A molecular switch integrates development and pathogen response signaling in plants.
Abstract
How plants coordinate developmental processes and environmental stress responses is a pressing question. Here, we show that Arabidopsis (Arabidopsis thaliana) Rho of Plants6 (AtROP6) integrates developmental and pathogen response signaling. AtROP6 expression is induced by auxin and detected in the root meristem, lateral root initials, and leaf hydathodes. Plants expressing a dominant negative AtROP6 (rop6DN) under the regulation of its endogenous promoter are small and have multiple inflorescence stems, twisted leaves, deformed leaf epidermis pavement cells, and differentially organized cytoskeleton. Microarray analyses of rop6DN plants revealed that major changes in gene expression are associated with constitutive salicylic acid (SA)-mediated defense responses. In agreement, their free and total SA levels resembled those of wild-type plants inoculated with a virulent powdery mildew pathogen. The constitutive SA-associated response in rop6DN was suppressed in mutant backgrounds defective in SA signaling (nonexpresser of PR genes1 [npr1]) or biosynthesis (salicylic acid induction deficient2 [sid2]). However, the rop6DN npr1 and rop6DN sid2 double mutants retained the aberrant developmental phenotypes, indicating that the constitutive SA response can be uncoupled from ROP function(s) in development. rop6DN plants exhibited enhanced preinvasive defense responses to a host-adapted virulent powdery mildew fungus but were impaired in preinvasive defenses upon inoculation with a nonadapted powdery mildew. The host-adapted powdery mildew had a reduced reproductive fitness on rop6DN plants, which was retained in mutant backgrounds defective in SA biosynthesis or signaling. Our findings indicate that both the morphological aberrations and altered sensitivity to powdery mildews of rop6DN plants result from perturbations that are independent from the SA-associated response. These perturbations uncouple SA-dependent defense signaling from disease resistance execution.
Rho of Plants (ROPs), also known as RACs (for clarity, the ROP nomenclature will be used throughout this article), comprise a plant-specific group of Rho family small G proteins. Like other members of the Ras superfamily of small G proteins, ROPs function as molecular switches, existing in a GTP-bound “on” state and a GDP-bound “off” state. In the GTP-bound state, ROPs interact with specific effectors that transduce downstream signaling or function as scaffolds for interaction with additional effector molecules (Berken and Wittinghofer, 2008). Conserved point mutations in the G1 (P loop) Gly-15 or the G3 (switch II) Gln-64, which abolish GTP hydrolysis, or the G1 Thr-20 or G4 Asp-121 that compromise GDP/GTP exchange, can form either constitutively active or dominant negative mutants, respectively (Feig, 1999; Berken et al., 2005; Berken and Wittinghofer, 2008; Sorek et al., 2010). Primarily based on studies with neomorphic mutants, ROPs have been implicated in the regulation of cytoskeleton organization and dynamics, vesicle trafficking, auxin transport and response, abscisic acid (ABA) response, and response to pathogens (Nibau et al., 2006; Yalovsky et al., 2008; Yang, 2008; Lorek et al., 2010; Wu et al., 2011; and refs. therein).
In Arabidopsis (Arabidopsis thaliana), there are 11 ROP proteins (Winge et al., 1997). Assigning specific functions to individual members of this family is difficult, however, because ROPs are functionally redundant. A ROP10 loss-of-function mutant was reported to be ABA hypersensitive (Zheng et al., 2002), displaying enhanced expression of tens of genes in response to ABA treatments (Xin et al., 2005). However, in the absence of exogenous ABA, gene expression in the rop10 mutant was similar to that in wild-type plants (Xin et al., 2005). Loss of leaf epidermis pavement cell polarity was reported for rop4 rop2-RNAi (for RNA interference) double mutant plants (Fu et al., 2005). Mild changes in pavement and hypocotyl cell structure and microtubule (MT) organization were reported for a rop6 loss-of-function mutant (Fu et al., 2009).
The involvement of ROPs in auxin-regulated development has been addressed in several studies (Wu et al., 2011). Ectopic expression of a dominant negative ROP2 (rop2DN) mutant under regulation of the 35S promoter resulted in a loss of apical dominance and a reduction in the number of lateral roots. In contrast, ectopic expression of constitutively active ROP2 (rop2CA) caused an increase in the number of lateral roots and an enhanced decrease in primary root length in response to auxin. Consistent with these findings, the expression of a constitutively active NtRAC1 in tobacco (Nicotiana tabacum) protoplasts induced the expression of auxin-regulated genes in the absence of auxin and promoted the formation of protein nuclear bodies containing components of the proteasome and COP9 signalosome (Tao et al., 2002, 2005; Wu et al., 2011). The ROP effector ICR1 (for interactor of constitutively active ROP1) regulates polarized secretion and is required for polar auxin transport (Lavy et al., 2007; Bloch et al., 2008; Hazak et al., 2010; Hazak and Yalovsky, 2010). In the root, local auxin gradients induce the accumulation of ROPs in trichoblasts at the site of future root hair formation (Fischer et al., 2006). Recently, it was shown that interdigitation of leaf epidermis pavement cells depends on Auxin-Binding Protein1 (ABP1)-mediated ROP activation (Xu et al., 2010). Taken together, these data indicate that ROPs are involved in both mediating the auxin response and facilitating directional auxin transport. It is still unclear, however, which ROPs function in these processes.
