A mutant of arginine biosynthesis uncovers a novel role for the amino acid and blue light in regulating root meristem function.
Abstract
Arginine is an essential amino acid necessary for protein synthesis and is also a nitrogen storage compound. The genes encoding the enzymes of arginine biosynthesis in plants are not well characterized and have mainly been predicted from homologies to bacterial and fungal genes. We report the cloning and characterization of the TUMOR PRONE5 (TUP5) gene of Arabidopsis (Arabidopsis thaliana) encoding an acetylornithine aminotransferase (ACOAT), catalyzing the fourth step of arginine biosynthesis. The free arginine content was strongly reduced in the chemically induced recessive mutant tup5-1, root growth was restored by supplementation with arginine and its metabolic precursors, and a yeast (Saccharomyces cerevisiae) ACOAT mutant was complemented by TUP5. Two null alleles of TUP5 caused a reduced viability of gametes and embryo lethality, possibly caused by insufficient Arg supply from maternal tissue. TUP5 expression is positively regulated by light, and a TUP5-green fluorescent protein was localized in chloroplasts. tup5-1 has a unique light-dependent short root phenotype. Roots of light-grown tup5-1 seedlings switch from indeterminate growth to determinate growth with arresting cell production and an exhausted root apical meristem. The inhibitory activity was specific for blue light, and the inhibiting light was perceived by the root. Thus, tup5-1 reveals a novel role of amino acids and blue light in regulating root meristem function.
Arg is one of the 20 standard amino acids necessary for the biosynthesis of peptides and proteins. Therefore, it is vital for the functioning of the cell. Arg is a basic amino acid with a very low carbon:nitrogen ratio (6:4), which makes it ideal for nitrogen storage (Llácer et al., 2008). Arg can represent a significant part of the stored nitrogen in protein or as free amino acid in seeds, bulbs, or other parts of plants (Micallef and Shelp, 1989). Arg can account for 11% of the nitrogen pool in Arabidopsis (Arabidopsis thaliana) seeds (VanEtten et al., 1963; Zonia et al., 1995). Besides this, Arg is a precursor of compounds acting in developmental processes. Together with Orn, it is used for the production of polyamines (Slocum, 2005). Furthermore, it is a source of nitric oxide, which plays an important role in germination, defense responses, hormonal signaling, root growth, and flowering (Crawford, 2006; Grün et al., 2006). Taken together, Arg and its metabolism are of central importance in plant biology, but the genes and enzymes of Arg biosynthesis are only known partially.
Arg biosynthesis has been well studied in bacteria and fungi (Cunin et al., 1986; Davis, 1986). The enzymes of plant Arg biosynthesis have partly been characterized biochemically (Shargool et al., 1988), but little is known about the genes encoding these enzymes (Slocum, 2005). First, Orn is synthesized from Glu in bacteria, fungi, and plants in a highly conserved Orn pathway not found in most animals. Arg is then synthesized from Orn by the enzymes of the urea cycle in the Arg pathway (Slocum, 2005). Orn biosynthesis (see the overview in Supplemental Fig. S2) starts with Glu, which, in a first step, is acetylated by the N-acetyl-Glu synthase or by N2-acetyl-Orn:Glu acetyltransferase to form N-acetyl-Glu, which is then phosphorylated in a second step by N-acetyl-Glu kinase to produce N-acetyl-Glu-5-P. The next step consists of the formation of N-acetyl-Glu-5-semialdehyde catalyzed by N-acetyl-Glu-5-P reductase. In the fourth step, an amino group is transferred from a Glu molecule to N-acetyl-Glu-5-semialdehyde by N2-acetyl-Orn aminotransferase (ACOAT), yielding N2-acetyl-Orn. Subsequently, the acetyl residue is either removed by N2-acetyl-Orn:Glu acetyltransferase in the cyclic pathway found in nonenteric bacteria, fungi, and plants (Shargool et al., 1988) or by a putative N2-acetyl-Orn deacetylase in the linear pathway found in enteric bacteria to obtain Orn (Slocum, 2005). In the sixth step, Orn transcarbamylase (OTC) catalyzes the transfer of a carbamoyl residue to Orn, yielding citrulline (Cit). Argininosuccinate is then formed from Cit and Asp by argininosuccinate synthase. In the last step, fumarate is cleaved from argininosuccinate by argininosuccinate lyase, yielding Arg (Slocum, 2005).
Only four Arabidopsis genes that encode three enzymatic steps of Arg biosynthesis have been studied by analyzing mutants or by testing their function biochemically. N-acetyl-Glu kinase activity was shown for At3g57560 (Chen et al., 2006). ven3 and ven6 carry mutations in the large and small subunits of the carbamoyl phosphate synthetase, respectively (Mollá-Morales et al., 2011). Carbamoyl phosphate and Orn are substrates of OTC that produce Cit (Slocum et al., 2000). A transfer DNA (T-DNA) insertion in the OTC gene caused an increased sensitivity to Orn (Quesada et al., 1999).
Not much is known about the putative functions of amino acids beyond their role as building blocks of proteins and storage molecules, and only a few reports indicate a role for them in plant growth and development. One such example is the characterization of a Trp biosynthesis mutant displaying a severe growth defect under high light by Last et al. (1991); another one is the description of a Lys biosynthesis mutant showing altered leaf morphology and mild dwarfism by Song et al. (2004). Amino acid metabolism might also influence root meristem maintenance, as well as root growth and development, as was indicated by mutants of Asp and His biosynthesis (Miesak and Coruzzi, 2002; Mo et al., 2006). It is well documented that endogenous factors controlling root growth, such as phytohormones, are often regulated by exogenous signals, for example, by water and nutrient conditions (López-Bucio et al., 2003; Monshausen and Gilroy, 2009), as well as light (Canamero et al., 2006; Tong et al., 2008). There is evidence that roots contain different photoreceptors, that they are able to perceive light of different wavelengths, and that this regulates root functions (Sakamoto and Briggs, 2002; Canamero et al., 2006; Galen et al., 2007; Tong et al., 2008; Costigan et al., 2011; Dyachok et al., 2011, and references therein). However, a connection between light perception and the control of root development involving amino acid homeostasis as an influencing factor has not yet been reported.
In this article, we describe the characterization of the mutant tumor prone5-1 (tup5-1), which was initially identified in a screen for mutants responding to low concentrations of auxin and cytokinin by callus formation (Riefler, 2001). We found that tup5-1 carries a mutation in the Arabidopsis ACOAT gene. The free Arg content was consistently reduced in tup5-1 compared with the wild type, and root growth was restored in the mutant by supplementation with Arg. An interesting feature of the tup5-1 mutant was a blue light-dependent defect in root meristem maintenance, the inhibiting light being perceived by the root. Our results showed that TUP5 has a crucial role in plant development and that it connects primary metabolism, plant development, and blue light signaling.
RESULTS
The Mutant tup5-1
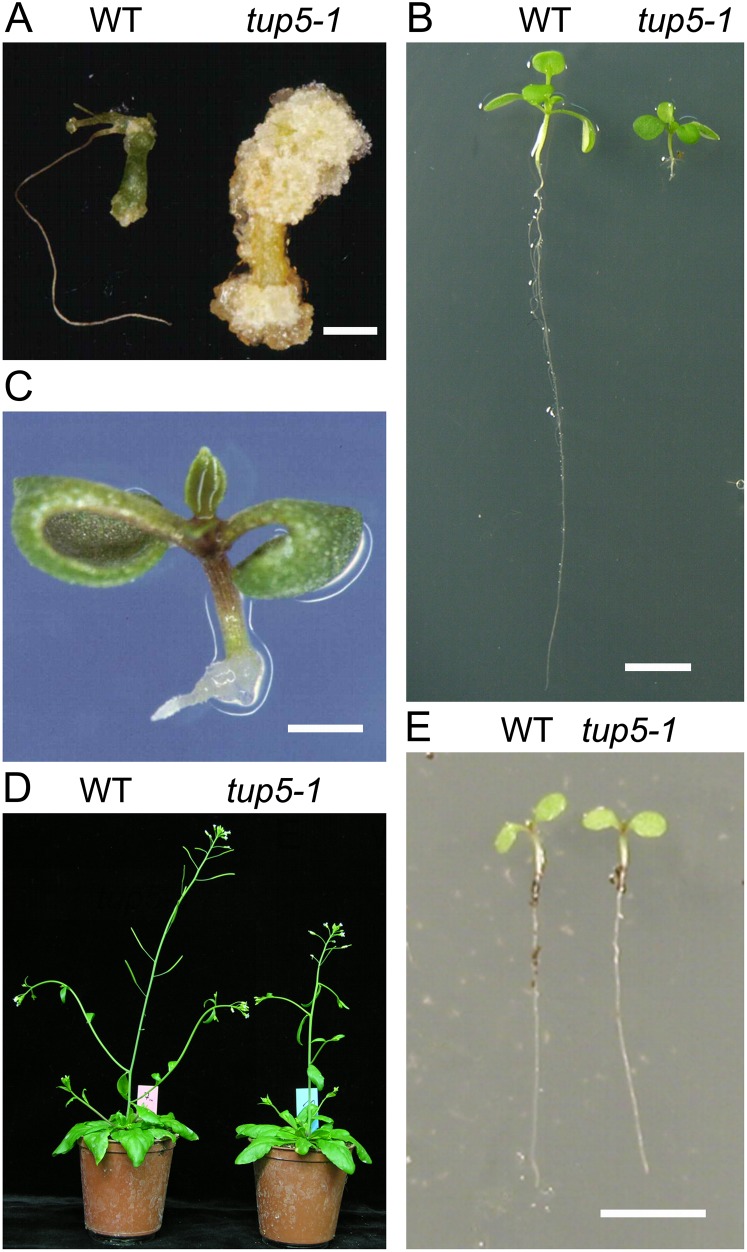
Initially, tup5-1 was found in a screen for mutants that react with higher sensitivity than the wild type to the phytohormones auxin and cytokinin by the formation of tumorous tissue. The recessive ethyl methanesulfonate mutant tup5-1 formed calli on hypocotyl and root explants when grown on medium containing low concentrations of the auxin α-naphthylacetic acid and the cytokinin isopentenyl adenine, which were not effective in the wild type (Fig. 1A; Riefler, 2001). Mutant seedlings had a very short root (no longer than 1–2 mm) when grown in vitro in light, and growth-arrested root primordia formed callus-like structures at the hypocotyl/root junction (Fig. 1, B and C; Riefler, 2001; Frémont, 2004). The shoot of in vitro-grown tup5-1 plants was also impaired, probably due to the root growth arrest (Fig. 1B). In contrast, the shoots of soil-grown tup5-1 mutants were morphologically similar to the wild type, flowered only approximately 2 d later and formed a normal root system (Fig. 1, D and E). This result indicated a potential role of light in inducing root growth inhibition in the tup5-1 mutants, which will be explored further below.
