Reducing abnormal isoaspartyl accumulation in seed nuclear proteome increases vigor and seed longevity.
Abstract
PROTEIN l-ISOASPARTYL METHYLTRANSFERASE (PIMT) is a widely distributed protein-repairing enzyme that catalyzes the conversion of abnormal l-isoaspartyl residues in spontaneously damaged proteins to normal aspartyl residues. This enzyme is encoded by two divergent genes (PIMT1 and PIMT2) in plants, unlike many other organisms. While the biological role of PIMT1 has been elucidated, the role and significance of the PIMT2 gene in plants is not well defined. Here, we isolated the PIMT2 gene (CaPIMT2) from chickpea (Cicer arietinum), which exhibits a significant increase in isoaspartyl residues in seed proteins coupled with reduced germination vigor under artificial aging conditions. The CaPIMT2 gene is found to be highly divergent and encodes two possible isoforms (CaPIMT2 and CaPIMT2′) differing by two amino acids in the region I catalytic domain through alternative splicing. Unlike CaPIMT1, both isoforms possess a unique 56-amino acid amino terminus and exhibit similar yet distinct enzymatic properties. Expression analysis revealed that CaPIMT2 is differentially regulated by stresses and abscisic acid. Confocal visualization of stably expressed green fluorescent protein-fused PIMT proteins and cell fractionation-immunoblot analysis revealed that apart from the plasma membrane, both CaPIMT2 isoforms localize predominantly in the nucleus, while CaPIMT1 localizes in the cytosol. Remarkably, CaPIMT2 enhances seed vigor and longevity by repairing abnormal isoaspartyl residues predominantly in nuclear proteins upon seed-specific expression in Arabidopsis (Arabidopsis thaliana), while CaPIMT1 enhances seed vigor and longevity by repairing such abnormal proteins mainly in the cytosolic fraction. Together, our data suggest that CaPIMT2 has most likely evolved through gene duplication, followed by subfunctionalization to specialize in repairing the nuclear proteome.
The generation and accumulation of spontaneously damaged proteins in seed due to aging or stresses often adversely affect their vigor and viability. Such damaged proteins are thought to arise primarily due to spontaneous covalent modifications of existing proteins. Among such covalent protein modifications, the conversion of l-aspartyl or asparaginyl residues to abnormal isoaspartyl (isoAsp) residues in proteins is quite prevalent among organisms (Mudgett et al., 1997; Clarke, 2003; Ogé et al., 2008). The formation of such isoAsp in proteins often leads to the loss of protein function and the consequent loss of cellular function (Johnson et al., 1987; Clarke, 2003). One enzyme that participates in repairing such damaged protein is PROTEIN l-ISOASPARTYL METHYLTRANSFERASE (PIMT; EC 2.1.1.77). This enzyme catalyzes the transfer of a methyl group from S-adenosyl-methionine (AdoMet) to the free α-carboxyl group of abnormal l-isoAsp residues as well as the β-carboxyl group of d-aspartyl residues (Lowenson and Clarke, 1992; Aswad et al., 2000). Such PIMT-catalyzed methylation can initiate their conversion back to normal l-aspartyl residues and eventually reestablish the protein’s inherent functions (Johnson et al., 1987; McFadden and Clarke, 1987).
PIMT has been considered as an ancient and highly conserved enzyme that is widely distributed in evolutionarily diverged organisms such as eubacteria, archaebacteria, protozoa, fungi, nematodes, mammals, and plants (Kagan et al., 1997; O’Connor, 2006).
The physiological role of PIMT as an aging-related protein-repairing enzyme has been established through various genetic and biochemical studies, particularly from bacterial and animal systems. For example, overaccumulation of PIMT in Escherichia coli is associated with high heat shock survival under mild heat stress conditions, while a PIMT-deficient E. coli mutant showed higher sensitivity toward oxidative stress. PIMT-deficient mice were shown to suffer with epileptic seizures and a reduced life span (Kim et al., 1997; Visick et al., 1998; Kindrachuk et al., 2003). In such PIMT-deficient mutants, isoAsp residues become overrepresented in proteins, suggesting that PIMT maintains a low level of isoAsp in proteins, thus combating the effect of aging or stress particularly in cells with low metabolic activity.
As in other organisms, PIMT activity is also detected in a wide range of plants (Mudgett et al., 1997; Thapar et al., 2001), although the PIMT gene has been cloned only from a few plant species, namely Arabidopsis (Arabidopsis thaliana), wheat (Triticum aestivum), and chickpea (Cicer arietinum). Interestingly, two differentially regulated PIMT coding genes (AtPIMT1 and AtPIMT2) have been reported in Arabidopsis (Xu et al., 2004; Dinkins et al., 2008), in contrast to bacterial or animal systems, where such enzymes are encoded by a single gene.
The role of PIMT is not well elucidated in higher plants. Based on the occurrence of high PIMT activity in quiescent and germinating seeds, the role of PIMT in seed vigor and longevity has been suggested (Mudgett and Clarke, 1993). Additionally, PIMT activity was shown to associate with extreme seed longevity in sacred lotus (Nelumbo nucifera; Shen-Miller et al., 1995). Recently, PIMT1 (AtPIMT1) has been shown to be involved in seed longevity and germination vigor in Arabidopsis (Ogé et al., 2008), although the biological role and significance of PIMT2 still remain elusive.
In this study, we have chosen chickpea, since it is a major source of human and domestic animal food, particularly in developing countries. Chickpea ranks third in food legume crop production in the world, with 96% cultivation in developing countries. The productivity of this grain legume is low, and it is further reduced by environmental stresses (Ahmad et al., 2005). Despite having an immense agronomic and nutritional importance, research in chickpea is very limited, probably due to the nonavailability of genome sequencing data and mutant resources. Furthermore, the lack of an efficient and dependable transformation protocol has largely restricted transgenic research in chickpea. Chickpea seeds have been shown to be sensitive to aging and deterioration over time, hence causing a major concern for chickpea seed storage and seedling establishment (Kapoor et al., 2010). Thus, a large effort is required to elucidate the underlying mechanism controlling seed vigor and viability of this important crop plant. Therefore, we attempted to study PIMT in this plant, as PIMT1 has previously been shown to be involved in seed vigor and longevity in Arabidopsis; however, the biological role of PIMT2 is still unknown in plants to date.
In our previous study, we reported CaPIMT1 from this plant (Verma et al., 2010). In this study, we examined the correlation of PIMT activity and isoAsp accumulation with reduced germination vigor upon accelerated aging in chickpea. Subsequently, we isolated the second gene for PIMT (CaPIMT2) and showed it to produce two transcripts (CaPIMT2 and CaPIMT2′) through alternative splicing. Such spliced isoforms were biochemically characterized and compared with the CaPIMT1 enzyme. As a step toward elucidating the physiological role of CaPIMT2 in chickpea, the expression pattern of this gene was analyzed in major organs, stresses, and hormone treatments. Subcellular localization was also determined through confocal visualization of stably expressed GFP-fused PIMT proteins in roots. Finally, to examine the role of CaPIMT2 in seed vigor and longevity, seed-specific overexpression lines were generated in Arabidopsis and further analyzed.
To our knowledge, this is the first functional characterization of a PIMT2 gene involved in maintaining seed vigor and longevity by repairing abnormal isoAsp residues in the seed nuclear proteome. This study also exemplifies the subfunctionalization of genes after duplication through the partitioning of gene expression and localization.
RESULTS
Accelerated Aging Affects PIMT Activity and Enhances isoAsp Content in Chickpea Seed
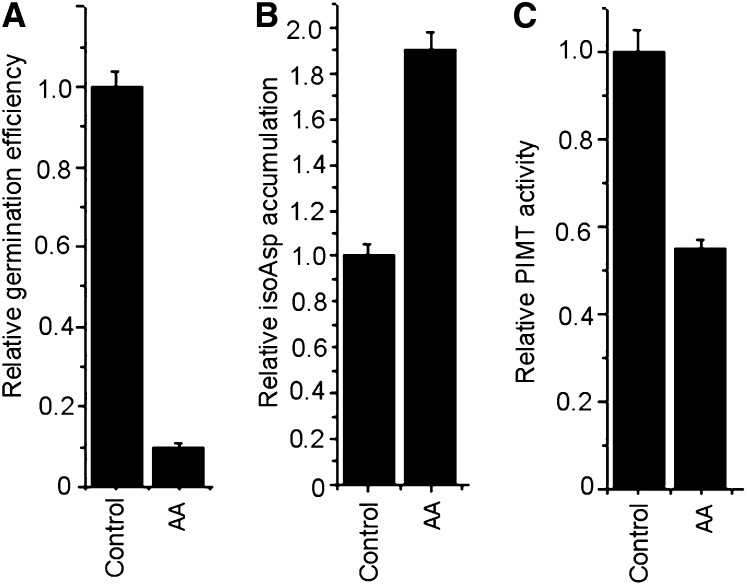
In higher plants, PIMT activity is mostly confined to seeds and has been shown to reduce abnormal isoAsp accumulation during aging or stress (Mudgett and Clarke, 1993; Mudgett et al., 1997; Ogé et al., 2008). In our previous study in chickpea, maximum PIMT activity was also recorded in dry and imbibed seeds but progressively decreased following radicle emergence (Verma et al., 2010). Here, we further investigated the isoAsp accumulation and PIMT activity in chickpea seed upon accelerated aging, since chickpea seed vigor is adversely affected by such stresses (Kapoor et al., 2010). To examine this, seeds were subjected to accelerated aging treatment and isoAsp accumulation was quantified. Subsequently, PIMT activity and germination percentage were also analyzed. The data revealed that seeds subjected to accelerated aging showed a significant reduction in germination percentage coupled with a significant increase in isoAsp residues in seed proteins and a substantial decrease in PIMT activity (Fig. 1). These data suggest that the PIMT activity might be required to maintain seed vigor in chickpea during aging.
Figure 1.
Effect of accelerated aging (AA) on germination (A), isoAsp accumulation (B), and PIMT activity (C) in chickpea seed. Seeds (moisture content, 22% ± 2%) were aged at 45°C and 100% relative humidity for 5 d. Values are the result of triplicate analyses. Error bars indicate sd.
CaPIMT2 from Chickpea Is Highly Divergent and Produces Two Different Transcripts through Alternative Splicing
Unlike other organisms, higher plants are reported to possess two PIMT coding genes (PIMT1 and PIMT2), and the PIMT1 gene from chickpea (CaPIMT1; accession no. GQ421817) has previously been reported from our laboratory (Verma et al., 2010). Hence, we attempted to isolate a second PIMT gene and complementary DNA (cDNA) from chickpea. To isolate the cDNA of the second PIMT gene (CaPIMT2), PCR was performed using degenerate primers, and eventually two different partial fragments were acquired. After sequencing, one fragment appeared to be CaPIMT1 while the other was found to be a paralog to CaPIMT1 (CaPIMT2). Next, to achieve the complete nucleotide sequence of CaPIMT2, both 3′ and 5′ RACE were performed. Besides the expected full-length PIMT2 transcript (CaPIMT2; accession no. JQ690076), another transcript (CaPIMT2′; accession no. JQ690077), differing only by 6 bp in the coding sequence, was also recovered. Both transcripts possess identical 5′ (35-bp) and 3′ (252-bp) untranslated regions, with 861- and 855-bp regions encoding the two protein isoforms CaPIMT2 (286 amino acids) and CaPIMT2′ (284 amino acids), respectively.