ROP function was linked to plant defense responses in several studies. In rice (Oryza sativa), OsRAC1 is a positive regulator of the hypersensitive response, possibly through interactions with the NADPH oxidase RbohB, Required for Mla12 Resistance, and Heat Shock Protein90 (Ono et al., 2001; Thao et al., 2007; Wong et al., 2007). Interestingly, other members of the rice ROP family, namely RAC4 and RAC5, are negative regulators of resistance to the rice blast pathogen Magnaporthe grisea (Chen et al., 2010). Similar to rice, when expressed in tobacco, dominant negative OsRAC1 suppressed the hypersensitive response (Moeder et al., 2005). In barley (Hordeum vulgare), several constitutively active ROP/RAC mutants and a MT-associated ROPGAP1 loss-of-function mutant enhanced susceptibility to the powdery mildew Blumeria graminis f. sp. hordei (Bgh). The activated ROP-enhanced susceptibility to Bgh was attributed to disorganization of the actin cytoskeleton and was shown to depend on Mildew Resistance Locus O (MLO; Schultheiss et al., 2002, 2003; Opalski et al., 2005; Hoefle et al., 2011). In barley, three ROP proteins, HvRACB, HvRAC1, and HvRAC3, were linked to both development and pathogen response (Schultheiss et al., 2005; Pathuri et al., 2008; Hoefle et al., 2011).
We have analyzed the function of the Arabidopsis AtROP6 (ROP6) by characterizing its expression pattern and its regulation by auxin and the phenotype of plants that express rop6DN under the regulation of its endogenous promoter. The utilization of the dominant negative mutant overcame functional redundancy, while expression under the regulation of the endogenous promoter enabled the analysis of ROP6 function in a developmental context. Phenotypic and gene expression analyses indicate that ROP6 functions in developmental, salicylic acid (SA)-dependent, and SA-independent defense response pathways.
RESULTS
ROP6 Expression Pattern and Subcellular Localization
Previously, we analyzed lipid modifications, membrane interaction dynamics, and the function of ROP6 in the regulation of cell polarity using the constitutively active rop6CA mutant expressed under the regulation of the cauliflower mosaic virus 35S promoter. Because of the functional redundancy of ROPs, the rop6 mutant phenotype is inseparable from that of wild-type plants. To overcome this difficulty and elucidate the specific function of ROP6, its promoter was isolated (Supplemental Fig. S1) and subcloned into the LhG4/pOp transcription transactivation system (Moore et al., 1998) to drive the expression of GUS, DsRed, GFP-ROP6, GFP-rop6CA, and GFP-rop6DN. To verify the expression pattern and phenotypes, three independent transgenic lines were analyzed for each expression construct.
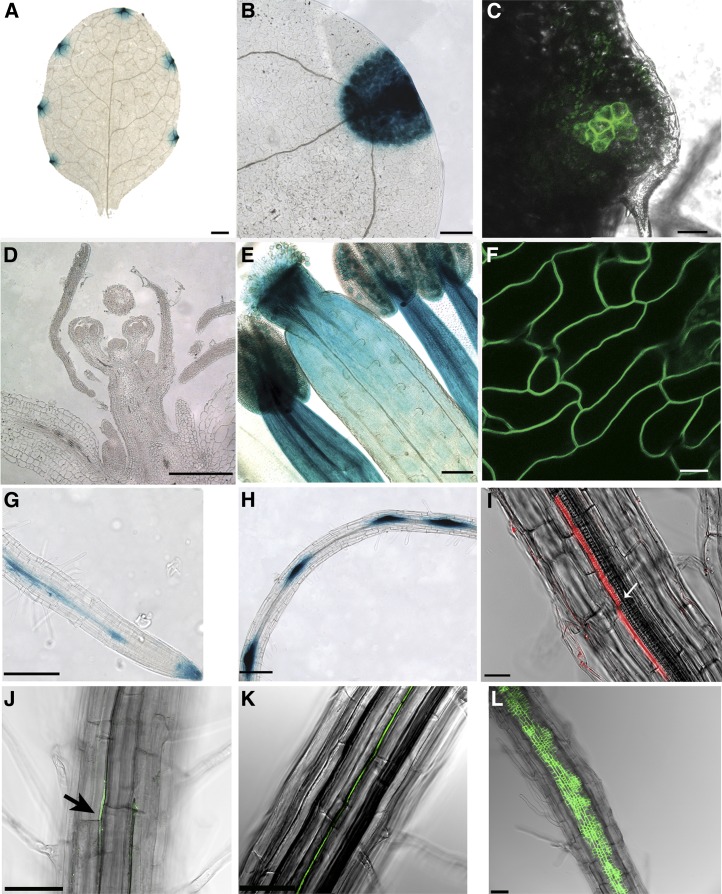
In rosette leaves, ROP6 expression was detected at the tip and in the hydathodes at the leaf margin (Fig. 1, A–C). In cauline leaves, however, expression was not confined to the leaf margins, and leaf epidermis pavement cells expressing a constitutively active GFP-rop6CA (rop6CA) were rectangular with few and shallow lobes (Fig. 1F). No expression could be detected in the inflorescence meristem and in young flowers (Fig. 1D). In mature flowers, strong expression was detected in stamen filaments, pollen grains, and stigma and weak expression was detected in ovules (Fig. 1E). In the primary root, expression was detected in the root tip, was absent from the cell division zone, and increased in the stele of the transition zone. In the root differentiation zone, expression increased in lateral root initials (LRI) and was already detected in lateral root founder cells in the pericycle (Fig. 1, G–I). The expression pattern of ROP6 in the root was further confirmed by whole-mount RNA in situ hybridization. Similar to expression detected with reporter genes fused to the ROP6 promoter, following hybridization with an antisense probe, ROP6 RNA was detected in the stele, LRI, and the tip of the emerging lateral root (Fig. 2).
