A landmark-based system quantifies root architecture using holistic trait capture methods.
Abstract
The architecture of plant roots affects essential functions including nutrient and water uptake, soil anchorage, and symbiotic interactions. Root architecture comprises many features that arise from the growth of the primary and lateral roots. These root features are dictated by the genetic background but are also highly responsive to the environment. Thus, root system architecture (RSA) represents an important and complex trait that is highly variable, affected by genotype × environment interactions, and relevant to survival/performance. Quantification of RSA in Arabidopsis (Arabidopsis thaliana) using plate-based tissue culture is a very common and relatively rapid assay, but quantifying RSA represents an experimental bottleneck when it comes to medium- or high-throughput approaches used in mutant or genotype screens. Here, we present RootScape, a landmark-based allometric method for rapid phenotyping of RSA using Arabidopsis as a case study. Using the software AAMToolbox, we created a 20-point landmark model that captures RSA as one integrated trait and used this model to quantify changes in the RSA of Arabidopsis (Columbia) wild-type plants grown under different hormone treatments. Principal component analysis was used to compare RootScape with conventional methods designed to measure root architecture. This analysis showed that RootScape efficiently captured nearly all the variation in root architecture detected by measuring individual root traits and is 5 to 10 times faster than conventional scoring. We validated RootScape by quantifying the plasticity of RSA in several mutant lines affected in hormone signaling. The RootScape analysis recapitulated previous results that described complex phenotypes in the mutants and identified novel gene × environment interactions.
Roots have a crucial impact on plant survival because of their major functions: anchorage of the plant in the soil, water and nutrient acquisition, and symbiotic interaction with other organisms (Den Herder et al., 2010). One important characteristic of root systems is the manner in which the primary and lateral roots comprise the superstructure or root architecture. Root architecture is an ideal system for studying developmental plasticity, as it continually integrates intrinsic and environmental responses (Malamy, 2005), which represents a vital and dynamic component of agricultural productivity (Lynch, 1995).
Root system architecture (RSA) is defined as the spatial configuration of the roots in their environment (Lynch, 1995). The complexity of RSA was initially appreciated several decades ago, and terms like morphology, topology, distribution, and architecture were often used to describe the nature of RSA (Fitter, 1987; Fitter and Stickland, 1991; Lynch, 1995). These early reports argued that simple traits like root mass are insufficient to describe roots, because they do not capture the spatial configuration of roots in the soil, which is critical to plant performance (Fitter and Stickland, 1991). Root systems are integrated organs that adopt specific architectures to maximal foraging of the heterogeneous soil environment in different ways (Fitter, 1987; Fitter and Stickland, 1991; Lynch, 1995). More recently, new approaches have incorporated the measurement of many individual developmental traits that together comprise RSA (De Smet et al., 2012; Dubrovsky and Forde, 2012). For example, one recent report identified three fundamental components of RSA in generating complex topologies, including the contribution of lateral axes to branching, the rate and path of growth of the axis, and the increase in root surface area (Topp and Benfey, 2012). Thus, RSA is an important and complex trait that requires convenient measurement methods for rapid screening of diverse plant mutants and genotypes.
With increasing research in RSA in the genetically tractable model plant Arabidopsis (Arabidopsis thaliana), the need for high-throughput methods of root phenotyping has dramatically increased over the years. Consequently, different methods and approaches have been developed in order to address this demand. Currently, three major approaches for phenotyping RSA are used (for review, see Zhu et al., 2011; De Smet et al., 2012). The first group of methods uses classical measures of RSA, which involve measurements of individual root traits. These methods often use software to manually draw the RSA onto digital two-dimensional images to quantify root length and number (Abramoff et al., 2004; http://www.machinevision.nl). These traditional methods provide the most accurate measurements of the root system but have a major disadvantage in being extremely time consuming.
The second group of methods utilizes advanced semiautomated software for RSA measurements like EZ-Rhizo (Armengaud et al., 2009). EZ-Rhizo also uses digital two-dimensional images of plants grown on vertical plates (similar to the classical methods above) but is faster and produces different traits and basic statistics. The method works best when root features do not physically overlap, but we have found root overlap to be common when working with Arabidopsis plants older than 10 d. Other recent programs also provide semiautomated analysis of RSA, including RootReader2D (http://www.plantmineralnutrition.net/rootreader.htm) and SmartRoot (Lobet et al., 2011). However, while completely automated detection is potentially the highest throughput, we found that the root surface detection step is frequently prone to failure when using both of these programs, even after considerable adjustment by the user, where root features are missed or background noise is incorrectly labeled as roots.
Finally, in a third group, recent developments include three-dimensional analysis of RSA of plants grown on transparent gel cylinders or in soil. The three-dimensional gel-based imaging approach is reported to be suitable for high-throughput phenotyping (Iyer-Pascuzzi et al., 2010). However, this approach requires special equipment, and imaging the root system of single plants can take 10 min (Iyer-Pascuzzi et al., 2010). X-ray computed tomography (Perret et al., 2007; Tracy et al., 2010) and magnetic resonance imaging (Van As, 2007) also provide highly detailed three-dimensional RSA analysis, but they require long scanning times and are extremely expensive and inaccessible. Most laboratories still utilize relatively convenient, inexpensive, and rapid two-dimensional phenotypic characterization of RSA, at least for initial screening purposes.
The aim of this work is to address the need for a simple method to measure many different aspects of root architecture for high-throughput laboratory screening of mutants and genotypes in Arabidopsis. Here, we describe a landmark-based allometric (size and shape) approach called RootScape, a user-friendly software platform that enables rapid, comprehensive, and integrative phenotyping of the RSA in Arabidopsis. Unlike recent methods that collect information on different root traits to describe the RSA, RootScape places user-defined root landmarks on a two-dimensional grid to measure root architecture as a single integrated root system. The method employs rapid manual placement of root system landmarks. This manual step avoids one of the most problematic steps in automated image analysis (recognition of the root surface), providing a simple tool that does not require image processing. This method uses simple, two-dimensional digital images of the root system and a 20-point landmark model created in AAMToolbox, a freely available MATLAB plugin. While in-depth developmental analysis of root systems will often require knowing the contribution of individual traits, RootScape is a rapid method to access the holistic contribution of many individual root traits to RSA and to capture the overall property of the spatial configuration of roots in the soil (Fitter and Stickland, 1991). To demonstrate its utility, we used RootScape to quantify the root plasticity of Arabidopsis plants (Columbia [Col-0]) grown on four different media and compared the RootScape results with conventional measurements of individual root traits captured using the Optimas6 image-analysis software or Image J (Abramoff et al., 2004). This analysis showed that by measuring integrative root traits using RootScape, we could capture the vast majority of the individual trait variation, as verified by multiple regression analysis. Additionally, we tested the ability of RootScape to quantify the plasticity response in Arabidopsis mutants defective in hormone signaling. For this analysis, wild-type Col-0 and three hormone signaling mutants (auxin-resistant4 [axr4], abscisic acid insensitive4 [abi4], and cytokinin response1 [cre1]) were treated with auxin, cytokinin, or abscisic acid (ABA) versus controls. Statistical analyses (ANOVA/multivariate ANOVA [MANOVA]) allowed us to confirm most of the previously known interactions of genotype with these distinct environments and to potentially identify novel ones. Thus, we demonstrate that RootScape can be used as a rapid and efficient approach for quantifying the plasticity of the RSA in mutant (or ecotype) backgrounds of Arabidopsis and can identify new conditional root phenotypes.
