An improved synthetic reporter monitors cytokinin signaling in planta.
Abstract
Cytokinins are classic plant hormones that orchestrate plant growth, development, and physiology. They affect gene expression in target cells by activating a multistep phosphorelay network. Type-B response regulators, acting as transcriptional activators, mediate the final step in the signaling cascade. Previously, we have introduced a synthetic reporter, Two Component signaling Sensor (TCS)::green fluorescent protein (GFP), which reflects the transcriptional activity of type-B response regulators. TCS::GFP was instrumental in uncovering roles of cytokinin and deepening our understanding of existing functions. However, TCS-mediated expression of reporters is weak in some developmental contexts where cytokinin signaling has a documented role, such as in the shoot apical meristem or in the vasculature of Arabidopsis (Arabidopsis thaliana). We also observed that GFP expression becomes rapidly silenced in TCS::GFP transgenic plants. Here, we present an improved version of the reporter, TCS new (TCSn), which, compared with TCS, is more sensitive to phosphorelay signaling in Arabidopsis and maize (Zea mays) cellular assays while retaining its specificity. Transgenic Arabidopsis TCSn::GFP plants exhibit strong and dynamic GFP expression patterns consistent with known cytokinin functions. In addition, GFP expression has been stable over generations, allowing for crosses with different genetic backgrounds. Thus, TCSn represents a significant improvement to report the transcriptional output profile of phosphorelay signaling networks in Arabidopsis, maize, and likely other plants that display common response regulator DNA-binding specificities.
The plant hormone cytokinin comprises a class of small, adenine-derived organic molecules that influence plant development and physiology in diverse contexts throughout the plant life cycle. Cytokinins initiate a multistep phosphorelay (MSP) signaling cascade by binding to and activating the cognate receptors, hybrid kinases with a cyclases/His kinases-associated sensory extracellular ligand-binding domain (Anantharaman and Aravind, 2001; Mougel and Zhulin, 2001). In Arabidopsis (Arabidopsis thaliana), these are encoded by the ARABIDOPSIS HIS KINASE2 (AHK2), AHK3, and AHK4 genes. Ligand binding triggers autophosphorylation at a conserved His residue in the receiver domain and subsequent transfer of the phosphoryl group to a conserved Asp residue in the attached transmitter domain. Besides the cytokinin receptors, eight other hybrid kinases are encoded by the Arabidopsis genome, including CYTOKININ INDEPENDENT1 (CKI1), which can potentially activate the MSP signaling network. From the Asp of the hybrid kinase, the phosphoryl group is passed on to one of five ARABIDOPSIS HIS PHOSPHOTRANSFER proteins and then to a nuclear ARABIDOPSIS RESPONSE REGULATOR (ARR), of which there are type-A, type-B, and type-C. Members of the type-B class bind to promoters of target genes via their Myb-like DNA-binding domain and activate transcription, while type-A and type-C ARRs inhibit signaling activity. At the same time, type-A ARRs are immediate-early target genes of activated type-B ARR proteins, which establishes a negative feedback loop to the signaling pathway (Werner and Schmülling, 2009; Argueso et al., 2010; Perilli et al., 2010; Bishopp et al., 2011a; Hwang et al., 2012).
Despite the apparent simplicity of the MSP signaling mechanism, the precise identification and functional characterization of the diverse signaling locales poses several challenges. First, the distribution of active cytokinin ligands in planta is difficult to determine. Cytokinins are produced by complex enzymatic biosynthetic pathways in different cellular compartments and are subject to long- and short-range transport and degradation (Werner et al., 2006; Hirose et al., 2008; Bishopp et al., 2011b). Although distribution patterns of cytokinins using monoclonal antibodies have been reported (Aloni et al., 2004, 2005), the available antibodies detect only a subset of active cytokinins, as well as inactive precursor forms (Eberle et al., 1986). Besides AHK2, AHK3, and AHK4, cytokinin-independent hybrid kinases, in particular CKI1 (Pischke et al., 2002; Hejátko et al., 2003, 2009; Deng et al., 2010), but potentially also the ethylene receptor ETR1 (Cho and Yoo, 2007; Hall et al., 2012) or AHK5 (Mira-Rodado et al., 2012; Pham et al., 2012), can activate the MSP network. The use of mutants is complicated because of redundantly acting signaling components, which require the generation of higher order mutants. For many gene families, these are difficult or impractical to generate due to the high number of genes involved, the lack of null mutants, or the close linkage of loci. Moreover, phenotypes caused by a loss of signaling are often pleiotropic or cause early lethality, which can mask functions of interest. In contrast to these difficulties, visualizing the transcriptional MSP output with a synthetic reporter reveals the sites of action during wild-type development. This information then allows focusing on the specific context for functional analyses, such as applying targeted genetic approaches or chemical and pharmacological treatments, and tracking the immediate consequences on the signaling output. The Myb-like DNA-binding domain of the 11 different type-B ARR family members is conserved, in particular, in the nine residues that were shown to make direct DNA contact (Hosoda et al., 2002). Accordingly, in vitro binding studies with the DNA-binding domains of different type-B ARRs identified very similar binding specificities, with the consensus sequence 5′-(A/G)GAT(C/T)-3′ (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003). This apparent similarity in the DNA binding specificity of the different type-B ARR family members was exploited to design a specific synthetic sensor, Two Component signaling Sensor (TCS), which is based on concatemeric 5′-(A/G)GAT(C/T)-3′ binding sites (Fig. 1, A–D; Müller and Sheen, 2008). In transgenic TCS::GFP plants, the GFP signal reflecting the signaling output pattern has facilitated describing novel cytokinin functions (Müller and Sheen, 2008; Bencivenga et al., 2012; Marsch-Martínez et al., 2012), as well as refining and deepening the understanding of existing cytokinin functions (Leibfried et al., 2005; Gordon et al., 2009; Zhao et al., 2010; Bielach et al., 2012; Chickarmane et al., 2012; Murray et al., 2012). Despite the documented value of TCS-controlled reporters, some limitations emerged, which motivated us to construct an improved version. First, TCS-induced expression is weak in certain developmental contexts where cytokinin signaling has a documented role, such as in the embryo sac (Pischke et al., 2002; Hejátko et al., 2003; Deng et al., 2010; Bencivenga et al., 2012), in the shoot (Gordon et al., 2009; Zhao et al., 2010; Chickarmane et al., 2012), and in the vasculature (Mähönen et al., 2000, 2006a, 2006b; Dello Ioio et al., 2008). Second, we observed that GFP expression becomes progressively reduced with increasing generations, such as in the root meristem of the seedling (Fig. 1B), presumably due to silencing effects triggered by the monotony of the repetitive sequence in TCS (Chan et al., 2005). Here, we present a superior version, TCS new (TCSn)::GFP, which exhibits higher sensitivity to cytokinin and MSP components in transient transfection assays (Fig. 2) and a much brighter GFP signal in most tissues analyzed (Figs. 3 and 4). Thus, the TCSn::GFP expression pattern reveals aspects of the MSP output that were not reported by TCS::GFP. Furthermore, GFP expression has been stable during propagation, indicating that unlike TCS::GFP, it does not easily get silenced.