Figure 1.
Root growth is arrested in tup5-1 when grown in vitro, but tup5-1 develops normally on soil. A, Hypocotyl explants of tup5-1 formed callus tissue when cultivated on medium containing low concentrations of α-naphthylacetic acid and isopentenyl Ade, while wild-type explants did not (explants after 6 weeks on medium). B, Root phenotype of 12-d-old wild-type and tup5-1 plants cultivated in vitro under standard light conditions. C, tup5-1 seedling (10 d old) grown in vitro displayed a short primary root and formed adventitious root primordia in a tumorous fashion. D, Shoot phenotype of soil-grown tup5-1 plants compared with the wild type. E, Roots of 5-d-old tup5-1 seedlings developed normally in soil. Bars = 0.2 mm (A), 5 mm (B) and (E), 1 mm (C). WT, Wild type.
TUP5 Encodes a Putative Acetyl-Orn Aminotransferase
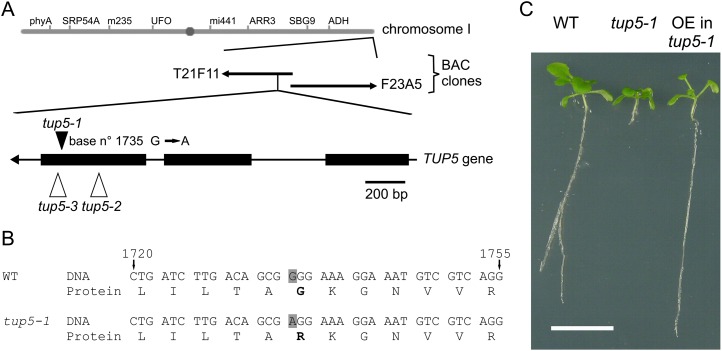
The tup5-1 mutation was located on the lower arm of chromosome I close to the telomere by marker-assisted gene mapping (Fig. 2A). No recombinants were found to further reduce the mapping interval of 88 kb containing 29 predicted genes. Therefore, 4- to 22-kb large subclones containing one to five predicted genes were generated from the two BAC clones T21F11 and F23A5 covering the tup5-1 mapping interval. These subclones were then transformed into tup5-1 plants to screen for root phenotype complementation. Three complementing BAC subclones contained only one predicted gene, At1g80600, in their common region. The tup5-1 allele showed as a single point mutation, a transition from G to A in the third exon of At1g80600 at base number 1735 of the unspliced sequence, leading to an altered amino acid sequence of the predicted protein (G424R; Fig. 2, A and B). Transformation of tup5-1 plants with the complementary DNA (cDNA) of TUP5 under control of the 35S promoter complemented the root phenotype (Fig. 2C), demonstrating that the mutation in At1g80600 caused the tup5-1 phenotype.
Figure 2.
Identification of the tup5-1 mutation and complementation of the phenotype. A, Genetic map position of the TUP5 gene on chromosome I and scheme of the TUP5 gene. The genomic sequence of TUP5 is composed of three exons (indicated as black boxes) and two introns (shown as lines between exons). The locus of the tup5-1 mutation is marked with a black arrowhead, and the positions of the two T-DNA insertions tup5-2 and tup5-3 are shown as white arrowheads. B, Nucleotide and amino acid sequences neighboring the tup5-1 mutation. The tup5-1 mutation (gray shade) is a G-to-A transition at base number 1735 of the genomic sequence leading to a Gly-to-Arg (G424R) exchange (marked in bold letters). C, Overexpression of TUP5 complements the root phenotype of tup5-1. From left to right: wild-type, tup5-1, P35S:TUP5 transgenic seedlings in tup5-1 background grown in vitro (12 d old). Bar = 1 cm. WT, Wild type. [See online article for color version of this figure.]
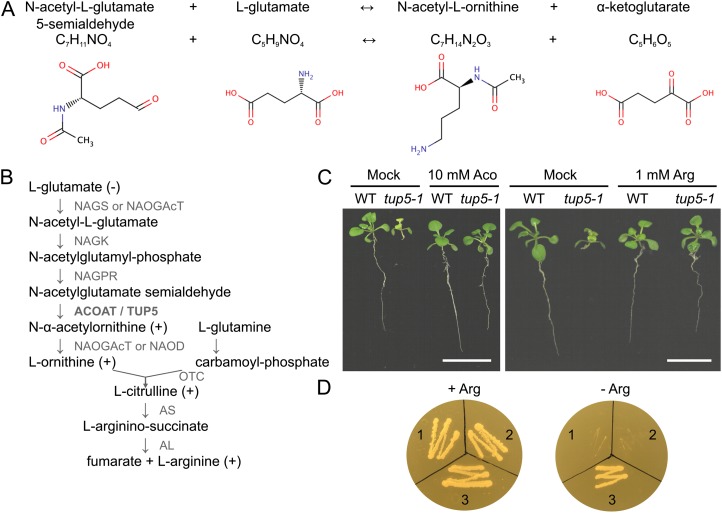
Protein sequence analysis identified TUP5 as a putative ACOAT (EC 2.6.1.11.). The protein is predicted to have a length of 457 amino acids and a Mr of 48.8. The enzyme catalyzes the transfer of an amino group from l-Glu to acetyl-l-Glu 5-semialdehyde, yielding α-ketoglutarate and acetyl-Orn (www.expasy.org/enzyme/2.6.1.11; Fig. 3A), using pyridoxal 5-P as a cofactor (Albrecht and Vogel, 1964). The synthesis of acetyl-Orn is the fourth step (from Glu) in de novo Arg biosynthesis (Slocum, 2005; Fig. 3B). Proteins with high sequence similarity to TUP5 are encoded by bacteria, fungi, and plants. A sequence alignment with different known and putative ACOATs showed that the amino acid mutated in tup5-1 (G424) was conserved in all enzymes from bacteria to higher plants, indicating that it is functionally relevant (Supplemental Fig. S1A). However, in silico analyses using different structure prediction programs have not been informative about the possible impact of the mutation on protein structure and catalytic activity (data not shown). The phylogenetic relationships of the seven aligned proteins correspond to the relationships between the source organisms. TUP5 of Arabidopsis is closely related to the Populus trichocarpa protein and farthest related to the bacterial and fungal ACOAT (Supplemental Fig. S1B).
Figure 3.
TUP5 encodes an acetyl-Orn aminotransferase. A, Chemical reaction catalyzed by ACOAT (skeletal formula of compounds from the Human Metabolome Database; http://www.hmdb.ca). B, Arg biosynthesis pathway. tup5-1 seedlings were supplemented with different compounds of the pathway to test for complementation of the short root phenotype. (−), the substance did not complement; (+), the substance complemented the root phenotype. The enzymes catalyzing the reaction steps of Arg biosynthesis are indicated in gray. NAGS, N-acetyl-Glu synthase; NAOGAcT, N2-acetyl-Orn Glu acetyltransferase; NAGK, N-acetyl-Glu kinase; NAGPR, N-acetyl-Glu-5-P reductase; NAOD, N2-acetyl-Orn deacetylase; AS, argininosuccinate synthase; AL, argininosuccinate lyase (modified from Slocum, 2005). C, Normal root development in tup5-1 when supplemented with acetyl-Orn (Aco) or Arg (14-d-old seedlings in vitro). Bar = 1 cm. WT, Wild type. D, TUP5 restores Arg autotrophy in the ACOAT-deficient yeast mutant Y37711. Left, transformed yeast on medium containing Arg; right, transformed yeast on medium lacking Arg. Y37711 transformed with 1, the empty yeast expression vector p423TEF; 2, vector containing the genomic Arabidopsis TUP5 sequence; and 3, vector containing the Arabidopsis TUP5 cDNA. [See online article for color version of this figure.]
The Root Phenotype of tup5-1 Can Be Complemented by Exogenous Supplementation of Arg
tup5-1 is probably impaired in the biosynthesis of the amino acids that are produced downstream of the ACOAT enzymatic reaction in the Arg biosynthetic pathway (Fig. 3B; Slocum, 2005). Consistent with this assumption, it has been possible to complement the short root phenotype of tup5-1 by supplementation of the medium with acetyl-Orn, l-Orn, l-Cit, and l-Arg (Fig. 3, B and C; data not shown). A number of other metabolites linked to Arg biosynthesis, including different polyamines and the nitric oxide donor sodium nitroprusside, were not effective in restoring the tup5-1 root phenotype, indicating that the mutant phenotype is specifically linked to Arg biosynthesis (Supplemental Fig. S2).