RESULTS
RootScape: Adapting Software Platform to Measure RSA
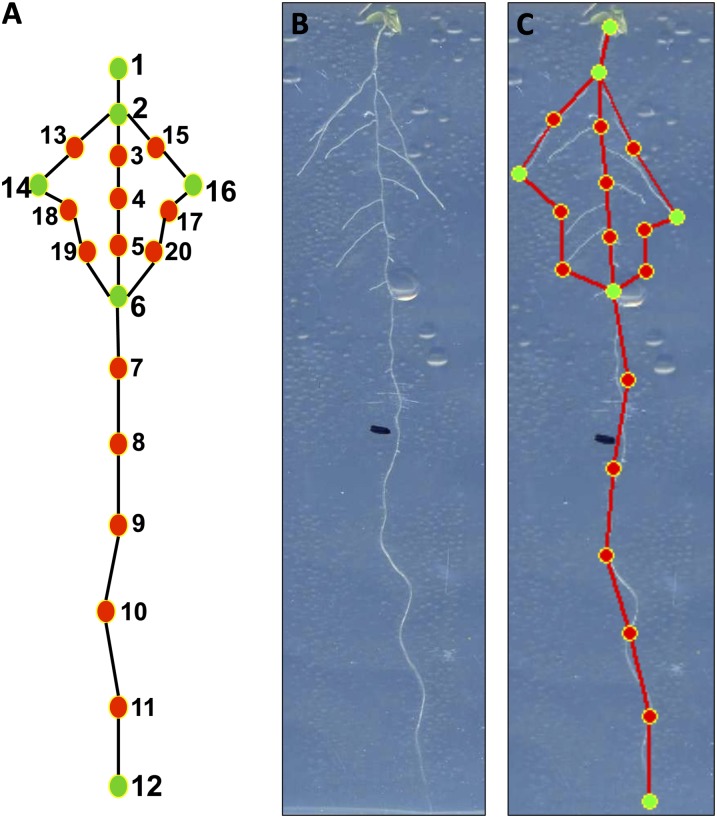
Allometric methods have been applied previously to measure plant organs such as leaves (Langlade et al., 2005; Bensmihen et al., 2008). To implement an allometric method for quantifying RSA, we created a 20-point landmark template (model) in the AAMToolbox software, a publicly available MATLAB plugin (http://cmpdartsvr1.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software) originally developed for face shape recognition in lip reading (Matthews et al., 2002). This 20-point template captures the main characteristics of the RSA. The six “primary” landmarks (green points in Fig. 1A) are defined by recognizable developmental landmarks on the root. These included four of the primary landmarks, placed on the transition between shoot and root (point 1), the position of the first lateral root (point 2), the position of the last lateral root (point 6), and the apex of the primary root (point 12; Fig. 1A). Two other primary landmarks (points 14 and 16) were placed at the apex of the lateral root that was farthest from the primary root along an axis perpendicular to the primary on each side (Fig. 1A). All primary landmark points of the template are placed manually at the defined developmental positions of each root sample (Supplemental Fig. S2). Secondary landmarks or semilandmarks (red points in Fig. 1) were defined as having a position between the primary landmarks. These secondary landmarks were also placed manually on the lateral root periphery to capture the corresponding shape and along the length of the primary root. Following manual placement of these secondary landmarks, the AAMToolbox software automatically spaces these secondary landmarks evenly between the primary landmarks (Supplemental Fig. S2). In a side-by-side comparison performed by the same researcher, this new, landmark-based method called RootScape was 5 to 10 times faster than individual measurements of root traits as performed using Optimas6 or Image J (Abramoff et al., 2004).
Figure 1.
RootScape, a landmark-based allometric model for integrative quantification of RSA. Twenty landmarks were used to describe the RSA: six primary landmarks (green; 1, 2, 6, 12, 14, and 16) placed at recognizable positions of the RSA and 14 secondary landmarks (red) spaced evenly between the primary landmarks. A, RootScape template of 20 landmarks. B, RSA of a 12-d-old seedling. C, RootScape applied on the same seedling.
The plant root system does not have a defined shape, compared with leaves, and the number of lateral roots is quite variable depending on the conditions or genotype. Thus, the constructed model of 20 landmarks represents the primary root as a line and lateral roots as a polygon, which is the shape that explains how the root system spans the surface of its growth environment on the plate. Root shapes that were captured by this model were aligned by translation according to the root shape centroids and translated to minimize the variance between corresponding landmarks. No size normalization of the root shapes was performed in this model, since the aim is to capture the variation in both the size and shape (allometry) of the root system in response to the hormonal treatments. This RootScape model of 20 landmarks yields 40 coordinate values (two per point) that define a 40-dimensional space in which each axis represents variation in one of the coordinate values. Each root shape can be characterized as a single point in this space, where all root shapes from the same treatment together define a “cloud” of points. The center of the cloud, corresponding to the mean root shape, is defined by the means of each of the 40 coordinate values.