Figure 1.
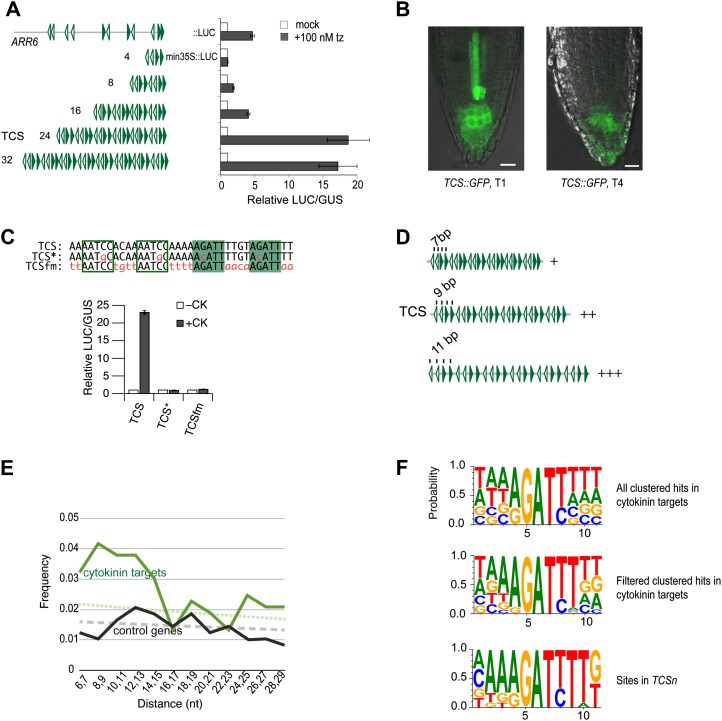
Optimization of TCS. A, A concatemer of 24 repeats of the 5′-(A/G)GAT(C/T)T-3′ binding motif caused the strongest cytokinin-dependent induction of a LUC reporter in transient transfection assays. B, Reduced GFP signal in the primary root meristem of a 5-d-old transgenic TCS::GFP seedling in the fourth generation (T4) compared with a primary transformant (T1). C, Similar to mutating nucleotides essential for in vitro binding of type-B ARRs (TCS*::LUC, where the asterisk indicates point mutation G→C), the mutation of flanking nucleotides (TCSfm::LUC) abolished cytokinin-dependent response of TCS::LUC. D, Scheme representing the quantitative effects of different phasings of core 5′-(A/G)GAT(C/T)T-3′ motifs. Phasing of 11 bp results in strongest reporter gene expression. E, The frequency of 5′-(A/G)GAT(C/T)T-3′ motifs with a distance of 7 to 15 bp in cytokinin target genes (top curve) is higher than expected (dotted line), while the same motifs are not significantly higher than expected in control genes (lower curve and dashed line). F, Sequence logos (Crooks et al., 2004) generated from the alignment of clustered 5′-(A/G)GAT(C/T)T-3′ motifs in cytokinin target genes as listed in Supplemental Table S2 (top) after filtering with 5′-A(A/G)GAT(C/T)T-3′ and 5′-A (A/G)GAT(C/T)TT-3′ (middle), and the alignment based on the 12 motifs used to construct TCSn as listed in Supplemental Table S4 (bottom). Filled or empty arrowheads (A and D) or boxes (C) indicate 5′-A (A/G)GAT(C/T)TT-3′ motifs on the forward or reverse DNA strand, respectively. Bars = 20 µm. [See online article for color version of this figure.]
Figure 2.
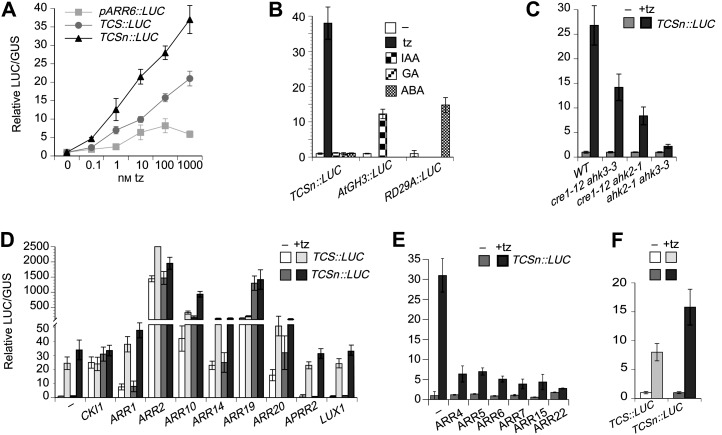
Sensitivity and specificity of TCSn::LUC in transient transfection assays. A, Induction of ARR6::LUC, TCS::LUC, and TCSn::LUC to increasing concentrations of transzeatin. B, TCSn::LUC is induced by transzeatin, but not by auxin, GA3, or abscisic acid. AtGH3::LUC and RD29A::LUC serve as positive controls for auxin and abscisic acid hormone induction, respectively (Müller and Sheen, 2008). C, Cytokinin-dependent induction of TCSn::LUC is compromised in ahk4 ahk3, ahk4 ahk2, and ahk2 ahk3 double mutant cells. cre1-12 is a mutant allele of AHK4 (Higuchi et al., 2004). D, Positive regulators of the MSP network induce TCSn::LUC expression. APRR2 and LUX have no effect. E, Type-A and type-C ARR attenuate cytokinin-dependent induction of TCSn::LUC. F, TCS::LUC and TCSn::LUC are induced in maize protoplasts by transzeatin. tz, Transzeatin; IAA, auxin; GA, GA3; ABA, abscisic acid.