Arabidopsis TUP5 Rescues an ACOAT-Deficient Yeast Mutant
To test whether TUP5 has the predicted function as an ACOAT, we examined its ability to restore Arg autotrophy in the ACOAT-deficient yeast (Saccharomyces cerevisiae) mutant strain Y37711. For this purpose, the genomic Arabidopsis TUP5 sequence and the TUP5 cDNA were cloned downstream of the strong constitutive promoter of the translation elongation factor1α (TEF) gene in yeast vector p423TEF (Mumberg et al., 1995). The yeast mutant transformed with the empty vector p423TEF or the same vector containing the genomic TUP5 sequence were unable to grow on medium lacking Arg (Fig. 3D). In contrast, the yeast mutant Y37711 expressing the TUP5 cDNA became Arg autotrophic (Fig. 3D), thus confirming the function of TUP5 as an ACOAT.
tup5-1 Is Arg Deficient and Shows a General Deregulation of Amino Acid Metabolism
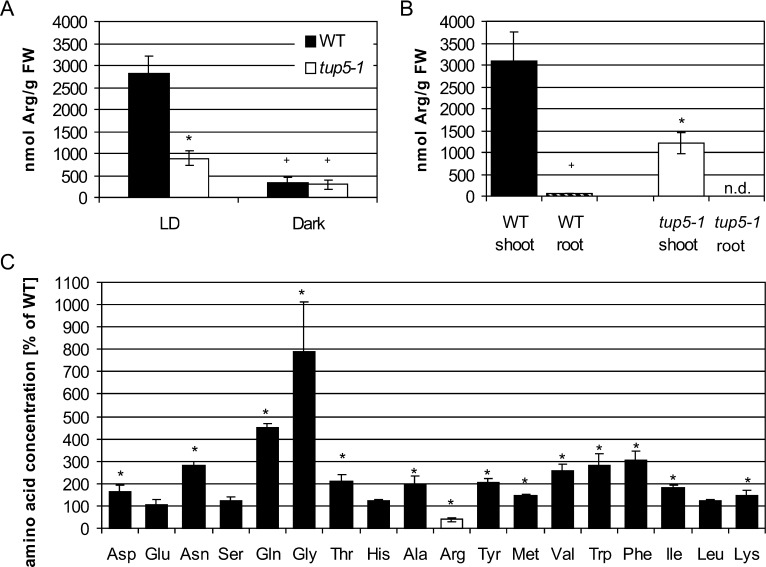
We measured the content of Arg and other amino acids and compared it with the wild-type content to analyze the consequences of the apparent block in Arg biosynthesis in the tup5-1 mutant. The content of free Arg in tup5-1 seedlings grown in vitro under long-day conditions was lowered to 31% of that of the wild type (Fig. 4A). Additional measurements in seedlings of different age (7, 11, and 17 d old) consistently showed a reduction of Arg content between 61% and 85% in tup5-1 (data not shown). In contrast, no difference in Arg content was found between tup5-1 and the wild type grown in darkness. Dark-grown seedlings of both genotypes showed a strong decrease of their Arg content (Fig. 4A).
Figure 4.
tup5-1 plants have a reduced Arg content and a generally deregulated amino acid metabolism. A, Free Arg content of wild-type and tup5-1 seedlings grown in long days (LD) or in darkness. n = 6. B, Free Arg content in wild-type shoots, wild-type roots, and tup5-1 shoots grown in the light. Wild-type shoot, n = 4; wild-type root, n = 3; tup5-1 shoot, n = 4. C, Free amino acid content of tup5-1 shoots compared with wild-type shoots. The concentration of each amino acid in tup5-1 is shown in percentage referring to the values measured in the wild type set as 100%. Arg content is shown as a white column. n = 4. Seedlings were 7 d old and grown in one-half-strength MS liquid culture (A) and 11 d old and grown on vertical plates of MS medium (B and C). Each sample was a pool of at least 30 seedlings. Data are shown as mean (±sd). Significant differences (calculated with Student’s t test) between wild-type and tup5-1 samples are marked with asterisks (*P < 0.05), while significant differences between standard light and other light conditions or tissues within a genotype are marked with a plus sign (+P < 0.05). FW, Fresh weight; WT, wild type; n.d., not determined.
The distribution of free Arg between root and shoot was unequal. Wild-type roots contained about 50 times less Arg than wild-type shoots (61 ± 11 compared with 3084 ± 673 nmol g–1 fresh weight; Fig. 4B). This was the most extreme difference in shoot/root distribution of all amino acids analyzed (Supplemental Fig. S3). The Arg content of tup5-1 shoots was approximately 39% of that of wild-type shoots (1214 ± 252 compared with 3084 ± 673 nmol g–1 fresh weight), which is similar to the difference found in whole seedlings.
We analyzed the content of 18 different amino acids to explore whether the disruption of Arg biosynthesis also affects the level of other amino acids. Out of these, the level of 13 amino acids was increased in 11-d-old seedlings (Fig. 4C). The strongest increases of up to approximately 800% and 400% compared with the wild type were found for Gly and Gln, respectively. Interestingly, only Asn and Gln were increased in tup5-1 in 17-d-old seedlings, while the level of all other amino acids was comparable to the wild type, except for Arg, which was again reduced (Supplemental Fig. S4; data not shown).
Subcellular Localization of TUP5
The TUP5 gene was fused at the 3′ end in frame to a GFP gene and positioned downstream of the 35S promoter to determine the subcellular localization of TUP5 (Karimi et al., 2005). The fusion gene complemented the tup5-1 root phenotype, indicating its functionality (data not shown). The subcellular localization of the TUP5-GFP fusion protein was analyzed by confocal laser scanning microscopy in leaf protoplasts obtained from stably transformed lines in wild-type background. The TUP5-GFP signal colocalized with the autofluorescence of chloroplasts, indicating a chloroplastic localization (Fig. 5). This is in good agreement with the database predictions of TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000) and Aramemnon (http://aramemnon.botanik.uni-koeln.de/; Schwacke et al., 2003).
Figure 5.
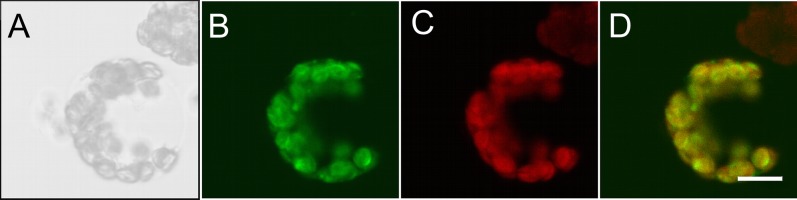
TUP5 is localized in chloroplasts. A, Protoplast of B, C, and D shown in bright field. B, Fluorescence signal of TUP5-GFP. C, Autofluorescence signal of the chloroplasts. D, Merged image of B and C showing an overlap (yellow) of TUP5-GFP and chloroplast signals. Bar = 10 µm. Leaf protoplasts were isolated from stably transformed Arabidopsis plants. Images were taken with a confocal laser scanning microscope.
Expression of the TUP5 Gene
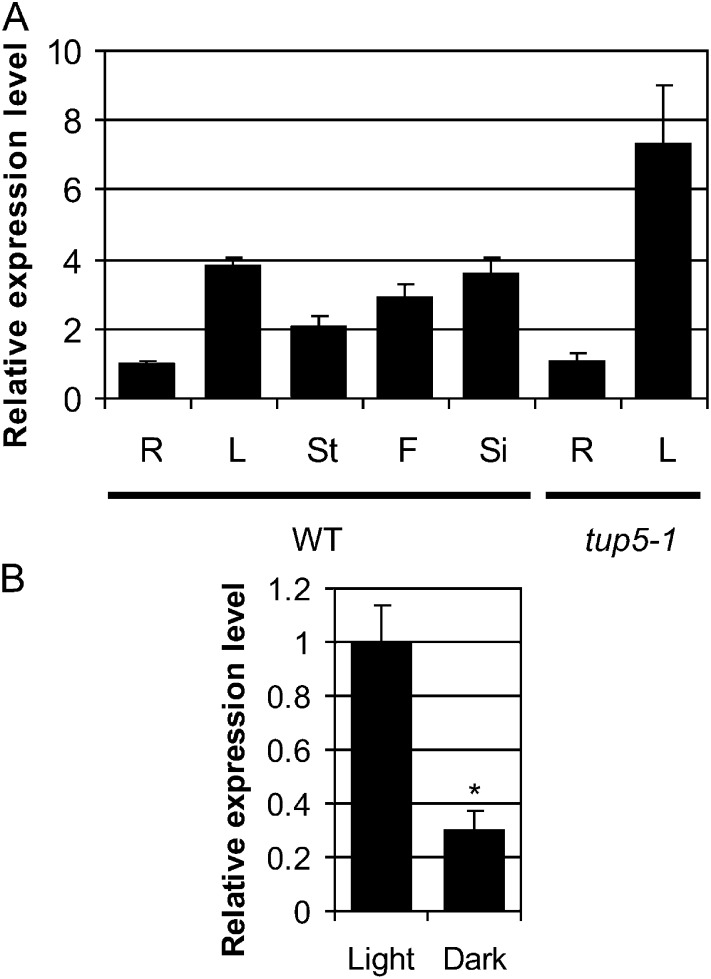
The expression of TUP5 was examined by quantitative real-time reverse transcription (RT)-PCR in different organs and under different light conditions. TUP5 was expressed in all organs tested (root, rosette leaves, stem, flowers, and siliques). The highest expression was found in rosette leaves and the lowest in roots (Fig. 6A). The ubiquitous expression of TUP5 is consistent with the results of microarray analyses found in databases such as the eFP browser and Genevestigator (Winter et al., 2007; Hruz et al., 2008).
Figure 6.
TUP5 transcript level in different organs and its dependency on light conditions. A, TUP5 and tup5-1 expression in different plant organs in wild-type and tup5-1 mutant plants. The transcript level was measured by quantitative real-time RT-PCR in roots (R) and rosette leaves (L) of 32-d-old plants and in stems (St), flowers (F), and siliques (Si) of 35-d-old plants grown in soil. Wild-type (WT) and tup5-1 root and rosette leaves were also analyzed. The differences were not significant, as calculated with Student’s t test. B, Effect of light on TUP5 expression in Arabidopsis wild-type seedlings. Seedlings were grown in long-day conditions for 4 d and subsequently exposed for 24 h to continuous light or darkness and then harvested. Transcript levels are given as relative values compared with the wild-type root tissue (A) or light-treated wild-type seedlings (B). Relative expression levels were normalized using At UBC10 (At5g53300) and At3g25800 as reference genes. Significance was calculated with Student’s t test (*P < 0.01).