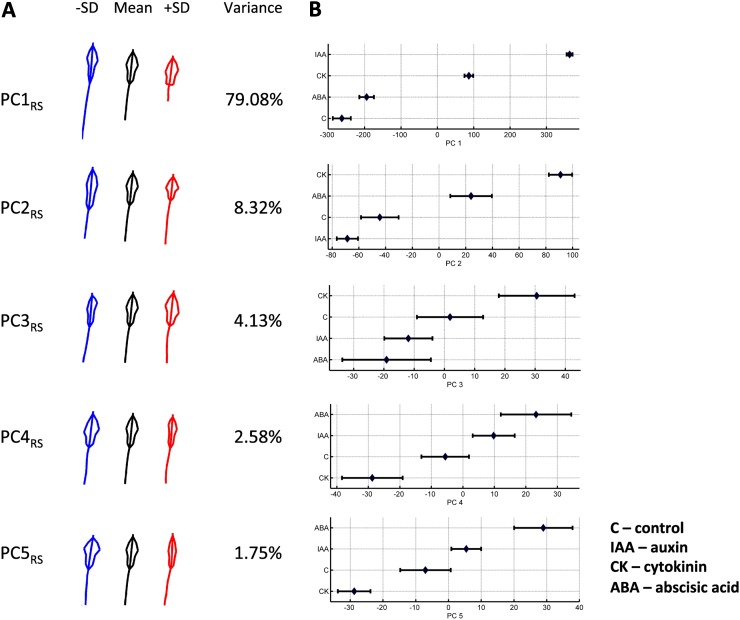
We used the RootScape template of 20 landmarks to quantify the RSA of Arabidopsis (Col-0) plants grown on four conditions, which consisted of control and three hormone treatments, auxin (indole-3-acetic acid [IAA]), cytokinin (CK), and ABA, with 24 to 25 plant replicates for each treatment condition. These treatment conditions were selected to create a variety of well-documented and stereotypical root architectures (Nibau et al., 2008). Plants were initially grown on a single growth medium and then transferred to either a control plate or one of the three hormone treatment conditions (for details, see “Materials and Methods” and Supplemental Fig. S1). After 5 d on the different hormonal treatments or control plates, the root system was imaged using a document scanner, and the unprocessed images were used for subsequent analysis using the 20-point allometric template of RootScape (Supplemental Fig. S2). This landmark data set was used to create the RSA morphospace and was named the “allometric Col-0 plasticity space.” These data were further processed by the AAMToolbox software in a principal component analysis (PCA) that captures the main trends of variation in the root architecture. This analysis revealed that five PCs captured more than 95% of the root variation of the total root shape and size variance in Col-0 genotype (Fig. 2A). Figure 2 shows a plot of the range of the RootScape-derived principal component (PC) values of the first five PCs to identify the treatments that are driving the extreme phenotypes in RSA (Fig. 2B). PC1RS of the allometric Col-0 model explains 79.08% of the total variance and mostly affects size, but it also affects the shape of the RSA and thus represents a major allometric trait. Low PC1RS values relate to having a longer primary root and a smaller polygon of lateral roots with a narrowed polygon base (Fig. 2A; Supplemental Video S1), which corresponds to plants grown in control medium or with ABA (Fig. 2B). High PC1RS values relate to shorter primary root and rectangle-like shape of the lateral root polygon (Fig. 2A; Supplemental Video S1). This extreme value for PC1RS corresponds to the IAA-treated plants (Fig. 2B). PC2RS accounts for 8.32% of the variance in RSA and affects mostly the shape of the polygon occupied by lateral roots. Low PC2RS values give a longer, rectangle-like polygon (Fig. 2A; Supplemental Video S1) and correspond to IAA-treated plants (Fig. 2B), whereas higher PC2RS values have a diamond-shaped lateral root polygon (Fig. 2A; Supplemental Video S1) and correspond to CK-treated plants (Fig. 2B). Together, PC3RS and PC4RS capture about 7% of the RSA variance and affect only the asymmetry of the lateral root polygon, which reflects the random variation in root shape among individual plants but does not represent any trends resulting from the hormone treatment (Fig. 2B). PC5RS accounts for only about 2% of the variance in RSA and mainly affects the lateral root polygon width (i.e. the length of the lateral roots that make the polygon shape). Negative PC5R values correspond to RSAs with a wider lateral root polygon, while a positive value corresponds to RSAs with a very narrow lateral root polygon (Fig. 2A; Supplemental Video S1).
Figure 2.
Variability of the root system shape and size of Arabidopsis Col-0 quantified with RootScape, as described by the five main PCs from the allometric Col-0 model. A, For each PC, the mean root shape outline is shown in black (middle), blue (left), and red (right); shapes are shown by varying the PC value minus or plus 1 sd (−SD and +SD). The percentage of variance of each PC is shown next to the shapes. B, PC values for the first five PCs are plotted on the x axis against the four different conditions. The mean of each of the four treatments is represented by a diamond, and the error bars display the variation observed for each group.
Comparison of PCs Captured by RootScape versus Individual Trait Measurements
As described above, RootScape quantifies the RSA as a series of integrative traits, where the phenotypic output is represented by PCs that each captures different aspects of the shape and size of RSA. We were interested in determining how these phenotypic outputs of RootScape are related to the classical method of RSA quantification, which measures individual traits. In order to answer this question, individual traits were first quantified using Optimas6 or Image J (Abramoff et al., 2004; for details, see “Materials and Methods” and Supplemental Text S1) and PCA was performed on the individual traits. This allowed us to compare how PCs derived from the individual traits (PCIT) relate to the PCs derived from RootScape (PCRS) measurements (Table I; Supplemental Text S1). The PCA analysis of the individual traits (Table II) revealed that several traits have a high contribution to PC1IT (e.g. root length ratio, root formation zone, branching density, growth of primary root). Next, we performed Pearson correlation analysis of the first five PCs of the individual trait analysis with the first five PCs derived from the RootScape analysis (Table I). PC1RS has a high and significant correlation (0.88) with PC1IT (Table I). Thus, we can conclude that individual traits such as root length ratio, root formation zone, and branching density are captured by PC1RS. Following this logic, PC2RS has the highest correlation (−0.7) with PC2IT (Table I), which mainly captures total root length (Table II). Note that the direction of PC axes is arbitrary, so either correlation or anticorrelation indicates a trend between a trait and a PC. PC3RS, PC4RS, and PC5RS correlate moderately, but significantly (0.35, −0.36, and −0.5, respectively), with the same PC from the individual trait analysis, PC3IT (Table I). The highest contribution in PC3IT is from the average lateral root length (Table I). Therefore, RootScape captures variation in lateral root length in PC3RS, PC4RS, and PC5RS. This correlation analysis shows that the RootScape allometric landmark-based method captures very similar root variation as measured by many individual traits. Thus, RootScape is able to capture information contributed by many individual traits, even though it measures RSA as one integrative trait.
Table I. Correlation matrix of the first five PCs of RootScape and the first five PCs of individual traits, followed by a variability contribution for each.
Sample size is represented by 24 to 25 plant replicates per treatment. Boldface values show highest significant correlations. ***P < 0.0001, **P < 0.001, *P < 0.01.
| Correlation | PC1RS (79%) | PC2RS (8%) | PC3RS (4%) | PC4RS (3%) | PC5RS (2%) |
|---|---|---|---|---|---|
| PC1IT (63%) | 0.884*** | −0.334** | 0.069 | −0.081 | −0.147 |
| PC2IT (22%) | −0.414*** | −0.669*** | 0.197 | −0.284* | −0.129 |
| PC3IT (11%) | 0.004 | 0.437*** | 0.347** | −0.364** | −0.497*** |
| PC4IT (2%) | 0.145 | 0.057 | 0.033 | 0.037 | −0.005 |
| PC5IT (1%) | −0.023 | 0.267* | −0.006 | 0.008 | −0.089 |
Table II. Loading matrix of PCA, scored by 10 individual traits.