Figure 1.
Expression patterns of AtROP6 in the shoot and the root. A, B, D, E, G, and H, pROP6>>GUS. C, F, J, K, and L, pROP6>>GFP-rop6CA. I, pROP6>>DsRed. A to C and F, In rosette leaves, ROP6 expression is primarily restricted to hydathodes, whereas in cauline leaves, it is detected throughout the epidermis. D, ROP6 expression could not be detected in the inflorescence meristem and young flowers. E, In mature flowers, expression is detected in stamen filaments and pollen grains, stigma, style, and weakly in ovules. G, In the root, expression is detected in the root cap, in and around the quiescent center; it is absent from the cell division and transition zone, and it reappears in the stele in the differentiation zone. H and I, Expression then drops again and reappears in LRI and founder cells. The arrow denotes a protoxylem file. J, Expression of ROP6 is induced by auxin and can be detected in pericycle cells as early as 3 h after induction. K, After 6 h, expression is spread to more cells in the pericycle. L, Twenty-four hours after induction, expression is spread throughout the pericycle and extensive cell divisions are detected. Bars = 100 μm (A), 50 μm (B, D, E, and G–L), and 20 μm (C and F). For more information, see Supplemental Figure S2.
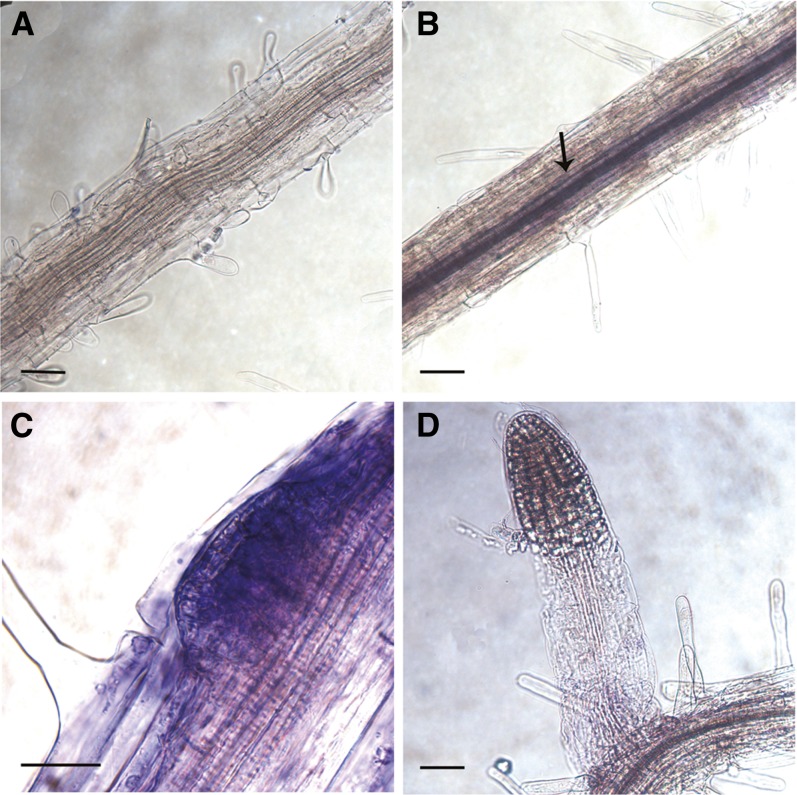
Figure 2.
Expression of ROP6 in the roots determined by whole-mount in situ hybridization. A, Hybridization with a ROP6 gene-specific sense probe, negative control. B to D, An antisense ROP6 gene-specific probe. Expression was detected in the stele (B, arrow, and D), in the LRI (C), and in the meristematic region of the mature lateral root (D). B, Lateral root founder cells prior to the first anticlinal division. C, Lateral root primordia emerging from the primary root. D, Young lateral root. Bars = 20 μm. [See online article for color version of this figure.]