Figure 3.
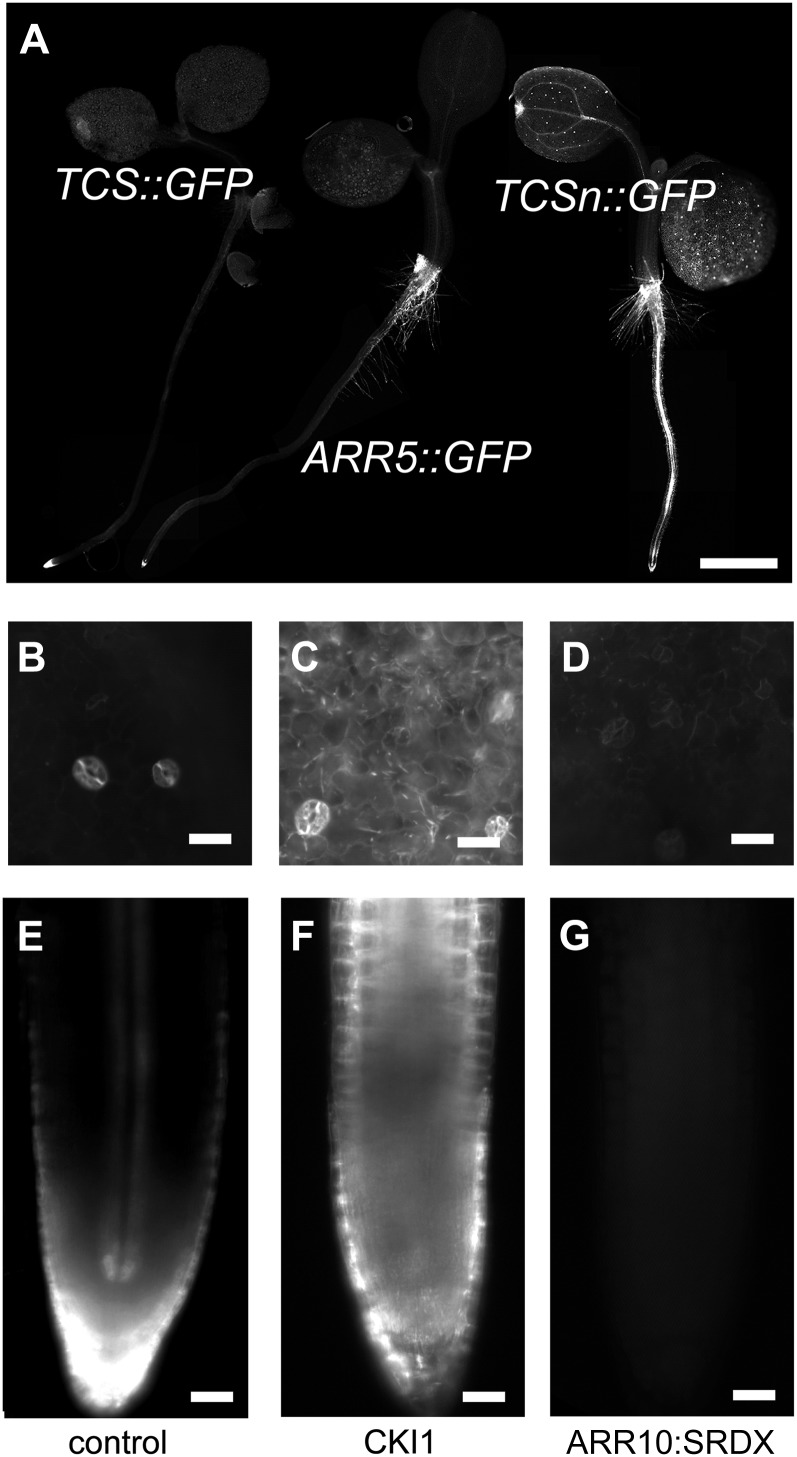
TCSn::GFP in the seedling. A, Compared with TCS::GFP and ARR5::GFP, TCSn::GFP exhibits strong GFP expression both in the root and shoot of the seedling. B to G, Induced overexpression of CKI1 (C and F) and ARR10:SRDX (D and G) causes ectopic activation or repression of TCSn::GFP, respectively, compared with controls (B and E) in the cotyledons (B–D) and the root meristem (E–G). Bars = 20 µm.
Figure 4.
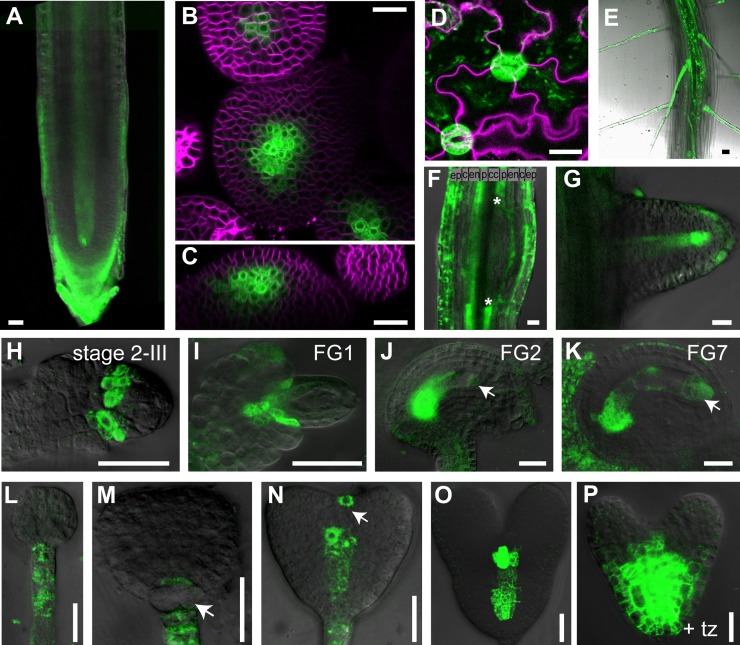
TCSn::GFP expression in different developmental contexts. A, Primary root meristem of 5-d-old seedling. B, Top view of shoot apical meristem. C, Side view of shoot apical meristem. D, Pavement cells and guard cells. E, Primary seedling root with root hairs. F, Lateral root primordium, early stage. Asterisks delineate lateral root primordium founder cells of pericycle that down-regulate MSP output. G, Emerging lateral root primordium. H, Ovule primordium after first mitotic division of megaspore mother cell stages, according to Schneitz et al. (1995). I to K, Embryo sac, stages according to Christensen et al. (1997). Arrows denote faint GFP signal in nuclei of embryo sac. L to P, Embryos. L, Globular stage. M, Transition stage, arrow denotes down-regulation of GFP in basal cell lineage. N, Heart stage, arrow denotes transient signal in the prospective shoot meristem. O, Late heart stage. P, Late heart stage, overnight incubation with 10 µm transzeatin. The signal from the membrane stain FM4-64 is shown in magenta. tz, Transzeatin; ep, epidermis; c, cortex; en, endodermis; p, pericycle cells; cc, central cylinder. Bars = 20 µm.