Boldface values show the highest contribution(s) to the component.
| Individual Trait | PC1IT | PC2IT | PC3IT | PC4IT | PC5IT |
|---|---|---|---|---|---|
| Root formation zone (RFZ) | −0.95 | 0.11 | 0.21 | −0.10 | 0.16 |
| Branching density (Bd) | 0.94 | 0.01 | −0.21 | 0.03 | 0.28 |
| Lateral root number (LR#) | 0.67 | 0.58 | −0.43 | −0.09 | −0.02 |
| Growth of primary root (P2) | −0.94 | 0.32 | −0.01 | −0.01 | 0.06 |
| Primary root length (P) | −0.89 | 0.43 | 0.01 | −0.13 | 0.01 |
| Lateral root length (LRI) | 0.56 | 0.17 | 0.79 | 0.16 | 0.03 |
| Length ratio (LR) | 0.98 | 0.09 | −0.10 | 0.01 | −0.01 |
| Total root length (TRL) | 0.19 | 0.95 | 0.21 | −0.14 | −0.02 |
| Total lateral root length (TLRL) | 0.85 | 0.49 | 0.17 | −0.03 | −0.03 |
| LR# in P2 | −0.60 | 0.65 | −0.28 | 0.37 | 0.001 |
| Variability (%) | 63.1 | 22.30 | 10.53 | 2.22 | 1.08 |
| Cumulative variability (%) | 63.1 | 85.40 | 95.94 | 98.16 | 99.24 |
Furthermore, we were interested in how the results obtained by RootScape could be explained (predicted) by the main PCs identified by the individual trait analysis. To answer this question, we used multiple linear regression analysis where each of the first five PC values from RootScape were taken as a response variable and the first five PC values from individual trait analysis were taken as the explanatory variables (Supplemental Table S1). We found that PC1RS can be explained almost completely (r2 = 0.97) by three PCs from the individual trait analysis (PC1IT, PC2IT, and PC4IT). PC2RS also has a high degree of predictability (r2 = 0.82) based on four PCs from the individual trait analysis (PC1IT, PC2IT, PC3IT, and PC5IT). PC3RS, PC4RS, and PC5RS can be explained to a modest extent by the first five PCs from the individual trait analysis (r2 = 0.17, 0.22, and 0.29), although only PC3IT shows significance for prediction. These results demonstrate that RootScape phenotypic outputs can be highly predicted by classical quantification of RSA measured using individual traits.
Reciprocally, we asked how much of the RSA variation captured with the conventional individual trait analysis can be explained by RootScape allometric measurements (Supplemental Table S2). PC1IT is largely explained by PC1RS, PC2RS, and PC5RS (r2 = 0.93). PC2IT and PC3IT can also be explained to a high degree (r2 = 0.75 and 0.69) where different PCs from RootScape have significant contributions. Thus, this analysis confirms that the simple and rapid allometric approach of RootScape has the power to predict the first three PCs from the individual trait analysis, which accounts for about 96% of the variation using that method. More generally, the reciprocal multivariate analysis shows that RootScape and individual trait analysis largely capture the same trends in variation of RSA. Thus, overall, the correlation and multivariate analyses show that RootScape is a rapid method that describes the same major trends in variation of RSA as more detailed and time-consuming analyses of individual traits.
Using RootScape to Characterize the Plasticity of RSA
Different phytohormones are known to exert specific effects on root architecture, with partially overlapping effects often due to cross talk between hormones (Bishopp et al., 2011; for review, see Depuydt and Hardtke, 2011). We used RootScape to characterize the plasticity of the Arabidopsis root system under hormone treatments. In addition, we measured the root architecture of well-characterized Arabidopsis mutants in hormone signaling to determine if RootScape could identify known phenotypes, including insensitivity to hormone treatments, which were the basis for the mutant screens. We also characterized all of these Arabidopsis mutants with a panel of hormone treatments to explore the ability of RootScape to rapidly characterize root architecture under a variety of conditions and to identify potential cross talk among hormones. We analyzed three Arabidopsis mutants known to affect root development due to signaling defects in auxin (axr4-1; Hobbie and Estelle, 1995), ABA (abi4-1; Signora et al., 2001), and CK (cre1-2; Inoue et al., 2001).
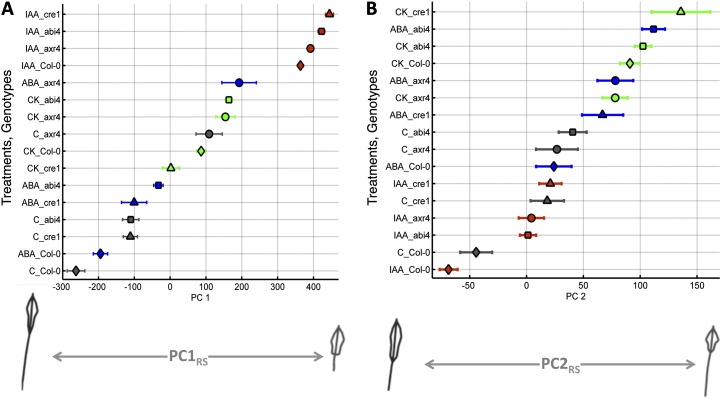
For each mutant genotype, 10 to 15 plant replicates were analyzed in each of the hormonal treatments, compared with controls. Growth conditions of the mutants were identical as for Col-0 (see “Materials and Methods”; Supplemental Fig. S1). The same RootScape allometric template of 20 landmarks was applied to each plant. In order to compare changes in root phenotype between the mutant lines and Col-0, we projected both the root architectural plasticity in Col-0 and the three mutant lines onto PC1RS and PC2RS axes (e.g. the allometric Col-0 plasticity space defined by RootScape measurements) and obtained phenotypic scores (PC values). By carrying out this projection, we were able to visualize the distribution of phenotypes along the phenotypic Col-0 plasticity “space” (Fig. 3).
Figure 3.
Range of PC values obtained for Col-0 (diamonds) and three mutants, abi4-1 (squares), axr4-1 (circles), and cre1-2 (triangles), on the four different media (control [C], gray; IAA, red; CK, green; and ABA, blue) for the allometric Col-0 model. The mean of each genotype based on the four treatments is represented by a different shape, and the bars represent the se within the observed group. A, PC1. B, PC2. Extreme root shapes of minus or plus 1 sd are shown on each side below the x axis.