The influence of white light on TUP5 expression was then studied because tup5-1 shows a light-dependent root phenotype. Seedlings were cultivated in standard conditions and then exposed to light or darkness for 24 h. The expression level of TUP5 in light-exposed seedlings was 3 times higher than in dark-exposed seedlings (Fig. 6B).
Effects of Changes in TUP5 Gene Dosage
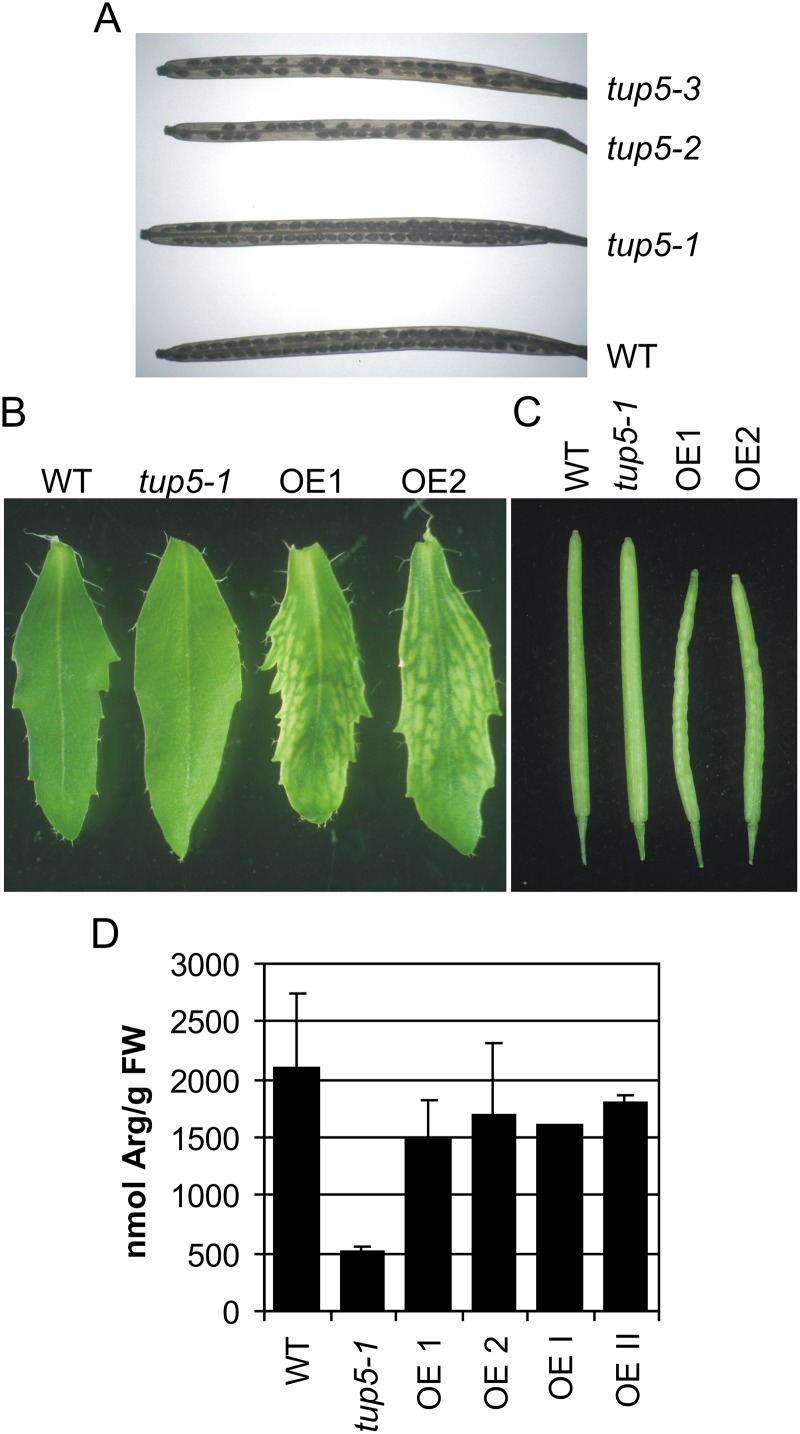
We analyzed the consequences of lower and higher TUP5 gene expression to gain a better understanding of the role of TUP5. Loss-of-function analysis was carried out with two T-DNA insertion alleles of the SALK collection named tup5-2 and tup5-3 (Alonso et al., 2003). Sequencing confirmed the sites of both insertions within the third exon of the TUP5 gene (Fig. 2A). F1 populations of heterozygous self-fertilized parents of both lines were screened by PCR for homozygous progeny, but no homozygous line was found among 118 F1 plants tested for tup5-2 and 62 plants tested for tup5-3. This indicated that the homozygous state might be lethal for both alleles. Inspection of cleared siliques of heterozygous tup5-2 and tup5-3 plants revealed empty spaces between seeds, indicating premature termination of seed development (Fig. 7A). Microscopy examination showed that these spaces in the siliques contained aborted ovules or very small embryos (Supplemental Fig. S5). The percentage of abortion was about 42% ± 4% in tup5-2 and 35% ± 7% in tup5-3, while in the wild type and tup5-1, only 1% to 2% of abortion was found (Supplemental Table S1).
Figure 7.
Developmental impairment caused by altered TUP5 dosage. A, Empty seed positions in siliques of heterozygous TUP5 T-DNA insertion lines compared with the wild type. Mature siliques were bleached with chloral hydrate. B, Cauline leaves of TUP5 overexpressing transgenic lines show interveinal chlorosis. C, Siliques of TUP5 overexpressing transgenic lines are shorter and wrinkled. D, Free Arg content of transgenic lines overexpressing TUP5 in wild-type and tup5-1 background compared with the wild type and tup5-1. FW, Fresh weight. The plants in B and C were cultivated on soil in a growth chamber under long-day conditions. Plants used for the analysis in D were cultivated in vitro for 17 d. OE1 and OE2, exemplary lines expressing 35S:TUP5 in the wild type; OEI and OEII, exemplary lines expressing 35S:TUP5 in tup5-1 background. WT, Wild type.
In addition, a meiotic drift was found in the F1 generation of heterozygous self-fertilized tup5-2 and tup5-3 lines. According to Mendel’s Laws, the ratio between wild-type and heterozygous F1 plants should be 1:2. Only 36% ± 10% of the plants in tup5-2 and 43% ± 11% in tup5-3 were heterozygous for mutated tup5 allele (Supplemental Table S2). All other plants contained the TUP5/TUP5 allele combination. Because of this strong underrepresentation of the T-DNA insertion alleles in the segregating progeny, the ratio between wild-type and heterozygous F1 plants was 1.7:1 in tup5-2 progeny and 1.3:1 in tup5-3 progeny.
Reciprocal crosses of tup5-2 and tup5-3 heterozygous plants with the wild type were carried out to find out whether embryo development, pollen, and/or ovule fertility were impaired. PCR analysis of F1 progenies showed that the T-DNA insertion allele was transmitted by both pollen and ovule. However, the transmission frequency in both cases was reduced: Instead of 50%, only approximately 24% to 42% (20 heterozygous plants of 82 plants [24%] in the case of wild type × tup5-3, 10 of 24 [42%] for wild type × tup5-2, and 18 of 48 plants [37%] in tup5-3 × wild type) of the F1 generation carried the T-DNA insertion alleles, indicating that the tup5-2 and tup5-3 alleles reduce male and female gametophytic fitness leading to meiotic drift. In addition, the low T-DNA allele frequency in the self-fertilized tup5-2 and tup5-3 lines and in the reciprocal crosses suggests an impairment of embryo development even in a heterozygous state. Taken together, null mutation of the TUP5 gene impairs gametophytic fitness and is lethal at early stages of embryo development.
Plants overexpressing TUP5 were produced by transformation of the wild type with the 35S:TUP5 gene used previously for complementation to test the consequences of increased TUP5 expression (Fig. 2C). TUP5 overexpression in the wild type caused two visible morphological changes: interveinal chlorosis in cauline leaves and the formation of wrinkled, bumpy siliques (Fig. 7, B and C). Interestingly, the Arg level was not increased above the wild type in cultivated 35S:TUP5 lines in vitro (Fig. 7D).
Influence of Light Conditions on tup5-1 Root Meristem Development
As described above, the root phenotype of tup5-1 mutants differed when plants were grown in vitro or on soil (Fig. 1, B, C, and E). The roots of tup5-1 seedlings grown on Murashige and Skoog (MS) medium in short-day conditions grew longer than the roots of tup5-1 seedlings cultivated under long-day conditions (data not shown). This indicated that day length, and therefore light, influenced root growth and not a component of the growth medium. The roots of tup5-1 seedlings of different ages cultivated in darkness consistently immediately stopped growth when they were shifted into light.
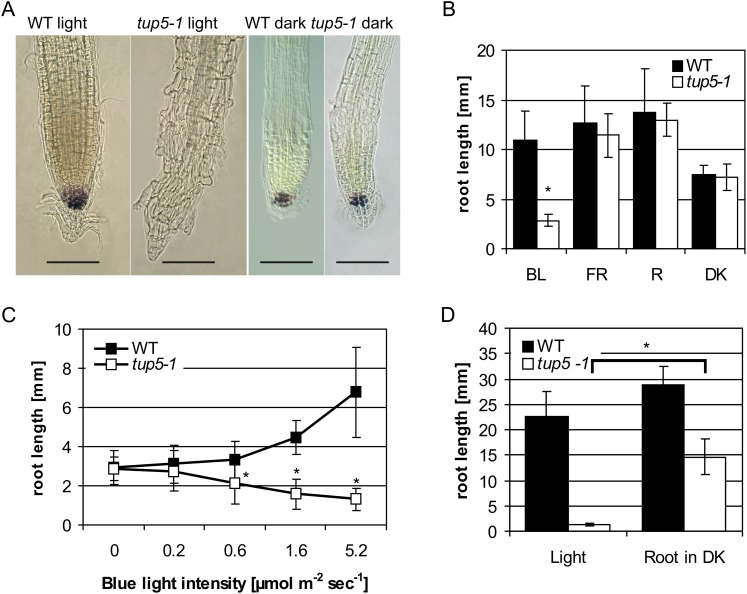
Microscopy analysis of the root tips of light-grown tup5-1 plants revealed that the meristem had lost its organized structure, the cells had differentiated, and starch granules were absent in the columella cells (Fig. 8A). In contrast, dark-grown tup5-1 seedlings had a root meristem similar to that of the wild type (Fig. 8A).
