Abstract
An evolutionary argument supports the conclusion that plants do not have G protein coupled receptors.
In the classic 1942 movie Casablanca, Vichy Police Captain Louis Renault obfuscated the truth by commanding his lieutenants to “round up the usual suspects,” knowing well that the culprit with the gun stood in plain view. Something similar has happened in the plant G protein field. This Scientific Correspondence was written to shed light on the source of misunderstanding and to preempt further confusion. Plant heterotrimeric G proteins are self-activating and therefore do not need and do not utilize G protein-coupled receptors (GPCRs). This conclusion was reached previously from biochemical analyses of plant G proteins (Johnston et al., 2007a; Urano et al., 2012); here, we buttress this point of view using an evolutionary argument. Proteins suspected as plant GPCRs were “rounded up” because they have the predicted topology of animal GPCRs and/or have been misannotated as such; however, these proteins are highly conserved in organisms that lack heterotrimeric G proteins. Therefore, they have functions unrelated to G-coupled signaling. Instead, the culprit protein standing in plain view is a receptor GTPase-accelerating protein (GAP), a receptor GAP called AtRGS1 (for regulator of G signaling).
GPCRS ARE RECEPTOR GEFS
In animals and fungi, GPCRs are cell surface receptors that perceive a wide spectrum of signals. The human genome encodes about 850 different GPCRs, which constitute the largest gene family (Nordström et al., 2011). They are involved in the perception of various external signals, like light, neurotransmitters, or peptide hormones/pheromones, even proteolytic activity. As the name designates, GPCRs are coupled to a cytoplasmic, membrane-tethered, heterotrimeric GTP-binding complex composed of a Gα subunit partnered to an obligate Gβγ dimer. Gα tightly binds GDP in the heterotrimeric deactivated complex. Upon receptor activation by its signal, the GPCR pries away the GDP from Gα and stabilizes the open state of the complex (Chung et al., 2011; Rasmussen et al., 2011), allowing GTP, which is in excess over GDP in animal cells, to bind Gα (Schneider and Seifert, 2010). Thus, GPCRs should be considered as receptor enzymes having Guanine Nucleotide Exchange Factor (GEF) activity. In other words, GPCRs are receptor GEFs. The GTP-bound form of Gα changes its conformation to activate downstream effectors that consequently alter cell behavior. Subsequently, the intrinsic GTP hydrolysis property of the Gα subunit returns Gα to the inactive GDP-bound form, allowing reassociation of the heterotrimer, the resting state of this complex. This hydrolysis “back reaction” is often stimulated by a GAP RGS protein.
The defining topological feature of GPCRs is the seven transmembrane-spanning (7TM) domain, and it is this topological feature that is its most conserved feature. However, conservation at the amino acid sequence level is poor among GPCRs, even within a single species. Unfortunately, the 7TM topology alone is often used as evidence to annotate divergent 7TM-encoding genes as GPCRs, although, as discussed below, methods that do not rely on sequence alignments have been used to assemble 7TM receptor candidate (Moriyama and Kim, 2006; Moriyama et al., 2006; Gookin et al., 2008; Lu et al., 2009).
The definitive test for a GPCR is GEF activity, but GEF activity has been demonstrated for only well-studied GPCRs. This is an onerous criterion for classification that has relaxed over time and with the deluge of new genomes to annotate.
PLANT G PROTEINS ARE SELF-ACTIVATING
In 2007, Francis Willard and colleagues (Johnston et al., 2007a) showed that the Arabidopsis (Arabidopsis thaliana) Gα subunit (AtGPA1) is the fastest known nucleotide-exchanging G protein, having an astonishingly 200 times faster rate than the typical animal Gα subunit (Jones et al., 2011a, 2011b, 2012). The spontaneous exchange property of AtGPA1 paired with its slow GTPase property indicates that AtGPA1 is 99% occupied by GTP in vitro. Evidence supports the conclusion that all plant Gα subunits spontaneously load GTP (Urano et al., 2012), and while this cycling property means that nearly all Gα is in its active form in the test tube, that is not the case in planta. Increasing the amount of GTP-bound AtGPA1 in plant cells confers “active” phenotypes in vivo (Ullah et al., 2001; Chen et al., 2003). This means that the GTP-bound form is the active Gα form in plant cells, just as it is in animal cells, and that an unknown element in plant cells must be controlling this active state. That element is not a GPCR.
