosines 421, 466, and 482 (Ammer and Weed, 2008; Daly, 2004). Using affinity precipitation assays with purified Src SH2, SH3, and tandem SH2/SH3 fusion proteins, we confirmed previous work (Okamura and Resh, 1995) that the Src SH2 domain is the only region on Src responsible for binding cortactin (supplementary material Fig. S1A). To determine whether the Src SH2 domain can interact with any of the primary cortactin phosphotyrosine residues, a commercial SH2 domain array was screened with cortactin peptides surrounding the two main Src phosphorylation targets (Y421 and Y466). Interestingly, neither phosphorylated nor unphosphorylated peptides interacted with the Src SH2 domain, although both peptides showed phospho-dependent binding to the SH2 domains of the tyrosine kinases Abl and Fyn (supplementary material Fig. S1B–D), both which target cortactin (Boyle et al., 2007; Huang et al., 2003). We pursued this finding utilizing GST-Src SH2 pull-down assays from lysates of epidermal growth factor (EGF)-stimulated cells. There was no difference between the amounts of cortactin precipitated with the Src GST-SH2 domain from EGF-stimulated cells compared to non-stimulated cells, even though stimulated cells showed increased cortactin tyrosine phosphorylation (Fig. 1A). We next evaluated the direct association of GST-Src SH2 with cortactin using far western analysis. Cells were transfected with FLAG-tagged constructs expressing full-length cortactin (FL) or cortactin containing phenylalanine substitutions at the dominant Src phosphorylation site (Y421F; Huang et al., 1998) or all three Src phosphorylation sites (triple tyrosine mutant; TYM). The Src SH2 domain bound cortactin at equivalent levels from starved or EGF-stimulated cells whether or not Src-targeted cortactin tyrosine residues were present (Fig. 1B). The same result was obtained by GST-Src SH2 affinity precipitation (Fig. 1C). To further confirm phosphotyrosine independence, we assayed bacterially-produced, non-phosphorylated recombinant cortactin by GST-SH2 affinity precipitation. A dose-dependent increase in cortactin binding by the Src SH2 domain was observed above saturated GST control levels (Fig. 1D). Constitutively active Src phosphorylated recombinant cortactin (supplementary material Fig. S2A) and GST-Src SH2 interacts with FAK Y397 in a phosphotyrosine-dependent manner (supplementary material Fig. S2B), indicating that the assayed proteins retained correct functionality.
Fig. 1.
Src SH2 binding to cortactin does not involve tyrosine phosphorylation and binds cortactin repeats 1 and 5. (A) GST and GST-Src SH2 affinity precipitation from MTLn3 cells evaluated for cortactin tyrosine phosphorylation. The ratio of phosphorylated cortactin levels and the normalized amounts of total precipitated cortactin are indicated. (B) Src SH2 far western analysis of FLAG-tagged recombinant wild-type and cortactin phosphorylation mutants. TYM, triple tyrosine mutant. (C) Affinity precipitation analysis of FLAG-tagged recombinant wild-type and cortactin phosphorylation mutants from extracts with GST-Src SH2 domain. (D) Affinity precipitation of non-phosphorylated, recombinant cortactin with GST and GST-Src SH2. Normalized intensity levels are shown relative to GST control. (E) GST-SH2 domain far western blotting of the cortactin NTA and repeats region. CT, C terminus; LC, IgG light chain; NT, N terminus. Arrows denote position of IgG heavy chain (HC) recognized by cross reactivity with secondary antibodies during the blotting process. Asterisks indicate the positions of recombinant cortactin proteins. (F) GST-SH2 far western analysis of cortactin deletion cortactin constructs. (G) Far western binding of the GST-Src SH2 domain to tandem cortactin repeat chimeric constructs.
The Src SH2 domain binds to cortactin repeat 1 and repeat 5
To identify the cortactin region responsible Src SH2 binding, we utilized a series of deletion mutants with the systematic removal of cortactin structural domains that independently retain their respective functions (Weaver et al., 2001; Weed et al., 2000) (supplementary material Fig. S3). Far western and affinity precipitation assays indicated that GST-Src SH2 bound to the N-terminal (NT) half of cortactin (residues 1–330) rather than the carboxyl terminal (CT) half that contains the sites of tyrosine phosphorylation (supplementary material Fig. S2C,D). Separation of the Arp2/3-binding N-terminal acidic (NTA) domain from the F-actin binding repeats domain indicated that the Src SH2 domain associated with the cortactin repeats region (Fig. 1E). Serial deletion of individual cortactin repeats beginning with repeat 3 and retaining the carboxyl terminus demonstrated a significant reduction of Src SH2 domain binding with removal of the 5th cortactin repeat (Fig. 1F). Deletion of repeat 5 in the context of the full-length cortactin protein failed to prevent SH2 binding (supplementary material Fig. S4A), suggesting that there is at least one additional repeat that binds the Src SH2 domain. To test this, we utilized a carboxyl terminal deletion series by far western analysis where each cortactin repeat was serially removed (supplementary material Fig. S4B). Src SH2 binding was observed in all constructs containing repeat 1. Since either set of deletion constructs cannot clearly determine that repeats 1 and 5 are the only repeats capable of Src SH2 binding, we created chimeric constructs by adding each individual cortactin repeat in tandem N-terminal to repeat 6 in the SH2-null binding R6-CT construct (supplementary material Fig. S3). Far western assays of these constructs unambiguously established cortactin repeat 1 and repeat 5 as individual interaction regions for Src SH2 domain binding (Fig. 1G).
Cysteine 112 and cysteine 246 mediate cortactin binding to the Src SH2 domain
Sequence alignment of each cortactin repeat identified cysteine 112 in repeat 1 and cysteine 246 in repeat 5 as residues that lacked significant homology with cortactin residues in the same position (Fig. 2A). To determine if these residues were responsible for their respective cortactin repeats to bind the Src SH2 domain, we mutated each cysteine individually to alanine in the full-length wild-type molecule (WT C112A and WT C246A) and in their respective dual tandem repeat 6 chimeras (R1R6 C112A and R5R6 C246A). Far western assays indicated that Src SH2 binding was retained in the WT C112A and WT C246A constructs but not by the R1R6 C112A and R5R6 C246A chimeric mutants (Fig. 2B,C). These data indicate that C112 and C246 function as separate, independent binding motifs for the Src SH2 domain. Given the duplicity of these SH2 binding sites, we mutated cysteine 112 and 246 to alanine in the WT cortactin construct (double cysteine mutant; DCM) and tested Src SH2 binding by far western analysis and affinity precipitation. Mutation of both cysteine residues abolished Src SH2 domain binding (Fig. 2D–E). These data indicate that cysteine 112 and 246 are required for Src SH2 association with cortactin.
Fig. 2.
Cortactin cysteines 112 and 246 are required for Src SH2 domain binding. (A) Alignment of cortactin repeats denoting C112 and C246. (B,C) Far western blotting of GST-Src SH2 domain with cortactin cysteine to alanine mutants. CT, C terminus; HC, IgG heavy chain; LC, IgG light chain; NT, N terminus; WT; full-length wild-type cortactin. Dashed line separates HC from chimeric cortactin proteins (asterisks) due to similar molecular weights. (D) Far western analysis of Src SH2 domain binding to the C112/C246A cortactin double cysteine mutant (DCM). (E) Affinity precipitation analysis of Src SH2 domain binding to the C112/C246A cortactin double cysteine mutant (DCM).