Figure 8.
Influence of light on tup5-1 root development. A, The root meristem of tup5-1 disappears in light-grown seedlings but not in dark-grown seedlings. Plants grown in light were 5 d old, and those grown in darkness were 6 d old. Starch granules were stained with Lugol solution. Bars = 100 µm. B, tup5-1 root growth is specifically inhibited by blue light. BL, blue light (445 nm), approximately 2 µmol m−2 s−1; FR, far-red light (724 nm), approximately 3.5 µmol m−2 s−1; R, red light (660 nm), 3.5 µmol m−2 s−1; DK, darkness. n = 18 to 26, except far-red light for the wild type, n = 5. Plants were treated with white light for 16 h to induce germination and then transferred to constant (24 h) monochromatic light exposure for 5 d. Student’s t test was used to compare the wild type and tup5-1 grown under the same light conditions; significance: *P < 0.001. C, tup5-1 root growth inhibition depends on the intensity of blue light irradiation. Seedlings were cultivated under various intensities of blue light (445 nm) for 6 d. *P < 0.05 as calculated by Student’s t test. D, The root growth inhibiting light is perceived in the root of tup5-1. Seedlings were cultivated for 28 d in vitro in light or with the roots kept in darkness. Mean ± sd of four to seven plants. Significance: *P < 0.001. WT, Wild type; DK, darkness. [See online article for color version of this figure.]
Root growth of tup5-1 was tested under various monochromatic light qualities to find out whether a specific part of the light spectrum causes this switch from an indeterminate growth to determinate growth. The inhibition of root growth in tup5-1 was induced by blue light but not by red or far red light (Fig. 8B). The root growth inhibition caused by blue light was dose dependent (Fig. 8C). The root length was not significantly different between tup5-1 and the wild type up to 0.2 µmol m−2 s−1, while a blue light intensity of 0.6 µmol m−2 s−1 or more inhibited root elongation in tup5-1. To test whether a known light signaling pathway is involved in mediating this effect, we combined by crossing tup5-1 with the following light receptor or light signaling mutants: the phytochrome apoprotein mutant phyB-1 (hy3-Bo64; Reed et al., 1993; Rockwell et al., 2006), the phytochrome chromophore biosynthesis mutant hy2-1 (Kohchi et al., 2001), the cryptochrome apoprotein mutant cry1 (hy4-1; Ahmad and Cashmore, 1993), and the bZIP transcription factor mutant hy5-1 (Oyama et al., 1997). Analysis of the double mutants in the F2 generation showed that none of these mutants suppressed the tup5-1 root phenotype (data not shown). Therefore, the light receptors or light signaling factors analyzed are not epistatic to tup5-1.
Wild-type and tup5-1 plants were grown in vitro in glass tubes with illumination of the whole plant or illumination of only the shoot to find out whether the perception of the inhibiting light takes place in the shoot or in the root. When grown in light, tup5-1 exhibited its typical severe root growth defect (Fig. 8D). In contrast, when only the shoots were illuminated, the root length of tup5-1 increased significantly. Although tup5-1 roots did not reach the length of wild-type roots, which was probably due to the imperfect darkening of the roots in the experimental setup, it can be concluded that the root itself perceives the inhibiting light signal causing the tup5-1 root phenotype.
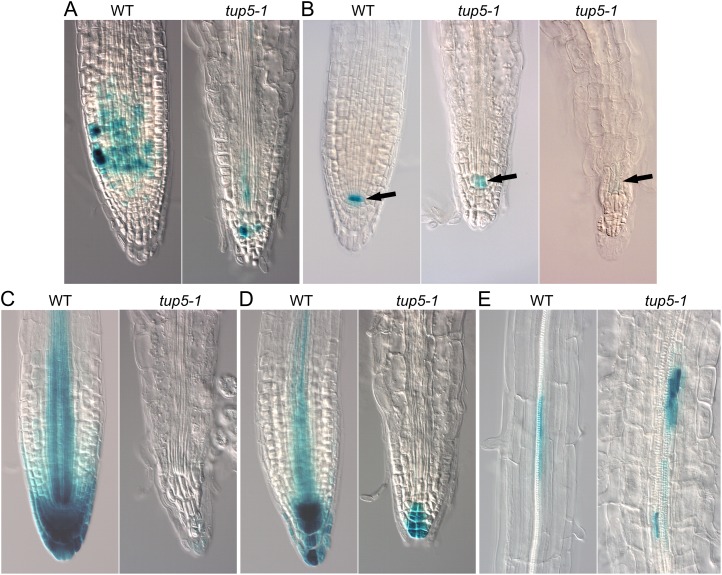
We studied the cell cycle marker gene CYCB1;1pro:GUS-DBox (Colón-Carmona et al., 1999) and QC25 (Sabatini et al., 1999), which specifically marks the quiescent center (QC), in the tup5-1 background to follow more closely the light-dependent switch to determinate growth and the subsequent loss of the root meristem. The meristem of tup5-1 roots grown in vitro in light showed a strongly reduced cell cycle-dependent GUS activity compared with the wild type. A weak signal was visible in only a few cells (Fig. 9A). The QC-dependent GUS activity was reduced in light-grown roots of tup5-1. When the meristematic structure was not yet completely lost, a light-blue color showed a decreased GUS activity in the QC cells (Fig. 9B). The shape of the QC cells was also changed in tup5-1: While the cells were flattened in the wild-type meristem, they stretched parallel to the root axis in tup5-1 (Fig. 9B). When the loss of meristematic structure progressed, the GUS activity in the QC of tup5-1 roots almost vanished (Fig. 9B). As tup5-1 was found in a screen for callus formation on low concentrations of cytokinin and auxin, we also examined the hormonal status in the differentiated tup5-1 root meristem using the cytokinin-sensitive marker ARR5:GUS (Sabatini et al., 1999; D’Agostino et al., 2000) and the auxin reporter DR5:GUS (Sabatini et al., 1999; D’Agostino et al., 2000). The GUS activity pattern in both cases was much weaker in tup5-1 than in the wild type or had almost vanished in the mutant root (Fig. 9, C and D). Nevertheless, DR5:GUS expression marking in pericycle cells at the beginning of lateral root primordia formation (Benková et al., 2003) was also observed in the short growth-arrested tup5-1 primary roots (Fig. 9E).
Figure 9.
Expression of different marker genes is altered in the tup5-1 root. A, Cell cycle-dependent GUS staining with the marker CYCB1;1pro::GUS-DBox is reduced in the primary root tip of tup5-1. B, GUS activity of the marker QC25 in the QC of the root apical meristem in tup5-1 primary roots decreases during meristem differentiation. Arrows point to the stained QC. C, Staining with the cytokinin-sensitive marker ARR5::GUS is strongly reduced in the primary root tip of tup5-1. D, Auxin-responsive DR5::GUS expression is decreased in the primary root tip of tup5-1. E, Auxin-responsive DR5::GUS expression indicating early events during the formation of lateral roots in the pericycle of the primary root is also found in tup5-1. All seedlings were 5 d old and grown in vitro. Photos were taken at a 200-fold magnification. WT, Wild type.
DISCUSSION
TUP5 Encodes a Plastid-Localized Acetyl-Orn Aminotransferase
We showed that the phenotype of tup5-1 is due to a point mutation in a gene encoding the Arabidopsis ACOAT required for Arg biosynthesis. The function of TUP5 as ACOAT is supported by the findings that tup5-1 is Arg deficient and that the root phenotype could be complemented by supplementation with acetyl-Orn, Orn, Cit, and Arg, which are all synthesized downstream of ACOAT, while other metabolites did not complement. Additionally, a yeast ACOAT mutant was complemented by heterologous expression of the Arabidopsis TUP5 gene. A TUP5-GFP fusion protein has been detected in chloroplasts. This is consistent with the database prediction of a plastidic localization of TUP5 and biochemical experiments showing acetyl-Orn aminotransferase activity in the plastid fraction of a soybean (Glycine max) suspension culture (Jain et al., 1987).
Mutation of TUP5 Causes a Reduction of Arg Content and Has Pleiotropic Effects on Amino Acid Homeostasis
The Arg content of light-grown tup5-1 plants was strongly reduced compared with the wild type. Interestingly, the Arg content was reduced in both the wild type and tup5-1 to a comparable level in darkness. Arg in the wild type showed the most extreme difference in shoot/root distribution of all amino acids analyzed. The root contained 50 times less Arg than the shoot, indicating a fine-tuned and sensitive metabolic control system of Arg content in the root. It could be that further reduction of this already low Arg level causes a slowdown of root growth and, eventually, its complete arrest. Overexpression of TUP5 in tup5-1 restored the Arg content to the wild-type level, but overexpression in the wild type did not cause accumulation of higher Arg concentrations, indicating that other biosynthetic steps are rate limiting.