RESULTS
Defining Relevant Parameters to Improve TCS Activity
To reliably and consistently monitor low-to-intermediate output levels of the MSP network in planta and to avoid transgene silencing, we sought to improve the current synthetic sensor TCS (Müller and Sheen, 2008). Its design is based on the in vitro-defined DNA consensus sequence 5′-(A/G)GAT(C/T)-3′, as recognized by type-B ARRs (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003). To identify parameters that affect the activity of TCS, derivatives were constructed with variations in the number of binding sites, phasing, and identity of flanking nucleotides. All of the resulting fragments were cloned upstream of the cauliflower mosaic virus minimal 35S promoter and transcriptionally fused to luciferase (LUC). The ability of these constructs to confer cytokinin-dependent transcriptional activation was experimentally tested in transient transfection assays of primary mesophyll protoplasts (Müller and Sheen, 2008). An oligonucleotide harboring four such bindings sites, separated by arbitrarily selected flanking nucleotides, represented the basic building block for TCS (Fig. 1C). Multimerization of this sequence fragment resulted in various derivatives with an increasing number of binding sites (Fig. 1A). The arrangement of binding sites was chosen to realize all possible orientations that two given motifs can have relative to each other: tandem, tail to tail, and head to head (Fig. 1, A and D). Minimal but robust activity was observed with eight binding sites, while 16 sites resulted in a sensitivity comparable to ARR6::LUC, a reporter based on the 5′ cis-regulatory region of ARR6 (Hwang and Sheen, 2001), a type-A ARR with 11 clustered 5′-(A/G)GAT(C/T)-3′ motifs in its promoter (Fig. 1A; Supplemental Table S2). A dramatic increase in cytokinin responsiveness occurred when the number of sites increased from 16 to 24. Such a sigmoidal response curve is indicative of synergistic interactions among activator binding sites (Carey, 1998). The addition of more sites did not stimulate the activity further (Fig. 1A). Thus, 24 binding sites were chosen for the final design, which was named TCS (Müller and Sheen, 2008). A variant of TCS, TCSfm (for flanking nucleotides mutated), harbors mutations in the nucleotides that flank the in vitro-defined core sequences and was expected to integrate cytokinin-dependent induction similar to TCS, recapitulating the results of in vitro binding studies. Notably, TCSfm is insensitive to cytokinin (Fig. 1C), indicating that the in vitro-defined core motif 5′-(A/G)GAT(C/T)-3′ is too short to support transcriptional activation in vivo. Other derivatives of TCS differ in the distance between binding sites (Fig. 1D). Compared to TCS with a 9-bp distance between the core motifs, 11 bp resulted in higher activity, while reducing the distance to 7 bp caused a substantial reduction in activity (Fig. 1D). Eleven base pairs correspond approximately to one helical turn of the DNA double helix in its common B configuration (Wang, 1979). In agreement with our findings, helical phasing has been shown to be an important parameter for the functionality of transcription factor binding motifs in individual genes (Bouallaga et al., 2000; Mack et al., 2000; D’Alonzo et al., 2002) and has also been observed at a global scale (Ioshikhes et al., 1999; Makeev et al., 2003). Based on these experiments, we reasoned that improving TCS could be achieved by using extended type-B ARR binding motifs and adjusting the phasing of motifs to 11 bp.