The expression pattern of ROP6 in rosette leaves and primary and lateral roots resembled the expression pattern of the auxin-sensitive synthetic promoter DR5 (Sabatini et al., 1999; Aloni et al., 2003; Benková et al., 2003), suggesting that ROP6 expression might be induced by auxin. Furthermore, the ROP6 promoter contains a TGTCTC auxin response element. To examine whether ROP6 expression is induced by auxin, 5-d-old seedlings were transferred to control or auxin-containing media. The GFP-rop6CA signal was detected in some pericycle cells as early as 3 h following transfer to an auxin-containing medium (10 μm naphthaleneacetic acid [NAA]; Fig. 1J). Six hours after transfer to auxin-containing medium, GFP-rop6CA was detected in many pericycle cells, whereas on control plates, it was detected only in a few cells (Fig. 1K). GFP-rop6CA was visible in many dividing pericycle cells 24 h after transfer to auxin (Fig. 1L). The results of the auxin induction experiments confirm that ROP6 expression is quickly induced by auxin during lateral root development. Furthermore, the identical expression pattern, which was detected when using GUS, DsRed, GFP-ROP6, GFP-rop6CA, or GFP-rop6DN (Figs. 1 and 3), and the fast (3-h) response to auxin treatments make it unlikely that the activation status of the transgenic ROP protein affected the AtROP6 expression pattern and the preliminary response to auxin.
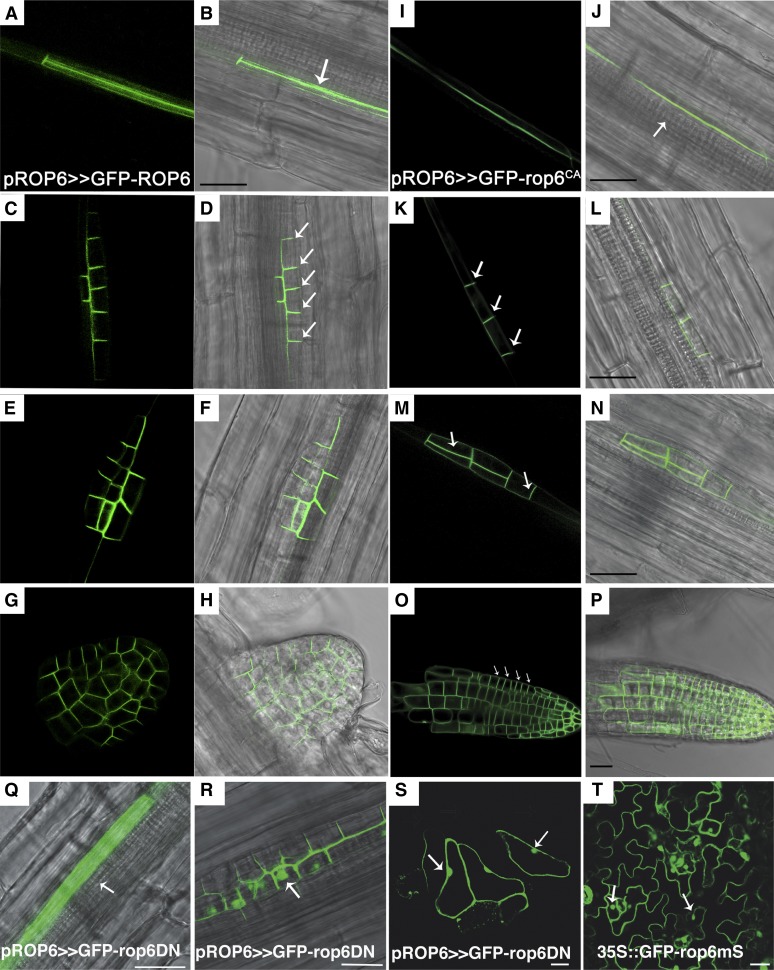
Figure 3.
Subcellular localization of GFP-ROP6, GFP-rop6CA, and GFP-rop6DN during lateral root development. Subcellular localizations of GFP-ROP6 (A–H) and GFP-rop6CA (I–P) were observed in developing lateral roots in pROP6>>GFP-ROP6 and pROP6>>GFP-rop6CA seedlings. A, B, I, and J, Lateral root founder cells. GFP-ROP6 and GFP-rop6CA were localized along the side of the cell adjacent to the xylem pole (J, arrow). C, D, K, and L, Stage I/II LRI. The majorities of the GFP-ROP6 and GFP-rop6CA proteins moved and were localized along anticlinal cell borders (arrows). E, F, M, and N, Stage II/III LRI. The majority of GFP-ROP6 and GFP-rop6CA were localized along anticlinal and periclinal cell borders (arrows) separating the newly formed outer and inner layers. G, H, O, and P, Emerged and mature lateral roots with organized cell files. The majority of the GFP-ROP6 and GFP-rop6CA proteins were concentrated on proximal anticlinal cell borders (arrows). In the meristematic zone, the protein was distributed more or less evenly around the cells. Q to S, GFP-rop6DN was observed in pROP6>>GFP-rop6DN seedlings in lateral root founder cells (Q), stage II/II LRI cells (R), and leaf epidermis pavement cells (S). The recruitment of GFP-rop6DN to the plasma membrane is compromised, and it accumulates in the cytoplasm and nuclei in roots (arrows). T, Nonprenylated GFP-rop6CA mutant accumulates in cytoplasm and nuclei (arrows) but, unlike GFP-rop6DN, does not affect cell polarity. Bars = 20 μm.
