Abstract
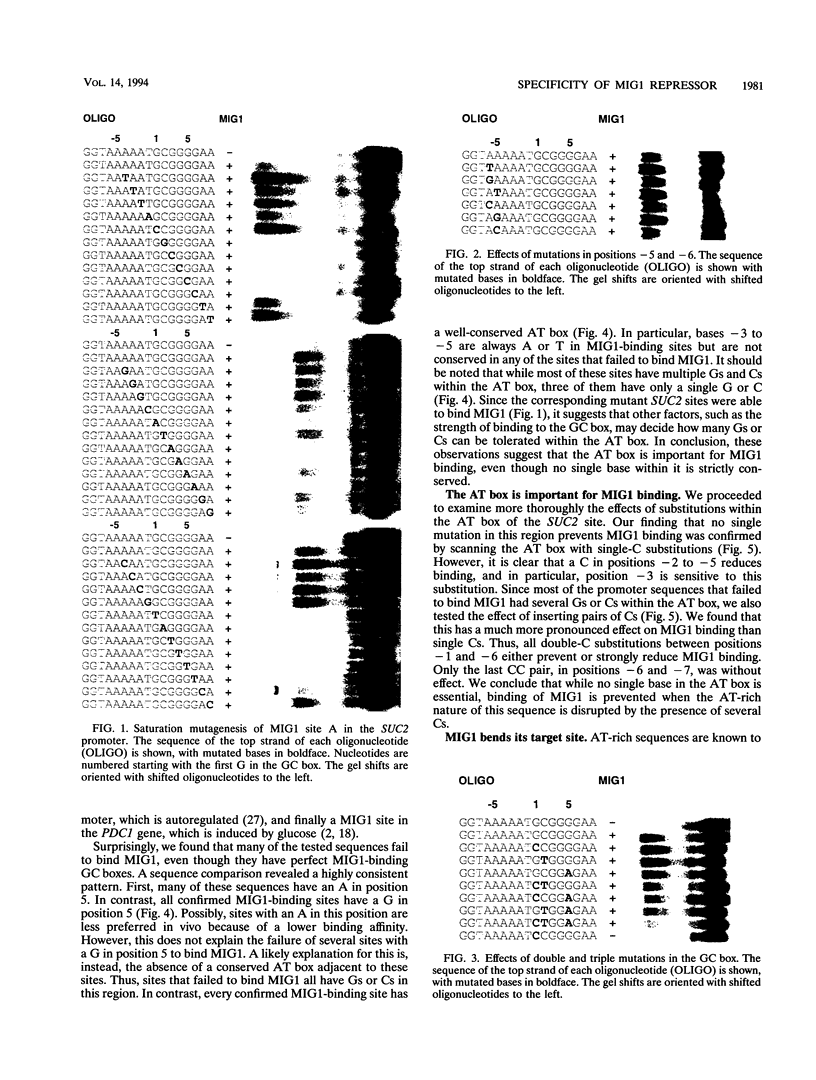
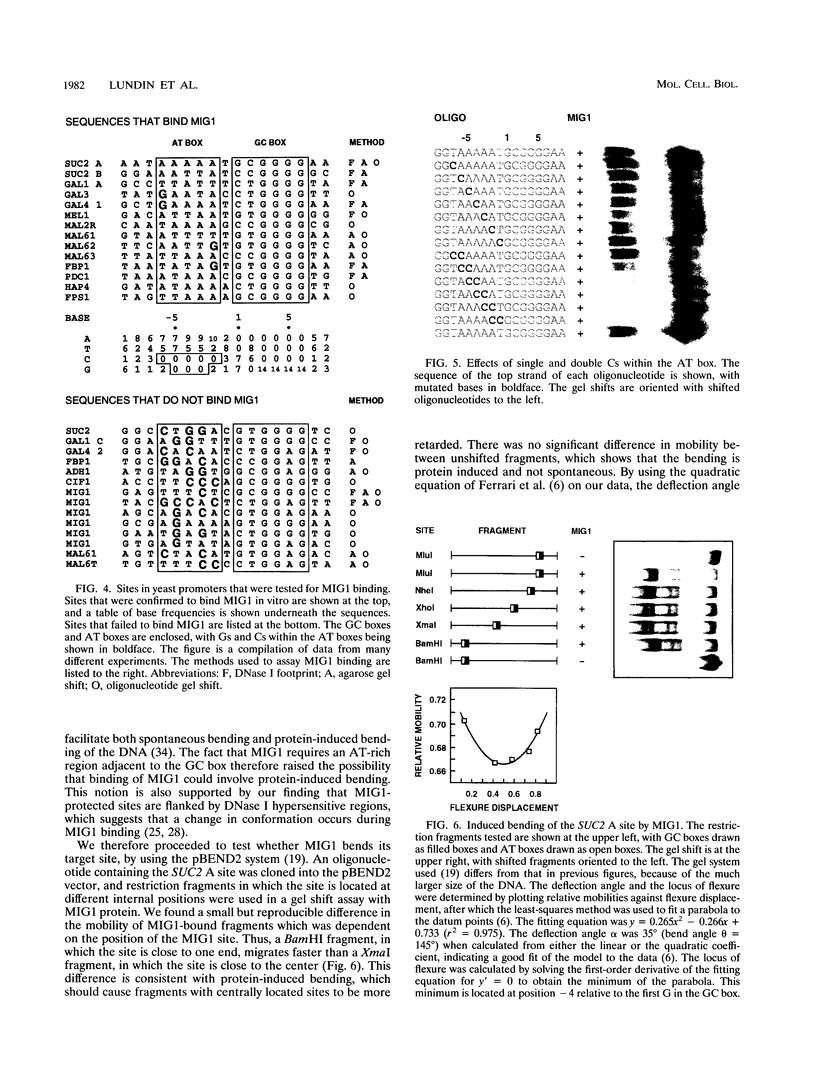
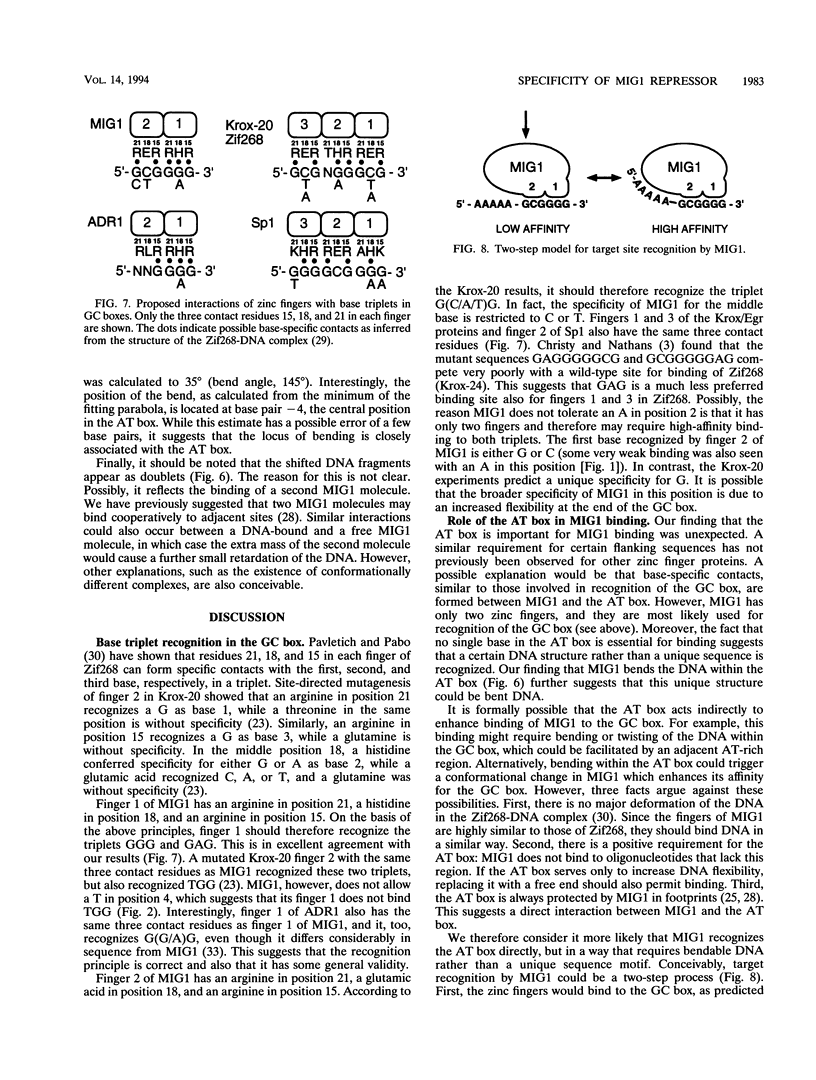
MIG1 is a zinc finger protein that mediates glucose repression in the yeast Saccharomyces cerevisiae. MIG1 is related to the mammalian Krox/Egr, Wilms' tumor, and Sp1 finger proteins. It has two fingers and binds to a GCGGGG motif that resembles the GC boxes recognized by these mammalian proteins. We have performed a complete saturation mutagenesis of a natural MIG1 site in order to elucidate its binding specificity. We found that only three mutations within the GC box retain the ability to bind MIG1: G1 to C, C2 to T, and G5 to A. This result is consistent with current models for zinc finger-DNA binding, which assume that the sequence specificity is determined by base triplet recognition within the GC box. Surprisingly, we found that an AT-rich region 5' to the GC box also is important for MIG1 binding. This AT box is present in all natural MIG1 sites, and it is protected by MIG1 in DNase I footprints. However, the AT box differs from the GC box in that no single base within it is essential for binding. Instead, the AT-rich nature of this sequence seems to be crucial. The fact that AT-rich sequences are known to increase DNA flexibility prompted us to test whether MIG1 bends DNA. We found that binding of MIG1 is associated with bending within the AT box. We conclude that DNA binding by a simple zinc finger protein such as MIG1 can involve both recognition of the GC box and flanking sequence preferences that may reflect local DNA bendability.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., McConnell D. J. Identification of an upstream activation site in the pyruvate decarboxylase structural gene (PDC1) of Saccharomyces cerevisiae. Curr Genet. 1988 Nov;14(5):405–412. doi: 10.1007/BF00521261. [DOI] [PubMed] [Google Scholar]