This analysis showed that auxin treatment mapped to one extreme of PC1RS, showing its strong effect on a suite of traits that varied dramatically among the treatments (Fig. 3A). We did not find any specific treatment grouping in the second PC (Fig. 3B). We next used PC values for the first five PCs to identify any genotype × treatment interactions. For this analysis, we performed a two-way ANOVA for each of the first five PCs, following the model PCRS = αgenotype + βtreatment + γgenotype × treatment + ε. There were significant differences due to genotype (PC1RS F = 70.84, P < 0.0001; PC2RS F = 23.31, P < 0.0001; PC4RS F = 11.26, P < 0.0001; PC5RS F = 10.25, P < 0.0001), treatment (PC1RS F = 343.37, P < 0.0001; PC2RS F = 54.15, P < 0.0001; PC3RS F = 3.06, P = 0.0288; PC5RS F = 16.52, P < 0.0001), and the interaction between genotype and treatment (PC1Col F = 16.04, P < 0.0001; PC2Col F = 2.82, P = 0.0036; PC3RS F = 5.63, P < 0.0001; PC4RS F = 6.16, P < 0.0001; PC5RS F = 3.09, P = 0.0015). Significant differences between the genotypes and treatments, when compared with the wild-type (Col-0) genotype and control treatment, are shown in Table III. RootScape measurements of the mutants and the wild type were able to recapitulate most of the known genotype × hormone interactions. For example, axr4, which was originally isolated as an auxin-resistant mutant (Hobbie and Estelle, 1995), displayed an interaction with IAA in PC1RS and PC4RS. In addition, axr4 showed an interaction with ABA in PC1RS, PC3RS, and PC4RS (Table III). These results support the variable resistance of axr4 to ABA, which was reported previously (Hobbie and Estelle, 1995). Additionally, another study reported cross talk between ABA and auxin signaling (axr4; Rock and Sun, 2005). The interaction of axr4 with cytokinin was present in all five PCRS (Table III) and fits with previous observations of variation in relative root elongation reported for axr4-1 at particular CK concentrations (Hobbie and Estelle, 1995). Thus, RootScape can identify many of the conditional phenotypes identified in the literature for the axr4 mutant.
Table III. Significant differences of the first five RootScape PC values comparing hormone treatments and mutant genotypes with control treatment and the wild-type genotype.
Only significant P values are shown (P < 0.05). *Indicative interaction.
| Groups |
P |
Interaction Reference | ||||
|---|---|---|---|---|---|---|
| PC1RS | PC2RS | PC3RS | PC4RS | PC5RS | ||
| Treatment (ABA) | <0.0001 | <0.0001 | <0.0001 | |||
| Treatment (CK) | <0.0001 | <0.0001 | ||||
| Treatment (IAA) | <0.0001 | <0.0001 | 0.0033 | |||
| Genotype (abi4) | 0.0006 | |||||
| Genotype (axr4) | <0.0001 | 0.0098 | <0.0001 | |||
| Genotype (cre1) | 0.0026 | 0.0133 | ||||
| Genotype (axr4) × treatment (IAA) | <0.0001 | <0.0001 | Hobbie and Estelle (1995) | |||
| Genotype (axr4) × treatment (ABA) | <0.0001 | <0.0001 | 0.0137 | Hobbie and Estelle (1995)*; Rock and Sun (2005)* | ||
| Genotype (axr4) × treatment (CK) | 0.0003 | 0.0088 | 0.0011 | <0.0001 | 0.0054 | Hobbie and Estelle (1995)* |
| Genotype (cre1) × treatment (CK) | 0.002 | 0.0149 | Inoue et al. (2001) | |||
| Genotype (cre1) × treatment (ABA) | 0.0226 | Inoue et al. (2001)* | ||||
| Genotype (cre1) × treatment (IAA) | 0.0004 | New | ||||
| Genotype (abi4) × treatment (CK) | 0.0091 | New | ||||
The RootScape analysis also uncovered an interaction of cre1-2 with all three hormonal treatments in at least one PCRS (Table III). First, the cre1-2 interaction with CK captured in PC1RS and PC5RS is consistent with previously published phenotypes for this mutant, fitting with its role as a CK receptor (Inoue et al., 2001). This analysis also identified an interaction of the cre1-2 mutation with ABA. This is reminiscent of another cre1 allele (cre1-1) that was shown to have lower root growth in response to low levels of ABA (Inoue et al., 2001). The abi4-1 mutant had only one interaction with CK in PC1 (Table III), which may be related to interactions found by another study in which CK treatment increased the transcript level of ABI4 (Shkolnik-Inbar and Bar-Zvi, 2010). Thus, RootScape is a sensitive and robust tool to characterize root architectural phenotypes using a rapid and simple protocol.
A Visual Representation of Phenotypic Plasticity Space
We have shown that RootScape can capture many of the individual traits that determine overall RSA. Furthermore, the PCA showed that suites of covarying traits can be summarized in single axes or PCs, capturing similar trends compared with the parallel analysis of individual traits. Thus, RootScape, in combination with PCA, offers a way to rapidly summarize the complex characteristics of RSA in a given genotype or background, as has been done for other traits (Adams, 2010). The first two PCs from RootScape capture about 87% of the total root shape variation for the allometric Col-0 plasticity space (Fig. 2). These PCs represent suites of individual root traits (Table I; Supplemental Table S1). The RootScape PC space, therefore, provides the opportunity to quantify and visualize trends in root architecture in two dimensions, providing a visual overview of phenotypic space.