Molecular modeling of cortactin C112 and C246 pentapeptides with the Src SH2 domain
Given the apparent unique binding requirements for cortactin to the Src SH2 domain, we conducted molecular modeling with cortactin C112 and C246 peptides to gain mechanistic insight into how the cortactin cysteine residues might interact with the SH2 domain. The SH2 domain of Src was obtained from the published crystal structure (Xu et al., 1997). As a positive control, a pentapeptide encompassing Src pY527 was docked to Src SH2. The peptide was predicted to dock in the phospho-tyrosine binding pocket in close proximity with the βB5 arginine 175 (Fig. 3A). The phosphotyrosine in the docked structure displayed the identical 2.72 Å distance between R175 and the pY527 phosphate group obtained by co-crystallization and NMR analysis (Waksman et al., 1992). Analogous cortactin and corresponding control pentapeptides, encompassing C112, A112, C246, and A246, docked in the presence the SH2/pY527 complex failed to produce sufficient binding energies predictive of cortactin binding. This suggested that the tested cortactin peptides might bind within the Src SH2 phosphotyrosine binding cavity. To test this, similar docking studies were conducted in the absence of the pY527 Src peptide. Under these conditions all cortactin peptides were predicted to bind within the Src SH2 phospho-tyrosine binding cleft (Fig. 3A). Interestingly, the cysteine/alanine residues within each cortactin peptide dock in close proximity to Src C185 in the βC3 position (C112; 5.83 Å, A112; 5.96 Å, C246; 3.56 Å, A246; 3.17 Å) (Fig. 3A). These data suggested the possibility that cortactin C112 or C246 might form a cystine bond with Src C185 to mediate Src-cortactin binding. The predicted distances between cortactin C112/246 and Src C185 are greater than the typical ∼2 Å distance for cystine bonds determined by Raman spectroscopy (Van Wart et al., 1973). This difference is attributed to the inability of the modeling program to construct disulfide bonds, as well as program-predicted deprotonation of all cysteines in the assay, imparting negative charges to C112/246 and Src C185. In spite of the introduced repulsive effects, the predicted binding energies of the C112/C246 cortactin peptides are lower than the A112/A246 peptides by 6–8 kcal/mole, suggesting a more favorable binding affinity for the cysteine-containing cortactin peptides (Fig. 3B). Although nearly 50% weaker than predicted pY527 peptide binding, the presence and location of cysteine at cortactin 112 and 246 within the Src SH2 binding pocket suggested a potentially favorable interaction with the Src SH2 domain. These data implicate Src C185 as a key residue within the SH2 domain responsible for cortactin binding through cystine bond formation.
Fig. 3.

Cysteine-containing cortactin peptides dock within the Src SH2 phosphotyrosine binding region. (A) Molecular modelling of the Src SH2 domain with phosphorylated Src and cortactin pentapeptides. Enlarged views show position of Src R175, Src C185 and the respective central Src or cortactin peptide residues. (B) Calculated binding energies for each peptide docking condition shown in (A).
Cystine bonding mediates Src SH2 binding to cortactin
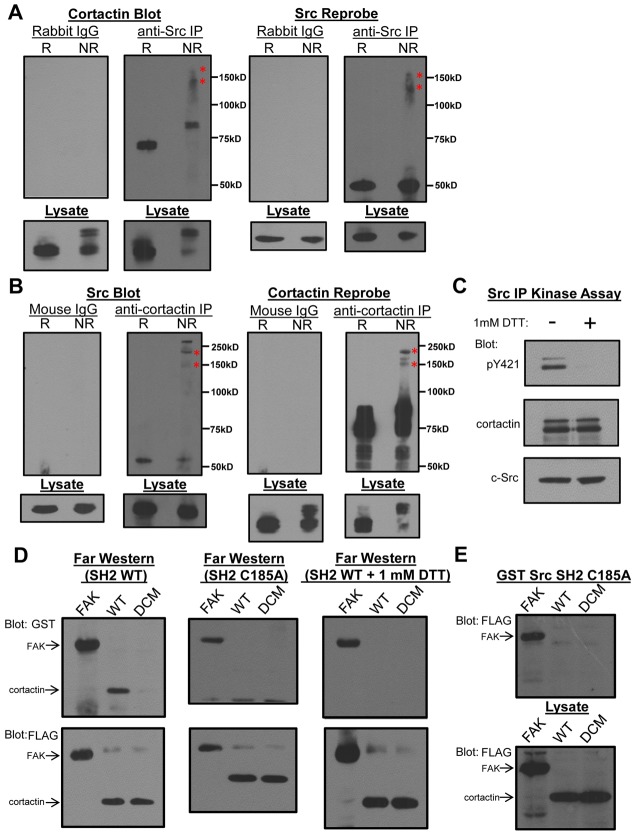
To evaluate if cystine bonding mediates Src binding to cortactin, the association of endogenous Src and cortactin was determined under differential redox conditions. Cortactin specifically co-immunoprecipitated with Src from UMSCC1 HNSCC cells migrated at the typical 70 kDa Mr when immune complexes were incubated with 2-mercaptoethanol and analyzed by SDS-PAGE and Western blotting (Fig. 4A). However, cortactin Mr was severely retarded when immunoprecipitates were prepared in the absence of reductant, banding as several higher molecular weight species at 90, 120 and 165 kDa (Fig. 4A). A similar pattern was observed for Src in cortactin immunoprecipitates (Fig. 4B), indicating that a subset of Src and cortactin complex under oxidative conditions required for cystine bonding. Cortactin phosphorylation by Src is also oxidation dependent, as incubation of Src immunoprecipitates with purified recombinant cortactin in the presence of dithiothreitol (DTT) prevents Src phosphorylation of cortactin Y421 (Fig. 4C).
Fig. 4.
Binding and phosphorylation of cortactin by Src is redox dependent and requires Src C185. (A) Co-immunoprecipitation (IP) of cortactin with Src followed by analysis under reducing (R) and non-reducing (NR) conditions. Asterisks denote equivalent bands in cortactin and Src immunoblots. (B) Co-immunoprecipitation of Src with cortactin followed by analysis under reducing (R) and non-reducing (NR) conditions. Asterisks denote equivalent bands in cortactin and Src immunoblots. (C) Phosphorylation of cortactin by Src in the absence and presence of DTT. (D) Far western analysis of GST-Src SH2 and C185A. (E) Affinity precipitation analysis of FAK and cortactin binding to GST-Src SH2 C185A.
Since Src C185 is predicted to be the key residue mediating Src-cortactin bonding, Src C185 was mutated to alanine (C185A) and the recombinant SH2 domain assayed for cortactin binding. C185A eliminated WT cortactin binding but did not perturb phosphotyrosine-dependent binding to FAK (Fig. 4D–E). Mutation of the arginine responsible for phosphotyrosine binding (R175) to alanine did not alter binding to WT or NT cortactin, verifying phosphotyrosine independence (supplementary material Fig. S5). Reduction of the Src SH2 domain with DTT did not impact binding to FAK but abolished cortactin binding (Fig. 4D).
To directly verify the existence of cystine bonding between Src C185 and cortactin C112/246, we incubated the GST-Src SH2 domain with saturating amounts of 7-mer peptides encompassing cortactin C112 and C246 in the absence of reducing agents. Control (GST-SH2 alone) and SH2-cortactin peptide mixtures were subjected to LC-MS/MS to identify cystine bonding between the Src SH2 domain and each cortactin peptide. The predicted trypsin digest product from the Src SH2 domain containing C185 is a 14-mer peptide with C185 in position four. The fragment composition and mass for the Src SH2 domain alone and cystine-bound to the digested C112 and C246 cortactin peptides are shown in Fig. 5A. Fragmentation and subsequent spectral sequence analysis indicated that cystine bonding occurred between Src C185 and cortactin C112 or C246, as evidenced by mass shifts of the b4 ions in the Src C185 peptide fragment in experiments containing cortactin C112 (Fig. 5B vs Fig. 5C) and C246 (Fig. 5B vs Fig. 5D) peptides. Analogous shifts were observed upon inspection of the y11 ions for each experimental condition (supplementary material Fig. S6). Complete sequence coverage of the SH2 domain fragment from the SH2-C112 and SH2-C246 experiments in the b- and y- planes yielded observed peptide masses identical or very close to the predicted mass at each ion parameter (Table 1). These data collectively indicate that Src C185 is capable of forming cystine bonds with cortactin C112 and C246 under oxidizing conditions, representing a novel mode of interaction between an SH2 domain and its respective target ligand.