NOT ALL 7TM PROTEINS ARE GPCRS
Possession of a 7TM domain does not justify a GPCR moniker. The insect odorant receptors, originally discussed as GPCRs, are ligand-gated cation channels with the N terminus inside the cell (Sato et al., 2008; Wicher et al., 2008). Further examples are the green algae light sensor, which is homologous to bacteriorhodopsin and functions as a light-activated channel (Nagel et al., 2002), the microbial type I rhodopsin that functions as an ion pump (Oesterhelt and Stoeckenius, 1971), the human and fungal adiponectin receptors, which have ceramidase activity (Kupchak et al., 2009; Villa et al., 2009), and the bacterial homolog hemolysin III, which has hemolysis activity (Baida and Kuzmin, 1996). In addition, some human genes annotated as orphan receptors are likely not GPCRs. Notably, the human GPR89 (NP_001091081), GPR107 (NP_001130029), GPR108 (NP_001073921), adiponectin receptors (NP_057083 and NP_078827), and GPR175 (NP_001129525) have no sequence similarity to any characterized GPCRs (Tang et al., 2005; Nordström et al., 2011), and there is no evidence that they function as GPCRs. As will be discussed later, plant proteins with homologies to these faux GPCRs discussed above and those with predicted 7TMs are still annotated as candidate GPCRs in the databases. Misannotation is one source of the plant GPCR problem.
PLANTS LACK GPCRS WITH ANIMAL AND FUNGAL HOMOLOGY
Mining genomes for divergent GPCRs is a daunting, if not impossible, task because GPCRs evolved at a rapid pace (Fredriksson and Schiöth, 2005). Therefore, in 2006, as a fresh approach to solve this problem, Etsuko Moriyama and colleagues avoided comparing sequences by using nonconventional algorithms (Hill et al., 2002) to assemble a set of 54 candidate Arabidopsis 7TM receptors (Moriyama et al., 2006). Two years later, this work was extended to rice (Oryza sativa) proteins (Gookin et al., 2008). Included in this set are G PROTEIN-COUPLED RECEPTOR1 (GCR1), 15 MILDEW-RESISTANCE LOCUS O (MLO) proteins, five HEPTAHELICAL PROTEIN (HHP) proteins, AtRGS1, TOBAMOVIRUS replication protein (TOM), and CANDIDATE GPCR (CAND) proteins. None of these are plant GPCRs for the reasons described below.
GCR1
Except for GCR1, no plant protein carries any vestige of GPCR homology. GCR1 homology to Dictyostelium discoideum cAMP Receptor1 (cAR1) lies in the third and fourth transmembrane spans and is weak at best. Even if GCR1 is homologous to cAR1, it is still not clear whether cAR1, or at least the ancestor of cAR1, was a GPCR. Furthermore, there is no biochemical proof that D. discoideum cAR1 has GEF activity, although there is indirect evidence showing that cAMP-induced FRET changes between Gα and Gβ subunits (Janetopoulos et al., 2001). In lieu of direct biochemical proof that cAR1 is a receptor GEF, we turn to evolution to assess its function. D. discoideum cAR1 may be the closest extant protein to the ancestor of animal GPCRs, but this ancestor was probably not a GPCR (Krishnan et al., 2012). cAR1 is extant broadly in eukaryotes, notably found in alveolata, red algae, and green algae, but each of these groups lacks a G protein signaling system; therefore, the cAR1 homologs are not likely to activate G proteins (Fig. 1; Supplemental Fig. S1). Based on our argument that the cAR1 ancestor from which GCR1 evolved was not a GPCR, we conclude that GCR1 does not activate G proteins.