Bioinformatic Analyses to Identify Relevant Type-B ARR Consensus Binding Sites in Vivo
To analyze the type-B ARR binding motifs as they occur in vivo, we analyzed the sequence of the 10 type-A ARR genes (ARR3–ARR9 and AAR15–AAR17). These genes represent the best-documented direct cytokinin target genes (D’Agostino et al., 2000; Taniguchi et al., 2007; Brenner et al., 2012). As a negative control, genes were randomly picked from a list of genes that showed stable expression irrespective of developmental stage, stress, and pharmacological or physiological treatments (Czechowski et al., 2005). Since the cis-regulatory sequence is typically found upstream, but can also be located the within transcribed sequence (Yant et al., 2010; Ritter et al., 2012), about 3 kb of the 5’-upstream sequence, as well as the transcribed sequence from each gene, was included in the analysis, totaling 45 kb of sequence for the 10 type-A ARR genes and 58 kb for the control genes (Supplemental Table S6). In each set, the number of hits to the 5′-(A/G)GAT(C/T)-3′ motif was counted. This motif has been shown to be indispensable for type-B ARR binding in vitro (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003) and for function in vivo (Ross et al., 2004; Müller and Sheen, 2008; Zhao et al., 2010; Liang et al., 2012). However, it is short and degenerate and thus occurs frequently by chance: on average, once in 108 bp of random DNA sequence. In the control genes, the frequency of motifs falls within a 95% confidence interval of a Poisson distribution, consistent with random frequency. By contrast, a hit is found every 86 bp in the cytokinin target genes. Such, or even higher, densities are very unlikely to occur by chance assuming Poisson distribution (P < 10–7), suggesting that a considerable fraction of the 5′-(A/G)GAT(C/T)-3′ motifs found in cytokinin target genes is functional in integrating cytokinin input (Supplemental Table S1). Next, distances and relative orientations of these motifs were analyzed. Specifically, we measured the distances between two given hits and sorted them into different size classes. The motif itself measures 5 bp; thus, the shortest distance between two hits is 6 bp. Because we were mainly interested in clustered hits, we did not resolve distances greater than an arbitrary 30 bp. The expected size distribution follows the function: F(n) = (1 – P)n – 1P, where P is the probability to find a hit (Basler, 2000). In the cytokinin target genes, distances from 6 to 30 bp are significantly overrepresented (X2 [1, n = 527] = 21.1, P < 0.00001), suggesting that these motifs are functional to support cooperativity of transcription factor binding in natural promoters, similar to what was observed in synthetic sequences (Fig. 1A). The observed enrichment concentrates to distances of 7 to 14 bp (Fig. 1E; Supplemental Table S3). When the relative orientation of clustered motifs was analyzed, no apparent bias either toward tandem or inverse orientations was detected (Supplemental Table S3). The analysis suggests that clustered 5′-(A/G)GAT(C/T)-3′ hits are significantly enriched in cytokinin target genes, and we used these hits to create an alignment. An additional three nucleotides flanking the core site were included, totaling 11 nucleotides, which was determined as the optimal phasing based on transient transfection experiments reported above (Fig. 1D). The resulting alignment, represented by a sequence logo (Crooks et al., 2004), revealed the tendency for conservation of nucleotides flanking the core (Fig. 1F). Specifically, sequences accompanied by a 5′ extension, 5′-A-3′ and/or a 3′ extension, 5′-T-3′ appear more frequently, similarly to previous findings (Rashotte et al., 2003; Taniguchi et al., 2007). We used this information to reduce the number of nonspecific motifs from the alignment and filtered the list of motifs using the sequences 5′-A(A/G)GAT(C/T)-3′ and 5′-(A/G)GAT(C/T)T-3′ (Supplemental Table S2). The resulting smaller set of binding motifs yielded a sequence logo with a AA(A/G)GAT(C/T)TT consensus (Fig. 1F). This consensus was also found enriched in the cytokinin target genes compared with the control genes (not shown), similar to previous studies (Brenner et al., 2012).
Based on the refined sequence logo, we created 12 sites, each slightly different from the other. Their alignment creates a sequence logo, which is similar to the natural sites (Fig. 1F; Supplemental Table S4). These synthetic sites were combined in random order to result in a synthetic sequence fragment that was repeated once to harbor 24 binding sites (Supplemental Table S5). Since clustered sites in cytokinin targets show no preference for a specific relative orientation, we preserved the relative site arrangement of TCS, which supports all possible orientations (Fig. 1A). This improved synthetic fragment was named TCSn. To summarize, the results obtained from transient transfection assays combined with bioinformatic analyses of bona fide cytokinin target genes allowed us to construct a new synthetic cytokinin promoter with an optimized number, spacing, and sequence of motifs, while also including variations to reflect the range of potential diversity among sites found in natural targets. At the same time, sequence variations avoid sequence monotony that could trigger the silencing of TCSn::GFP in transgenic plants.
TCSn Specifically Integrates MSP Activity
Using transient transfection experiments of mesophyll protoplasts, TCSn::LUC was subjected to various assays to determine its sensitivity and specificity to MSP signaling (Fig. 2). Its sensitivity to cytokinin was higher compared with TCS::LUC or ARR6::LUC (Fig. 2A). Similar to TCS (Müller and Sheen, 2008), TCSn did not cause transcription of LUC upon incubation with the auxin indole-3-acetic acid, GA3, or abscisic acid (Fig. 2B). Compared with wild-type cells, cytokinin-dependent expression of TCSn::LUC is compromised in cells that are mutated in two out of the three cytokinin receptor genes AHK2, AHK3, and AHK4 (Fig. 2C). Cotransfection with positively acting signaling components, including CKI1 and type-B ARRs, stimulated TCSn::LUC expression. Notably, overexpression of ARR10, ARR19, and ARR20 caused markedly stronger induction of TCSn::LUC than of TCS::LUC, while the remaining type-B ARRs showed similar induction of both reporters. By contrast, ARABIDOPSIS PSEUDO-RESPONSE REGULATOR2 (APRR2) and LUX ARRHYTHMO (LUX), both of which share a very similar DNA-binding domain with type-B ARRs (Hwang et al., 2002; Helfer et al., 2011), do not activate TCS nor TCSn transcription (Fig. 2D). As expected, cotransfection with type-A ARRs attenuated cytokinin-dependent induction (Fig. 2E). Next, we tested the reporters in maize (Zea mays) mesophyll protoplasts. As could be predicted from the similarity of the type-B response regulators between monocots and dicots (Chu et al., 2011), TCS::LUC and TCSn::LUC expression is activated by cytokinin in maize protoplasts (Fig. 2F). In summary, TCSn performs superior to TCS using protoplast transient assays, while retaining its specificity.
TCSn-Directed GFP Signals in the Seedling Are Brighter Than ARR5- or TCS-Controlled Reporters and Depend on MSP Signaling
Next, we analyzed the GFP expression pattern in plants that were transformed with a TCSn::GFP construct. Overall, TCSn::GFP expression levels are higher than TCS::GFP or ARR5::GFP, another frequently used cytokinin reporter (Fig. 3A). To address whether TCSn::GFP expression in planta is also controlled by MSP signaling, we transiently overexpressed proteins that either dominantly activate or repress MSP signaling. Compared with steady mutants, this approach avoids lethality issues and secondary effects. CKI1 induces MSP activity constitutively independent of cytokinins (Hwang and Sheen, 2001; Hejátko et al., 2009). Thus, CKI1 expression was ubiquitously induced for 30 h in 3-d-old seedlings. Consequently, both in the main root tip and in epidermal cells of the cotyledons, ectopic and ubiquitous signaling activity is observed (Fig. 3, C and F), compared with the control (Fig. 3, B and E). By contrast, TCSn-dependent expression is affected by a dominant-negative version of the type-B ARR10. Specifically, ubiquitously expressed ARR10:SRDX caused a loss of the endogenous expression domains (Fig. 3, D and G). This result is in agreement with previous experiments where type-B ARRs harboring a chimeric repressor domain suppress phosphorelay signaling (Heyl et al., 2008; Müller and Sheen, 2008). Thus, ectopic activation of MSP signaling in planta induces TCSn::GFP expression, while dominant-negative interference with MSP output causes loss of the TCSn::GFP domains, indicating that TCSn specifically integrates MSP output.