To examine the possibility of more transcript variants and the location of the 5′ end of PIMT2 RNAs, a primer extension experiment was also conducted. Primer extension analysis also identified only about 520 cDNA products corresponding to CaPIMT2 and CaPIMT2′ (Supplemental Fig. S1).
To check the existence of PIMT proteins in vivo, western-blot analysis was carried out in chickpea seed proteins using CaPIMT2 and PIMT-specific antibody. The results revealed only one immunoreactive band corresponding to CaPIMT2 and CaPIMT2′ protein when PIMT2-specific antibody was used (Fig. 2). As expected, two immunoreactive proteins corresponding to CaPIMT1 and CaPIMT2s (CaPIMT2 and CaPIMT2′) were observed when PIMT-specific antibody was used (Fig. 2). Apart from these, an immunoreactive band of 10 kD was also detected, which is possibly nonspecific, as no PIMT transcript corresponding to such low-mass protein has been identified in primer extension or RACE analysis.
Figure 2.
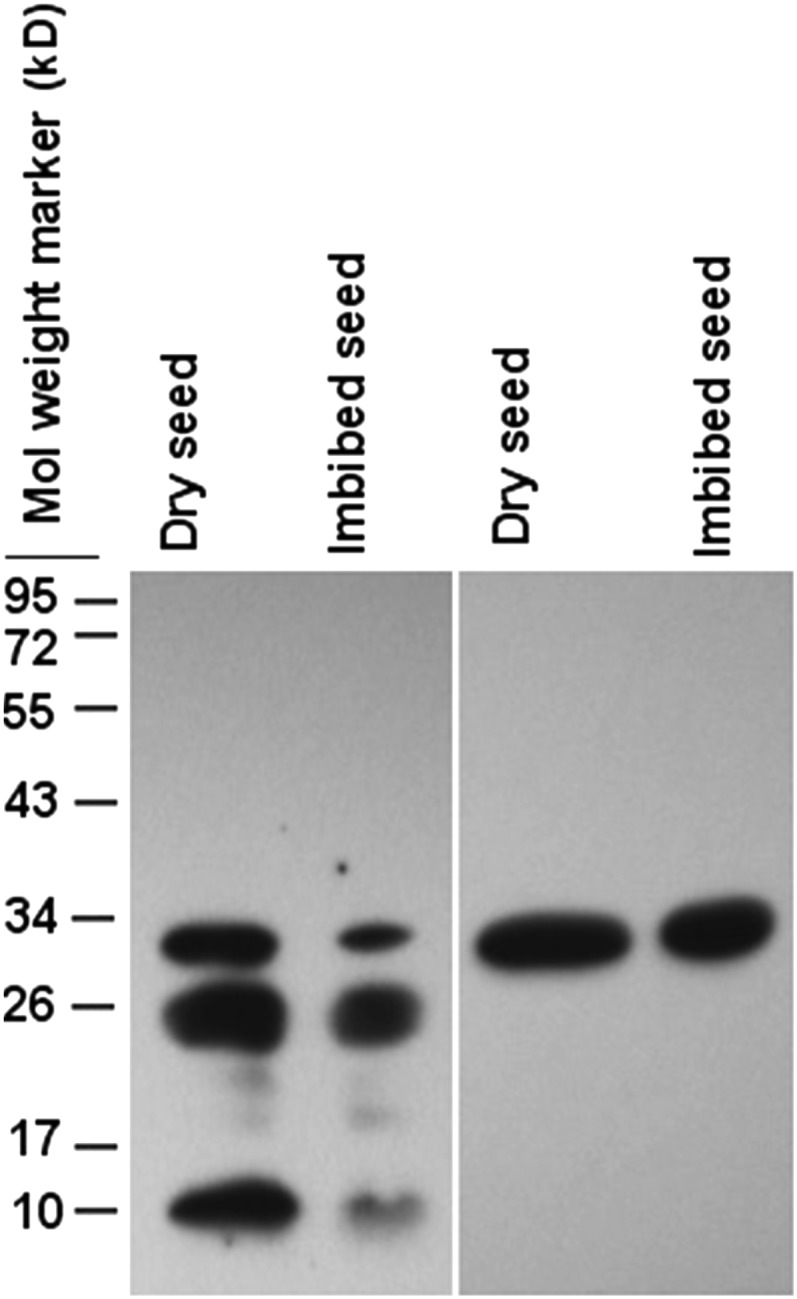
Western-blot analysis of PIMT proteins in dry and imbibed seeds. Approximately 50 μg of total proteins was separated by 12% SDS-PAGE and probed with anti-PIMT (left panel) or anti-CaPIMT2 (right panel) antibody.
Both CaPIMT2 and CaPIMT2′ polypeptide sequences contain five conserved domains (preregion I, TISAPHMHA; region I, ALDVGSGTG/ALDVGS–G, where boldface text signifies the presence of these two amino acids in CaPIMT2 and absence in the splice variant CaPIMT2′; region II, APYDAIHV; region III, QLKPGGRMVI; and postregion III, VRYPLTS [Kagan et al., 1997]), like other PIMT proteins; however, the CaPIMT2′ isoform lacks two important amino acids (Gly and Thr) in region I (ALDVGS–G) due to the absence of 6 bp in its coding region. Interestingly, region I (ALDVGSGTG) is important for the catalytic activity of enzymes utilizing AdoMet, where Gly is one of the key active-site amino acids (Smith et al., 2002). In addition, both polypeptides possess a unique 56-amino acid N-terminal extension that is absent in CaPIMT1 protein (Fig. 3A).
Figure 3.
(Figure continues on following page.)
A comparison of amino acid sequences of chickpea PIMTs with Arabidopsis and wheat PIMTs is shown in Figure 3A. Sequence comparisons among these plant PIMTs revealed identities ranging between 60% and 65%. When the 56-amino acid N-terminal extension was excluded in this analysis, CaPIMT2 was 81% identical to CaPIMT1. Moreover, this 56-amino acid N-terminal extension displayed no homology with any other proteins, even with the N-terminal extension of AtPIMT2′.
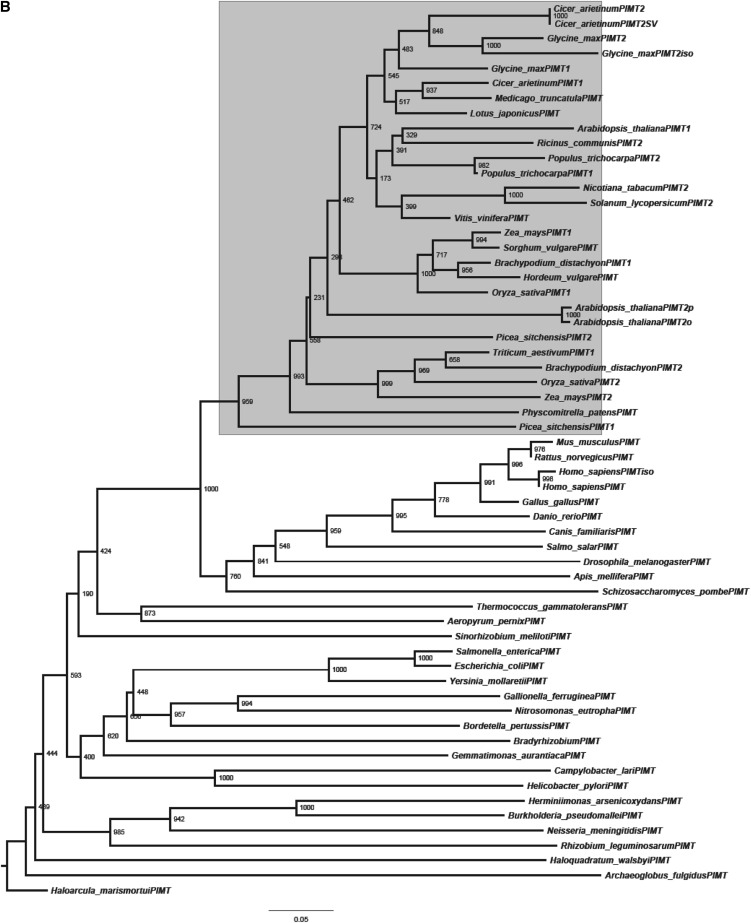
A phylogenetic tree was also constructed to analyze the evolutionary relationship of CaPIMT proteins with other PIMTs (Fig. 3B). The analysis supports the ubiquitous distribution of PIMTs with higher divergence among bacteria than eukaryotes, as suggested previously (Kagan et al., 1997; O’Connor, 2006). Among eukaryotes, plant PIMT proteins clustered together in one subgroup and are distinct from other subgroups of eukaryotes. Additionally, legume PIMTs that form a separate clade within plant and chickpea PIMTs were found to be closer to soybean (Glycine max) and Medicago spp. than others. The phylogenetic analysis also highlighted the sequence divergence between PIMT1 and PIMT2 proteins that exists in plants. From genomic information, it is now clearly evident that higher plants have duplicated PIMT genes, although in a few plants, only a single PIMT gene has been identified, possibly due to incomplete genome annotation. To analyze the intron-exon boundaries and to verify the possibility of alternative splicing, the genomic sequence of CaPIMT2 was cloned and analyzed. Sequence comparison of CaPIMT1 and CaPIMT2 genes revealed strong divergence in their introns and flanking sequences in contrast to exons. The CaPIMT2 (accession no. JQ690075) gene spans about 3,210 bp and contains four exons and three introns. Interestingly, the 6-bp difference in the coding region of CaPIMT2′ appears to be the result of alternative 3′ splice site selection of the second intron (Supplemental Fig. S2).
The CaPIMT2 Enzyme Exhibits Similar But Distinct Biochemical Characteristics to CaPIMT1
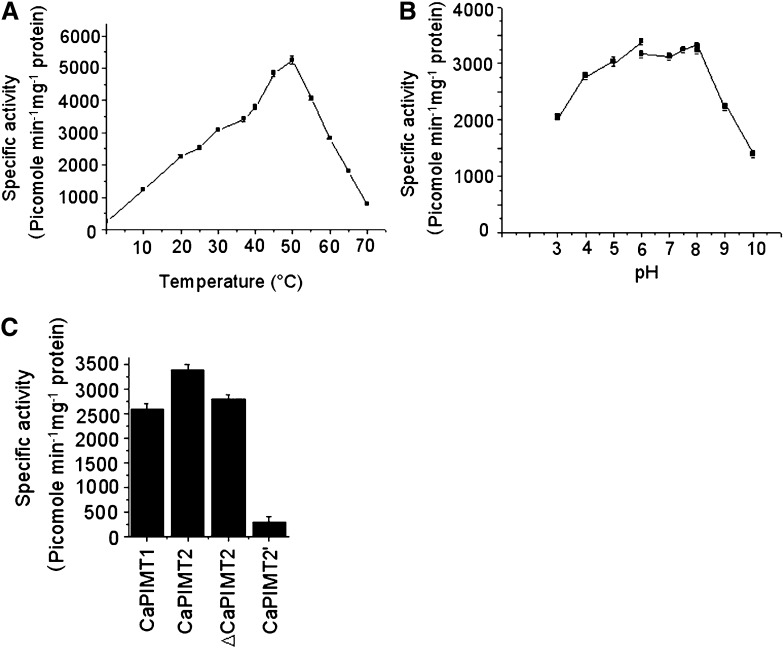
Since we observed significant differences in amino acid sequences between CaPIMT1 and CaPIMT2, particularly due to the N-terminal extension, we were interested in studying the enzymatic properties of CaPIMT2. To do that, the protein was expressed with a C-terminal hexahistidyl tag in E. coli. The protein was found to accumulate predominantly in the particulate fraction (Supplemental Fig. S3), which was solubilized using 8 m urea buffer and was subsequently purified to near homogeneity through nickel-charged affinity chromatography. Purified protein was used to determine the biochemical characteristics of the enzyme. The enzyme showed better activity at high assay temperatures, and maximum activity was recorded at 50°C. However, at temperatures above 60°C, PIMT activity showed a sharp decline (Fig. 4A). The pH optimum for the enzyme was examined over a broad range of pH values from 3 to 10 using three different buffers. Unlike CaPIMT1, the enzyme showed significant activity even in acidic conditions at pH 3 to 6, although maximum enzyme activity was detected at pH 6 to 8. However, the enzyme activity was significantly inhibited above pH 9.5 (Fig. 4B). Subsequently, using Lineweaver-Burk plots, the Km and Vmax values for the enzyme for both peptide and AdoMet were determined and compared with those of CaPIMT1 (Verma et al., 2010). The data are summarized in Table I. The CaPIMT2 enzyme was found to have lower Km but higher Vmax values particularly for peptide and overall higher enzymatic activity than CaPIMT1 (Fig. 4C).