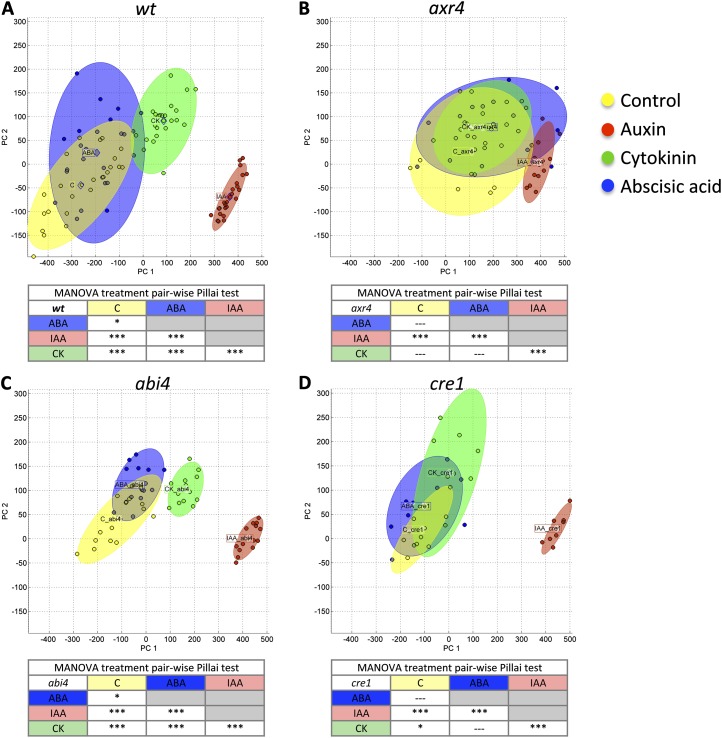
To compare the root phenotypes of the various Arabidopsis hormone signaling mutants, we plotted PC1RS against PC2RS values of root shapes for each genotype (Col-0, axr4, abi4, and cre1) on four treatment media (control, IAA, CK, and ABA; Fig. 4). By plotting PC1RS against PC2RS values, one can visually assess how the plasticity of the RSA is affected in different genotypes (Fig. 4). For example, as reported previously (Hobbie and Estelle, 1995; Depuydt and Hardtke, 2011) and shown by ANOVA above, the change of root plasticity of axr4 (Fig. 4B) compared with the wild type (Fig. 4A) is visually apparent in the first two PCs of RootScape (87% variation). When wild-type (Col-0) root plasticity space is compared with abi4 (Figs. 3C and 4A), it is evident that the overall trend is maintained. In this case, the appropriate analysis to test whether root architecture is significantly different in the two-dimensional PC space is multivariate ANOVA (MANOVA; Pillai test). We used MANOVA and corrected for multiple pairwise testing by applying a Bonferroni adjustment (α = 0.008). This MANOVA detects differences in the plasticity space occupied by all the different treatments applied to abi4. Mapping the individual samples on the two-dimensional PC space shows that the variability within treatments is much reduced in abi4 without reducing the differences among treatments. This leads to the observation that the abi4 mutant appears to constrain variability within all treatments; such a trait might not have been a part of quantitative measurements in the screening process but becomes apparent with visualization. This shows how this visual representation of RSA space can help screen for complex phenotypes that could be followed up with subsequent quantitative analysis.
Figure 4.
Visualization and MANOVA quantification of RSA phenotypic plasticity in genotypes. Distribution of PC1+ and PC2 values is shown for wild-type (wt) Col-0, axr4-1, abi4-1, and cre1-2 on four different treatments for the allometric Col-0 model quantified by RootScape. The mean of each treatment is shown by a colored diamond, and ellipsoids in the same color represent clouds around the replicates. A, Wild type. B, axr4-1. C, abi4-1. D, cre1-2. A MANOVA summary table for pairwise comparison is shown below each plot (Pillai test with Bonferroni adjustment). *P < 0.008, ***P < 0.00008.
DISCUSSION
This paper presents RootScape, a rapid method for the allometric and integrative quantification of RSA applied to Arabidopsis. The RootScape method uses a 20-point landmark template created in AAMToolbox (a MATLAB plugin), which can rapidly and accurately characterize RSA variation in different genetic backgrounds or treatments. RSA variation was generated experimentally by supplying Arabidopsis plants grown in full Murashige and Skoog (MS) medium with three different hormones known to affect different aspects of root development (IAA, CK, and ABA). The landmark data from the root templates applied to this data set were then used in a PCA. These PC values were then used for correlation and multiple regression analyses in order to compare how the RootScape method is related to standard measurements of individual root traits. This analysis showed that RootScape is able to recapitulate very similar trends of variation and is able to capture trends exhibited by nearly all individual traits. Additionally, we tested three mutant lines in hormone signaling known to affect root development and validated the RootScape technique by recapitulating previously characterized phenotypes and potentially describing new aspects of mutant phenotypes.
In a typical analysis with RootScape, a user could apply simple criteria for comparing root architectures, such as a significant difference in one or several PCs, between wild-type and mutant samples. Examination of components can identify the root system properties that are changing along a PC.
The RootScape template of 20 landmarks is meant to quantify RSA as a comprehensive or holistic phenotype (see “Results”). However, it might be argued that the template can have a different (or more detailed) set of landmarks to enable a more detailed quantification of RSA. In such a case, a new template can easily be created that is designed to record specific aspects of root architecture. The current 20-landmark model design captures intuitive and well-established features of the RSA with a minimal number of landmarks, permitting high-throughput phenotyping. The secondary landmarks add sensitivity to detect a range of phenotypes, such as bending of the root in agravitropic mutants. However, we note that the primary intention of RootScape is rapid screening of RSA of Arabidopsis seedlings grown vertically on agar plates. It may not capture all possible defects in root architecture. For example, the RootScape method does not measure the diameter of roots, and subtle bending or waving phenotypes may be missed.
We show that the first PC captured by RootScape, PC1RS, has the highest correlation with the first PC derived from measurements of individual traits (PC1IT), which mainly captures traits that are related to or functions of primary root length (e.g. root length ratio, growth of the primary root, root formation zone, and branching density). The second PC2RS is significantly correlated with PC2IT, in which total root length and lateral root number have major contributions. We note that RootScape does not explicitly measure traits like lateral root number or branching density, but it appears that these traits correlate with other root features that RootScape does effectively capture. RootScape also effectively captures other traits that are based on ratios of two important root traits, for example, length ratio (total lateral root length to primary root length). The important point is that RootScape captures trends in the variation of root architecture that are described by standard measurements, even if they were not directly measured. The first and second PCs captured by RootScape significantly correlate with more than one PC of individual traits, signifying the integrative nature of RSA captured by RootScape.
One interesting finding is that lateral root length is captured by three different RootScape PCs (PC3RS, PC4RS, and PC5RS). The RootScape template was designed to capture the set of lateral roots that grew farthest away from the primary root axes. In addition, RootScape measures the extent of lateral root outgrowths along the primary root. This design appeared to effectively capture both the length of the lateral roots and much of the shape of the space covered by lateral roots on the two-dimensional plate. This enabled us to measure different ways in which lateral root length influences root architecture, as multivariate analysis showed that this trait contributed to three different RootScape-derived PCs. These findings are consistent with earlier opinions that simple traits like the length of roots and total root mass are unable to explain the RSA variation and complexity, while spatial configuration and topology have more crucial impacts on RSA (Fitter and Stickland, 1991). Correlation and multiple regression analyses indicate that the first two PCs of RootScape are correlated with more than one PC from the individual traits. This shows that PCA analysis on RootScape groups the covariation among traits somewhat differently than PCA derived from an individual trait analysis, as can be visualized in PC “walks” (Supplemental Video S1) that represent the variation in root forms along a PC. Thus, RootScape measures the spatial configuration of roots to provide a new view of RSA, one that adheres to the concept that roots are integrated organs (Fitter, 1987; Fitter and Stickland, 1991; Lynch, 1995).