The content of most other amino acids was increased in tup5-1 mutants, although to a different extent at different developmental stages. The increased content of Asn and Gln, two amino acids that play a major role in nitrogen storage and transport (Lam et al., 1995), may partially compensate for the absence of Arg as a nitrogen sink. Accumulation of surplus amino groups in these storage pools in the tup5-1 mutant might also be caused by a reduced protein biosynthesis capacity due to the lack of Arg as a rate-limiting compound. The dynamic changes in the concentrations of numerous other amino acids support the notion of cross-pathway regulation of different amino acid biosynthesis pathways in plants. This mechanism has been shown to exist in yeast (Hinnebusch, 1992) and may also cause a general deregulation of free amino acid content in plants if one amino acid is not produced sufficiently (Guyer et al., 1995; Noutoshi et al., 2005). Indeed, a general amino acid metabolic deregulation with a global increase of free amino acid content was also found in other mutants, for example, in albino and pale green10, which is disrupted in His biosynthesis (Noutoshi et al., 2005), Phe insensitive growth1-1 (Voll et al., 2004), and Gln dumper1 (Pilot et al., 2004). Alternatively, the increased free amino acid content in tup5-1 might be due to protein degradation caused by deleterious effects due to the lack of Arg (Noctor et al., 2002). Together, the strong effect of a single partial loss-of-function mutation in the Arg biosynthesis pathway on amino acid homeostasis underpins that a tight regulatory network controls a delicate balance that is not yet fully understood.
TUP5 Is Required for Gametogenesis and Seed Development
Analysis of two tup5 functional null alleles revealed the absolute requirement of Arg synthesis for proper male and female gametogenesis, as well as early seed development. Meiotic drift, as indicated by reduced transmission frequencies of the tup5-2 and tup5-3 insertion alleles in reciprocal crosses with the wild type, is likely due to a dysfunction of ovules and pollen grains carrying this genotype. The reduced fitness of gametes carrying a TUP5 null allele indicates that they are not exclusively cross-fed from surrounding tissues but need a certain degree of Arg autotrophy (Muralla et al., 2011). However, survival of some gametes also shows that maternal tissue ensures at least limited Arg supply to ovules and pollen grains.
Similarly, it has not been clear so far if Arabidopsis embryos need to be Arg autotrophic or whether their demand can be covered by maternal tissue. The knowledge of how amino acid supply of developing seeds takes place is still incomplete for Arabidopsis (Baud et al., 2008). All amino acids, but mainly Gln, Glu, Asp, Asn, Ser, and Ala, are transported from source tissues of the mother plant to the seed via the phloem (Sanders et al., 2009). The amino acids can be converted to other amino acids in the seed coat (maternal tissue) or in the embryo (Baud et al., 2008; Sanders et al., 2009). Early failure of seed development in tup5 nulls indicated that these need their own Arg biosynthesis and are not exclusively nourished by the maternal tissue. It is not entirely clear from our analysis which tissue of the developing seed is primarily impaired by the lack of Arg and causes growth arrest, but the embryo is most likely affected. Different amino acid biosynthesis mutants showing embryo lethal phenotypes have been described, including mutants in His (Muralla et al., 2007), Lys (Song et al., 2004; Hudson et al., 2006), and Pro biosynthesis genes (Székely et al., 2008). Meinke (1991) and Bryant et al. (2011) also pointed out that embryonic development is often interrupted in mutants defective in essential housekeeping genes, including those of amino acid biosynthesis.
tup5-1 Mutants Show an Unusual Blue Light-Dependent Root Growth Inhibition
Probably the most intriguing aspect of the tup5-1 mutant phenotype is the blue light-dependent switch from indeterminate growth of the root to a determinate growth phase. Importantly, the root growth arrest is not simply due to the arrest of cell cycling, but the apical root meristem is completely used up. This implies that central functions relevant for the maintenance of the QC and the surrounding initials are lost and, thus, critically depend on Arg in the presence of light.
We are not aware of any other mutant showing a blue light-dependent inhibition of root growth. This phenotype also distinguishes tup5-1 from other Arabidopsis mutants impaired in amino acid biosynthesis or metabolism showing defects in root growth, such as the hpa1 His biosynthesis mutant (Mo et al., 2006), the mutant Rm 57 disturbed in the Lys biosynthetic pathway (Sarrobert et al., 2000), and two mutants of the genes encoding Asp aminotransferase AspAT2, aat2-2 and aat2-T (Schultz et al., 1998; Miesak and Coruzzi, 2002). The His biosynthesis mutant hpa1 resembles tup5-1 insofar as it has a short root when grown in vitro, but there is no obvious phenotype in the aerial part and hpa1 mutants are fully fertile plants (Mo et al., 2006). Mutants for several Trp biosynthetic pathway genes are generally impaired in development under high-light but not under low-light conditions (Last et al., 1991; Radwanski et al., 1996). It has been suggested that the residual activity of the mutant enzyme might be able to cover the lower Trp demand of slow-growing plants in low light (Last et al., 1991). In contrast, the roots and shoots of soil-grown tup5-1 plants grown under standard light develop normally, though high metabolic activity can be expected under these conditions. This argues for specifically light-induced changes in the root tissue.
One possibility is a light-dependent change in protein stability or catalytic activity of the mutated ACOAT (TUP5G424R). The molecular environment of root plastids is presumably different from that of green tissue chloroplasts; therefore, some blue light-regulated proteins might interfere with TUP5 in the illuminated root, or proteins could be lacking due to the influence of light. It could also be hypothesized that shoot Arg production is sufficient even with the mutated protein and that the shoot exports Arg into the root when the latter is grown in dark, but not when grown in light. However, although significantly higher Arg concentrations are present in the shoot compared with the root (Fig. 4B), it is not clear yet whether Arg long-distance transport normally plays an important role in Arabidopsis. Glu, Gln, Asp, and Asn were found to be the predominant amino acids in phloem sap and xylem exudates and are therefore presumably the major nitrogen transport molecules (Lam et al., 1995). Other amino acids are assumed to be mainly synthesized from these transport amino acids at the site of requirement (Lea and Miflin, 1980).
The root growth arrest in tup5-1 is specifically caused by blue light and very low fluence rates are sufficient to trigger the developmental switch. Light is encountered by roots on the soil surface where roots emerge from germinating seeds and in the upper soil layers. Light penetrates several millimeters through soil and also reaches plant roots through the vascular tissue (Tester and Morris, 1987; Kasperbauer and Hunt, 1988; Sun et al., 2003). The very low effective light fluence rates that act on tup5-1 indicate a physiological relevance and suggest an as yet unknown connection between amino acid biosynthesis, light perception, and root development. Roots contain red and blue light photoreceptors that may perceive light directly (Sakamoto and Briggs, 2002; Galen et al., 2007; Costigan et al., 2011; Dyachok et al., 2011, and references therein) and also respond to UV-B (Tong et al., 2008). Our genetic analyses have largely excluded several known components from light signaling pathways (CRY1, PHYB, HY2, and HY5) to operate in the TUP5 pathway, but not all known light receptors that could have a function in light perception by roots have been tested. Other candidate proteins include cry2, though cryptochrome effect on root elongation was perceived in the shoot according to a study of Canamero et al. (2006). Phototropin phot1 was also found to have a role in blue light perception of the root where it mainly regulates directional growth (Galen et al., 2007; Kutschera and Briggs, 2012). phyA can absorb blue light and could thus also be a potential receptor in the signaling pathway regulating TUP5 (Lin, 2000). Intriguingly, recent work has demonstrated that phyA and phot1 interact functionally at the plasma membrane, which further increases the variety of light signaling pathways (Jaedicke et al., 2012). In addition, it cannot be ruled out that an unknown receptor is involved in the light regulation of TUP5.
Interestingly, three mutants linked to UV light perception and light stress have a light-dependent short root phenotype comparable to tup5-1. The rus1 and rus2 mutants have short roots under UV-B light and the inhibiting light is perceived by the root (Tong et al., 2008; Leasure et al., 2009). pdx1-3 mutants missing a pyridoxal synthase necessary for the production of the TUP5 cofactor pyridoxal-5′-P show a light-dependent root growth arrest. However, it has not been tested whether the root perceives the inhibiting light and whether a specific wavelength is involved (Titiz et al., 2006; Havaux et al., 2009). Interestingly, a screen for second site suppressor mutants of the rus phenotype identified mutations in the ASP AMINOTRANSFERASE2 gene, and adding pyridoxine (vitamin B6) to the growth medium of rus mutants partly rescued root growth (Leasure et al., 2011). This illustrates that as yet unknown pathways connecting UV-B light sensitivity, vitamin B6 homeostasis, and amino acid biosynthesis exist.
Our data show that for a complete understanding of root meristem function, essential metabolites, such as Arg, need to be considered in addition to plant hormones and transcription factors that are generally in the focus of root meristem research (Perilli et al., 2012). The relevance of additional poorly considered factors in meristem regulation is corroborated by the recent report that the metabolic cofactor thiamine has an important role during the proliferation of stem and initial cell populations in the shoot meristem (Woodward et al., 2010). The tup5-1 mutant will be an important tool to investigate the mechanisms that link Arg to known regulators maintaining indeterminate root meristem development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
tup5-1 was isolated from an ethyl methanesulfonate-mutagenized population of Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 glabra (Riefler, 2001). The T-DNA insertional mutants tup5-2 (N878354) and tup5-3 (N875828) and the light signaling pathway mutants phyB-1 (N69; Reed et al., 1993; Rockwell et al., 2006), hy5-1 (N71; Oyama et al., 1997), cry1 (N70; Ahmad and Cashmore, 1993), and hy2-1 (N68; Kohchi et al., 2001) were obtained from the Nottingham Arabidopsis Stock Center (http://arabidopsis.info/) and the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm). tup5-1 was combined by crossing with the marker gene lines CYCB1;1pro:GUS-DBox, QC25, ARR5:GUS, and DR5:GUS provided by Peter Doerner, Sabrina Sabatini, and Joe Kieber, respectively (Colón-Carmona et al., 1999; Sabatini et al., 1999; D’Agostino et al., 2000).