TCSn::GFP Expression Patterns Correlate with Known Cytokinin Functions and Also Reveal New Functions
A detailed analysis of the TCSn::GFP expression patterns in different tissues revealed that the GFP activity is consistent with documented cytokinin functions. For example, during ovule primordia formation, TCSn-directed GFP expression, similar to TCS::GFP (Bencivenga et al., 2012), localizes to the basal part of the funiculus (Fig. 4I). However, while TCS::GFP expression levels are below the detection level at later stages, TCSn::GFP expression is visible till female gametophyte stage 7 of ovule development (Fig. 4K). A very weak signal is also detected in the nuclei of the female gametophyte nuclei at the micropylar pole of the embryo sac (Figure 4, J and K, arrows). During embryogenesis, TCSn::GFP expression is detected in the suspensor and later in the hypophysis. GFP expression is down-regulated in progenitors of the hypophysis defining the basal cell lineage, while a weak signal is detected in the lens-shaped cell, similar to TCS::GFP (Müller and Sheen, 2008). Notably, expression levels in suspensor and suspensor-derived cells is much weaker compared with TCS::GFP. By contrast, provascular cells and the prospective cells of the shoot meristem exhibit distinct and bright GFP signals (Fig. 4, N and O), which potentially allow for addressing novel cytokinin functions in these contexts. As expected, application of exogenous cytokinin leads to increased and expanded GFP expression (Fig. 4P). Expression in the columella cells of the root meristem in the seedling (Fig. 4A), in the vasculature of root (Figs. 3, A and E, and 4A), in the lateral root primordia (Fig. 4, F and G), in root hairs (Fig. 4E), in the shoot meristem (Fig. 4, B and C), in the shoot vasculature (Fig. 3A), and in pavement cells and guard cells (Fig. 4D) are qualitatively very similar to TCS::GFP (Müller and Sheen, 2008; Bielach et al., 2012; Chickarmane et al., 2012), but much stronger. Thus, in addition to revealing the peaks of cytokinin output, TCSn::GFP shows intermediate to low levels of signaling output as well. Furthermore, no reduction in GFP levels has been observed after three generations of selfing of transgenic plants (data not shown), indicating that unlike TCS::GFP, TCSn::GFP is not subject to transgene silencing.
DISCUSSION
Cytokinins activate a MSP network in target cells, which culminates in transfer of phosphoryl groups to type-B response regulators, nuclear proteins that specifically bind to DNA and activate transcription of selected target genes. Concatemeric binding motifs combined with a minimal promoter and transcriptionally fused with LUC or GFP resulted in a reporter that specifically mediates MSP output in vivo (Müller and Sheen, 2008). Ideally, activity mediated by a synthetic promoter reflects the pure and universal transcriptional output profile of the signaling activity, devoid of tissue-specific aspects or unrelated signaling input. Indeed, TCS-dependent expression patterns of GFP or GUS were useful in monitoring the specific sites of phosphorelay signaling output in different tissues, which guided the discovery of previously unknown cytokinin functions (Müller and Sheen, 2008; Bencivenga et al., 2012; Marsch-Martínez et al., 2012) and refined existing models of cytokinin function (Gordon et al., 2009; Zhao et al., 2010; Bielach et al., 2012; Chickarmane et al., 2012; Murray et al., 2012). However, TCS-mediated expression in planta is low in many contexts where MSP is known to be important, which motivated us to revise the design of TCS by optimizing and extending the binding motifs for the type-B ARRs. Furthermore, to counteract the silencing, we introduced sequence variations in nonessential nucleotides, which broke the monotony of the repetitive TCS sequence. These modifications resulted in TCSn, which, compared with TCS, demonstrates higher sensitivity to cytokinin (Fig. 2A) and a more balanced response to different type-B family members (Fig. 2D).
In transgenic plants, these improvements translate into increased GFP activity in all tissues analyzed (Figs. 3A and 4, A–K, N, and O), except for the suspensor and suspensor-derived cells of the embryo (Fig. 4, L–N). Qualitatively, the expression patterns of TCS::GFP and TCSn::GFP in planta are very similar during, for example, ovule primordia formation, embryogenesis, lateral root development, shoot meristem function, and vasculature formation (Figs. 3A and 4). The increased sensitivity of TCSn::GFP renders additional aspects of phosophorelay readout visible, which remained below the level of detection with TCS::GFP. Specifically, the expression domain in the shoot meristem is broader (Fig. 4, B and C; Gordon et al., 2009; Chickarmane et al., 2012), a GFP signal in pavement cells becomes visible (Fig. 4D), and a transient signal in the shoot meristem of the embryo at the heart stage is observed (Fig. 4N; Müller and Sheen, 2008). The apparent resistance against silencing will allow the crossing of the TCSn::GFP line in various genetic backgrounds with a reduced risk of decreased or variable GFP expression in the progeny. This will facilitate the analysis of phosphorelay signaling in many contexts, including the little-known roles in root hair development (Fig. 4E) or the emerging role in pavement (Fig. 4D; Li et al., 2012) and guard cells (Fig. 4D; Desikan et al., 2008; Mira-Rodado et al., 2012), among others.
Ideally, a synthetic reporter integrates the activities of all transcription factors involved in relaying the signal without bias toward specific family members. The 11 members of the type-B ARR family differ in their inherent activity levels, both in transient assays and in planta (Hwang and Sheen, 2001; Sakai et al., 2001; Imamura et al., 2003; Hass et al., 2004; Tajima et al., 2004; Heyl et al., 2008; Müller and Sheen, 2008; Kim et al., 2012; Liang et al., 2012). However, no reference exists for a given family member or relevant combinations thereof, and consequently, it is not clear how the ideal synthetic reporter would respond. Thus, while all type-B ARRs tested activate TCS and TCSn (Fig. 2D), we cannot exclude that the reporters may exhibit some bias toward specific type-B ARRs. For example, the low expression levels in the suspensor mediated by TCSn, compared with TCS, might reflect the better binding conditions for the set of type-B ARRs expressed in the suspensor, which facilitates integration of phosphorelay response; while in all other tissues, TCSn appears to provide better conditions.