We generated experimental RSA variation by supplementing wild-type Arabidopsis (Col) with three different hormones, all known to affect root development. One of the hormones, auxin, has a dramatic effect on the root phenotype, inhibiting new outgrowth of the primary root and stimulating the initiation of new lateral roots while inhibiting their elongation. It is possible that because of this strong effect, the RSA variation created in our experimental design will be driven by auxin treatment. This appears to be the case, since PC1RS captures mainly variations in primary root length or overall size. However, this trend of capturing a size effect in the first PC is observed in separate landmark-based quantifications of leaf shape (Langlade et al., 2005; Bensmihen et al., 2008). We have also used the RootScape method for RSA quantification on 69 Arabidopsis ecotypes (U. Rosas, A. Cibrian-Jaramillo, D. Ristova, J. Banta, M. Gifford, A.H. Fan, R.W. Zhou, G.J. Kim, G. Krouk, K.D. Birnbaum, M.D. Purugganan, and G.M. Coruzzi, unpublished data) in one environment and found that the first PC is also driven by the size effect, even if no auxin treatment is applied.
Testing the RSA plasticity space in hormone signaling mutants with RootScape followed by two-way ANOVA confirmed most of previously reported phenotypes and hormone interactions in the mutant lines. For example, axr4 was isolated for its resistance to auxin treatment and also shows variable resistance to ABA (Hobbie and Estelle, 1995). In the same study (Hobbie and Estelle, 1995), axr4 was reported to be sensitive to a particular kinetin concentration. Here, we were able to use RootScape to confirm the interaction of axr4 with auxin (PC1RS and PC4RS), with ABA (PC1RS, PC3RS, and PC4 RS), and with CK (PC1RS–PC5RS). In addition, cre1 was originally isolated as having reduced sensitivity to cytokinin in the inhibitory effect on primary root growth (Inoue et al., 2001). We confirmed this interaction of cre1 with CK (PC1RS and PC5RS). Additionally, our two-way ANOVA results indicate interactions of cre1 with auxin (PC1RS) and ABA (PC3RS) and an interaction of abi4 with CK (PC1RS). These findings support the results from the correlation and multiple regression analyses that RootScape is able to recapitulate very similar trends in RSA variation to classical measurements of individual root traits. In addition, RootScape can potentially identify new interactions of genotypes with treatments. We identified two new genotype × environment interactions (cre1 × IAA, abi4 × CK) as well as three previously identified interactions (axr4 × ABA, axr4 × CK, cre1 × ABA; Hobbie and Estelle, 1995; Inoue et al., 2001). This result demonstrates that RootScape can be used as a sensitive screening tool to explore the RSA plasticity space of mutant lines (alleles or ecotypes) exposed to different environments in Arabidopsis. Furthermore, with ANOVA and/or MANOVA analysis, this plasticity of RSA and sensitivity can be visualized and quantified.
The experimental design presented here provides quantification of RSA at a single time point, at 12 d after germination. However, there is a need to observe the dynamics of root development, as recently advised (De Smet et al., 2012; Wells et al., 2012). In a pilot experiment, we tested if RootScape is able to distinguish RSA changes in different developmental stages of the Col-0 genotype grown on a single medium, 1 mm KNO3. In this analysis, PC110vs.14 showed a significant difference between day 10 and day 14 (Supplemental Fig. S3; Supplemental Text S1), confirming that RootScape can also be used for the quantification of RSA dynamics.
RSA differs between species by an enormous degree. We have developed RootScape, and its current template, using Arabidopsis as a model system. To be able to apply RootScape to other species, it will be necessary to adapt an optimal landmark template. This could potentially be a complex task, since the diversity of RSA is vast among ecotypes of the same species and even more diverse among different species. The current 20-landmark template developed for Arabidopsis is suitable for the quantification of any root system that has a dominant primary root. Therefore, we tested this 20-point template on a pilot set of Medicago truncatula ecotypes. Five M. truncatula ecotypes (Longi, A17, 2HA, Gaerta, and Tribu) were grown on 1 mm KNO3 for 14 d, and RSA was quantified with RootScape using the current template. This pilot study on a small number of ecotypes and replicates reveals a set of PCs that capture differences in RSA between the ecotypes captured mainly in the first five PCs (Supplemental Fig. S4; Supplemental Text S1).
In summary, RootScape, a rapid, cost-effective method to capture RSA allometry, is appropriate for studying and quantifying a wide range of questions in root biology in the model Arabidopsis and can be adapted to other species. We demonstrate that RootScape is able to quantify and recapitulate known root phenotypes from the literature. Characterization of RSA plasticity among different genotypes (e.g. mutants, alleles, or ecotypes) on a range of treatments could also be investigated by the same analysis. These examples support the notion of a potentially wide application of RootScape in different areas of high-throughput root studies in plants.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) genotypes used in this study were Col-0, axr4-1 (Hobbie and Estelle, 1995), abi4-1 (Signora et al., 2001), and cre1-2 (Inoue et al., 2001). Medicago truncatula ecotypes used were 2HA, Gaerta, Longi (M. truncatula var longispina), Tribu (M. truncatula ssp. tribuloides), and A17.
Experimental Design and Treatments
To generate variation in the RSA, we first grew 15 seeds per plate of the Col-0 wild type on full MS basal medium (M 5524; Sigma) with 0.1% Suc as a carbon source, 0.05% MES sodium salt (Sigma), pH 5.7, and 1% Bacto Agar (BD) for 7 d. Seeds were first surface sterilized, plated on 10- × 10-cm2 plates (without tape or Parafilm), and then stratified for 3 to 5 d in the dark at 4°C. Plates were set up vertically in a Percival growth incubator (Intellus) at 22°C with a 16-h/8-h light/dark cycle and a light intensity of 50 μmol m−2 s−1. After 7 d on each plate, five out of the 15 most uniform plants were chosen and transferred onto a new plate with different media as described in Supplemental Figure S1: full MS control or full MS supplemented with 500 nmol of IAA (Sigma), 500 nmol of CK (Sigma), or 1 µmol of ABA (Sigma). Each experiment was performed twice, with a total of 24 to 25 plants per treatment. The same procedure was applied for the mutant genotypes, where we carried out two experimental replicates and used a total of 10 to 15 plant replicates per genotype per treatment. Five days after the transfer of the 7-d-old plants to different media, plates were scanned (Epson Perfection V350 Photo) at 300 dots per inch and images were obtained.
Individual Trait Analysis of RSA
We scored 12 individual root traits quantified as described previously (Remans et al., 2006; Ruffel et al., 2011; Dubrovsky and Forde, 2012). Briefly, we used Optimas6 software (MediaCybernetics) or Image J (Abramoff et al., 2004) that allows one to completely draw the root system and export the main numerical values. We note that Optimas software is no longer commercially available, however, Image J (Abramoff et al., 2004) and other freely available software give the same results. We measured primary root length on the transfer day, primary root length growth after 5 d from the transfer, lateral root number, lateral root number in primary root, root branching zone, root formation zone, and average lateral root length. Other traits are obtained from these main traits by applying appropriate calculations (for details, see Supplemental Text S1 and Supplemental Fig. S1). Using this analysis, we quantified Col-0 plants (24–25 plants per treatment) in four different treatments (control, IAA, CK, and ABA) and later performed PCA in order to compare with RootScape.