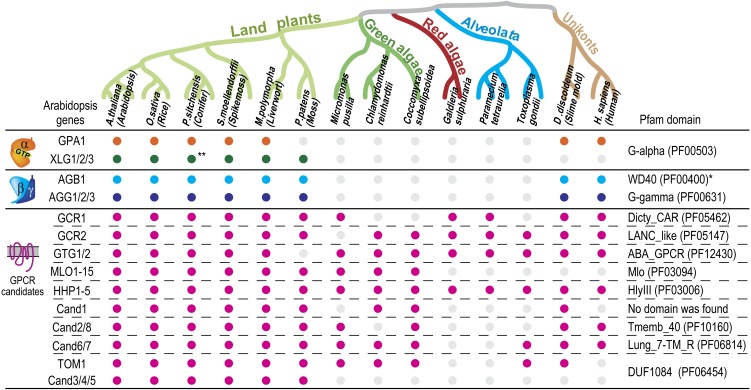
Figure 1.
Gene conservation of G protein components and plant GPCR candidates. Genes homologous to Gα, Gβ, Gγ, and plant GPCR candidates were identified as mentioned in Supplemental Materials and Methods S1. The Pfam domain was determined using Arabidopsis genes shown on the left of the table. Color dots indicate gene conservation. Phylogenetic trees for GPCR candidates are available in Supplemental Materials and Methods S1. *WD40 (PF00400) contains Gβ and other proteins possessing WD40 repeats. **Picea sitchensis XLG is currently not registered in the National Center for Biotechnology Information database but found in EST data for Picea glauca. [See online article for color version of this figure.]
Other reasons that preclude GPCR functionality for GCR1 have been discussed (Johnston et al., 2008). In addition, genetic epistasis shows that GCR1 and G proteins act independently in at least some signaling pathways. GCR1 was reported to interact physically with AtGPA1, but we have not been able to confirm that result (J. Huang and A.M. Jones, unpublished data), and deep screens for G protein and GCR1 partners have yet to suggest a GCR1-Gα interaction.
MLO
In 2002, Ralph Panstruga and colleagues showed, using loss-of-function mutations, that the fungal resistance role for MLO proteins is independent of G proteins (Kim et al., 2002). One might argue that this finding does not exclude coupling to G proteins, since the endogenous function of MLOs is unknown; however, the evidence to date does not suggest that MLO proteins regulate the activation state of G proteins. Epistasis analysis does not indicate that G proteins and MLOs share the same signaling pathway, which is consistent with the conclusion that MLOs are not coupled to G proteins. It should be noted that among the entire set of candidate plant GPCRs, only barley (Hordeum vulgare) MLO1 was confirmed biochemically to have a 7TM domain (Devoto et al., 1999); as such, we emphasize that we are only refuting the existence of plant GPCRs (receptor GEFs), not plant 7TM proteins.
HHP1 to HHP5
HHPs were proposed as GPCR candidates based on their similarity to human progestin and adipoQ receptors (PAQRs; Tang et al., 2005; Gookin et al., 2008). However, human PAQRs have no homology to GPCRs (Tang et al., 2005); rather, they have significant similarity to hemolysin III (Pfam:PF03006; Baida and Kuzmin, 1996), with a topology unlike GPCRs (Yamauchi et al., 2003). While PAQRs stimulate inhibitory G protein pathways (Thomas et al., 2006, 2007; Thomas, 2008), they do so by acting as ceramidases (Kupchak et al., 2009; Villa et al., 2009), which produce sphingolipids (Moussatche and Lyons, 2012). Sphingolipids are well-known ligands for GPCRs (Spiegel and Milstien, 2003) and, hence, the root of this HHP confusion.
GCR2 and GPCR-TYPE G Proteins
Finally, although neither GCR2 nor GPCR-TYPE G (GTG) protein were retrieved by 7TMR search engines, these proteins were originally misannotated as Arabidopsis GPCRs in GenBank and accepted into the plant biology community without a healthy dose of skepticism. GCR2 shares sequence similarity with a cytoplasmic protein homologous to the prokaryotic enzyme lanthionine synthase (Bauer et al., 2000; Mayer et al., 2001). Despite that, Liu et al. (2007) proclaimed GCR2 to be a GPCR, and subsequently, the data from the original publication were quickly refuted (Gao et al., 2003, 2008; Johnston et al., 2007b; Illingworth et al., 2008). Careful examination of topological predictions for GTG1 and GTG2 indicate that these proteins have eight or nine transmembrane domains; eight would be consistent with the authors’ own split-ubiquitin yeast (Saccharomyces cerevisiae) complementation data (Pandey et al., 2009). GTGs are most likely Golgi ion transporters, based on their homologous animal counterparts (Maeda et al., 2008), consistent with GTG localization to the Golgi apparatus (Jaffé et al., 2012).