To further explore possible function(s) of ROP6 in lateral roots, we examined its subcellular localization in the developing LRI. In lateral root founder cells in the pericycle, either GFP-ROP6 or GFP-rop6CA was localized in the cell at the side that is closer to the xylem pole (Fig. 3, A, B, I, and J). At later stages of lateral root development, GFP-ROP6 and GFP-rop6CA were localized such that they aligned the cell division planes (Fig. 3, C–P) and were absent from the side of the cell pointing toward the lateral root tip (Fig. 3, E–H, M, and N). This polar localization suggests that ROP6 relays a special signal in LRI cells. Similarly, it has been found that during stomata development in maize (Zea mays), the type I ROPs ROP2 and ROP9 enhance the polarization of asymmetric cell divisions (Humphries et al., 2011). Together, these data suggest that ROP-dependent polarization could be critical for asymmetric cell divisions. In contrast to GFP-ROP6 and GFP-rop6CA, the dominant negative GFP-rop6DN accumulated in the cytoplasm and nuclei in root and leaves (Fig. 3, Q–S), resembling the subcellular distribution of the nonprenylated GFP-rop6C195S mutant (Fig. 3T). Hence, nucleotide exchange is required for the recruitment of ROPs to the plasma membrane.
The Phenotype of rop6DN Plants
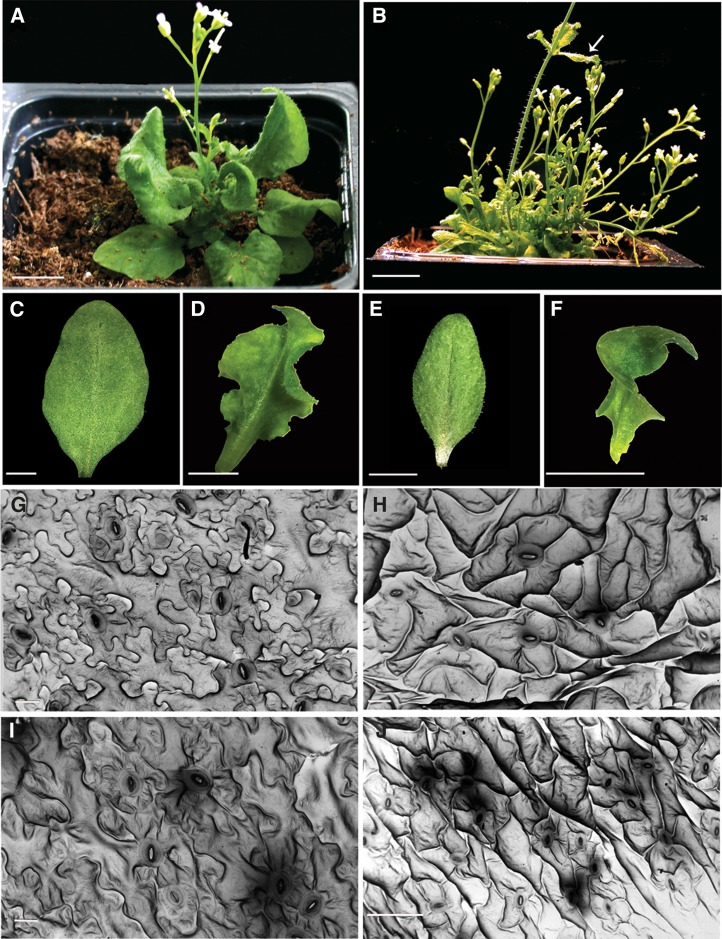
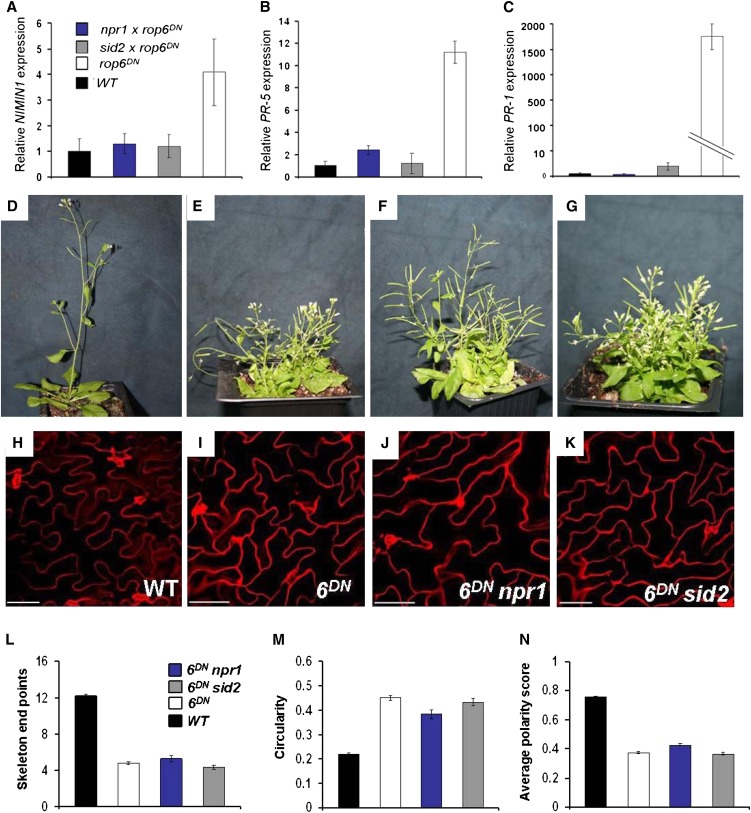
Next, we examined the phenotype of the pROP6>>GFP-rop6DN (rop6DN) plants (Fig. 4). The rop6DN plants were small and developed multiple inflorescence stems (Figs. 4, A and B, and 7, D and E). The rosette leaves of the rop6DN plants were curled, and their cauline leaves were twisted (Fig. 4, A–F). The stronger effect of rop6DN on the structure of cauline leaves compared with rosette leaves was closely correlated with its expression levels in the two leaf types (Fig. 1, A and F). Analysis of the cell structures in rosette (Figs. 4, G and H, and 7, H and I) and cauline leaves (Fig. 4, I and J) revealed that pavement cells in the rop6DN