RootScape, Integrative and Allometric Quantification of RSA
The same plants quantified by the individual trait analysis were also measured by RootScape. Three insertion mutations (axr4-1, cre1-2, and abi4-1; 10–15 plants per treatment per genotype) were quantified by this method. Twenty landmarks (six primary and 14 secondary) were fitted along the length of the primary (main) and lateral (secondary) roots by placing the landmarks at key and recognizable positions of RSA: the start and end of the primary root, the first and last lateral roots on the main root, and the widest points of lateral roots on each side of the main root (Fig. 1). The PCA model of root shape and size was created from the 20-point model in the wild-type Col-0 treatment data set. In this wild-type plasticity space, we projected the model shapes for the three mutant lines and obtained PC values for later statistical analysis. Models were generated using version 1 of the AAMToolbox MATLAB plugin, available free at http://cmpdartsvr1.cmp.uea.ac.uk/wiki/BanghamLab/index.php/Software. The user manual can be downloaded at the following link:http://lemur.cmp.uea.ac.uk/Research/cbg/Documents/Bangham-Coen-Group/AAMToolbox/AAMToolbox.htm.
Procrustes Alignment, Normalization, and Representation of Shapes
For each analyzed root, root outlines were normalized using the Procrustes method, where each root shape is transformed to a mean shape using iterative translation and rotation of the landmark data generating superimposition that minimizes the aberrations from the overall mean root shape (Bensmihen et al., 2008).
Statistical Analysis
One-way ANOVA for treatment in the wild-type Col-0 data set for the 12 individual traits was performed in JMP 9. In JMP, we also performed two-way ANOVA, PCA of individual traits, correlation, and multiple regression analysis. RSA shape and size images (RootScape) were obtained in AAMToolbox in MATLAB. PCA for RootScape was also performed in AAMToolbox in MATLAB. Pairwise MANOVA comparison was done in R.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Experimental design of the study.
Supplemental Figure S2. Application of 20-landmark template of RootScape.
Supplemental Figure S3. Quantification of developmental stages with RootScape.
Supplemental Figure S4. Quantification of M. truncatula with RootScape.
Supplemental Table S1. Multiple regression of each of the first five principal components of RootScape.
Supplemental Table S2. Multiple regression of each of the first five principal components of individual trait analysis.
Supplemental Video S1. Variability of the root system shape and size of Arabidopsis Col-0 quantified with RootScape.
Supplemental Text S1. RootScape template.
Acknowledgments
We thank Lawrence Hobbie for providing axr4-1 seeds and Thomas Schmülling and Michael Riefler for providing cre1-2 seeds. We thank Miriam L. Gifford for providing seeds of M. truncatula ecotypes. We thank Daniel Tranchina for guidance on multiple regression analysis and Miriam L. Gifford and Amy Marshall Colon for helpful comments and suggestions.
Glossary
- RSA
root system architecture
- Col-0
Columbia
- ABA
abscisic acid
- MANOVA
multivariate ANOVA
- IAA
indole-3-acetic acid
- CK
cytokinin
- PCA
principal component analysis
- PC
principal component
- MS
Murashige and Skoog
References
- Abramoff MD, Magelhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Adams DC. (2010) Parallel evolution of character displacement driven by competitive selection in terrestrial salamanders. BMC Evol Biol 10: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Hanna AI, Langlade NB, Micol JL, Bangham A, Coen ES. (2008) Mutational spaces for leaf shape and size. HFSP J 2: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Benková E, Helariutta Y. (2011) Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr Opin Plant Biol 14: 10–16 [DOI] [PubMed] [Google Scholar]
- Den Herder G, Van Isterdael G, Beeckman T, De Smet I. (2010) The roots of a new green revolution. Trends Plant Sci 15: 600–607 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Hardtke CS. (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21: R365–R373 [DOI] [PubMed] [Google Scholar]
- De Smet I, White PJ, Bengough AG, Dupuy L, Parizot B, Casimiro I, Heidstra R, Laskowski M, Lepetit M, Hochholdinger F, et al. (2012) Analyzing lateral root development: how to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Forde BG. (2012) Quantitative analysis of lateral root development: pitfalls and how to avoid them. Plant Cell 24: 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter AH. (1987) An architectural approach to the comparative ecology of plant-root systems. New Phytol 106: 61–77 [Google Scholar]
- Fitter AH, Stickland TR. (1991) Architectural analysis of plant-root systems. 2. Influence of nutrient supply on architecture in contrasting plant-species. New Phytol 118: 383–389 [Google Scholar]
- Hobbie L, Estelle M. (1995) The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J 7: 211–220 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao YL, Belcher H, Harer J, Weitz JS, Benfey PN. (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlade NB, Feng XZ, Dransfield T, Copsey L, Hanna AI, Thébaud C, Bangham A, Hudson A, Coen E. (2005) Evolution through genetically controlled allometry space. Proc Natl Acad Sci USA 102: 10221–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Pagès L, Draye X. (2011) A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 157: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Matthews I, Cootes TF, Bangham JA, Cox S, Harvey R. (2002) Extraction of visual features for lipreading. IEEE Trans Pattern Anal Mach Intell 24: 198–213 [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Perret JS, Al-Belushi ME, Deadman M. (2007) Non-destructive visualization and quantification of roots using computed tomography. Soil Biol Biochem 39: 391–399 [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD, Sun X. (2005) Crosstalk between ABA and auxin signaling pathways in roots of Arabidopsis thaliana (L.) Heynh. Planta 222: 98–106 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H. (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Topp CN, Benfey PN. (2012) Growth control of root architecture. In A Altman, PM Hasegawa, eds, Plant Biotechnology and Agriculture. Elsevier, London, UK, pp 373–386 [Google Scholar]
- Tracy SR, Roberts JA, Black CR, McNeill A, Davidson R, Mooney SJ. (2010) The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. J Exp Bot 61: 311–313 [DOI] [PubMed] [Google Scholar]
- Van As H. (2007) Intact plant MRI for the study of cell water relations, membrane permeability, cell-to-cell and long distance water transport. J Exp Bot 58: 743–756 [DOI] [PubMed] [Google Scholar]
- Wells DM, French AP, Naeem A, Ishaq O, Traini R, Hijazi HI, Bennett MJ, Pridmore TP. (2012) Recovering the dynamics of root growth and development using novel image acquisition and analysis methods. Philos Trans R Soc Lond B Biol Sci 367: 1517–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Ingram PA, Benfey PN, Elich T. (2011) From lab to field, new approaches to phenotyping root system architecture. Curr Opin Plant Biol 14: 310–317 [DOI] [PubMed] [Google Scholar]