Fig. 5.
Src C185 forms a cystine bond with cortactin C112 and C246. (A) Sequence of the predicted Src SH2 domain tryptic fragment containing C185. Predicted cystine bonding between Src C185 and the cortactin C112 and C246 tryptic peptides with predicted masses are shown below. (B–D) Extracted ion chromatogram (left) and ion fragmentation spectra (right) from tandem LC-MS/MS of the GST-Src SH2 domain (B), the GST-SH2 domain with the cortactin C112 peptide (C) and the GST-SH2 domain with cortactin C246 peptide (D). Spectra were enlarged to indicate the position of the Src C185 b4 ion (boxed in red).
Table 1.
Observed and predicted ion masses of GST-Src-SH2, GST-Src-SH2 + cortactin C112 and GST-Src-SH2 + cortactin C246 tryptic peptides
| GST-Src SH2 | GST-Src SH2 + C112 Peptide (HCSQV) | GST-Src SH2 + C246 Peptide (CALG) | |||||||||||||||
| Ion | Exp. Da | Obs. Da | Ion | Exp. Da | Obs. Da | Ion | Exp. da | Obs. Da | Ion | Exp. Da | Obs. Da | Ion | Exp. Da | Obs. Da | Ion | Exp. Da | Obs. Da |
| b1 | 57.02 | ---- | y1 | 146.11 | 147.11 | b1 | 57.02 | ---- | y1 | 146.11 | ---- | b1 | 57.02 | ---- | y1 | 146.11 | 147.11 |
| b2 | 128.06 | 129.07 | y2 | 217.14 | 218.15 | b2 | 128.06 | 129.07 | y2 | 217.14 | 218.15 | b2 | 128.06 | 129.07 | y2 | 217.14 | 218.15 |
| b3 | 291.12 | 292.13 | y3 | 331.19 | 332.19 | b3 | 291.12 | 292.13 | y3 | 331.19 | 332.19 | b3 | 291.12 | 292.13 | y3 | 331.19 | 332.19 |
| b4 | 394.13 | 395.14 | y4 | 446.21 | 447.22 | b4 | 965.36 | 965.36* | y4 | 446.21 | 447.22 | b4 | 755.29 | 755.28* | y4 | 446.21 | 447.21 |
| b5 | 507.22 | 508.22 | y5 | 593.28 | 594.29 | b5 | 1078.45 | 1078.44* | y5 | 593.28 | 594.29 | b5 | 868.37 | 868.36* | y5 | 593.28 | 594.28 |
| b6 | 594.25 | ---- | y6 | 708.31 | 709.32 | b6 | 1165.47 | 1165.48* | y6 | 708.31 | 709.32 | b6 | 955.40 | 955.38* | y6 | 708.31 | 709.31 |
| b7 | 693.32 | 694.32 | y7 | 795.34 | 796.35 | b7 | 1264.55 | 1264.55* | y7 | 795.34 | 796.35 | b7 | 1054.47 | 1054.46* | y7 | 795.34 | 796.34 |
| b8 | 780.35 | 781.36 | y8 | 894.41 | 895.42 | b8 | 1351.58 | 1351.58* | y8 | 894.41 | 895.42 | b8 | 1141.50 | 1141.50* | y8 | 894.41 | 895.40 |
| b9 | 895.38 | ---- | y9 | 981.44 | 982.45 | b9 | 1466.61 | 1466.60* | y9 | 981.44 | 982.45 | b9 | 1256.53 | 1256.51* | y9 | 981.44 | 982.44 |
| b10 | 1042.44 | 1043.45 | y10 | 1094.52 | 1095.53 | b10 | 1613.67 | 1613.67* | y10 | 1094.52 | 1095.53 | b10 | 1403.60 | 1403.58* | y10 | 1094.52 | 1095.52 |
| b11 | 1157.47 | 1158.48 | y11 | 1197.53 | 1198.54 | b11 | 1728.70 | 1728.70* | y11 | 1768.76 | 1768.76* | b11 | 1518.62 | 1518.63* | y11 | 1558.69 | 1558.68* |
| b12 | 1271.51 | 1272.52 | y12 | 1360.60 | 1361.60 | b12 | 1842.74 | ---- | y12 | 1931.83 | 1931.83* | b12 | 1632.67 | 1632.66* | y12 | 1721.75 | ---- |
| b13 | 1342.55 | 1343.56 | y13 | 1431.63 | 1432.64 | b13 | 1913.78 | 1913.78* | y13 | 2002.86 | ---- | b13 | 1703.71 | 1703.70* | y13 | 1792.79 | ---- |
| b14 | 1470.65 | ---- | y14 | 1488.66 | ---- | b14 | 2041.88 | ---- | y14 | 2059.89 | ---- | b14 | 1831.80 | ---- | y14 | 1849.81 | ---- |
Non-bold numerals indicate expected ion masses; bold numerals indicate observed ion masses; the asterisks indicate the peptide modified masses.
Cortactin C112 and C246 are required for cortactin tyrosine phosphorylation and Src-based cellular processes
To evaluate the impact of cortactin C112/C246 on biochemical and cellular functions involving Src, we initially determined the ability of Src to phosphorylate cortactin DCM. While expression of cortactin WT with WT Src (c-Src) in murine fibroblasts lacking Src, Yes, and Fyn (SYF) demonstrated robust cortactin Y421 phosphorylation, expression of cortactin DCM with c-Src resulted in a complete lack of cortactin tyrosine 421 phosphorylation (Fig. 6A). Expression of cortactin DCM in MTLn3 cells resulted in diminished Y421 phosphorylation on par with the phosphorylation-null cortactin TYM construct (Fig. 6B), indicating that cortactin C112/C246 are essential for Src-mediated cortactin phosphorylation. We next analyzed the effect of cortactin DCM on cell migration and adhesion, two processes that involve Src activation and downstream cortactin tyrosine phosphorylation. Cortactin DCM expression in 1483 head and neck squamous cell carcinoma (HNSCC) cells with stable endogenous cortactin knockdown (supplementary material Fig. S7A) failed to rescue adhesion and migration to levels similar to control or WT cortactin, and were equivalent to knockdown (sh) and TYM expressing cells (Fig. 6C–D). Finally, we analyzed how cortactin DCM affects invadopodia formation and extracellular matrix degradation, processes dependent on Src and cortactin. Control (Ctl) OSC19 HNSCC cells spontaneously form numerous invadopodia that focally degrade labeled gelatin matrices. Invadopodia were completely absent in cortactin knockdown (sh) cells (Fig. 6E) as previously shown (Artym et al., 2006; Clark et al., 2007). Re-expression of WT cortactin rescued the knockdown phenotype, restoring matrix degradation to control levels. However, rescue of cortactin sh cells with cortactin TYM resulted in the formation of invadopodia structures with limited matrix degradation ability. These structures are likely pre-invadopodia as previously described (Kelley et al., 2010a; Oser et al., 2009). Rescue of cortactin sh cells with cortactin DCM failed to restore invadopodia/pre-invadopodia formation and gelatin degradation. These results indicated that the genesis of pre-invadopodia requires initial binding of Src to cortactin (Fig. 6E). The DCM protein correctly localizes within lamellipodia (Fig. 6E), suggesting that the C112/246A mutations do not deleteriously impact cortactin structure or alter proper cortactin subcellular localization. C112/246A does not significantly alter binding to F-actin as determined by F-actin co-sedimentation assays (supplementary material Fig. S7B,C). Collectively these data indicate that cystine-mediated Src binding is required for Src phosphorylation of cortactin and regulation of cellular events required for pro-motile and invasive activity.