- Christy B., Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991 Nov;11(11):5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Analysis of URSG-mediated glucose repression of the GAL1 promoter of Saccharomyces cerevisiae. Genetics. 1992 Feb;130(2):295–304. doi: 10.1093/genetics/130.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gancedo J. M. Carbon catabolite repression in yeast. Eur J Biochem. 1992 Jun 1;206(2):297–313. doi: 10.1111/j.1432-1033.1992.tb16928.x. [DOI] [PubMed] [Google Scholar]
- Gogos J. A., Hsu T., Bolton J., Kafatos F. C. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992 Sep 25;257(5078):1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- González M. I., Stucka R., Blázquez M. A., Feldmann H., Gancedo C. Molecular cloning of CIF1, a yeast gene necessary for growth on glucose. Yeast. 1992 Mar;8(3):183–192. doi: 10.1002/yea.320080304. [DOI] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber D. A., Buckler A. J. WT1: a novel tumor suppressor gene inactivated in Wilms' tumor. New Biol. 1992 Feb;4(2):97–106. [PubMed] [Google Scholar]
- Herschman H. R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Jacobs G. H. Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J. 1992 Dec;11(12):4507–4517. doi: 10.1002/j.1460-2075.1992.tb05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kellermann E., Hollenberg C. P. The glucose-and ethanol-dependent regulation of PDC1 from Saccharomyces cerevisiae are controlled by two distinct promoter regions. Curr Genet. 