CAND and TOM1
Gookin et al. (2008) used whole-proteome analysis and reported several other Arabidopsis GPCR candidates, including CAND proteins (Supplemental Figs. S4–S6), unfortunately not to be confused with other Arabidopsis proteins of the prototype abbreviation but a different name, CULLIN-ASSOCIATED AND NEDDYLATION-DISSOCIATED (Zhang et al., 2008). The authors reported interaction of several CAND proteins with AtGPA1, using a yeast complementation assay (Gookin et al., 2008). CAND6 and CAND7 are homologous to human GPR107 and GPR108, and CAND2 and CAND8 are similar to human GPR175/TPRA40 (Vassilatis et al., 2003; Aki et al., 2008; Nordström et al., 2011). Notably, these human proteins possess no sequence similarity to GPCRs and are now classified into non-GPCR domain families (Fig. 1). These authors also proposed that TOM1 and the distant homologs (CAND3/CAND4/CAND5) were candidate plant GPCRs (Supplemental Fig. S4), although there is no homology between these proteins and GPCRs. TOM proteins have a domain of unknown function (DUF1084) not found in animal genomes.
THE FUNCTIONS OF CANDIDATE PLANT GPCRS PREDATE THE ORIGIN OF G PROTEINS
Figure 1 shows the distribution of genes encoding G protein subunits (Gα, Gβ, and Gγ) and GPCR candidates. Gα, Gβ, and Gγ genes are lacking within certain evolutionary clades, such as red and green algae and alveolata (Anantharaman et al., 2011). The GPCR candidates described above are present in the clades lacking Gα, Gβ, and Gγ genes (Fig. 1; Supplemental Figs. S1–S7). Under the neutral theory of molecular evolution (Kimura, 1968), DNA sequences are mutated randomly and gradually lose their original signature because there is no evolutionary pressure for synonymous mutations to be restored to the original value (Nei, 2005). This is not the case for nonsynonymous mutations. For these, evolutionary constraint is not only applied by the intrinsic molecular function (e.g. catalytic core residues of enzymes) but also by other molecules (i.e. binding surfaces with ligands, proteins, or DNA; Temple et al., 2010). For example, where we see that Gα, Gβ, and Gγ subunits are independently deleted within certain evolutionary clades (Anantharaman et al., 2011), the loss of the collective group is correlated (Anantharaman et al., 2011). In other words, once a genome loses one of the three subunits, there is little genetic constraint to keep the other two genes. On the other hand, when proteins do not evolve rapidly after the loss of a hypothesized protein partner, there is some other constraint. For example, proteins like the candidate plant GPCRs discussed above did not evolve much in the absence of G proteins (in certain organisms), indicating that these proteins have evolutionary constraints that are unrelated to G signaling.
PLANT G PROTEIN CYCLING
The evidence indicates that the regulation of plant G protein cycling is at the hydrolysis step, not the GPCR-requiring nucleotide-exchange step. That means that either a GAP (i.e. an RGS protein) or a GDP-dissociation protein (GDI) is regulating the active state of plant G proteins. A GDI serving this job makes more sense. Assuming that GTP levels in plant cells are in excess of GDP, uncontrolled consumption of GTP promoted by an RGS protein just to keep G protein cycling in the inactive state is energy expensive. Logic dictates that there must be a GDI, rather than a GAP, because GDIs simply “hold” the G protein in its GDP-bound active state and do not promote nucleotide consumption, as do the GAPs. Another reason a GDI makes sense is that not all plants have RGS proteins; cereals and some lower plants have self-activating G proteins but lack a canonical RGS protein (Urano et al., 2012). For these species, we speculate that a switchable (e.g. ligand-regulated) GDI serves the purpose of regulating the plant G protein activation state.