leaves had fewer lobes and indentations. Quantitative analysis of cell structure, using a value designated the average polarity score, which is based on the ImageJ tools “circularity” and “Skeleton end points” (Sorek et al., 2010, 2011), shows that the rop6DN cells are significantly less polar compared with wild-type cells (P ≤ 0.001, ANOVA, Tukey-Kramer test; Fig. 7, L–N).
Figure 4.
Phenotype of pROP6>>GFP-rop6DN plants. A and B, A rop6DN plant with folded rosette leaves. Note the loss of apical dominance in the rop6DN plant, characterized by the development of many short adventitious inflorescence stems. Folded cauline leaves are highlighted (arrow). C and D, Rosette leaves of wild-type (C) and rop6DN (D) plants. E and F, Cauline leaves of wild-type (E) and rop6DN (F) plants. G to J, Scanning electron microscopy images of epidermal cells from rosette leaves (G and H) and cauline leaves (I and J) from wild-type (G and I) and rop6DN (H and J) plants. Note the loss of lobes in the epidermal cells of rop6DN plants. Bars = 1 cm (A and B), 0.5 cm (C–F), and 10 μm (G–J). [See online article for color version of this figure.]
Figure 7.
PR gene expression and phenotypes of rop6DN npr1 and rop6DN sid2 double mutants. A to C, Expression analysis of NIMIN1 (A), PR-5 (B), and PR-1 (C) by qPCR in wild-type (WT), rop6DN, rop6DN npr1, and rop6DN sid2 plants. For each gene, its relative expression level in the wild type was taken as 1. Note the suppression of expression of each of the three genes to wild-type levels in the rop6DN npr1 and rop6DN sid2 double mutants. D to G, Plants of the wild type (D), rop6DN (E), rop6DN npr1 (F), and rop6DN sid2 (G). H to K, Abaxial epidermis pavement cells. Bars = 20 μm. L to N, Quantitative parameters of epidermal cell structure. L, Number of lobes. M, Circularity. N, Average polarity score. Note that the rop6DN single mutant and the rop6DN npr1 and rop6DN sid2 double mutants have the same plant and cell structures. [See online article for color version of this figure.]