1988 Oct;14(4):337–344. doi: 10.1007/BF00419991. [DOI] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Kulmburg P., Mathieu M., Dowzer C., Kelly J., Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993 Mar;7(6):847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- Mercado J. J., Vincent O., Gancedo J. M. Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 1991 Oct 7;291(1):97–100. doi: 10.1016/0014-5793(91)81112-l. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T., Charnay P. Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res. 1992 Aug 25;20(16):4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992 Oct 25;20(20):5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Cook G. R., Bradbury E. M., Gottesfeld J. M. Transcription factor IIIA induced bending of the Xenopus somatic 5S gene promoter. Nature. 1989 Aug 10;340(6233):487–488. doi: 10.1038/340487a0. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Entian K. D. Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol. 1991 Mar;173(6):2045–2052. doi: 10.1128/jb.173.6.2045-2052.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Mutations in the zinc fingers of ADR1 that change the specificity of DNA binding and transactivation. Mol Cell Biol. 1992 Jun;12(6):2784–2792. doi: 10.1128/mcb.12.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Trumbly R. J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992 Jan;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- el-Baradi T., Pieler T. Zinc finger proteins: what we know and what we would like to know. Mech Dev. 1991 Nov;35(3):155–169. doi: 10.1016/0925-4773(91)90015-x. [DOI] [PubMed] [Google Scholar]