Fig. 6.
Cortactin C112 and 246 are essential for phosphotyrosine-based cortactin function. (A) Analysis of Src-mediated cortactin Y421 phosphorylation in cortactin wild-type (WT) and double cysteine mutant (DCM) proteins following reintroduction of Src into SYF cells. Ratio indicates pY421 phosphorylation to total cortactin levels. (B) Analysis of cortactin WT, triple tyrosine mutant (TYM) and DCM tyrosine phosphorylation in MTLn3 cells. Asterisks show the position of the 80 kDa and 85 kDa cortactin forms. (C) Quantitation of ECIS cell adhesion assays (n = 3) in control (Ctl) and shRNA (sh) knockdown 1483 cells re-expressing the indicated FLAG-cortactin constructs. (D) Quantitation of ECIS cell migration assays (n = 3) in control (Ctl) and shRNA (sh) knockdown 1483 cells re-expressing the indicated FLAG-cortactin constructs. Bars in (C,D) represent residual standard error from one-way ANOVA without the intercept. ***, p≤0.001. (E) OSC19 cells transfected with the indicated FLAG-cortactin constructs were assayed for invadopodia formation and matrix degradation (n = 50 cells from three independent experiments). Bars represent residual standard error from one-way ANOVA without the intercept. ***, p≤0.001. Arrows indicate invadopodia and regions of degraded matrix. Scale bar = 20 mm.
Discussion
While SH2 domains have been shown to interact with ligands through phosphorylation-dependent and -independent mechanisms, binding to phosphotyrosine residues within target proteins is the predominant mode of interaction, having been characterized at the structural and thermodynamic levels for numerous ligand/domain pairs (Pawson, 2004; Schlessinger and Lemmon, 2003). SH2 domains are present in proteins that mediate most cellular functions and provide a wide combinatorial variety for selective intracellular signal transfer (Liu et al., 2006). The identification of a cystine-based, tyrosine phosphorylation-independent interaction between the phosphotyrosine binding interface of the Src SH2 domain with cortactin increases the potential ability for Src to interact with substrate ligands in a previously unrealized manner, expanding the repertoire and complexity of Src SH2 domain interactions in signal transduction.
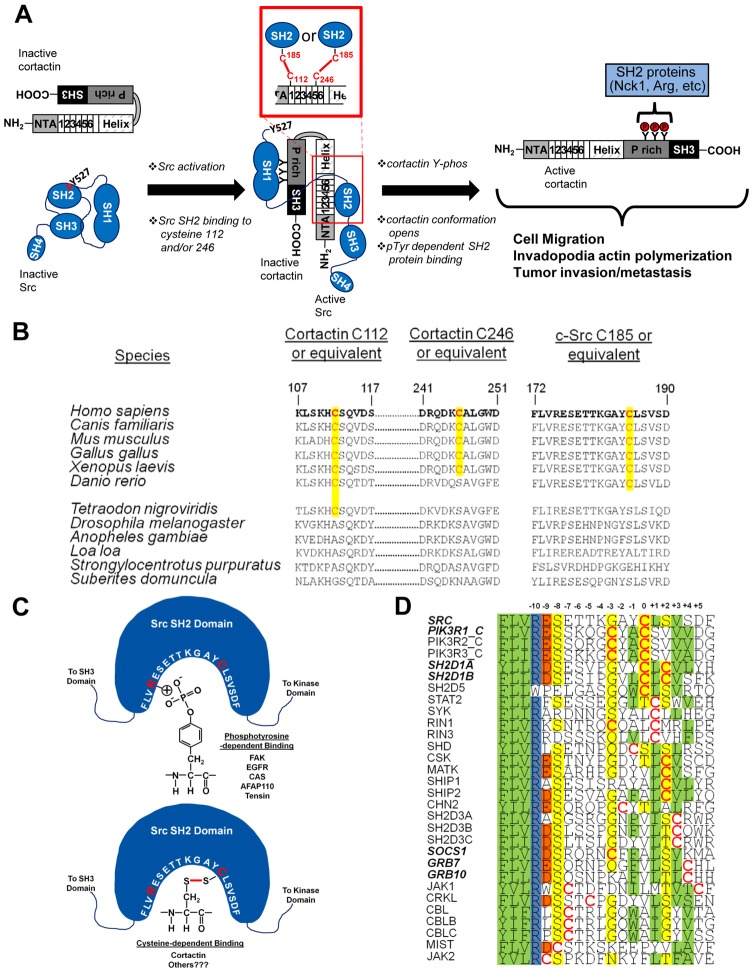
Evidence from our analysis of Src SH2 domain binding to cortactin indicates that Src C185 forms a cystine bond with C112 and/or C246 in the cortactin repeats region that is critical for mediating cortactin binding, tyrosine phosphorylation, and downstream cellular events. When bound to tyrosine phosphorylated ligands, C185 lies in close proximity to the phosphate group within the binding pocket, where the intrinsic repulsive nature of the deprotonated C185 facilitates release of pY527 from the SH2 domain, assisting in relieving the kinase from the autoinhibited state (Bradshaw et al., 1999). Our data provides an additional and alternative function for Src C185 in docking to cortactin, where cystine bonding to C112 or C246 mediates association with activated Src. Structural studies indicate cortactin exists as a partially globular protein, where the repeats are in a paraordered molten globule state that is highly dynamic in solution (Cowieson et al., 2008; Shvetsov et al., 2009). Biophysical and biochemical analysis indicates that the carboxyl terminus folds back onto the N-terminal region, suggesting that the protein resides in a “closed” conformation as supported by previously observed conformational isomers (Campbell et al., 1999; Huang et al., 1997). C112 and C246 can be crosslinked to each other, suggesting they are exposed and either lie in close proximity or are dynamically brought together (Cowieson et al., 2008; Shvetsov et al., 2009). The relatively unstructured nature of the cortactin repeats therefore provides the flexibility and accessibility of C112 and C246 to freely dock with the Src SH2 domain, allowing cystine bonding to occur. The nearness of the cortactin repeats region to the carboxyl terminal target tyrosine residues would allow for SH2-directed Src binding to the cortactin N-terminus, followed by subsequent processive phosphorylation of Y421/466/482 within the P-rich domain (Head et al., 2003). In conjunction with Erk1/2 phosphorylation at S405/418 (Campbell et al., 1999; Kelley et al., 2010b), cortactin is predicted to assume an open conformation, exposing the phosphotyrosine residues for association with SH2 containing adaptor proteins (Tehrani et al., 2007) or Abl-family kinases (Lapetina et al., 2009) (Fig. 7A). The net effect of Src-based phosphorylation would enhance actin dynamics through adaptor protein interactions or by maintaining activation of SH2-bound Abl or Arg, promoting phosphorylation of cortactin and neighboring target proteins (Lapetina et al., 2009; Oser et al., 2010). This model could be regulated by additional binding interactions and modifications that involve the cortactin repeats region. While mutation of the C112/246 Src docking sites does not alter F-actin binding, other modifications to the cortactin repeats region do modulate F-actin binding, including binding of phosphatidylinositol 4,5-bisphosphate (He et al., 1998), acetylation of the cortactin repeats (Zhang et al., 2007), and phosphorylation of S113 in the first repeat by PAK (Webb et al., 2006). Whether these events impact the association of Src with cortactin remains to be determined.
Fig. 7.