Organization of the Cytoskeleton in Wild-Type and rop6DN Plants
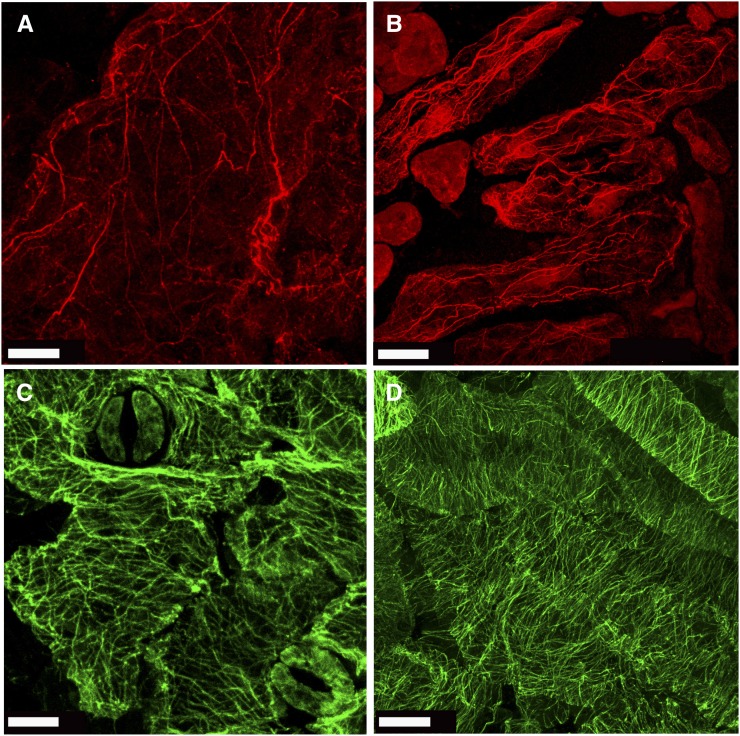
ROPs have been implicated in cytoskeleton organization and dynamics, suggesting that the phenotype of rop6DN plants could be associated with impaired actin and MT functions. Therefore, we examined the organization of MT and actin in wild-type and rop6DN plants by immunostaining the cytoskeleton in rosette leaves (Fig. 5). In wild-type pavement cells, F-actin was organized in thin filaments and a few thicker bundles that were oriented at different angles. In addition, diffuse fluorescence likely corresponding to fine F-actin or G-actin could be seen (Fig. 5A; Supplemental Fig. S3, A–C). In comparison, in the rop6DN pavement cells, F-actin was primarily organized in thick bundles, many of which were arranged parallel to the long cell axis, and little diffuse fluorescence could be seen (Fig. 5B; Supplemental Fig. S3, E–H). The MTs were arranged at different angles in wild-type pavement cells (Fig. 5C; Supplemental Fig. S3, I–L), while in rop6DN pavement cells, they were arranged mainly in parallel arrays, perpendicular to the long axis of cells (Fig. 5D; Supplemental Fig. S3, M–P). Taken together, the immunostains demonstrate that both actin and MT were differentially organized in the rop6DN plants.
Figure 5.
Immunostaining showing actin and MT organization in Col-0 wild-type and GFP-rop6DN plants. Actin (red) and MTs (green) are shown in wild-type Col-0 (A and C) and GFP-rop6DN (B and D) leaf epidermal cells. Bars = 10 μm. [See online article for color version of this figure.]
rop6DN-Associated Changes in Gene Expression
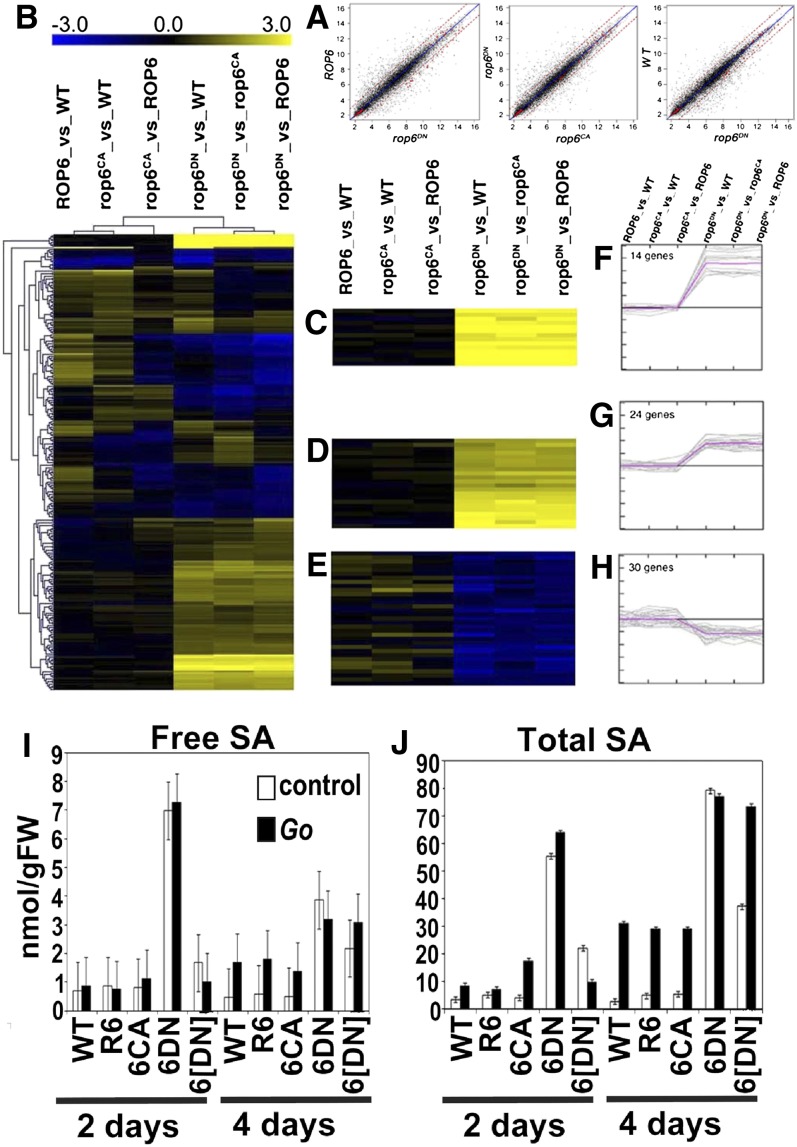
The obvious developmental perturbations of rop6DN plants prompted us to examine whether the phenotype of the rop6DN plants is associated with changes in gene expression. Gene expression in rop6DN, wild-type, pROP6>>GFP-ROP6 (ROP6), and pROP6>>GFP-rop6CA (rop6CA) plants (Supplemental Fig. S2) was analyzed using microarrays. Entire seedlings were used in the microarrays. To identify differentially expressed genes, we used the “rank product” method. The rank product analysis gives an estimate of false discovery rate while providing flexible means to assign a significance level to each gene (Breitling et al., 2004). Differentially expressed genes are listed in Supplemental Tables S1 to S5. Pairwise plots showing relative gene expression levels revealed that the expression of many genes was changed in rop6DN seedlings relative to wild-type, ROP6, and rop6CA plants (Fig. 6A). Gene expression data were analyzed using hierarchical (Fig. 6B) and K-means (Fig. 6, C–H) clustering algorithms to identify genes with similar expression patterns. The clustering analyses revealed a group of 14 genes that were strongly induced in rop6DN plants (Fig. 6, C and F), a group of 24 genes that were mildly up-regulated (Fig. 6, D and G), and a group of 30 genes that were suppressed in rop6DN plants relative to wild-type, ROP6, or rop6CA transgenic plants (Fig. 6, E and H). Functional categorization of the differentially expressed genes by Gene Ontology (Martin et al., 2004) revealed that many of the up-regulated genes in the rop6DN plants were reminiscent of SA-dependent defense responses (Supplemental Tables