Figure 4.
Biochemical characterization of the CaPIMT2 enzyme. A, Effect of temperature on the activity of purified recombinant CaPIMT2 enzyme. The specific activity was measured at different temperatures ranging from 0°C to 70°C. B, Effect of pH on the activity of purified recombinant CaPIMT2. The specific activity was measured at 37°C using different buffers (pH 3–6, citrate phosphate buffer; pH 6–8, sodium phosphate buffer; pH 8–10, 2-amino-2-methyl-1,3-propanediol buffer). C, Comparison of enzyme activity among CaPIMT1, CaPIMT2, ΔCaPIMT2 (CaPIMT2 lacking the 56-amino acid N terminus), and CaPIMT2′. Approximately 5 μg of purified protein was assayed in each case. Error bars in all graphs indicate se from three independent experiments.
Table I. Comparative analysis of CaPIMT1 and CaPIMT2 enzyme characteristics.
To examine whether the unique 56-amino acid N terminus of the CaPIMT2 protein has any contribution to its higher enzymatic activity, the CaPIMT2 deletion protein lacking the 56-amino acid N terminus (ΔCaPIMT2) was similarly expressed and purified. Purified protein was used to determine its activity, which was compared with that of CaPIMT2. As shown in Figure 4C, ΔCaPIMT2 exhibited slightly lower activity than the full-length protein.
Furthermore, we investigated the catalytic activity of CaPIMT2′, since this isoform lacks important active-site residues due to alternative splicing. To do that, recombinant protein was similarly expressed in E. coli and purified to homogeneity. Purified protein was assayed for PIMT activity, and a remarkably reduced activity was observed in contrast to CaPIMT2 or CaPIMT1 protein (Fig. 4C). These results were confirmed in multiple replicates with varying enzyme concentrations.
CaPIMT2 Is Differentially Expressed and Regulated
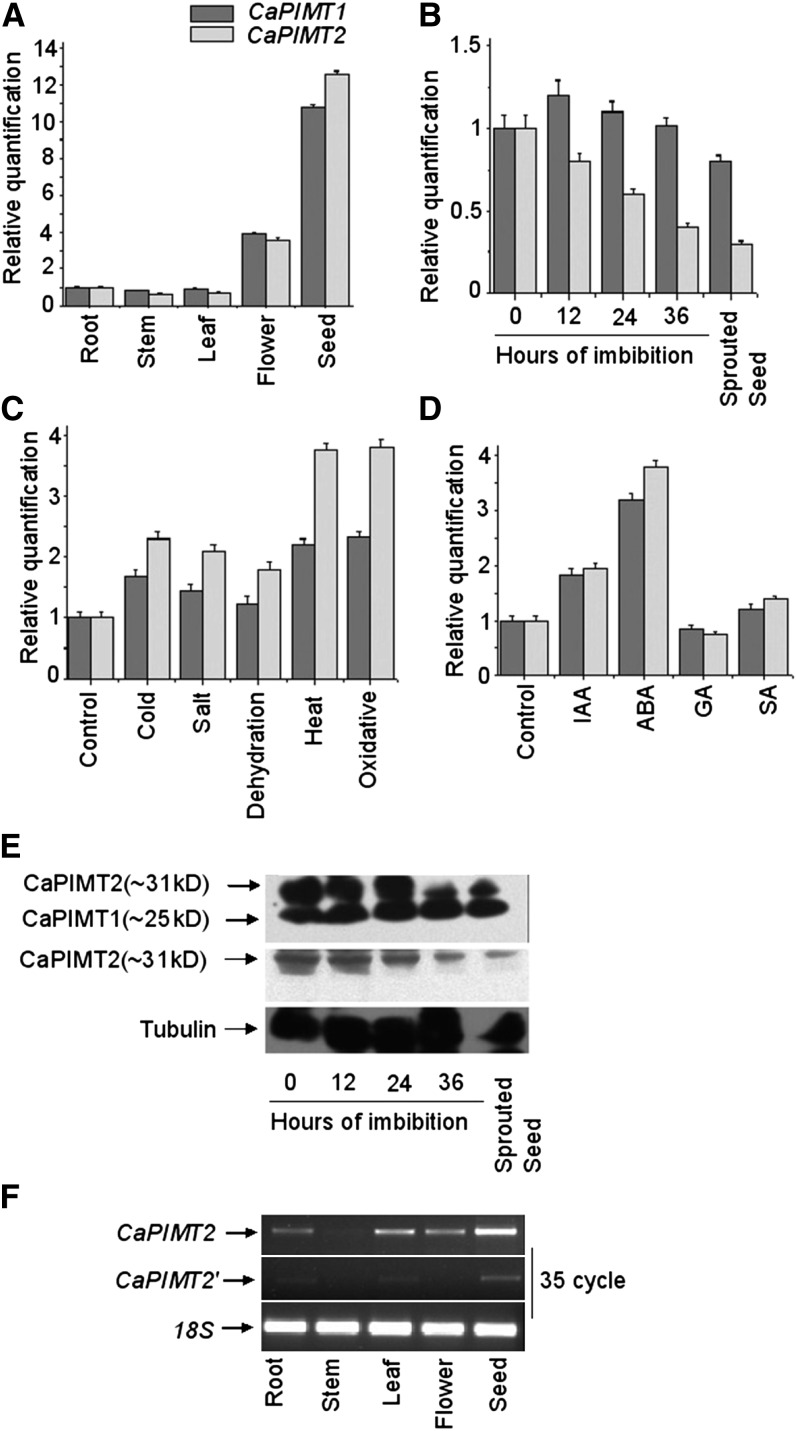
As a step toward elucidating the role of CaPIMT2 in chickpea, the transcript accumulation across different organs was examined along with CaPIMT1 through quantitative real-time PCR. The data showed that the expression pattern of CaPIMT2 was fairly similar to that of CaPIMT1, since CaPIMT2 transcript also accumulated predominantly in the seed followed by the flower and was also noticeable in leaf, shoot, and root tissues (Fig. 5A). However, it is interesting that the abundance of CaPIMT2 transcript appeared lower than PIMT1 in almost all organs, except in dry seed, where transcript accumulation was found to be slightly more than the CaPIMT1 transcript. Furthermore, the transcript abundance of these two genes was examined during the course of seed germination. RNA was extracted from dry, imbibed, and sprouted seeds, and subsequently, real-time PCR was performed. The results revealed that CaPIMT2 transcript progressively declined with imbibition period and was significantly reduced in sprouted seeds. However, in the case of CaPIMT1, transcript accumulation was observed to be the same or a little greater during initial imbibition periods but significantly reduced in sprouted seeds (Fig. 5B).
Figure 5.
Expression analysis of CaPIMT1 and CaPIMT2. A to D, Quantitative real-time PCR analysis of CaPIMT1 and CaPIMT2 transcript abundance in different organs (A), during germination (B), under different stresses (C), and in the presence of different hormones (D). Total RNA from each sample was reverse transcribed and subjected to real-time PCR analysis. The relative expression value of each gene was normalized to an endogenous control (18S or β-actin) and calculated using the ΔΔCT method (Applied Biosystems). Values are the result of triplicate analyses of two biological replicates. Error bars indicate sd. IAA, Indole-3-acetic acid; SA, salicylic acid. E, Western-blot analysis of CaPIMT1 and CaPIMT2 in dry, imbibed, and sprouted seeds. Approximately 50 μg of total proteins was separated by 12% SDS-PAGE and probed with anti-PIMT (top panel) or anti-CaPIMT2 (middle panel) antibody. A similar blot was also probed with anti-tubulin antibody (bottom panel). F, Transcript analysis of CaPIMT2 and CaPIMT2′ in different organs through semiquantitative real-time PCR.
To confirm whether such a pattern of transcript accumulation during the course of germination is also reflected in protein level, we carried out western-blot analysis using PIMT- and CaPIMT2-specific antibodies. Initially, the specificity of these antibodies was verified using purified E. coli-expressed proteins (Supplemental Fig. S4). Proteins were extracted from dry, imbibed, and sprouted seeds, and western blotting was performed using these antibodies. The result from this analysis was found to be fairly similar to the transcript accumulation in seeds, where the CaPIMT2 protein level was high in dry seeds and progressively decreased with imbibition, although the difference was more evident at 36 h of imbibition and in sprouted seeds. On the contrary, CaPIMT1 protein level was fairly similar throughout dry, imbibed, and sprouted seeds (Fig. 5E).
Previously, PIMT activity and CaPIMT1 transcript accumulation were shown to increase in chickpea seedlings challenged with various environmental stress conditions (Verma et al., 2010). Hence, we were also interested to examine the CaPIMT2 transcript profile under such situations. The results showed that CaPIMT2 transcript accumulated about 4-fold more relative to the control under all stress conditions tested (Fig. 5C). Interestingly, stress-exposed seedlings accumulated more CaPIMT2 transcript than CaPIMT1, indicating that such stresses preferentially induce the production of CaPIMT2 transcript over CaPIMT1. Next, to investigate the effect of various hormones on CaPIMT1 and CaPIMT2 gene expression, seedlings were treated with hormones, and subsequently, transcript levels were examined through quantitative real-time PCR. The results showed that both CaPIMT1 and CaPIMT2 transcript accumulations were significantly greater in abscisic acid (ABA)-treated seedlings than in the control, although CaPIMT2 was seen to be more influenced by ABA. In the presence of exogenous GA, both CaPIMT1 and CaPIMT2 transcript accumulations were found to be the same or little reduced relative to the control plant. Slightly enhanced accumulation of these transcripts was also observed in the presence of indole-3-acetic acid and salicylic acid (Fig. 5D). These results were confirmed through several independent experiments.
Transcript accumulation of CaPIMT2′ across the organs was also analyzed. To do that, we designed specific primers for both CaPIMT2 and CaPIMT2′. Initially, PCR specificity was checked using respective plasmids (Supplemental Fig. S5). CaPIMT2′ transcript accumulation was observed only in root, leaf, and seed with extremely low abundance (Fig. 5F).