Model of cysteine-mediated interactions in cortactin regulation. (A) Model of cysteine-based cortactin activation and phosphorylation by Src. (B) Phylogenetic co-conservation of cortactin C112/246 and Src C185. Conserved cysteines are in red and the homologous positions highlighted in yellow. (C) Cartoon representation of Src SH2 binding to phosphotyrosine and cysteine residues. Src amino acids 172–191 within the SH2 domain binding pocket are in white; interacting arginine 175 and cysteine 185 residues are in red. Cystine bonding is indicated as a red line. Phosphotyrosine and cystine binding ligands are listed. (D) Alignment of cysteine-containing SH2 domains. Domains known to bind ligands in a phosphotyrosine-independent manner are in bold italics. Cysteine residues are in red. Shading: green; hydrophobic, blue; positively charged, red; negatively charged, yellow; polar.
Given the unprecedented nature of SH2 domains utilizing cystine bonding for binding ligands, we conducted a phylogenetic analysis of sequences containing cortactin C112/C246 and Src C185 in species ranging from Homo sapiens to Suberites domuncula (Fig. 7B). This analysis shows that the cortactin cysteine 112 equivalent appears in accord with the equivalent Src C185 residue in Danio rerio and is conserved throughout higher species. Cortactin C246 first appears in Xenopus laevis and is present in all higher organisms, collectively indicating co-conservation of the Src and cortactin cysteines in vertebrates.
There is mounting evidence to date that oxidative-based cystine and disulfide bonding occurs within and between multiple different cytoplasmic proteins during conditions of cytoplasmic oxidative stress as a mechanism that serves to regulate protein functionality (Cumming, 2009; Cumming et al., 2004; Cumming and Schubert, 2005). Src activity can be regulated by reactive oxygen species (ROS) (Sun and Kemble, 2009), which have a broad impact on multiple cellular processes that utilize oxidative signaling (Giannoni et al., 2010). Specifically, ROS-induced cysteine thiol oxidation of Src C245 in the SH2 domain and C487 in the kinase domain has been proposed to sustain Src activity by cystine bonding between these residues, maintaining Src in an open conformation to promote kinase activity (Giannoni et al., 2005). The proximity of Src C185 to cortactin C112/246 docked within the SH2 phosphotyrosine binding cleft provides the molecular setting that allows for a similar redox-mediated oxidation and consequential cystine bonding between the two proteins. Oxidative regulation of Src-cortactin binding is supported by increased Src-based cortactin phosphorylation in cells treated with hydrogen peroxide (Li et al., 2000). The requirement for cortactin C112/246 in tumor cell motility, adhesion, and invadopodia formation is in line with oxidative Src regulation utilized in these processes (Giannoni et al., 2010; Weaver, 2009). Furthermore, work in invadopodia has shown that localized ROS production generated by the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (Nox) system occurs through association of the invadopodial protein Tks5 with the Nox component p22phox (Diaz et al., 2009). Tks5 generated ROS promotes invadopodia formation and generates a feed forward loop through ROS-induced Src activity by suppression of the Src inactivating phosphatase PTP-PEST (Diaz et al., 2009) and potentially by sustaining cystine-based Src activity as described. ROS production in invadopodia would therefore facilitate oxidation of Src C185 and cortactin C112/246, promoting cystine bonding and maintenance of the Src-cortactin complex at sites of invadopodia formation. Since pre-invadopodia can form in the absence of cortactin phosphorylation (Oser et al., 2009), we speculate that the initial event for pre-invadopodia formation involves oxidative-based cystine binding of Src to cortactin. Once established, successive cycles of Src activity coupled with dynamic cortactin phosphorylation/dephosphorylation promotes establishment of SH2 protein-based actin regulatory complexes to propagate actin filament production and ECM matrix degradation (Kelley et al., 2010a; Oser et al., 2009). While recent work in MD-MBA-231 cells suggests that Arg, and not Src, is responsible for regulating cortactin phosphorylation during invadopodia formation (Mader et al., 2011), our data in HNSCC cells suggests that the tyrosine-independent binding event between Src and cortactin is the essential trigger for pre-invadopodia production. Determination of whether these differences are tumor specific remains to be resolved.
Cystine formation between Src and cortactin must be transient and reversible, since the Src SH2 domain does not stably interact with cortactin as opposed to other phospho-tyrosine ligands (Cobb et al., 1994; van Damme et al., 1997). Also, dynamic cycling of Src kinase activity is required for invadopodia maturation (Kelley et al., 2010b; Oser et al., 2009). Cytoplasmic glutathione likely plays a part in providing the reductive counterbalance to Tks5-generated ROS production, providing the necessary redox balance to generate and reduce thiolate anions at pH levels present in invadopodia (Cumming et al., 2004; Magalhaes et al., 2011). Another possibility is that Src and cortactin binding may be downregulated through the action of thiol reductases, although the subcellular localization of these enzymes has not been extensively evaluated.
Collectively our data indicates that the Src SH2 phosphotyrosine binding cleft is capable of phosphotyrosine and cystine dependent and independent interactions. The dual specificity of this SH2 subregion may be utilized to increase the selectivity between different classes of Src substrates (Fig. 7C) and may warrant reinvestigation of Src SH2-targeting compounds designed to react with C185 as alternative therapeutic strategy to the current class of kinase targeted Src inhibitors being evaluated as anti-cancer therapeutics in clinical trials (Charifson et al., 1997). Ablating C185 reactivity, while retaining phosphotyrosine binding, would selectively impair the Src-cortactin axis without impeding essential phosphorylation-based SH2 domain-ligand interactions. This rationale may be warranted in HNSCC, where Src activating epidermal growth factor receptor (EGFR) and cortactin are frequently amplified and overexpressed, corresponding with poor patient outcome (Ang et al., 2002; Rodrigo et al., 2000).
Database analysis indicates that Src is the only SH2 domain-containing cytoplasmic tyrosine kinase in its class that harbors a cysteine residue within the SH2 domain (Fig. 7D). However, there are thirty additional SH2 domains (25% of the total known number) that contain cysteine residues within the phosphotyrosine binding region in close proximity to the Src βC3 (Fig. 7D). While untested, this indicates that SH2 domains containing cysteine residues within this region may broadly utilize cystine bonding to select ligands. This alternate SH2 signaling mode may be commonplace in conditions of high intracellular ROS due to environmental stress (e.g.; heat, ionizing radiation, ultraviolet light), as well as in hypoxic tumors. Alternatively, cystine-based SH2 signaling may serve as a “backup” mode of preserving domain-ligand binding to ensure that essential survival signals are mediated in conditions of low ATP availability, where tyrosine kinase signaling might be compromised (i.e.; Warburg effect). While future investigation is required to examine these possibilities, our results demonstrating cystine bonding between Src and cortactin represent a new paradigm for SH2 domain function in mediating domain-based signal transduction.
Materials and Methods
Cell culture
MTLn3 cells were maintained in alpha-minimal essential medium (α-MEM) (Mediatech) with 10% FBS (Hyclone) in a 5% CO2 humidified atmosphere. SYF, 293T, 1483, and OSC19 cell lines were maintained in Dulbecco's modified Eagles medium (DMEM) (Mediatech) with 10% FBS.
Generation of plasmids
Cortactin truncation, Y421, and TYM mutants were previously described (Head et al., 2003; Weed et al., 2000). Tandem repeat chimeric mutants were generated by subcloning each individual repeat containing flanking BamH1 and Xho1 restriction sites with the Xho1/EcoR1 R6-CT fragment into BamH1/EcoR1 digested pcDNA3 FLAG-2AB. CFP-Src was produced by subcloning the Src containing Xho1/BamH1 fragment from GFP-Src (a gift from Margaret Frame) into pECFP-N1 (Clontech). FAK WT and Y397F were subcloned into FLAG-2AB using BamH1 and EcoR1 restriction sites. Site-directed mutagenesis of cortactin constructs was performed using the QuickChange II™ Site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's protocol.