“LOUIS, I THINK THIS IS THE BEGINNING OF A BEAUTIFUL FRIENDSHIP”
The (mis)annotation of a plant protein as a GPCR in a database prompts an irresistible urge to order the mutants from the stock center, phenotype them, and submit the data set for a quick publication, all while riding on the coattails of Nobel laureates who discovered the original and bona fide GPCRs in animal cells. Similarly, obtaining a topological prediction of a 7TM domain in a plant protein should not make us want to “play it again, Sam.” We simply point out that plants do not need, and therefore do not use, animal-like GPCRs to control the active state of heterotrimeric G proteins. Instead of embracing the animal GPCR paradigm, our collective research effort would be more productive if we focused on the mechanism of G cycle regulation in plants in the absence of GPCRs.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenies of AtGCR1, AtGCR2 or AtGTG homologous proteins.
Supplemental Figure S2. Phylogeny of HHP family proteins.
Supplemental Figure S3. Phylogeny of MLO family proteins.
Supplemental Figure S4. Phylogeny of TOM and Cand3 (At3G599090.1) family proteins.
Supplemental Figure S5. Phylogeny of lung 7TM receptor (AtCand6/7) family proteins.
Supplemental Figure S6. Phylogenies of AtCand1 (At1G57680) or AtCand2 (At3G05010) homologous proteins.
Supplemental Figure S7. Phylogeny and multiple sequence alignment of canonical and extra-large Gα proteins.
Supplemental Materials and Methods S1. Data collection and phylogeny construction.
Glossary
- GPCR
G protein-coupled receptor
- GAP
GTPase-accelerating protein
- 7TM
seven transmembrane-spanning
- PAQRs
progestin and adipoQ receptors
- GDI
GDP-dissociation protein
References
- Aki T, Funakoshi T, Nishida-Kitayama J, Mizukami Y. (2008) TPRA40/GPR175 regulates early mouse embryogenesis through functional membrane transport by Sjögren’s syndrome-associated protein NA14. J Cell Physiol 217: 194–206 [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Abhiman S, de Souza RF, Aravind L. (2011) Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 475: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baida GE, Kuzmin NP. (1996) Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta 1284: 122–124 [DOI] [PubMed] [Google Scholar]
- Bauer H, Mayer H, Marchler-Bauer A, Salzer U, Prohaska R. (2000) Characterization of p40/GPR69A as a peripheral membrane protein related to the lantibiotic synthetase component C. Biochem Biophys Res Commun 275: 69–74 [DOI] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731 [DOI] [PubMed] [Google Scholar]
- Chung KY, Rasmussen SGF, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. (2011) Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature 477: 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P. (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274: 34993–35004 [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Schiöth HB. (2005) The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol 67: 1414–1425 [DOI] [PubMed] [Google Scholar]
- Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW. (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang S, Asami T, Chen J-G. (2008) Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein alpha subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol 49: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Gookin TE, Kim J, Assmann SM. (2008) Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice, and poplar: computational prediction and in-vivo protein coupling. Genome Biol 9: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. (2002) G protein-coupled receptors in Anopheles gambiae. Science 298: 176–178 [DOI] [PubMed] [Google Scholar]
- Illingworth CJ, Parkes KE, Snell CR, Mullineaux PM, Reynolds CA. (2008) Criteria for confirming sequence periodicity identified by Fourier transform analysis: application to GCR2, a candidate plant GPCR? Biophys Chem 133: 28–35 [DOI] [PubMed] [Google Scholar]
- Jaffé FW, Freschet G-EC, Valdes BM, Runions J, Terry MJ, Williams LE. (2012) G protein-coupled receptor-type G proteins are required for light-dependent seedling growth and fertility in Arabidopsis. Plant Cell 24: 3649–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C, Jin T, Devreotes P. (2001) Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science 291: 2408–2411 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen J-G, Siderovski DP, Jones AM, Willard FS. (2007a) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104: 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Temple BR, Chen J-G, Gao Y, Moriyama EN, Jones AM, Siderovski DP, Willard FS. (2007b) Comment on “A G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid.” Science 318: 914–, author reply 914. [DOI] [PubMed] [Google Scholar]