Promoter Sequences of CaPIMT1 and CaPIMT2 Are Highly Divergent
To understand the molecular basis of the differential expression of these two genes, 5′ upstream regulatory sequences of each gene were isolated and cloned through genome-walking PCR using a kit (Clontech). Nucleotide sequences of the promoters of these two genes are presented in Supplemental Figure S6. Sequence comparison revealed that promoter sequences are highly diverged, indicating a different transcriptional regulatory mechanism for each gene. Subsequently, upstream sequences were screened for putative cis-acting regulatory elements using various programs (PLACE, Cister, PlantCARE). The results are summarized in Supplemental Table S1. However, both promoters contain many similar putative cis-regulatory elements in different positions, and few cis-regulatory elements specific for the CaPIMT1 or CaPIMT2 promoter have also been noticed. A cis-regulatory element (TGACGTVMAMY) for high-level seed-specific expression has also been observed in both cases. In addition, both the promoters contain ABRE (ACGTG) and WBOXATNPRI (TTGAC) cis-elements for ABA- and salicylic acid- or biotic stress-induced gene expression, respectively. Interestingly, repeated MYC recognition sites (CANNTG) or MYB1 recognition sites (WAACCA), known to act as cis-acting elements for drought-induced expression, were present only in the CaPIMT2 promoter region. The presence of such similar but distinct cis-regulatory elements thus explains the differential expression of these two genes.
CaPIMT1 and CaPIMT2 Exhibit Differential Subcellular Localization
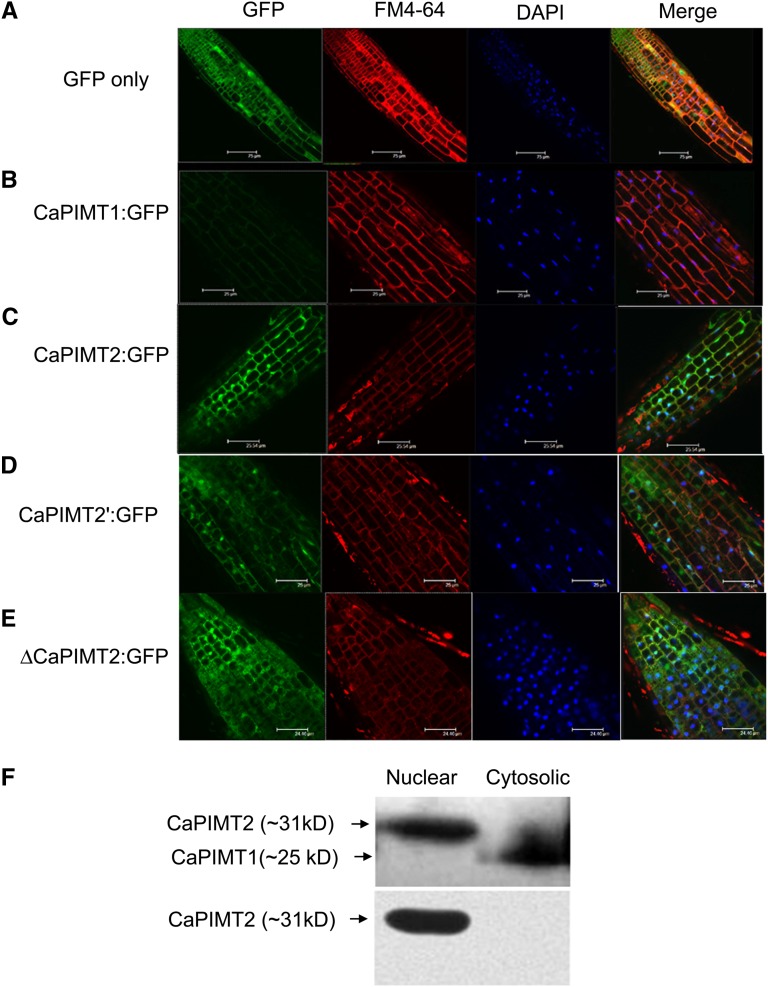
Previous reports based on the transiently expressed GFP fusion proteins revealed differential subcellular localization of AtPIMT1 and AtPIMT2s (Xu et al., 2004; Dinkins et al., 2008). Interestingly, AtPIMT2 transcriptional start sites were shown to target different subcellular compartments. In this study, we attempted to determine the subcellular localization of CaPIMT proteins (CaPIMT1, CaPIMT2, and CaPIMT2′) through confocal visualization of stably expressed GFP-fused proteins in Arabidopsis.
To do this, CaPIMT1, CaPIMT2, and CaPIMT2′ cDNAs were cloned under the control of the 35S promoter to express constitutively as C-terminal GFP-fused proteins, and transgenic lines were generated. Initially, to check whether the GFP-fused PIMT proteins (CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP) are functional in their respective transgenic Arabidopsis lines, PIMT activity was analyzed in these transgenic lines. The results showed that all GFP-fused PIMT proteins were functional, as PIMT transformed lines (CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP) exhibited enhanced PIMT activity as compared with the wild type or the GFP-only transformed line (Supplemental Fig. S7). Subsequently, the localizations of all GFP-fused PIMT proteins (CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP) were examined in roots of respective transgenic Arabidopsis seedlings through confocal imaging microscopy. The results are presented in Figure 6. As expected, in control Arabidopsis plants, GFP alone was distributed in nucleus and cytosol in the root (Fig. 6A). However, the distribution of CaPIMT1:GFP was observed predominantly in the cytosol but also detectable in the plasma membrane (Fig. 6B). On the contrary, both CaPIMT2:GFP and CaPIMT2′:GFP transformed plants presented a clear view of nuclear localization apart from its GFP fluorescence in the plasma membrane (Fig. 6, C and D; Supplemental Fig. S8). Furthermore, to investigate any possible role of the 56-amino acid N terminus of CaPIMT2 in determining such localization, we examined the localization of GFP-fused CaPIMT2 protein lacking the 56-amino acid N terminus (ΔCaPIMT2). The results showed that CaPIMT2 lacking the N terminus also localized in the nucleus and membrane (Fig. 6E). These data indicate that the 56-amino acid N terminus has no apparent role in determining the subcellular localization of CaPIMT2. The localization of CaPIMT proteins in the nucleus and membrane was confirmed by respective tracking dyes.
Figure 6.
Subcellular localization of CaPIMT1, CaPIMT2, CaPIMT2′, and ΔCaPIMT2 (CaPIMT2 lacking the 56-amino acid N terminus). A to E, Localization of GFP, CaPIMT1:GFP, CaPIMT2:GFP, CaPIMT2′:GFP, and ΔCaPIMT2:GFP in roots of Arabidopsis transgenic plants. Fluorescence images were taken using a confocal-laser scanning microscope. The plasma membrane and nuclei were stained by FM4-64 and 4′,6-diamino-phenylindole (DAPI), respectively. F, Western blot of nuclear and cytosolic proteins of 24-h-imbibed chickpea seed. Approximately 50 μg of nuclear or cytosolic protein was separated by 12% SDS-PAGE and probed with anti-PIMT (top panel) or anti-CaPIMT2 (bottom panel) antibody.
To confirm the nuclear localization of CaPIMT2 further, cell fractionation followed by western blotting was employed. Using the Qproteome nuclear protein kit (Qiagen), nuclear and cytosolic proteins were extracted from imbibed chickpea seeds. Subsequently, proteins were separated by 12% SDS-PAGE, and western-blot analysis was carried out using PIMT- and CaPIMT2-specific antibodies. As shown in Figure 6F, PIMT2 protein was found only in the nuclear fraction, while CaPIMT1 protein was found in the cytosolic fraction.
Seed-Specific Overexpression of CaPIMT1 and CaPIMT2 Genes in Arabidopsis Enhances Seed Vigor and Longevity by Reducing Abnormal isoAsp Accumulation Predominantly in Cytosolic and Nuclear Proteins, Respectively
PIMT1 from Arabidopsis was shown to be involved in enhancing germination vigor and longevity (Ogé et al., 2008); however, the involvement of PIMT2 in any such trait in seeds remains unexplored. Furthermore, in our previous experiment, we observed that dry seeds accumulate rather more PIMT2 transcript and protein than PIMT1 in chickpea. This observation prompted us to study the involvement of CaPIMT2 in seed vigor and longevity.
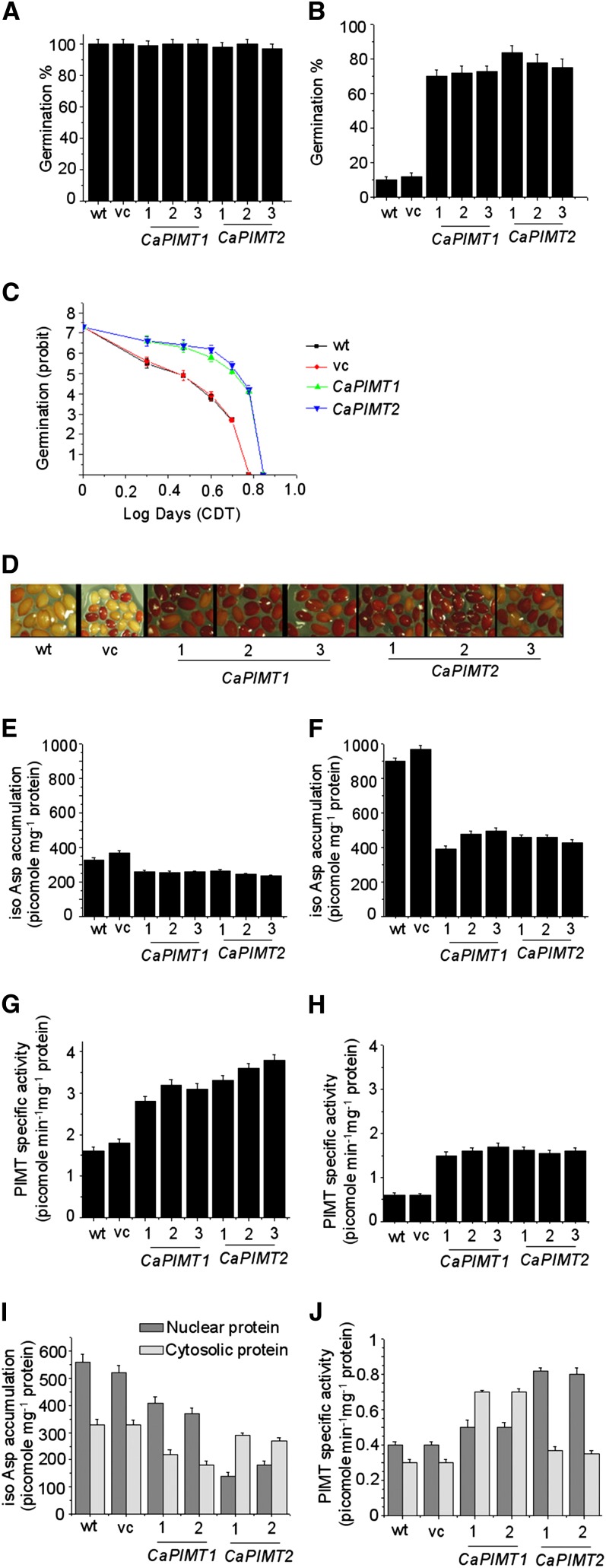
To investigate this, both CaPIMT1 and CaPIMT2 were overexpressed in seeds using the napin promoter in Arabidopsis. Seed-specific PIMT transcript accumulation was examined through real-time PCR, and a significant quantity of each respective CaPIMT transcript was observed in CaPIMT1 and CaPIMT2 transformants. Based on this analysis, three independent homozygous T3 lines for each overexpressed CaPIMT gene were selected and subsequently used to assess their germination vigor and longevity. To evaluate the seed vigor and longevity, a control deterioration test (CDT) was employed, since sensitivity or resistance of seeds to CDT has been successfully used for the rapid assessment and prediction of seed vigor and longevity in various studies (Delouche and Baskin, 1973; TeKrony, 1995; Lanteri et al., 1996; Prieto-Dapena et al., 2006; Ogé et al., 2008). Hence, to assess the seed vigor and longevity, seeds were subjected to CDT (0–7 d), and subsequently, the germination percentage was scored. Under normal conditions, CaPIMT1 and CaPIMT2 transformed seeds as well as control (empty vector transformed or wild-type) seeds showed no differences with respect to germination percentage, although seeds from each line progressively deteriorated as a function of the duration of the CDT. After 4 d of CDT, control seeds exhibited poor germination (10%–14%), in contrast to CaPIMT1 and CaPIMT2 transformed seeds, where, remarkably, 80% to 90% germination was observed (Fig. 7, A and B). To estimate the number of days of CDT required for a decline to 50% germination (LD50), seed survival curves fitted with probit analysis from germination data were generated (Fig. 7C; Supplemental Fig. S9). The results showed that the control seeds (wild-type LD50 = 2.50 ± 0.16 d; vector control LD50 = 2.55 ± 0.18 d) reached 50% mortality (unable to germinate) significantly earlier than the seeds from CaPIMT1 (LD50 = 3.85 ± 0.24 d) and CaPIMT2 (LD50 = 4.05 ± 0.26 d) transgenic lines. However, only slender differences of LD50 were observed between CaPIMT1 and CaPIMT2 transgenic lines.