Virus production and generation of stable cell lines
Stable knockdown of cortactin in 1483 and OSC19 cells was achieved by lentiviral transduction using short hairpin lentiviral constructs from Open Biosystems (1483: TRCN0000040275, OSC19: TRCN0000040275 and TRCN0000040273). Complete cortactin knockdown in OSC19 cells was achieved by subsequent transfection of cortactin-targeting siRNA (ON-TARGETplus SMARTpool catNO. L-010508-00-0020, Dharmacon).
Confocal microscopy and gelatin degradation assay
Immunofluorescent labeling, confocal microscopy, preparation of fluorescently-labeled gelatin coated coverslips, and ECM degradation assays were conducted as previously described (Ammer et al., 2009).
Peptide synthesis and SH2 domain array screening
Peptides were synthesized by Macromolecular Resources at Colorado State University. Screening of Transignal™ SH2 Domain Arrays (Panomics Cat. NO. MA3040) was conducted using 1.0 mg of each cortactin peptide according to the manufacturer's protocol.
Fusion protein purification
Purification of recombinant proteins were performed as described previously (Okamura and Resh, 1995).
Cell transfection, western blotting, and immunoprecipitation
Plasmid transfection, western blotting, and immunoprecipitation were performed as described (Rothschild et al., 2006).
Far western assays
Initially, cells transfected with FLAG-tagged cortactin plasmids were lysed and immunoprecipitated using EZ-View anti-FLAG resin (Invitrogen). Immune complexes were separated by SDS-PAGE, transferred to nitrocellulose membranes and proteins renatured in TBS-Tween containing 5% nonfat milk. Far western binding with Src SH2 domain constructs was conducted essentially as described (Shin et al., 2004), except 2 h incubations with 50–200 µg of fusion protein were used. In reduction experiments (Fig. 4D), 1 mM DTT was added to the incubation solution containing the GST-Src SH2 domain.
Affinity precipitation
SH2 domain precipitations were conducted as described (Okamura and Resh, 1995), except 100–200 µg of recombinant fusion protein and 1.0–1.5 mg of cell lysate were used for each assay.
Electric cell-substrate impedance sensing (ECIS)
ECIS was performed as described (Rothschild et al., 2006).
F-actin cosedimentation assay
F-actin cosedimentation assays were performed as described (Weed et al., 2000).
Statistical analysis
For standard errors (d-SE), variances were pooled using residual standard error from one-way ANOVA without the intercept. Delta-method approximation was used for all confidence bands (Fig. 6). Transformations performed in ANOVA used standard statistical model evaluation tools.
Molecular modeling
Docking studies were performed using the program eHiTS (SymBioSys Toronto, CA). Prior to docking, the hydrogens were added to the crystal structure PDBID: 1FMK (Xu et al., 1997) of the Src SH2 domain, solvated, and ions to neutralize the charge were added. The entire structure was then relaxed by running molecular dynamics (300 ps) using Amber (ver. 10) (Case et al., 2005). The cortactin peptides were built de novo in an extended conformation using Insight II (Accelrys, San Diego, CA) and each allowed to relax through 2500 steps of steepest descent molecular mechanics. The resulting structures were then docked. To obtain binding energies, molecular dynamics simulations were performed for 300 ps for: 1) each cortactin peptide in complex with the Src SH2 receptor, with docking results used as the starting structure; 2) the Src SH2 receptor alone and 3) each cortactin ligand alone. The resulting average potential energies calculated from the molecular dynamics simulation were then used to calculate the binding energy for each cortactin ligand to the SH2 receptor as the energy of the complex minus the energy of the Src SH2 receptor alone minus the energy of the cortactin ligand alone. Parameters for the phosphotyrosine were used (Homeyer et al., 2006).
Mass spectrometry
Purified GST-Src SH2 domain was incubated alone or mixed with a 10-fold molar excess of cortactin C112 peptide (SKHCSQV) or C246 peptide (QDKCALG) (Anaspec) in TBS (pH 7.2) for 2 h at room temperature. GST-Src SH2 samples (40 µg) were subsequently digested with trypsin (Promega) at a 1∶50 (wt/wt) ratio in 1 mM Tris, 150 mM NaCl (TBS), pH 7.2 overnight at 37°C. Digested samples were frozen at −80°C and lyophilized to remove solvents. Tryptic peptides were separated using an Acquity UPLC® System (Waters). Samples were separated using a reverse-phase Acquity BEH C18 1.7 mm, 1.0×50 mm column (Waters) directly coupled to a Waters Synapt® G2 HDMS mass spectrometer. Data were acquired using time of flight (TOF) MSE mode, which provides a comprehensive catalog of information for both precursor and fragment ions in a single analysis. A customized database was created using the GST-Src SH2 sequence, and the resulting precursor masses and MS/MS spectra were searched against the database using Biopharmalynx 3.1 (Waters). To evaluate cortactin C112 and C246 peptide binding to the Src SH2 domain, custom post-translational modifications were specified in Biopharmalynx based on monoisotopic masses for the C112 and C246 peptides binding at cysteine residues to Src C185 through disulfide bond formation. Extracted ion chromatograms (XIC) were obtained using MassLynx 2.2 (Waters).
Accession numbers for phylogenetic sequence analysis
Respective GenBank accession numbers for cortactin and Src are as follows: Homo sapiens AAH08799.1, NP_005408.1; Canis familiaris XP_851317.1, XP_865870.1; Mus musculus AAA19689.1, AAX90616.1; Gallus gallus Q01406.1, NP_990788.2; Xenopus laevis BAB79435.1, AAH45134.1; Danio rerio NP_001004121.1, AAI65380.1; Tetraodon nigroviridis CAF92908.1, CAG10364.1; Drosophila melanogaster NP_524426.2, NP_001189051.1; Anopheles gambiae XP_557457.3, XP_316537.2; Loa loa XP_003142854.1, EFO14749.1; Strongylocentrotus purpuratus AAD08655.1, ACI14304.1; Suberites domuncula CAC80140.1, AAT67598.1.
SH2 domain alignments
Twenty amino acids within the phosphotyrosine binding pocket of the Src SH2 domain beginning with the FLVRES sequence were aligned relative to Src C185, which was used as the zero residue reference point. SH2 domain sequence data were obtained from the University of Chicago Nash Laboratory SH2 domain database (http://sh2.uchicago.edu/clustalalignment.html).
Antibodies
Antibodies for immunoblotting were as follows: anti-cortactin (4F11), 1∶1000; anti-pY421 cortactin (Invitrogen), 1∶1000; anti-FLAG and anti-GST (Millipore), 1∶1000; anti-GFP (JL8; Clontech), 1∶1000; anti-phosphotyrosine (BD Transduction), 1∶1000. Antibodies for immunofluorescent labeling were used as described (Ammer et al., 2009).
Supplementary Material
Acknowledgments
We thank Mike Schaller, members of the Weed laboratory and MBR Cancer Center for advice and suggestions, M. Auble for early experimental help, and B. Robke for peptide synthesis. Imaging experiments were performed in the West Virginia University Microscope Imaging Facility. The West Virginia University Microscope Imaging Facility is supported in part by the Mary Babb Randolph Cancer Center and the National Institutes of Health [grant number P30 RR032138/GM103488].