Our experiments using an increasing number of binding elements in synthetic promoters suggest cooperative binding of the type-B ARRs (Fig. 1A). The statistical analysis of cytokinin target genes revealed an enrichment of clustered binding motifs, indicating that cooperative binding occurs in vivo as well. Further support for cooperative binding of type-B ARRs comes from the recent finding that ARR18 can homomerize in planta (Veerabagu et al., 2012). In prokaryotes, DNA-binding response regulators have been shown to dimerize and oligomerize (for review, see Galperin, 2006). Thus, in plants, an analogous model could be realized.
In summary, the improved sensor TCSn will allow a detailed study of MSP signaling in various developmental contexts, shedding new light on plant development and physiology.
MATERIALS AND METHODS
Plant Growth and Treatments
Seedlings were germinated on vertical agar plates containing 1% (w/v) Suc, 0.8% (w/v) phytagar, and one-half-strength Murashige and Skoog medium with a 16-h/8-h photoperiod at 120 μmol m–2 s–1 and 21°C. After 3 d of growth on vertical plates, seedlings were transferred to liquid medium plates containing 1% (w/v) Suc, one-half-strength Murashige and Skoog medium, 2mm MES, pH 5.7, and 2% (v/v) ethanol for transgene induction in 12-well plates, sealed with parafilm, and incubated for 30 h before recording GFP fluorescence. Plants that needed to be grown to adulthood were kept in the greenhouse with a 16-h/8-h photoperiod at 150 μmol m–2 s–1 and 21°C during the light period and 18°C during the dark period. Plants used for protoplast transient transfection assays were grown with a 12-h/12-h photoperiod at 90 μmol m–2 s–1 and 21°C during the light period and 18°C during the dark period. Embryo in vitro cultivation and hormone treatments with transzeatin were performed as described (Müller and Sheen, 2008).
Plant Materials and Reporter and Effector Constructs
The TCS::GFP reporter line and the expression plasmids used in transient transfection assays were previously described (Hwang and Sheen, 2001; Yoo et al., 2007; Müller and Sheen, 2008). LUX, APRR2, and ARR19 coding sequences were cloned into the expression vector as described (Hwang and Sheen, 2001). The ARR5::GFP reporter is composed of 2.3 kb of upstream regulatory sequence transcriptionally fused to GFP that localizes to the endoplasmatic reticulum (Ottenschläger et al., 2003) in the binary vector pCB302 conferring Basta resistance (Xiang et al., 1999). Plants carrying the inducible transgene RIBOSOMAL PROTEIN S5A::AlcR/AlcA::ARR10:SRDX are described in Müller and Sheen (2008). For the 35S::AlcR/AlcA::CKI1 transgene, the DM7 vector conferring kanamycin resistance (Müller and Sheen, 2008) was modified with a ligation-independent cloning adaptor annealed into linearized DM7 vector, digested with EcoRI and SalI, to allow ligation of a CKI1 PCR fragment, amplified from genomic ecotype Columbia DNA, which covers the CKI1 locus from start to stop codons (see Supplemental Table S5 for sequences of the oligonucleotides). The TCSn plasmids TCSn::GFP and TCSn::LUC differ from the TCS variants (Müller and Sheen, 2008) by the synthetic promoter sequence. Plasmid and oligonucleotide sequences are provided in Supplemental Table S5. All plasmids have been sequenced to ensure no unwanted mutations have been introduced during cloning.
Phenotypic Analysis and Microscopy
GFP expression patterns (Fig. 3) were recorded using a Leica DM6000 microscope equipped with epifluorescence and a Leica DFC350FX camera, or with a Leica SP2 laser scanning confocal microscope (Fig. 4). Micrographs of whole seedlings in Figure 3A were assembled from individual pictures using Adobe Photoshop Creative Suite 4. The lipophilic dye FM4-64 (Molecular Probes) was used at a concentration of 10 µg mL–1 to demarcate cell membranes in Figure 4, B to D. Imaging for Figure 4, B and C was done using a Zeiss 510 Meta laser scanning confocal microscope with a 63× air-water dipping lens using the multitracking mode.
Transient Expression in Protoplasts
Protoplast isolation and transfection experiments were performed as reported (Sheen, 1990; Yoo et al., 2007). All protoplast experiments were performed in duplicates, and independent biological replicates yielded similar results.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Summary of 5′-(A/G)GAT(T/C)-3′ hits in cytokinin target genes, control genes, and random sequence.
Supplemental Table S2. List of clustered 5′-NNN(A/G)GAT(T/C)NNN-3′ hits in cytokinin target genes and control genes.
Supplemental Table S3. Distance and orientation of clustered hits in cytokinin target genes and control genes.
Supplemental Table S4. Sites used for TCSn.
Supplemental Table S5. Plasmid and oligonucleotide sequences.
Supplemental Table S6. Gene sequences used for the bioinformatic analyses.
Acknowledgments
We thank Ueli Grossniklaus (University of Zurich) for helpful discussions, financial support, and critical reading of the manuscript; Marco Celio (University of Fribourg), Jean-Pierre Métraux (University of Fribourg), and Jen Sheen (Harvard Medical School) for financial support; Tatsuo Kakimoto (Osaka University) for ahk mutant seeds; and Eva Benková (Institute of Science and Technology Austria), Ari Pekka Mähönen (University of Helsinki), and Jan Hejátko (Masaryk University) for testing TCSn::GFP.