Figure 7.
Characterization of CaPIMT1 and CaPIMT2 transformed Arabidopsis seeds. Results from three independent transformed lines (1, line 1; 2, line 2; 3, line 3) of each are shown here. Eight-week-old seeds were subjected to CDT for 0 to 7 d. Germination frequency was evaluated after each day of treatment. Germination was scored after 5 d of imbibition at 22°C. A and B, Germination percentage of wild-type (wt), empty vector (vc), and CaPIMT1 and CaPIMT2 transformed Arabidopsis seeds before (A) and after (B) CDT (4 d). C, Seed survival curves (based on a seed’s ability to germinate) of CaPIMT1 and CaPIMT2 transformed Arabidopsis seed. Germination (probit) was plotted against the logarithmic time scale (days) of CDT. Only one representative independent line of each transgenic plant is shown here, and values are the result of triplicate analyses. D, Viability of seeds in the wild type and respective transformed lines after CDT (4 d). Seed viability was analyzed using TZ staining, and dark red staining indicates viable seeds. E to H, isoAsp accumulation (E and F) and PIMT activity (G and H) in seeds of the wild type and respective transformed lines before (E and G) and after (F and H) CDT (4 d). I and J, isoAsp accumulation (I) and PIMT activity (J) of the nuclear or cytosolic fraction of seed proteins in the wild type and respective transformed lines after CDT (4 d). Approximately 50 μg of crude protein was used for PIMT assay or isoAsp quantification in each case. Values are the result of triplicate analyses. Error bars indicate sd.
To check whether this improved seed germination after CDT is correlated with the potential seed viability in transgenic lines, a tetrazolium (TZ) assay was performed (Berridge et al., 1996). TZ precipitates to red-colored 2,3,5-triphenyl formazan by the activity of dehydrogenases present in the living cells. Transformed seeds along with control seeds were soaked in a solution of 2,3,5-triphenyl tetrazolium chloride before and after CDT. As expected under normal conditions, CaPIMT1, CaPIMT2, and control seeds were uniformly stained a dark red, while after control deterioration, only CaPIMT1 and CaPIMT2 transformed seeds exhibited such staining, in contrast to control seeds, which remained unstained or were stained pale red (Fig. 7D). These data clearly indicate that wild-type or empty vector transformed seeds subjected to control deterioration exhibited higher mortality while seeds overexpressing CaPIMT1 or CaPIMT2 exhibited less mortality, thus demonstrating the role of CaPIMT1 and CaPIMT2 in maintaining seed viability during aging or seed storage. To ascertain further whether such a phenotype is correlated with increased activity of PIMT and the consequent reduced accumulation of isoAsp residues in seed protein, PIMT activity and isoAsp content were quantified in seeds under normal conditions as well as after CDT. As expected, CaPIMT1- and CaPIMT2-overexpressing dry seeds exhibited enhanced PIMT activity and reduced accumulation of isoAsp in the proteome as compared with control seeds prior to imbibition (Fig. 7, E–H). Even though elevated levels of isoAsp were observed in both control and transgenic seeds after CDT, isoAsp accumulation was found to be restricted to 350 to 450 picomoles (pmol) mg−1 protein in CaPIMT1 or CaPIMT2 transgenic lines. On the contrary, wild-type or vector transformed lines accumulated detrimental isoAsp contents up to 900 pmol mg−1 protein after CDT (Fig. 7F).
To confirm this finding further, seed vigor and viability were also examined in GFP-fused PIMT transgenic lines (CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP), and reasonably similar results were observed (Supplemental Fig. S10). However, as compared with CaPIMT1:GFP and CaPIMT2:GFP transformed lines, the CaPIMT2′:GFP transformed line exhibited significantly less improved seed vigor, probably due to its reduced methyltransferase activity. We also noted that the improvement of seed vigor in 35S cauliflower mosaic virus promoter-driven GFP-fused PIMT transformed lines was less than in napin promoter-driven PIMT transformed lines. In each case, enhancement of seed vigor and longevity of transgenic lines were correlated well with their PIMT activity and isoAsp accumulation. To check whether these phenotypes are due to high PIMT activity but not due to the effect of an increased abundance of the overexpressed protein that could act as a molecular shield to prevent protein aggregation in the cells during stress, we carried out a similar experiment using Arabidopsis transgenic seeds overexpressing chickpea myoinositol 1-phosphate synthase (MIPS; unrelated to PIMT). The results clearly showed that the MIPS-overexpressing line did not show any such phenotypes (Supplemental Fig. S11) like PIMT-overexpressing lines and thus refute the above possibility of molecular shields but the phenotype is due to enhanced PIMT activity.
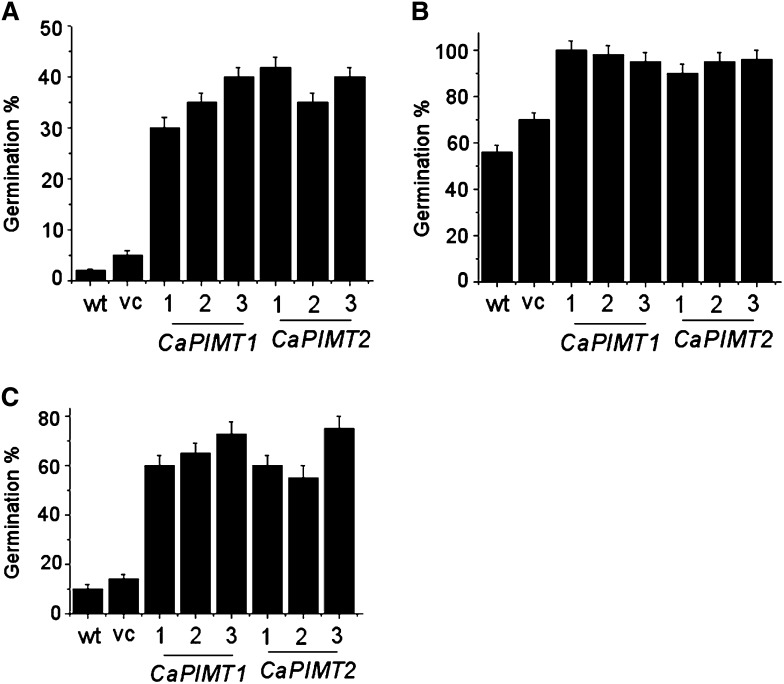
We further investigated isoAsp accumulation and PIMT activity in nuclear and cytosolic fractions of seed proteins in CaPIMT1 and CaPIMT2 transformed lines, since CaPIMT2 was observed to be localized particularly in the nucleus while CaPIMT1 was found to be cytosolic (Fig. 6). Cytosolic and nuclear protein fractions were extracted from seeds of respective transformed lines, and subsequently, PIMT activity and isoAsp content were quantified. Differential accumulations of isoAsp content and PIMT activity were observed in CaPIMT1- and CaPIMT2-overexpressing seeds. In the case of CaPIMT1, higher PIMT activity and a consequent reduced accumulation of isoAsp in the proteome were observed in the cytosolic fraction as compared with the nuclear fraction, while CaPIMT2 transformants exhibited higher PIMT activity and lower isoAsp content predominantly in the nuclear fraction (Fig. 7, I and J). Nuclear and cytosolic fractions were confirmed by western-blot analysis using the nuclear protein marker histone H3 antibody (Supplemental Fig. S12). Seed vigor also implies the ability to complete germination under widely variable environmental conditions. Hence, germination percentage under various stress situations was evaluated. In normal conditions, CaPIMT1 and CaPIMT2 transformed seeds and control seeds showed no differences in their germination percentage, while significant differences were observed at 200 mm sodium chloride, 0.5 μm paraquat, and polyethylene glycol (−0.5 megapascals). In each case, both CaPIMT1 and CaPIMT2 transformed seeds showed higher percentages of germination over wild-type or empty vector transformed seeds (Fig. 8).
Figure 8.
Analysis of germination efficiency of wild-type (wt), empty vector (vc), and CaPIMT1 and CaPIMT2 transformed Arabidopsis seeds under various stress conditions. A, Polyethylene glycol (−0.5 megapascals). B, Sodium chloride (200 mm). C, Paraquat (0.5 μm). Values are the result of triplicate analyses. Error bars indicate sd.
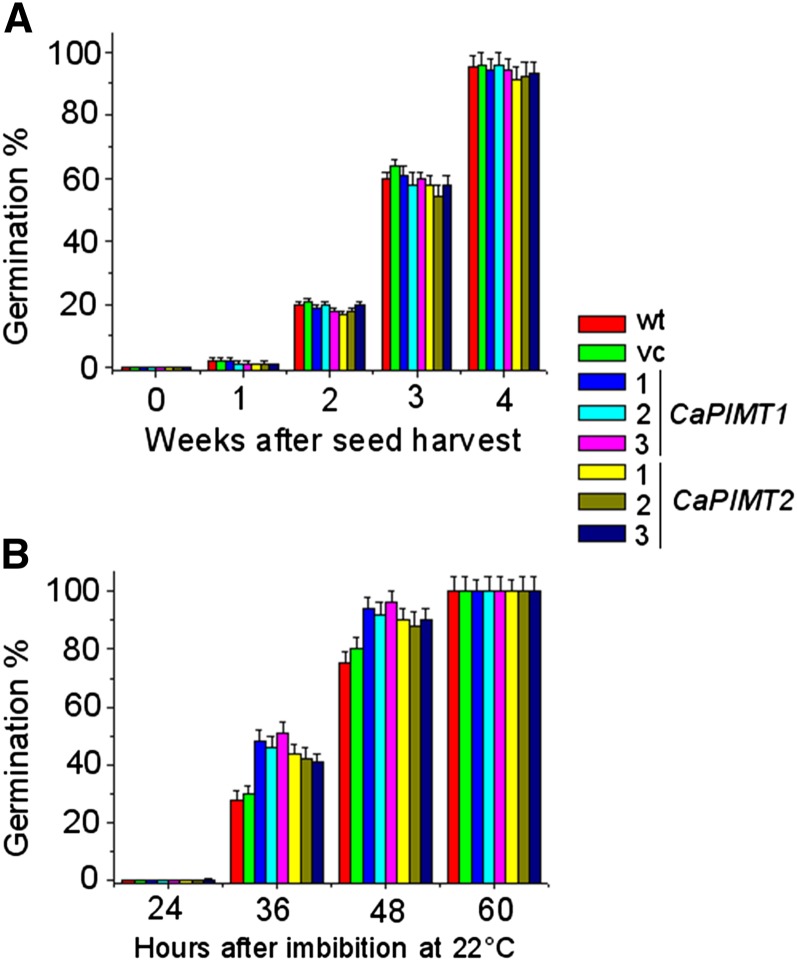
We further assessed the dormancy and speed of germination in these transgenic seeds, particularly to check whether the expression of PIMT caused any changes in seed dormancy or speed of germination. To assess dormancy, freshly harvested and 1- to 4-week-old dry seeds were tested for germination, and no significant change of dormancy was observed among control, CaPIMT1, and CaPIMT2 transformed seeds. Freshly harvested and 1-week-old seeds from wild-type, vector control, CaPIMT1, and CaPIMT2 transgenic lines showed almost no germination; however, after additional weeks of dry storage, the germination frequency of control and transgenic seeds was found to be increased in a fairly similar manner. After 4 weeks of dry storage (after ripening), wild-type, vector control, CaPIMT1, and CaPIMT2 transformed seeds showed more than 90% germination (Fig. 9A). A control experiment was also performed with these seeds, which showed nearly 100% germination after a cold treatment. To assess the germination speed in normal conditions, 8-week-old dry seeds were imbibed at 22°C and germination was scored at different hours of imbibition. The results showed that CaPIMT1 and CaPIMT2 transformed seeds completed germination faster than the wild type or vector control. CaPIMT1 and CaPIMT2 transformed seeds completed germination of less than 45% to 50% after 36 h of imbibition, while the wild type or vector control showed only about 30% germination. After 48 h of imbibition, more than 90% of CaPIMT1 and CaPIMT2 transformed seeds completed germination, while only 75% to 80% germination was observed in the case of wild-type or vector control seeds (Fig. 9B).