Footnotes
Funding
This work was supported by National Institutes of Health [grant numbers DE014364, DE014578 and RR16440 to S.A.W.]; the Mary Babb Randolph Cancer Center; and by the West Virginia University Department of Neurobiology and Anatomy. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.121046/-/DC1
References
- Ammer A. G., Weed S. A. (2008). Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton 65, 687–707 10.1002/cm.20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammer A. G., Kelley L. C., Hayes K. E., Evans J. V., Lopez–Skinner L. A., Martin K. H., Frederick B., Rothschild B. L., Raben D., Elvin P.et al. (2009). Saracatinib impairs head and neck squamous cell carcinoma invasion by disrupting invadopodia function. J. Cancer Sci. Ther. 1, 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang K. K., Berkey B. A., Tu X., Zhang H. Z., Katz R., Hammond E. H., Fu K. K., Milas L. (2002). Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 62, 7350–7356 [PubMed] [Google Scholar]
- Artym V. V., Zhang Y., Seillier–Moiseiwitsch F., Yamada K. M., Mueller S. C. (2006). Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66, 3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- Bae J. H., Lew E. D., Yuzawa S., Tomé F., Lax I., Schlessinger J. (2009). The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 138, 514–524 10.1016/j.cell.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougnères L., Girardin S. E., Weed S. A., Karginov A. V., Olivo–Marin J. C., Parsons J. T., Sansonetti P. J., Van Nhieu G. T. (2004). Cortactin and Crk cooperate to trigger actin polymerization during Shigella invasion of epithelial cells. J. Cell Biol. 166, 225–235 10.1083/jcb.200402073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S. N., Michaud G. A., Schweitzer B., Predki P. F., Koleske A. J. (2007). A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr. Biol. 17, 445–451 10.1016/j.cub.2007.01.057 [DOI] [PubMed] [Google Scholar]
- Bradshaw J. M., Mitaxov V., Waksman G. (1999). Investigation of phosphotyrosine recognition by the SH2 domain of the Src kinase. J. Mol. Biol. 293, 971–985 10.1006/jmbi.1999.3190 [DOI] [PubMed] [Google Scholar]
- Brown M. T., Cooper J. A. (1996). Regulation, substrates and functions of src. Biochim. Biophys. Acta 1287, 121–149 [DOI] [PubMed] [Google Scholar]
- Burnham M. R., Bruce–Staskal P. J., Harte M. T., Weidow C. L., Ma A., Weed S. A., Bouton A. H. (2000). Regulation of c-SRC activity and function by the adapter protein CAS. Mol. Cell. Biol. 20, 5865–5878 10.1128/MCB.20.16.5865-5878.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. H., Sutherland R. L., Daly R. J. (1999). Signaling pathways and structural domains required for phosphorylation of EMS1/cortactin. Cancer Res. 59, 5376–5385 [PubMed] [Google Scholar]
- Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr, Onufriev A., Simmerling C., Wang B., Woods R. J. (2005). The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charifson P. S., Shewchuk L. M., Rocque W., Hummel C. W., Jordan S. R., Mohr C., Pacofsky G. J., Peel M. R., Rodriguez M., Sternbach D. D.et al. (1997). Peptide ligands of pp60(c-src) SH2 domains: a thermodynamic and structural study. Biochemistry 36, 6283–6293 10.1021/bi970019n [DOI] [PubMed] [Google Scholar]
- Clark E. S., Whigham A. S., Yarbrough W. G., Weaver A. M. (2007). Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67, 4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- Cobb B. S., Schaller M. D., Leu T. H., Parsons J. T. (1994). Stable association of pp60src and pp59fyn with the focal adhesion-associated protein tyrosine kinase, pp125FAK. Mol. Cell. Biol. 14, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosen–Binker L. I., Kapus A. (2006). Cortactin: the gray eminence of the cytoskeleton. Physiology (Bethesda) 21, 352–361 10.1152/physiol.00012.2006 [DOI] [PubMed] [Google Scholar]
- Cowieson N. P., King G., Cookson D., Ross I., Huber T., Hume D. A., Kobe B., Martin J. L. (2008). Cortactin adopts a globular conformation and bundles actin into sheets. J. Biol. Chem. 283, 16187–16193 10.1074/jbc.M708917200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming R. C. (2009). Analysis of global and specific changes in the disulfide proteome using redox two-dimensional polyacrylamide gel electrophoresis. Methods Mol. Biol. 476, 160–174 10.1007/978-1-59745-129-1_12 [DOI] [PubMed] [Google Scholar]
- Cumming R. C., Schubert D. (2005). Amyloid-beta induces disulfide bonding and aggregation of GAPDH in Alzheimer's disease. FASEB J. 19, 2060–2062 [DOI] [PubMed] [Google Scholar]
- Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., Schubert D. (2004). Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem. 279, 21749–21758 10.1074/jbc.M312267200 [DOI] [PubMed] [Google Scholar]
- Daly R. J. (2004). Cortactin signalling and dynamic actin networks. Biochem. J. 382, 13–25 10.1042/BJ20040737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz B., Shani G., Pass I., Anderson D., Quintavalle M., Courtneidge S. A. (2009). Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2, ra53 10.1126/scisignal.2000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsberger B., Stewart B., Tatarov O., Edwards J. (2010). Is Src a viable target for treating solid tumours? Curr. Cancer Drug Targets 10, 683–694 10.2174/156800910793605802 [DOI] [PubMed] [Google Scholar]
- Giannoni E., Buricchi F., Raugei G., Ramponi G., Chiarugi P. (2005). Intracellular reactive oxygen species activate Src tyrosine kinase during cell adhesion and anchorage-dependent cell growth. Mol. Cell. Biol. 25, 6391–6403 10.1128/MCB.25.15.6391-6403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni E., Taddei M. L., Chiarugi P. (2010). Src redox regulation: again in the front line. Free Radic. Biol. Med. 49, 516–527 10.1016/j.freeradbiomed.2010.04.025 [DOI] [PubMed] [Google Scholar]
- Guarino M. (2010). Src signaling in cancer invasion. J. Cell. Physiol. 223, 14–26 [DOI] [PubMed] [Google Scholar]
- He H., Watanabe T., Zhan X., Huang C., Schuuring E., Fukami K., Takenawa T., Kumar C. C., Simpson R. J., Maruta H. (1998). Role of phosphatidylinositol 4,5-bisphosphate in Ras/Rac-induced disruption of the cortactin-actomyosin II complex and malignant transformation. Mol. Cell. Biol. 18, 3829–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head J. A., Jiang D., Li M., Zorn L. J., Schaefer E. M., Parsons J. T., Weed S. A. (2003). Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14, 3216–3229 10.1091/mbc.E02-11-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer N., Horn A. H., Lanig H., Sticht H. (2006). AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 12, 281–289 10.1007/s00894-005-0028-4 [DOI] [PubMed] [Google Scholar]
- Huang C., Ni Y., Wang T., Gao Y., Haudenschild C. C., Zhan X. (1997). Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J. Biol. Chem. 272, 13911–13915 10.1074/jbc.272.21.13911 [DOI] [PubMed] [Google Scholar]
- Huang C., Liu J., Haudenschild C. C., Zhan X. (1998). The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 273, 25770–25776 10.1074/jbc.273.40.25770 [DOI] [PubMed] [Google Scholar]
- Huang J., Asawa T., Takato T., Sakai R. (2003). Cooperative roles of Fyn and cortactin in cell migration of metastatic murine melanoma. J. Biol. Chem. 278, 48367–48376 10.1074/jbc.M308213200 [DOI] [PubMed] [Google Scholar]
- Kelley L. C., Ammer A. G., Hayes K. E., Martin K. H., Machida K., Jia L., Mayer B. J., Weed S. A. (2010a). Oncogenic Src requires a wild-type counterpart to regulate invadopodia maturation. J. Cell Sci. 123, 3923–3932 10.1242/jcs.075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. C., Hayes K. E., Ammer A. G., Martin K. H., Weed S. A. (2010b). Cortactin phosphorylated by ERK1/2 localizes to sites of dynamic actin regulation and is required for carcinoma lamellipodia persistence. PLoS ONE 5, e13847 10.1371/journal.pone.0013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. R., Fang Y., Mahon E. S., Anderson D. H. (2000). Using a phage display library to identify basic residues in A-Raf required to mediate binding to the Src homology 2 domains of the p85 subunit of phosphatidylinositol 3′-kinase. J. Biol. Chem. 275, 36450–36456 10.1074/jbc.M004720200 [DOI] [PubMed] [Google Scholar]