Unless stated otherwise, plants were grown at 22°C under long-day conditions (16-h light/8-h dark) on soil in a greenhouse (light intensity: 150–300 µmol m−2 s−1; natural light supplemented with high-pressure sodium vapor lamps, type SON-T, 2,000K) or in vitro in a growth chamber (light intensity: 150 µmol m−2 s−1). In vitro cultivation was on solid medium containing 1× MS salts (Duchefa Biochemie), 0.1 g L−1 myo-inositol, 0.5 g L−1 MES, 20 mg L−1 thiamine, 1 mg L−1 nicotinic acid, 1 mg L−1 pyridoxine, 1 mg L−1 biotin, 3% (w/v) Suc, and 0.9% (w/v) agar, pH 5.7, or in liquid medium containing 0.5× MS salts, 0.5 g L−1 MES, and 1% (w/v) Suc, pH 6 (modified from Murashige and Skoog, 1962).
Gene Mapping and BAC Subcloning
Homozygous tup5-1 plants were crossed with plants of the ecotype Landsberg erecta. The mapping was done with approximately 2,300 homozygous mutant plants of the F2 generation, as described by Krupková et al. (2007). No more recombinants were found at a mapping interval of 88 kb. Therefore, DNA fragments of the two BAC clones T21F11 (Choi et al., 1995) and F23A5 (Mozo et al., 1998) covering the tup5-1 mapping interval were used for complementation and gene identification.
Plasmid Construction for Plant Transformation
To generate subclones from the BAC clones T21F11 and F23A5, these were cut with appropriate restriction enzymes, separated by gel electrophoresis, the appropriate bands purified from the gel using QIAEX II gel extraction kit or QIAquick gel extraction kit (Qiagen), and cloned into vector pCB302 (Xiang et al., 1999). The resulting plasmids were used for transformation of tup5-1.
The 35S:TUP5 construct was produced using the TUP5 cDNA clone U15579 provided by Arabidopsis Biological Resource Center (Yamada et al., 2003). The cDNA was recombined into the vector pB2GW7 (Karimi et al., 2005) with LR clonase (Invitrogen) according to the manufacturer’s protocol.
To generate the P35S:TUP5-GFP gene fusion, the cDNA of TUP5 was amplified from clone U15579 using the gene-specific primers 5′-ATGGCGTCTCTTAGCCAAATC-3′ (forward) and 5′-ATCAAGCGCAGTCAAATTTT-3′ (reverse; Yamada et al., 2003). The resulting PCR product was used as a template to add 12 bp of Gateway attachment sites at the 5′ ends with the primers 5′-AAAAAGCAGGCTATGGCGTCTCTTAGCCAAATC-3′ (forward) and 5′-AGAAAGCTGGGTCATCAAGCGCAGTCAAATTTT-3′ (reverse). Finally, the full-length attachment sites were added in a PCR reaction with the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ (forward) and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′ (reverse). The resulting PCR product was inserted into the donor vector pDONR221 via recombination to create an entry clone for further use. The resulting entry clone was recombined with pB7FWG2 (Karimi et al., 2005), which inserts the TUP5 gene between the 35S promoter and the GFP gene creating the P35S:TUP5-GFP fusion.
The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101:pMP90 (Koncz and Schell, 1986), which was used for Arabidopsis transformation (Clough and Bent, 1998). Transformants were selected on medium containing the appropriate antibiotics.
Heterologous Complementation with TUP5 of a Yeast Mutant Defective in ACOAT
The yeast (Saccharomyces cerevisiae) strain with the accession number Y37711 (genotype: BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YOL140w::kanMX4/YOL140w::kanMX4) in which the ACOAT gene YOL140w (ARG8) is deleted was provided by Euroscarf. The genomic sequence and the cDNA of TUP5 were amplified with the primers 5′-CGGGATCCATGGCGTCTCTTAGCCAAATC-3′ (forward) and 5′-ACGCGTCGACTCAATCAAGCGCAGTCAAAT-3′ (reverse), generating a BamHI restriction site at the 5′ end and a SalI restriction site at the 3′ end for plasmid construction. The PCR products were inserted into the yeast vector p423TEF by directional cloning using the BamHI and SalI sites (Mumberg et al., 1995). The resulting plasmid was transformed into Y37711 by the lithium acetate method (Gietz and Schiestl, 1995; Bürkle et al., 2005). Yeast transformants were first selected for His autotrophy and ampicillin resistance. Transformed yeast strains were grown on synthetic defined medium containing 20 mg Arg L−1 or no Arg to test Arg autotrophy.
Gene Expression Analysis
Total RNA was purified from Arabidopsis tissue by the TRIzol method, as described by Brenner et al. (2005). cDNA synthesis, quantitative real-time RT-PCR, and data analysis were performed as described by Werner et al. (2010) using 7500 FAST Software_v2.0.1 (Applied Biosystems). In addition to UBC10, we used At3g25800 as a reference gene for normalization (Czechowski et al., 2005). TUP5 was amplified with the primers 5′-TCGGTGTGACTCCTGACAT-3′ (forward) and 5′-ACTGCACACAAGAGGGCT-3′ (reverse). The reference gene At3g25800 was amplified with the primers 5′-CCATTAGATCTTGTCTCTCTGCT-3′ (forward) and 5′-GACAAAACCCGTACCGAG-3′ (reverse) for gene expression analyses.
Subcellular Localization of TUP5
One to three leaves from stably transformed P35S:TUP5:GFP transgenic Arabidopsis lines were treated according to the protoplast isolation protocol described by Damm and Willmitzer (1988). The protoplasts were analyzed for subcellular localization of the TUP5-GFP fusion protein with a Leica TCS SP2 confocal laser scanning microscope. An excitation wavelength of 488 nm and a filter of 510 to 550 nm were used for the analysis of GFP fluorescence signal and an excitation wavelength of 488 nm and a filter of 610 to 680 nm for the autofluorescence signal of chloroplasts.
GUS Staining and Light Microscopy
The plant material was treated following the GUS staining protocol described by Krupková et al. (2007). The tissue was cleared and mounted according to Malamy and Benfey (1997). Microscopy was carried out with an Axioskop 2 plus with AxioCam ICc3 (Zeiss).
Measurement of Free Amino Acids
Seedlings were cultivated in liquid one-half-strength MS medium for 7 d under long-day conditions at standard light intensity (150 µmol m−2 s−1), at low light intensity (3 µmol m−2 s−1), or in darkness. After harvest, seedlings (approximately 100 mg per sample) were washed once with 5 mL sterile aqua bidest., frozen in liquid nitrogen, and stored at −80°C until extraction. Shoot and root samples were harvested from 11-d-old seedlings grown under long-day conditions on vertical plates containing 1× MS medium. The samples were extracted, derivatized, and analyzed as described by Rinder et al. (2008).
Sequence Comparison and Phylogenetic Analysis
The protein sequence of TUP5 was used for a search for homologous genes using BLAST search (Altschul et al., 1997). The amino acid sequences of six homologous proteins from different species were aligned with TUP5 using ClustalW 1.83 (http://www.ch.embnet.org/software/ClustalW.html; Larkin et al., 2007), and shading of conserved amino acids in the alignment was carried out with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). The fraction of sequences that had to agree for shading was set at 0.9. The phylogenetic analyses were conducted using MEGA5 (Tamura et al., 2011). The phylogenetic tree was revised with CorelDRAW 12.
Prediction of Subcellular Localization
The prediction of subcellular localization was carried out using TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000) and Aramemnon (http://aramemnon.botanik.uni-koeln.de/; Schwacke et al., 2003).
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: TUP5/WIN1 (AT1G80600), Populus trichocarpa unknown gene ABK95025.1, rice (Oryza sativa ssp. japonica cultivar group; GI:51854368), putative ACOAT, Physcomitrella patens ssp. patens (GI:162664563), predicted protein ACOAT family, Chlamydomonas reinhardtii ACOAT (GI:159480034), yeast ACOAT (GI:151945496), and Escherichia coli ACOAT (GI:242378886).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment and phylogenetic analysis of known ACOATs and highly similar genes from different species.
Supplemental Figure S2. Metabolites of the Arg biosynthesis pathway and related compounds tested for a complementing effect on the tup5-1 root phenotype.
Supplemental Figure S3. Free amino acid content differs between wild-type shoots and roots.
Supplemental Figure S4. Content of free Gln and Asn in 35S:TUP5 transgenic lines.
Supplemental Figure S5. Aborted seeds in siliques of heterozygous tup5-2 plants.
Supplemental Table S1. Percentage of aborted seeds in siliques of wild-type, tup5-1, heterozygous tup5-2, and tup5-3 plants.
Supplemental Table S2. Segregation analysis of heterozygous self-fertilized tup5-2 and tup5-3 F1 populations.
Acknowledgments
We thank Prof. Elmar Hartmann, Prof. Tilman Lamparter, and Dr. Alexander Repp (all formerly at Freie Universität Berlin) for giving us access to their monochromatic light experimental facility and also for helpful discussions. We thank Dr. Rainer Höfgen, Dr. Stephan Krüger, Dr. Hans-Michael Hubberten, and Dr. Mariusz Bromke (Max Planck Institute for Molecular Plant Physiology, Golm) for access to HPLC analyses and for great help and advice on measuring amino acid concentrations. We thank Prof. Bernhard Grimm and Dr. Christina Kühn (Institute of Biology, Humboldt University Berlin) for access to the confocal microscope and Prof. Klaus Harter and Dr. Virtudes Mira-Rodado (Center for Plant Molecular Biology, Eberhard Karls Universität Tübingen) for the opportunity to carry out the dose-response experiments with blue light. We thank Peter Doerner, Sabrina Sabatini, and Joe Kieber for providing the marker lines CYCB1;1pro:GUS-DBox, QC25, ARR5:GUS, and DR5:GUS, respectively. We acknowledge Euroscarf (Frankfurt) for providing the yeast mutant Y37711 and the Nottingham Arabidopsis Stock Center and Arabidopsis Biological Resource Center for providing various Arabidopsis mutant lines and clone U15579. We also thank Dr. Wolfram Brenner, Ulrike Deppe, Dr. Alexander Heyl, Dr. Eva Krupková, Prof. Wolfgang Schuster, and other members of the Institute of Biology/Applied Genetics for help and advice.