Figure 9.
A, Comparison of seed dormancy among wild-type (wt), empty vector (vc), and CaPIMT1 and CaPIMT2 transformed Arabidopsis seeds. Seeds were stored for 0 (fresh), 1, 2, 3, and 4 weeks after harvest and assayed for germination at 22°C. Germination was scored after 7 d of imbibition. B, Comparison of the speed of germination among wild-type (wt), empty vector (vc), and CaPIMT1 and CaPIMT2 transformed Arabidopsis seeds. Seeds were stored for 8 weeks after harvest and assayed for germination at 22°C. Germination was monitored and scored at different hours of imbibition. In each case, values are the result of triplicate analyses. Error bars indicate sd.
DISCUSSION
PIMT is a protein-repairing enzyme and is distributed through all phylogenetic domains. In higher plants, PIMT activity is particularly high in seeds and is reportedly enhanced in response to abiotic stresses in several plant species, including chickpea (Mudgett and Clarke, 1993, 1994; Verma et al., 2010). Interestingly, this protein-repairing enzyme is encoded by two genes in plants (PIMT1 and PIMT2), in contrast to bacterial or animal systems, where PIMT is encoded by a single gene. The PIMT1 gene from Arabidopsis has recently been shown to be involved in enhancing germination vigor and longevity (Ogé et al., 2008). However, the biological role and physiological significance of the second PIMT gene is unknown. In this study, we have investigated the molecular regulation, physiological role, and significance of the second PIMT gene in chickpea. We have chosen this particular plant not only because of its adaptation to harsh environmental conditions but also due to its immense agricultural importance.
Using degenerate primers and RACE strategies, we have identified a highly divergent second PIMT gene (CaPIMT2) that encodes two possible isoforms (CaPIMT2 and CaPIMT2′) differing by two amino acids (Gly and Thr) in the region I catalytic domain (Fig. 3A). This unique deletion of two amino acids is a result of alternative 3′ splice site selection in the second intron (Supplemental Fig. S2). Despite the fact that various AtPIMT2 splice variants have been reported previously, they were shown to arise either through alternative 5′ or 3′ splice site selection in the first intron, resulting in no such unique changes in their conserved domains. The CaPIMT2 gene was found to have four exons and three introns, similar to other reported plant PIMTs, including CaPIMT1. Phylogenetic analysis and sequence divergence between CaPIMT1 and CaPIMT2, particularly in their non-coding, 5′ and 3′ flanking regions, suggest that they might have arisen due to gene duplication like other plant PIMTs (O’Connor, 2006). Furthermore, we investigated the enzymatic properties of CaPIMT2, which exhibited reasonably similar but distinct biochemical properties to other PIMTs (Villa et al., 2006; Verma et al., 2010). CaPIMT2 appeared to be enzymatically more active and showed a higher Vmax but slightly lower Km for peptide than CaPIMT1 (Table I). In addition, CaPIMT2 showed significant activity at acidic pH (Fig. 4B), as opposed to CaPIMT1, which is mostly active in alkaline pH. Interestingly, alkaline pH favors isoAsp formation, so very little isoAsp formation from asparaginyl residues is expected in acidic conditions but continues to form from aspartyl residues even under acidic conditions (Patel and Borchardt, 1990). This result proposes that CaPIMT1 and CaPIMT2 are optimized to be active over a range of pH values where l-isoAsp formation occurs. Furthermore, deletion analysis of the CaPIMT2 enzyme suggests that the 56-amino acid N terminus may somehow contribute to the higher activity of CaPIMT2. Remarkably, the CaPIMT2′ splice variant exhibited extremely reduced activity (Fig. 4C), which could be explained by the deletion of active-site amino acid Gly (Smith et al., 2002). However, why chickpea plants require such a PIMT with extremely reduced methyltransferase activity remains to be demonstrated.
Earlier studies have shown that PIMT1 and PIMT2 are differentially expressed in Arabidopsis, and PIMT2 in particular is more influenced by developmental stage, ABA, and abiotic stresses (Xu et al., 2004). In this study, we also observed that CaPIMT1 and CaPIMT2 are differentially regulated in chickpea. While CaPIMT2 transcript and protein accumulation were found to progressively decline upon imbibition in seeds, CaPIMT1 exhibited fairly steady expression levels during the early phase of germination (Fig. 5, B and E). Similar observations were also reported from Arabidopsis, where AtPIMT2 transcript abundance declines upon imbibition and AtPIMT1 transcript remains the same (Xu et al., 2004). These expression patterns indicate that PIMT1 and PIMT2 are differentially regulated during seed dormancy and the germination period and may have a distinct role in these processes. In addition, CaPIMT2 appears to be more influenced by abiotic stresses and ABA than CaPIMT1 (Fig. 5, C and D). This differential expression pattern of these two genes could be explained by the divergence of their 5′ regulatory sequences.
Our subcellular localization study demonstrated that CaPIMT1 predominantly localized in cytosol while both isoforms of CaPIMT2 were primarily observed in nucleus. Previously, AtPIMT1 was also shown to be cytosolic and AtPIMT2 as primarily nucleus localized, although various splice variants and transcriptional start site variants were shown to have adopted different subcellular localizations, including mitochondria and plastid (Xu et al., 2004; Dinkins et al., 2008). PIMT activity and protein have been reported to localize to the nucleus in other organisms as well (O’Connor, 1987; O’Connor and Germain, 1987). Furthermore, histone H2B was shown as a nucleus-localized major target protein for PIMT repair in rat PC12 cells (Young et al., 2001). Additionally, the expression of GFP-fused CaPIMT1 and CaPIMT2 was visibly associated with plasma membrane. We were not surprised to see PIMT protein localize to the membrane, since increased levels of isoAsp residues were reported in erythrocyte membrane proteins and PIMT activity was suggested to play an important role for maintaining the structural integrity (Ingrosso et al., 2000). Moreover, the structural and functional integrity of membranes in seeds is a prerequisite to achieve high vigor and viability. Our result also suggests that the 56-amino acid N terminus has no apparent role in determining the subcellular localizations of CaPIMT2 isoforms. Together, our data and previous reports suggest that PIMT1 and PIMT2 are distributed to different subcellular compartments in order to cooperatively maintain a functional cellular proteome.
The accumulation of isoAsp in seeds due to aging or stressful environments was shown to adversely affect their vigor and viability (Mudgett et al., 1997; Ogé et al., 2008). Such damage needs to be repaired for normal and vigorous germination. Recently, Ogé et al. (2008) showed that PIMT1 overaccumulation can reduce the detrimental isoAsp residues in seed proteins, resulting in increased seed longevity and germination vigor. Here, we demonstrated the participation of PIMT2 along with PIMT1 in enhancing seed vigor and longevity through seed-specific overexpression in Arabidopsis (Fig. 7). Further analysis revealed that CaPIMT2 particularly reduces deleterious isoAsp accumulation in nuclear proteins while CaPIMT1 reduces such isoAsp predominantly in the cytosolic fraction (Fig. 7, I and J). Our results fit well with previous reports where PIMT substrates were shown to localize in various subcellular compartments, including the nucleus and cytosol in Arabidopsis seeds (Chen et al., 2010). These studies further substantiate that PIMT proteins are distributed in various subcellular compartments to repair various target proteins that may be essential for preserving the longevity and high germination ability of seed. Apart from the repair mechanisms, seed possesses a broad range of systems like protection and detoxification to maintain seed vigor and longevity (Clerkx et al., 2004; Rajjou et al., 2008; Rajjou and Debeaujon, 2008). The accumulation of antioxidant components like tocopherols, tocotrienols, flavonoids, and a number of antioxidative enzymes was shown to be important in maintaining seed vigor and viability (Bailly, 2004). The contribution of testa to seed longevity was also shown by various studies (Debeaujon et al., 2000; Pourcel et al., 2005). Desiccation tolerance and seed longevity in orthodox seeds were also shown to be associated with the presence and abundance of specific proteins like late embryogenesis abundant proteins or heat shock proteins (Blackman et al., 1995; Wehmeyer and Vierling, 2000). The ability of seed-specific overexpression of such protective proteins (heat shock factor/heat shock protein) to improve seed longevity has also been reported (Prieto-Dapena et al., 2006). Importantly, heat shock protein and late embryogenesis abundant proteins have been identified as AtPIMT1 substrates in Arabidopsis seed. These findings raise the intriguing possibility that PIMT repairs such antioxidative and protective proteins, which are involved in detoxification and protection mechanisms in seed and thus contribute to seed vigor and longevity. The identification of more PIMT substrates would authenticate such an assumption and also address the underlying mechanisms of PIMT-mediated improvement of seed vigor and longevity. Overall, seed vigor and longevity were shown to depend on a wide range of physical, chemical, molecular, and genetic factors, yet PIMT-mediated protein repair contributes substantially to improve seed vigor and longevity. Furthermore, our work provides evidence of the participation of both PIMT1 and PIMT2 in PIMT-mediated improvement of seed vigor and longevity.
CONCLUSION
In conclusion, our data strongly suggest that CaPIMT1 and CaPIMT2 are two divergent genes that differentially express and whose product localizations in chickpea combine to play discrete though overlapping roles, where CaPIMT2 likely evolved through gene duplication followed by subfunctionalization to specialize in repairing nucleus-localized abnormal proteins. We also identified a novel splice variant (CaPIMT2′) that encodes an extremely low-active PIMT enzyme. Finally, our research findings of a particular enhancement of seed vigor and longevity upon seed-specific overexpression of PIMT can be of great agricultural interest and might be exploited for long-term seed storage and germplasm conservation.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
Chickpea (Cicer arietinum ‘BGD72’) seeds were germinated and seedlings were grown with or without stresses according to the method described by Kaur et al. (2008).