- Kirkbride K. C., Sung B. H., Sinha S., Weaver A. M. (2011). Cortactin: A multifunctional regulator of cellular invasiveness. Cell Adh. Migr. 5, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. A., Anderson D., Moran M. F., Ellis C., Pawson T. (1991). SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 252, 668–674 10.1126/science.1708916 [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Cowburn D. (1997). Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 26, 259–288 10.1146/annurev.biophys.26.1.259 [DOI] [PubMed] [Google Scholar]
- Lapetina S., Mader C. C., Machida K., Mayer B. J., Koleske A. J. (2009). Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J. Cell Biol. 185, 503–519 10.1083/jcb.200809085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu J., Zhan X. (2000). Tyrosine phosphorylation of cortactin is required for H2O2-mediated injury of human endothelial cells. J. Biol. Chem. 275, 37187–37193 10.1074/jbc.M005301200 [DOI] [PubMed] [Google Scholar]
- Liao Y. C., Si L., deVere White R. W., Lo S. H. (2007). The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 176, 43–49 10.1083/jcb.200608015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. A., Jablonowski K., Raina M., Arcé M., Pawson T., Nash P. D. (2006). The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol. Cell 22, 851–868 10.1016/j.molcel.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Machida K., Thompson C. M., Dierck K., Jablonowski K., Kärkkäinen S., Liu B., Zhang H., Nash P. D., Newman D. K., Nollau P.et al. (2007). High-throughput phosphotyrosine profiling using SH2 domains. Mol. Cell 26, 899–915 10.1016/j.molcel.2007.05.031 [DOI] [PubMed] [Google Scholar]
- Mader C. C., Oser M., Magalhaes M. A., Bravo–Cordero J. J., Condeelis J., Koleske A. J., Gil–Henn H. (2011). An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71, 1730–1741 10.1158/0008-5472.CAN-10-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes M. A., Larson D. R., Mader C. C., Bravo–Cordero J. J., Gil–Henn H., Oser M., Chen X., Koleske A. J., Condeelis J. (2011). Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195, 903–920 10.1083/jcb.201103045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Knapp S. (2009). Out of the box binding determines specificity of SH2 domain interaction. Structure 17, 1040–1041 10.1016/j.str.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Nakagawa H. (1989). A protein tyrosine kinase involved in regulation of pp60c-src function. J. Biol. Chem. 264, 20886–20893 [PubMed] [Google Scholar]
- Okamura H., Resh M. D. (1995). p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J. Biol. Chem. 270, 26613–26618 10.1074/jbc.270.44.26613 [DOI] [PubMed] [Google Scholar]
- Oser M., Yamaguchi H., Mader C. C., Bravo–Cordero J. J., Arias M., Chen X., Desmarais V., van Rheenen J., Koleske A. J., Condeelis J. (2009). Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol. 186, 571–587 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M., Mader C. C., Gil–Henn H., Magalhaes M., Bravo–Cordero J. J., Koleske A. J., Condeelis J. (2010). Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123, 3662–3673 10.1242/jcs.068163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. (2004). Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 10.1016/S0092-8674(03)01077-8 [DOI] [PubMed] [Google Scholar]
- Pawson T., Gish G. D., Nash P. (2001). SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11, 504–511 10.1016/S0962-8924(01)02154-7 [DOI] [PubMed] [Google Scholar]
- Rodrigo J. P., García L. A., Ramos S., Lazo P. S., Suárez C. (2000). EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin. Cancer Res. 6, 3177–3182 [PubMed] [Google Scholar]
- Rothschild B. L., Shim A. H., Ammer A. G., Kelley L. C., Irby K. B., Head J. A., Chen L., Varella–Garcia M., Sacks P. G., Frederick B.et al. (2006). Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 66, 8017–8025 10.1158/0008-5472.CAN-05-4490 [DOI] [PubMed] [Google Scholar]
- Schaller M. D., Hildebrand J. D., Shannon J. D., Fox J. W., Vines R. R., Parsons J. T. (1994). Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 14, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J., Lemmon M. A. (2003). SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12 10.1126/stke.2003.191.re12 [DOI] [PubMed] [Google Scholar]
- Shin N. Y., Dise R. S., Schneider–Mergener J., Ritchie M. D., Kilkenny D. M., Hanks S. K. (2004). Subsets of the major tyrosine phosphorylation sites in Crk-associated substrate (CAS) are sufficient to promote cell migration. J. Biol. Chem. 279, 38331–38337 10.1074/jbc.M404675200 [DOI] [PubMed] [Google Scholar]
- Shvetsov A., Berkane E., Chereau D., Dominguez R., Reisler E. (2009). The actin-binding domain of cortactin is dynamic and unstructured and affects lateral and longitudinal contacts in F-actin. Cell Motil. Cytoskeleton 66, 90–98 10.1002/cm.20328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J.et al. (1993). SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 10.1016/0092-8674(93)90404-E [DOI] [PubMed] [Google Scholar]
- Sun G., Kemble D. J. (2009). To C or not to C: direct and indirect redox regulation of Src protein tyrosine kinase. Cell Cycle 8, 2353–2355 10.4161/cc.8.15.9225 [DOI] [PubMed] [Google Scholar]
- Tehrani S., Tomasevic N., Weed S., Sakowicz R., Cooper J. A. (2007). Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. USA 104, 11933–11938 10.1073/pnas.0701077104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme H., Brok H., Schuuring–Scholtes E., Schuuring E. (1997). The redistribution of cortactin into cell-matrix contact sites in human carcinoma cells with 11q13 amplification is associated with both overexpression and post-translational modification. J. Biol. Chem. 272, 7374–7380 10.1074/jbc.272.11.7374 [DOI] [PubMed] [Google Scholar]
- Van Wart H. E., Lewis A., Scheraga H. A., Saeva F. D. (1973). Disulfide bond dihedral angles from Raman spectroscopy. Proc. Natl. Acad. Sci. USA 70, 2619–2623 10.1073/pnas.70.9.2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G., Kominos D., Robertson S. C., Pant N., Baltimore D., Birge R. B., Cowburn D., Hanafusa H., Mayer B. J., Overduin M.et al. (1992). Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature 358, 646–653 10.1038/358646a0 [DOI] [PubMed] [Google Scholar]
- Weaver A. M. (2009). Regulation of cancer invasion by reactive oxygen species and Tks family scaffold proteins. Sci. Signal. 2, pe56 10.1126/scisignal.288pe56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. (2001). Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 10.1016/S0960-9822(01)00098-7 [DOI] [PubMed] [Google Scholar]
- Webb B. A., Zhou S., Eves R., Shen L., Jia L., Mak A. S. (2006). Phosphorylation of cortactin by p21-activated kinase. Arch. Biochem. Biophys. 456, 183–193 10.1016/j.abb.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000). Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151, 29–40 10.1083/jcb.151.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Harrison S. C., Eck M. J. (1997). Three-dimensional structure of the tyrosine kinase c-Src. Nature 385, 595–602 10.1038/385595a0 [DOI] [PubMed] [Google Scholar]
- Xu W., Doshi A., Lei M., Eck M. J., Harrison S. C. (1999). Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell 3, 629–638 10.1016/S1097-2765(00)80356-1 [DOI] [PubMed] [Google Scholar]
- Yaffe M. B. (2002). Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 3, 177–186 10.1038/nrm759 [DOI] [PubMed] [Google Scholar]
- Yeatman T. J. (2004). A renaissance for SRC. Nat. Rev. Cancer 4, 470–480 10.1038/nrc1366 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yuan Z., Zhang Y., Yong S., Salas–Burgos A., Koomen J., Olashaw N., Parsons J. T., Yang X. J., Dent S. R.et al. (2007). HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213 10.1016/j.molcel.2007.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.