Glossary
- MSP
multistep phosphorelay
References
- Aloni R, Langhans M, Aloni E, Dreieicher E, Ullrich CI. (2005) Root-synthesized cytokinin in Arabidopsis is distributed in the shoot by the transpiration stream. J Exp Bot 56: 1535–1544 [DOI] [PubMed] [Google Scholar]
- Aloni R, Langhans M, Aloni E, Ullrich CI. (2004) Role of cytokinin in the regulation of root gravitropism. Planta 220: 177–182 [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. (2001) The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci 26: 579–582 [DOI] [PubMed] [Google Scholar]
- Argueso CT, Raines T, Kieber JJ. (2010) Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Basler K. (2000) EMBO Gold Medal 1999. Waiting periods, instructive signals and positional information. EMBO J 19: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L. (2012) The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24: 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A, Podlesáková K, Marhavý P, Duclercq J, Cuesta C, Müller B, Grunewald W, Tarkowski P, Benková E. (2012) Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell 24: 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Benková E, Helariutta Y. (2011a) Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr Opin Plant Biol 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vatén A, Help H, El-Showk S, Scheres B, Helariutta K, Mähönen AP, Sakakibara H, Helariutta Y. (2011b) Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol 21: 927–932 [DOI] [PubMed] [Google Scholar]
- Bouallaga I, Massicard S, Yaniv M, Thierry F. (2000) An enhanceosome containing the Jun B/Fra-2 heterodimer and the HMG-I(Y) architectural protein controls HPV 18 transcription. EMBO Rep 1: 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. (1998) The enhanceosome and transcriptional synergy. Cell 92: 5–8 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD. (2007) ETHYLENE RESPONSE 1 histidine kinase activity of Arabidopsis promotes plant growth. Plant Physiol 143: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZX, Ma Q, Lin YX, Tang XL, Zhou YQ, Zhu SW, Fan J, Cheng BJ. (2011) Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet Mol Res 10: 3316–3330 [DOI] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN. (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. (2002) Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem 277: 816–822 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, Yang WC, Liang Y, Zuo J. (2010) Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Horák J, Chaban C, Mira-Rodado V, Witthöft J, Elgass K, Grefen C, Cheung MK, Meixner AJ, Hooley R, et al. (2008) The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS ONE 3: e2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle J, Arnscheidt A, Klix D, Weiler EW. (1986) Monoclonal antibodies to plant growth regulators. III. Zeatinriboside and dihydrozeatinriboside. Plant Physiol 81: 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188: 4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE. (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al. (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J, Pernisová M, Eneva T, Palme K, Brzobohatý B. (2003) The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol Genet Genomics 269: 443–453 [DOI] [PubMed] [Google Scholar]
- Hejátko J, Ryu H, Kim GT, Dobesová R, Choi S, Choi SM, Soucek P, Horák J, Pekárová B, Palme K, et al. (2009) The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 21: 2008–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmülling T. (2008) The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol 147: 1380–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T. (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J. (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. (2003) In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 122–131 [DOI] [PubMed] [Google Scholar]
- Ioshikhes I, Trifonov EN, Zhang MQ. (1999) Periodical distribution of transcription factor sites in promoter regions and connection with chromatin structure. Proc Natl Acad Sci USA 96: 2891–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ryu H, Cho YH, Scacchi E, Sabatini S, Hwang I. (2012) Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR2 attenuates signaling output in two-component circuitry. Plant J 69: 934–945 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Li H, Xu T, Lin D, Wen M, Xie M, Duclercq J, Bielach A, Kim J, Reddy GV, Zuo J, et al. (2012) Cytokinin signaling regulates pavement cell morphogenesis in Arabidopsis. Cell Res 23: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Wang X, Hong S, Li Y, Zuo J. (2012) Deletion of the initial 45 residues of ARR18 induces cytokinin response in Arabidopsis. J Genet Genomics 39: 37–46 [DOI] [PubMed] [Google Scholar]
- Mack CP, Thompson MM, Lawrenz-Smith S, Owens GK. (2000) Smooth muscle alpha-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ Res 86: 221–232 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. (2006a) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. (2006b) Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol 16: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Makeev VJ, Lifanov AP, Nazina AG, Papatsenko DA. (2003) Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res 31: 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martínez N, Ramos-Cruz D, Irepan Reyes-Olalde J, Lozano-Sotomayor P, Zúñiga-Mayo VM, de Folter S. (2012) The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J 72: 222–234 [DOI] [PubMed] [Google Scholar]
- Mira-Rodado V, Veerabagu M, Witthöft J, Teply J, Harter K, Desikan R. (2012) Identification of two-component system elements downstream of AHK5 in the stomatal closure response of Arabidopsis thaliana. Plant Signal Behav 7: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel C, Zhulin IB. (2001) CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci 26: 582–584 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JA, Jones A, Godin C, Traas J. (2012) Systems analysis of shoot apical meristem growth and development: integrating hormonal and mechanical signaling. Plant Cell 24: 3907–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S. (2010) The molecular basis of cytokinin function. Curr Opin Plant Biol 13: 21–26 [DOI] [PubMed] [Google Scholar]
- Pham J, Liu J, Bennett MH, Mansfield JW, Desikan R. (2012) Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol 194: 168–180 [DOI] [PubMed] [Google Scholar]
- Pischke MS, Jones LG, Otsuga D, Fernandez DE, Drews GN, Sussman MR. (2002) An Arabidopsis histidine kinase is essential for megagametogenesis. Proc Natl Acad Sci USA 99: 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ. (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter DI, Dong Z, Guo S, Chuang JH. (2012) Transcriptional enhancers in protein-coding exons of vertebrate developmental genes. PLoS ONE 7: e35202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EJ, Stone JM, Elowsky CG, Arredondo-Peter R, Klucas RV, Sarath G. (2004) Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J Exp Bot 55: 1721–1731 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A. (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hülskamp M, Pruitt RE. (1995) Wild-type ovule development in Arabidopsis thaliana; a light microscope study of cleared whole mount tissue. Plant J 7: 731–749 [Google Scholar]
- Sheen J. (1990) Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T. (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Veerabagu M, Elgass K, Kirchler T, Huppenberger P, Harter K, Chaban C, Mira-Rodado V. (2012) The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J 72: 721–731 [DOI] [PubMed] [Google Scholar]
- Wang JC. (1979) Helical repeat of DNA in solution. Proc Natl Acad Sci USA 76: 200–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Köllmer I, Bartrina I, Holst K, Schmülling T. (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8: 371–381 [DOI] [PubMed] [Google Scholar]
- Werner T, Schmülling T. (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU. (2010) Hormonal control of the shoot stem-cell niche. Nature 465: 1089–1092 [DOI] [PubMed] [Google Scholar]