Glossary
- ACOAT
N2-acetyl-Orn aminotransferase
- OTC
Orn transcarbamylase
- RT
reverse transcription
- MS
Murashige and Skoog
- QC
quiescent center
- cDNA
complementary DNA
- T-DNA
transfer DNA
- ACOAT
acetylornithine aminotransferase Cit: citrulline
References
- Ahmad M, Cashmore AR. (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Albrecht AM, Vogel HJ. (1964) Acetylornithine delta-transaminase. Partial purification and repression behavior. J Biol Chem 239: 1872–1876 [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L. (2008) Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113, doi/10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L, Meyer S, Dortay H, Lehrach H, Heyl A. (2005) In vitro recombination cloning of entire cDNA libraries in Arabidopsis thaliana and its application to the yeast two-hybrid system. Funct Integr Genomics 5: 175–183 [DOI] [PubMed] [Google Scholar]
- Canamero RC, Bakrim N, Bouly J-P, Garay A, Dudkin EE, Habricot Y, Ahmad M. (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224: 995–1003 [DOI] [PubMed] [Google Scholar]
- Chen YM, Ferrar TS, Lohmeier-Vogel EM, Morrice N, Mizuno Y, Berenger B, Ng KK, Muench DG, Moorhead GB. (2006) The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetyl glutamate kinase. J Biol Chem 281: 5726–5733 [DOI] [PubMed] [Google Scholar]
- Choi S, Creelman RA, Mullet JE, Wing RA. (1995) Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World 2: 17–20 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Costigan SE, Warnasooriya SN, Humphries BA, Montgomery BL. (2011) Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis. Plant Physiol 157: 1138–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM. (2006) Mechanisms for nitric oxide synthesis in plants. J Exp Bot 57: 471–478 [DOI] [PubMed] [Google Scholar]
- Cunin R, Glansdorff N, Piérard A, Stalon V. (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50: 314–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ. (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm B, Willmitzer L. (1988) Regeneration of fertile plants from protoplasts of different Arabidopsis thaliana genotypes. Mol Gen Genet 213: 15–20 [Google Scholar]
- Davis RH. (1986) Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev 50: 280–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB. (2011) SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell 23: 3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Frémont N. (2004) Molekulare und phänotypische charakterisierung der lichtabhängigen Wurzelmutante tup5 in Arabidopsis thaliana. PhD thesis. Freie Universität Berlin, Berlin [Google Scholar]
- Galen C, Rabenold JJ, Liscum E. (2007) Functional ecology of a blue light photoreceptor: effects of phototropin-1 on root growth enhance drought tolerance in Arabidopsis thaliana. New Phytol 173: 91–99 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. (1995) Transforming yeast with DNA. Methods Mol Cell Biol 5: 255–269 [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J. (2006) Nitric oxide and gene regulation in plants. J Exp Bot 57: 507–516 [DOI] [PubMed] [Google Scholar]
- Guyer D, Patton D, Ward E. (1995) Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc Natl Acad Sci USA 92: 4997–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Ksas B, Szewczyk A, Rumeau D, Franck F, Caffarri S, Triantaphylidès C. (2009) Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. (1992) General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae In EW Jones, JR Pringle, JB Broach, eds, The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, Vol 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 319–414 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AO, Singh BK, Leustek T, Gilvarg C. (2006) An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 140: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaedicke K, Lichtenthäler AL, Meyberg R, Zeidler M, Hughes J. (2012) A phytochrome-phototropin light signaling complex at the plasma membrane. Proc Natl Acad Sci USA 109: 12231–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain JC, Shargool PD, Chung S. (1987) Compartmentation studies on enzymes of ornithine biosynthesis in plant cells. Plant Sci 51: 17–20 [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kasperbauer M, Hunt P. (1988) Biological and photometric measurement of light transmission through soils of various colors. Bot Gaz 149: 361–364 [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. (2001) The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Krupková E, Immerzeel P, Pauly M, Schmülling T. (2007) The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J 50: 735–750 [DOI] [PubMed] [Google Scholar]
- Kutschera U, Briggs WR. (2012) Root phototropism: from dogma to the mechanism of blue light perception. Planta 235: 443–452 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Coruzzi G. (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR. (1991) Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase β genes. Plant Cell 3: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Miflin BJ. (1980) Transport and metabolism of asparagine and other nitrogen compounds within the plant. In BJ Miflin, ed, Amino Acids and Derivatives, Vol 5. Academic Press, New York, pp 569–607 [Google Scholar]
- Leasure CD, Tong H, Yuen G, Hou X, Sun X, He Z-H. (2009) ROOT UV-B SENSITIVE2 acts with ROOT UV-B SENSITIVE1 in a root ultraviolet B-sensing pathway. Plant Physiol 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure CD, Tong H-Y, Hou X-W, Shelton A, Minton M, Esquerra R, Roje S, Hellmann H, He Z-H. (2011) root uv-b sensitive mutants are suppressed by specific mutations in ASPARTATE AMINOTRANSFERASE2 and by exogenous vitamin B6. Mol Plant 4: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. (2000) Plant blue-light receptors. Trends Plant Sci 5: 337–342 [DOI] [PubMed] [Google Scholar]
- Llácer JL, Fita I, Rubio V. (2008) Arginine and nitrogen storage. Curr Opin Struct Biol 18: 673–681 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Meinke DW. (1991) Embryonic mutants of Arabidopsis thaliana. Dev Genet 12: 382–392 [Google Scholar]
- Micallef BJ, Shelp BJ. (1989) Arginine metabolism in developing soybean cotyledons: I. Relationship to nitrogen nutrition. Plant Physiol 90: 624–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesak BH, Coruzzi GM. (2002) Molecular and physiological analysis of Arabidopsis mutants defective in cytosolic or chloroplastic aspartate aminotransferase. Plant Physiol 129: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Zhu Q, Li X, Li J, Zeng Q, Rong H, Zhang H, Wu P. (2006) The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol 141: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollá-Morales A, Sarmiento-Mañús R, Robles P, Quesada V, Pérez-Pérez JM, González-Bayón R, Hannah MA, Willmitzer L, Ponce MR, Micol JL. (2011) Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J 65: 335–345 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Gilroy S. (2009) The exploring root: root growth responses to local environmental conditions. Curr Opin Plant Biol 12: 766–772 [DOI] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Shizuya H, Altmann T. (1998) Construction and characterization of the IGF Arabidopsis BAC library. Mol Gen Genet 258: 562–570 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D. (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. (2007) Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol 144: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for plant growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Noctor G, Novitskaya L, Lea PJ, Foyer CH. (2002) Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J Exp Bot 53: 939–945 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Shinozaki K. (2005) ALBINO AND PALE GREEN 10 encodes BBMII isomerase involved in histidine biosynthesis in Arabidopsis thaliana. Plant Cell Physiol 46: 1165–1172 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Di Mambro R, Sabatini S. (2012) Growth and development of the root apical meristem. Curr Opin Plant Biol 15: 17–23 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VPM, Frommer WB. (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from Hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. (1999) OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett 461: 101–106 [DOI] [PubMed] [Google Scholar]
- Radwanski ER, Barczak AJ, Last RL. (1996) Characterization of tryptophan synthase alpha subunit mutants of Arabidopsis thaliana. Mol Gen Genet 253: 353–361 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M. (2001) Identifizierung und Charakterisierung der tumor prone Mutanten von Arabidopsis thaliana und Atckx-Mutanten. PhD thesis. Eberhard-Karls-Universtität Tübingen, Tuebingen, Germany [Google Scholar]
- Rinder J, Casazza AP, Hoefgen R, Hesse H. (2008) Regulation of aspartate-derived amino acid homeostasis in potato plants (Solanum tuberosum L.) by expression of E. coli homoserine kinase. Amino Acids 34: 213–222 [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su Y-S, Lagarias JC. (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M. (2009) AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J 59: 540–552 [DOI] [PubMed] [Google Scholar]
- Sarrobert C, Thibaud M-C, Contard-David P, Gineste S, Bechtold N, Robaglia C, Nussaume L. (2000) Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J 24: 357–367 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Hsu M, Miesak B, Coruzzi GM. (1998) Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 149: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I, Kunze R. (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shargool PD, Jain JC, McKay G. (1988) Ornithine biosynthesis, and arginine biosynthesis and degradation in plant cells. Phytochemistry 27: 1571–1574 [Google Scholar]
- Slocum RD. (2005) Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol Biochem 43: 729–745 [DOI] [PubMed] [Google Scholar]
- Slocum RD, Nichols HF, III, Williamson CL. (2000) Purification and characterization of Arabidopsis ornithine transcarbamoylase (OTCase), a member of a distinct and evolutionarily-conserved group of plant OTCases. Plant Physiol Biochem 38: 279–288 [Google Scholar]
- Song JT, Lu H, Greenberg JT. (2004) Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yoda K, Suzuki M, Suzuki H. (2003) Vascular tissue in the stem and roots of woody plants can conduct light. J Exp Bot 54: 1627–1635 [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo Á, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Morris C. (1987) The penetration of light through soil. Plant Cell Environ 10: 281–286 [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB. (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48: 933–946 [DOI] [PubMed] [Google Scholar]
- Tong H, Leasure CD, Hou X, Yuen G, Briggs W, He Z-H. (2008) Role of root UV-B sensing in Arabidopsis early seedling development. Proc Natl Acad Sci USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten CH, Miller RW, Wolff IA, Jones Q. (1963) Amino acid composition of seeds from 200 angiospermous plant species. J Agric Food Chem 11: 399–410 [Google Scholar]
- Voll LM, Allaire EE, Fiene G, Weber AP. (2004) The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 136: 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JB, Abeydeera ND, Paul D, Phillips K, Rapala-Kozik M, Freeling M, Begley TP, Ealick SE, McSteen P, Scanlon MJ. (2010) A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell 22: 3305–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Zonia LE, Stebbins NE, Polacco JC. (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107: 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]