For hormone treatment, 6-d-old seedlings were treated with 100 µm ABA, indole-3-acetic acid, salicylic acid, or GA (Sigma) for 5 h as described by Syam Prakash and Jayabaskaran (2006).
Arabidopsis (Arabidopsis thaliana) Columbia ecotype was used for Arabidopsis transformation and was grown in a plant growth facility center (Conviron) maintained at 22°C ± 2°C with a 16/8-h light (200 μmol m−2 s−1)/dark cycle for general growth and seed harvesting unless otherwise mentioned.
Arabidopsis Transformation and Transgenic Analysis
To generate PIMT-overexpressing plants, full-length CaPIMT1 and CaPIMT2 cDNAs were cloned into pCAMBIA 2301 plant expression vector under the control of the napin promoter for seed-specific expression. For subcellular localization studies, cDNAs were fused with GFP and cloned into pCAMBIA 2301 under the control of the 35S promoter.
Transgenic plants were obtained by Agrobacterium tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998). Transformed plants were selected by kanamycin resistance on Murashige and Skoog agar plates.
Preparation of Protein Extracts: Total, Cytosolic, and Nuclear Proteins
Total crude proteins were extracted from seeds or other samples following the method described by Thapar et al. (2001) with minor modification. The nuclear and cytosolic proteins were extracted using the Qproteome nuclear protein kit (Qiagen). Dry or imbibed seeds (chickpea or Arabidopsis) were powdered using liquid nitrogen, and then powdered seed materials (approximately 20 mg) were used for cytosolic and nuclear protein extraction following the protocol supplied with the kit (Qiagen). Protein concentrations were determined using a protein assay kit (Bio-Rad) following the manufacturer’s protocol.
PIMT Assay
The PIMT assay was performed based on the vapor diffusion method as described by Mudgett and Clarke (1993) with minor modifications. The reaction involves the transfer of a radiolabeled methyl group from [3H]AdoMet to an isoAsp-containing methyl-accepting peptide [Val-Tyr-Pro(l-isoAsp)-His-Ala] (Peptron). The 30-μL reaction was allowed to continue at 37°C for 1 h and then stopped by quenching with 30 μL of freshly prepared 0.2 n NaOH and 1% SDS, which results in hydrolysis of the methyl ester to methanol. Radioactivity was measured using a liquid scintillation counter. The procedure was described previously in detail (Verma et al., 2010).
Antibody Preparation and Western-Blot Analysis
Two antigenic peptides corresponding to PIMT protein and CaPIMT2-specific protein were custom synthesized. For the general PIMT peptide, the conserved preregion I was selected (PIGYNATISAPHMHATC), while for CaPIMT2-specific peptide, the unique N-terminal region (PFFQNPNFLTGNTc) was selected.
Peptide was linked to Keyhole Limpet Hemocyanin and used to immunize white rabbits. Serum was collected, affinity purified (IMGENEX, India), and stored frozen in aliquots until use. Tubulin and histone H3 antibodies were purchased from Sigma.
Protein extracts (50 μg) for each sample were separated by 12% SDS-PAGE and then blotted onto nitrocellulose membrane. The blot was probed with the appropriate primary antibody followed by horseradish peroxidase-conjugated secondary antibody (GE Healthcare). As a loading control, tubulin was detected using an anti-tubulin antibody (Sigma) following the same procedure. Bands were detected according to the corresponding manufacturer’s instructions.
Isolation and Molecular Cloning of the PIMT2 Gene and cDNA from Chickpea
Total RNA was isolated, and subsequently, cDNA was made as described previously (Verma et al., 2010). Degenerate primers were used to amplify partial cDNA sequences, and subsequently, 3′ and 5′ RACE were performed on the basis of the partial cDNA sequences using a RACE kit (Invitrogen). Furthermore, full-length cDNA for CaPIMT2s was isolated and cloned into the pJET1.2/blunt cloning vector (Fermentas) and subsequently sequenced. Primers used in this study are listed in Supplemental Table S2.
The genomic sequence of CaPIMT2 was amplified through PCR using total genomic DNA and specific primers. The PCR product was cloned into pJET1.2/blunt cloning vector (Fermentas) and subsequently sequenced.
Primer Extension Assay
The primer extension assay was conducted using a kit (Primer Extension System AMV reverse transcriptase; Promega) following the kit protocol. The 21-nucleotide sequence after the splice site of the second intron was used as a primer to end label ([γ-32P]ATP, 3,000 Ci mmol−1; Perkin-Elmer). Total RNA (10 μg) from leaf, seedling, and seed was used for this reaction and hybridized with end-labeled primer at 60°C for 2 h. Primer extension products were analyzed on a 16- × 18-cm denaturing polyacrylamide gel containing 10% acrylamide (19:1 acrylamide:bis), 7 m urea, and 1× Tris-borate/EDTA buffer. ϕX174DNA/HinfI dephosphorylated marker was prepared as described in the kit manual. The gel was wrapped in plastic wrap and exposed to x-ray film overnight at −70°C.
Phylogenetic Analysis
PIMT protein sequences from various organisms of different taxonomic groups were derived from BLAST searches at the National Center for Biotechnology Information and genome databases (listed in Supplemental Table S3). Multiple sequence alignment for PIMT proteins was generated using ClustalX version 2.0.10 (Larkin et al., 2007) incorporating local pairwise alignment. The neighbor-joining method was conducted for construction of a phylogenetic tree with 1,000 replicates defined as the bootstrap values, and the final tree was created in FigTree version 1.3.1 (available at http://tree.bio.ed.ac.uk/software/figtree/).
Isolation of CaPIMT1 and CaPIMT2 Promoter Sequences
Promoter sequences were isolated by genome walking using the Genome Walker kit (Clontech) following the manufacturer’s protocol. Genome-walking PCR was performed on a genomic DNA library using gene-specific primers and the adaptor primers provided by the kit. Amplified fragments were then cloned into pJET1.2/blunt cloning vector and sequenced.
Quantitative Real-Time PCR
Total RNA from various samples of chickpea or Arabidopsis was isolated as described earlier (Kaur et al., 2008). The concentration of RNA was determined at 260 nm on a NanoDrop1000 spectrophotometer (Thermo Scientific).
Two micrograms of RNA was treated with DNaseI (Ambion) and further used for cDNA preparation using the ABI cDNA preparation kit following the manufacturer’s instruction (Applied Biosystems). A 1:20 dilution of the cDNA was used for quantitative real-time PCR in a 20-µL reaction with 10 pm suitable primers as described by Kaur et al. (2008). The expression of 18S or β-actin was used as an endogenous control. Primers used in this study are listed in Supplemental Table S2.
Bacterial Overexpression and Purification of Recombinant CaPIMT2s
The coding regions of CaPIMT2, CaPIMT2′, and ΔCaPIMT2 were subcloned into pET-23d vector (Novagen) and used for expression in the Escherichia coli host strain BL-21 (DE3) pLysS. Transformed cells were grown in Luria-Bertani medium at 37°C up to A600 of 0.5 and induced by 0.5 mm isopropylthio-β-galactoside for 6 h at 25°C. The particulate fraction of expressed protein was solubilized in 8 m urea, and subsequently, urea-solubilized protein was dialyzed as described by Majee et al. (2004). Solubilized and dialyzed proteins were purified to near homogeneity using nickel-charged affinity columns (GE Healthcare) following the manufacturer’s protocol.
Seed Germination Assays
Wild-type and all transgenic lines were grown in identical controlled conditions as described above. Seeds (from mature brown siliques) were harvested on the same day from all plants and then stored in the dark under dry conditions at room temperature (24°C ± 2°C) for at least 8 weeks before being used for the germination assay. However, to evaluate dormancy, freshly harvested and 1- to 4-week-old seeds were used (Léon-Kloosterziel et al., 1996).
For the seed germination assay, triplicates of 100 seeds were analyzed under controlled culture room conditions (22°C ± 2°C with a 16/8-h light/dark cycle). Seeds were surface sterilized with 20% (v/v) commercial bleach with 0.1% (v/v) Triton X-100 and rinsed with sterilized distilled water at least five times. Sterilized seeds were plated in one-half-strength Murashige and Skoog agar medium (Sigma) or aqueous agar medium (0.6% [w/v] agar, 10 mm MES [Sigma], pH 5.7) and kept in the dark at 4°C for 2 d or as mentioned in the figure legends before being allowed to germinate in controlled culture room conditions. To evaluate germination under stress conditions, seeds were plated with sodium chloride (200 mm), polyethylene glycol (19.5 g per 100 mL), and paraquat (0.5 μm). Seed germination was also evaluated without sterilization on two layers of Whatman No. 1 filter paper moistened with distilled, deionized water particularly to asses the dormancy and speed of germination. Seed germination was determined to be completed when the radicle protruded beyond the testa and was assessed until 5 or 7 d after imbibition as mentioned in the figure legends. In all experiments, at least three independent transformant lines were evaluated independently. For each line, seeds were collected from more than three plants, and analysis was done using 100 seeds in three replicates.
Accelerated Aging and CDT
Seeds used in these experiments were harvested on the same day from all plants grown in identical conditions as described above. Experiments were carried out using 100 8-week-old seeds in three replicates, and at least three independent transformant lines were evaluated independently. For each line, seeds were collected from more than three plants. Accelerated aging was performed according to the method described by Delouche and Baskin (1973) with minor modification. Dry seeds were rehydrated to increase the moisture content before their exposure to high temperature and humidity. Moisture content was increased to 22% ± 2% by imbibition in water for 1 h, and then seeds were air dried before incubating at 45°C in 100% relative humidity for 0 to 5 d. For CDT, moisture content was also increased to 22% ± 2% by imbibition, and then seeds were dried and placed in sealed bags before incubating at 45°C for 0 to 7 d (Prieto-Dapena et al., 2006). Treated seeds were further used to evaluate the viability, germination percentage, PIMT activity, or isoAsp accumulation.
TZ Assay
The TZ assay was performed as described by Wharton (1955). Seeds were soaked in a 1% solution of 2,3,5-triphenyl tetrazolium chloride (Sigma) and incubated in darkness at 30°C for 48 h (Wharton, 1955). TZ precipitates to red-colored 2,3,5-triphenyl formazan by the activity of dehydrogenases present in the live cells. As a result, viable seeds containing live cells stain red and nonviable or dead seeds remain unstained. Thus, the viability of seeds can be interpreted by the staining pattern and the color intensity. Heat-killed seeds (incubated at 100°C for 1 h) were used as a negative control.
Estimation of LD50 and Statistical Analysis
Germination data of the CDT were used for probit analysis on a logarithmic time scale (days) to estimate LD50 (Finney and Stevens, 1948; Finney, 1952; Prieto-Dapena et al., 2006). The germination percentage was initially converted to probit (Finney, 1952) and then plotted against the logarithmic time scale (days) and fitted to a line of regression (Microcal Origin). LD50 was determined by taking the inverse of the log of the concentration associated with probit 5. In each case, LD50 was independently assessed in three independent lines, and thereafter, LD50 values were averaged.
Student’s t test was applied to the data. Differences of germination percentage and other values between the transgenic and control plants were considered significant at P < 0.05.