- Johnston CA, Willard MD, Kimple AJ, Siderovski DP, Willard FS. (2008) A sweet cycle for Arabidopsis G-proteins: recent discoveries and controversies in plant G-protein signal transduction. Plant Signal Behav 3: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Duffy JW, Machius M, Temple BRS, Dohlman HG, Jones AM. (2011a) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci Signal 4: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Jones AM, Temple BRS, Dohlman HG. (2012) Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Gα proteins. Proc Natl Acad Sci USA 109: 7275–7279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BRS, Jones AM, Dohlman HG. (2011b) Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem 286: 13143–13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P. (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1968) Evolutionary rate at the molecular level. Nature 217: 624–626 [DOI] [PubMed] [Google Scholar]
- Krishnan A, Almén MS, Fredriksson R, Schiöth HB. (2012) The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS ONE 7: e29817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupchak BR, Garitaonandia I, Villa NY, Smith JL, Lyons TJ. (2009) Antagonism of human adiponectin receptors and their membrane progesterone receptor paralogs by TNFalpha and a ceramidase inhibitor. Biochemistry 48: 5504–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. (2007) A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Lu G, Wang Z, Jones AM, Moriyama EN. (2009) 7TMRmine: a Web server for hierarchical mining of 7TMR proteins. BMC Genomics 10: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Ide T, Koike M, Uchiyama Y, Kinoshita T. (2008) GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat Cell Biol 10: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Mayer H, Bauer H, Breuss J, Ziegler S, Prohaska R. (2001) Characterization of rat LANCL1, a novel member of the lanthionine synthetase C-like protein family, highly expressed in testis and brain. Gene 269: 73–80 [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Kim J. (2005) Protein family classification with discriminant function analysis. In JP Gustafson, ed, Data Mining the Genomes: 23rd Stadler Genetics Symposium. Kluwer Academic/Plenum Press, New York, pp 121–132 [Google Scholar]
- Moriyama EN, Strope PK, Opiyo SO, Chen Z, Jones AM. (2006) Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol 7: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, Lyons TJ. (2012) Non-genomic progesterone signalling and its non-canonical receptor. Biochem Soc Trans 40: 200–204 [DOI] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296: 2395–2398 [DOI] [PubMed] [Google Scholar]
- Nei M. (2005) Selectionism and neutralism in molecular evolution. Mol Biol Evol 22: 2318–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJV, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB. (2011) Independent HHsearch, Needleman-Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol 28: 2471–2480 [DOI] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233: 149–152 [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Schneider EH, Seifert R. (2010) Sf9 cells: a versatile model system to investigate the pharmacological properties of G protein-coupled receptors. Pharmacol Ther 128: 387–418 [DOI] [PubMed] [Google Scholar]
- Spiegel S, Milstien S. (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 4: 397–407 [DOI] [PubMed] [Google Scholar]
- Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, Funk WD. (2005) PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol 61: 372–380 [DOI] [PubMed] [Google Scholar]
- Temple B, Jones C, Jones A. (2010) Evolution of a signaling nexus constrained by protein interfaces and conformational states. PLoS Comp Biol 6: e1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. (2008) Characteristics of membrane progestin receptor α (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front Neuroendocrinol 29: 292–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Dressing G, Pang Y, Berg H, Tubbs C, Benninghoff A, Doughty K. (2006) Progestin, estrogen and androgen G-protein coupled receptors in fish gonads. Steroids 71: 310–316 [DOI] [PubMed] [Google Scholar]
- Thomas P, Pang Y, Dong J, Groenen P, Kelder J, de Vlieg J, Zhu Y, Tubbs C. (2007) Steroid and G protein binding characteristics of the seatrout and human progestin membrane receptor α subtypes and their evolutionary origins. Endocrinology 148: 705–718 [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Young JC, Im K-H, Sussman MR, Jones AM. (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM. (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, et al (2003) The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci USA 100: 4903–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa NY, Kupchak BR, Garitaonandia I, Smith JL, Alonso E, Alford C, Cowart LA, Hannun YA, Lyons TJ. (2009) Sphingolipids function as downstream effectors of a fungal PAQR. Mol Pharmacol 75: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, et al. (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423: 762–769 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ito H, Quint M, Huang H, Noël LD, Gray WM. (2008) Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci USA 105: 8470–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]