Quantification of isoAsp Content
The protein samples were digested by a denaturing digestion method. Fifty micrograms of vacuum-dried protein sample was resuspended in 20 µL of denaturing digestion buffer (50 mm Tris-Cl, pH 8.0, 8 m urea, and 5 mm dithiothreitol). Next, 60 µL of 50 mm Tris-Cl, pH 8.0, was added to dilute urea to 2 m, and then 10 µL of 0.025 µg µL−1 trypsin was added. The mixture was incubated at 37°C for 2 h before 0.01 volume of protease inhibitors was added to stop proteolysis. The digested samples were kept at −20°C or used directly for isoAsp detection. A blank reaction was performed in parallel without lyophilized protein sample. Subsequently, isoAsp content was quantified using the Isoquant Iso-Asp detection kit (Promega) and radiolabeled [3H]AdoMet (370 gigabecquerel mmol−1; Perkin-Elmer) according to the kit’s protocol with minor modifications.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CaPIMT1, GQ421817; CaPIMT2 coding sequence, JQ690076; CaPIMT2′, JQ690077; CaPIMT2 gene, JQ690075.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primer extension analysis of CaPIMT2.
Supplemental Figure S2. Nucleotide sequence of the CaPIMT2 gene and its deduced amino acid sequence.
Supplemental Figure S3. Bacterial expression of CaPIMT2, ΔCaPIMT2, and CaPIMT2′.
Supplemental Figure S4. Specificity of the PIMT antibodies used in western-blot analysis of CaPIMT1 and CaPIMT2 proteins.
Supplemental Figure S5. Specificity of the primers used in semiquantitative PCR analysis of CaPIMT2 and CaPIMT2′ transcripts.
Supplemental Figure S6. Sequences of the putative promoter regions of CaPIMT1 (A) and CaPIMT2 (B) genes.
Supplemental Figure S7. PIMT activity in seed of GFP, CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP transformed lines.
Supplemental Figure S8. Subcellular localization of CaPIMT2.
Supplemental Figure S9. Seed survival curves (based on a seed’s ability to germinate) of CaPIMT1 and CaPIMT2 transformed Arabidopsis seed.
Supplemental Figure S10. Viability and germination percentage of seeds in wild-type GFP, CaPIMT1:GFP, CaPIMT2:GFP, and CaPIMT2′:GFP transformed seeds before and after CDT.
Supplemental Figure S11. Viability and germination percentage of seeds in the wild type and MIPS transformed lines (as a negative control) before and after CDT.
Supplemental Figure S12. Western-blot analysis of nuclear and cytosolic fractions of seed protein with histone H3 antibody.
Supplemental Table S1. Summary of putative cis-regulatory elements in CaPIMT1 and CaPIMT2 promoter regions.
Supplemental Table S2. List of primers used in this study.
Supplemental Table S3. List of PIMT sequences used in the phylogenetic analysis.
Acknowledgments
We thank Dr. Susmita Maitra Majee and Brinderjit Singh Buttar (University of Delhi, South Campus) for helping us to make the phylogenetic tree of PIMT proteins.
Glossary
- ABA
abscisic acid
- isoAsp
isoaspartyl
- AdoMet
S-adenosyl-methionine
- CDT
control deterioration test
- LD50
number of days of control deterioration test required for a decline to 50% germination
- TZ
tetrazolium
- cDNA
complementary DNA
References
- Ahmad F, Gaur PM, Croser J (2005) Chickpea (Cicer arietinum L.). In R Singh, P Jauhar, eds, Genetic Resources, Chromosome Engineering and Crop Improvement: Grain Legumes, Vol 1. CRC Press, Boca Raton, FL, pp 187–217 [Google Scholar]
- Aswad DW, Paranandi MV, Schurter BT. (2000) Isoaspartate in peptides and proteins: formation, significance, and analysis. J Pharm Biomed Anal 21: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Bailly C. (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14: 93–107 [Google Scholar]
- Berridge MV, Tan AS, McCoy KD, Wang R. (1996) The biochemical and cellular basis of cell proliferation assays that use tetrazolium salts. Biochemica 4: 725–732 [Google Scholar]
- Blackman SA, Obendorf RL, Leopold AC. (1995) Desiccation tolerance in developing soybean seeds: the role of stress proteins. Physiol Plant 93: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Nayak N, Majee SM, Lowenson J, Schäfermeyer KR, Eliopoulos AC, Lloyd TD, Dinkins R, Perry SE, Forsthoefel NR, et al. (2010) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J Biol Chem 285: 37281–37292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev 2: 263–285 [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, Blankestijn-DeVries H, Ruys GJ, Groot SPC, Koornneef M. (2004) Genetic differences in seed longevity of various Arabidopsis mutants. Physiol Plant 121: 448–461 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delouche YC, Baskin C. (1973) Accelerated ageing techniques for predicting the relative storability of seeds lots. Seeds Sci Technol 1: 427–452 [Google Scholar]
- Dinkins RD, Majee SM, Nayak NR, Martin D, Xu Q, Belcastro MP, Houtz RL, Beach CM, Downie AB. (2008) Changing transcriptional initiation sites and alternative 5′- and 3′-splice site selection of the first intron deploys Arabidopsis protein isoaspartyl methyltransferase2 variants to different subcellular compartments. Plant J 55: 1–13 [DOI] [PubMed] [Google Scholar]
- Finney DJ (1952) Probit Analysis. Cambridge University Press, Cambridge, UK [Google Scholar]
- Finney DJ, Stevens WL. (1948) A table for the calculation of working probits and weights in probit analysis. Biometrika 35: 191–201 [PubMed] [Google Scholar]
- Ingrosso D, D’Angelo S, di Carlo E, Perna AF, Zappia V, Galletti P. (2000) Increased methyl esterification of altered aspartyl residues in erythrocyte membrane proteins in response to oxidative stress. Eur J Biochem 267: 4397–4405 [DOI] [PubMed] [Google Scholar]
- Johnson BA, Langmack EL, Aswad DW. (1987) Partial repair of deamidation-damaged calmodulin by protein carboxyl methyltransferase. J Biol Chem 262: 12283–12287 [PubMed] [Google Scholar]
- Kagan RM, McFadden HJ, McFadden PN, O’Connor C, Clarke S. (1997) Molecular phylogenetics of a protein repair methyltransferase. Comp Biochem Physiol B Biochem Mol Biol 117: 379–385 [DOI] [PubMed] [Google Scholar]
- Kapoor N, Arya A, Siddiqui MA, Amir A, Kumar H. (2010) Seed deterioration in chickpea (Cicer arietinum L.) under accelerated aging. Asian J Plant Sci 9: 158–162 [Google Scholar]
- Kaur H, Shukla RK, Yadav G, Chattopadhyay D, Majee M. (2008) Two divergent genes encoding L-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ 31: 1701–1716 [DOI] [PubMed] [Google Scholar]
- Kim E, Lowenson JD, MacLaren DC, Clarke S, Young SG. (1997) Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc Natl Acad Sci USA 94: 6132–6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J, Parent J, Davies GF, Dinsmore M, Attah-Poku S, Napper S. (2003) Overexpression of L-isoaspartate O-methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. J Biol Chem 278: 50880–50886 [DOI] [PubMed] [Google Scholar]
- Lanteri S, Nada E, Balleti P, Quagliotti L, Bino RJ. (1996) Effects of controlled deterioration and osmoconditioning on germination and nuclear replication in seeds of pepper (Capsicum annuum L.). Ann Bot (Lond) 77: 591–597 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, van de Bunt GA, Zeevaart JA, Koornneef M. (1996) Arabidopsis mutants with a reduced seed dormancy. Plant Physiol 110: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenson JD, Clarke S. (1992) Recognition of D aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D aspartyl protein methyl transferase: implication for the repair hypothesis. J Biol Chem 276: 5985–5995 [PubMed] [Google Scholar]
- Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC, Das KP, Majumder AL. (2004) A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. J Biol Chem 279: 28539–28552 [DOI] [PubMed] [Google Scholar]
- McFadden PN, Clarke S. (1987) Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins. Proc Natl Acad Sci USA 84: 2595–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. (1993) Characterization of plant L-isoaspartyl methyltransferases that may be involved in seed survival: purification, cloning, and sequence analysis of the wheat germ enzyme. Biochemistry 32: 11100–11111 [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. (1994) Hormonal and environmental responsiveness of a developmentally regulated protein repair L-isoaspartyl methyltransferase in wheat. J Biol Chem 269: 25605–25612 [PubMed] [Google Scholar]
- Mudgett MB, Lowenson JD, Clarke S. (1997) Protein repair L-isoaspartyl methyltransferase in plants: phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiol 115: 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM. (1987) Regulation and subcellular distribution of a protein methyltransferase and its damaged aspartyl substrate sites in developing Xenopus oocytes. J Biol Chem 262: 10398–10403 [PubMed] [Google Scholar]
- O’Connor CM (2006) Protein L-isoaspartyl, D-aspartyl O-methyl transferase: catalysts of protein repair. In SG Clarke, F Tamanoi, eds, The Enzymes, Vol XXIV. Elsevier, Amsterdam, pp 385–433 [Google Scholar]
- O’Connor CM, Germain BJ. (1987) Kinetic and electrophoretic analysis of transmethylation reactions in intact Xenopus laevis oocytes. J Biol Chem 262: 10404–10411 [PubMed] [Google Scholar]
- Ogé L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. (2008) Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20: 3022–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Borchardt RT. (1990) Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm Res 7: 703–711 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J. (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142: 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Debeaujon I. (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. C R Biol 331: 796–805 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghazi M, Job C, Job D. (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 148: 620–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Miller J, Mudgett MB, Schopf JW, Clarke S, Berger R. (1995) Exceptional seed longevity and robust growth: ancient sacred lotus from China. Am J Bot 82: 1367–1380 [Google Scholar]
- Smith CD, Carson M, Friedman AM, Skinner MM, Delucas L, Chantalat L, Weise L, Shirasawa T, Chattopadhyay D. (2002) Crystal structure of human L-isoaspartyl-O-methyl-transferase with S-adenosyl homocysteine at 1.6-A resolution and modeling of an isoaspartyl-containing peptide at the active site. Protein Sci 11: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syam Prakash SR, Jayabaskaran C. (2006) Expression and localization of calcium-dependent protein kinase isoforms in chickpea. J Plant Physiol 163: 1135–1149 [DOI] [PubMed] [Google Scholar]
- TeKrony DM (1995) Accelerated aging. In HA van de Venter, ed, Seed Vigour Testing Seminar. International Seed Testing Association, Zurich, pp 53–73 [Google Scholar]
- Thapar N, Kim AK, Clarke S. (2001) Distinct patterns of expression but similar biochemical properties of protein l-isoaspartyl methyltransferase in higher plants. Plant Physiol 125: 1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Singh A, Kaur H, Majee M. (2010) Protein L-isoaspartyl methyltransferase1 (CaPIMT1) from chickpea mitigates oxidative stress-induced growth inhibition of Escherichia coli. Planta 231: 329–336 [DOI] [PubMed] [Google Scholar]
- Villa ST, Xu Q, Downie AB, Clarke SG. (2006) Arabidopsis protein repair l-isoaspartyl methyltransferase: predominant activities at lethal temperatures. Physiol Plant 128: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick JE, Cai H, Clarke S. (1998) The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol 180: 2623–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E. (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton MJ. (1955) The use of tetrazolium test for determining the viability of seeds of the genus Brassica. Proc Int Seed Test Assoc USA 20: 81–88 [Google Scholar]
- Xu Q, Belcastro MP, Villa ST, Dinkins RD, Clarke SG, Downie AB. (2004) A second protein l-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol 136: 2652–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Carter WG, Doyle HA, Mamula MJ, Aswad DW. (2001) Structural integrity of histone H2B in vivo requires the activity of protein L-isoaspartate O-methyltransferase, a putative protein repair enzyme. J Biol Chem 276: 37161–37165 [DOI] [PubMed] [Google Scholar]