S1–S5; Supplemental Fig. S4). SA-mediated disease resistance is a mechanism of induced defense that confers protection against a broad spectrum of microorganisms. SA-mediated pathogen defense requires the accumulation of SA, which serves as a signaling molecule leading to the induction of pathogenesis-related proteins that contribute to pathogen resistance (Durrant and Dong, 2004; Conrath, 2006). The frequency of SA-associated genes among the up-regulated genes is statistically significant (P ≤ 0.05) compared with their random up-regulation (Supplemental Table S4). Furthermore, expression analysis of the SA-associated genes in Genevestigator (http://www.genevestigator.ethz.ch; Hruz et al., 2008) showed that their expression is primarily associated with biotic defense responses and not with other stress responses. Examination of the gene expression patterns of all the genes that were differentially expressed in rop6DN plants revealed that most of them are subject to regulation by biotic stress. Hence, the microarray study suggested that expression of the dominant negative rop6DN induces a constitutive SA-mediated defense response and that most if not all of the observed changes in gene expression can be explained by activated SA signaling.
Figure 6.
Microarray analysis of gene expression in pROP6>>GFP-rop6DN seedlings and SA measurements. A, Pairwise plots showing relative gene expression levels in rop6DN versus the wild type (WT), rop6DN versus rop6CA, and rop6DN versus ROP6. The dashed red lines denote 2-fold change in gene expression levels. B, Hierarchical clustering showing the pattern of expression in rop6CA versus the wild type, rop6CA versus ROP6, ROP6 versus the wild type, rop6DN versus the wild type, rop6DN versus rop6CA, and rop6DN versus ROP6. C to H, K-means clustering showing a cluster of 14 genes that were strongly induced (C and F), a cluster of 24 genes that were mildly induced (D and G), and a cluster of 30 genes that were suppressed (E and H). Yellow color indicates an increase in gene expression, and blue indicates a decrease. I and J, Free (I) and total (J) SA levels in control and G. orontii-inoculated (Go) plants 2 and 4 d post spore inoculation. 6DN, GFP-rop6DN plants; 6[DN], GFP-rop6DN plants with low expression levels and weak phenotype; FW, fresh weight. Note the high levels of free and total SA in noninoculated rop6DN plants.
The microarray data required validation of the gene expression pattern by an independent method and raised questions that required additional experimental work. The questions that we asked were as follows. (1) Are the levels of SA higher in the rop6DN plants compared with the controls, as would be expected from the gene expression analysis? (2) Is the developmental phenotype of the rop6DN plants associated with a constitutive systemic acquired resistance response? (3) Are the rop6DN plants more resistant to pathogens as a result of the constitutive systemic acquired resistance response?
rop6DN and the SA Response
Validation of the microarray data was carried out by real-time quantitative reverse transcription PCR (qPCR) analysis of three genes that displayed strong differential expression in the rop6DN plants and are often used as markers for SA-dependent defense responses: (1) NIMIN1 (for nonexpresser of PR genes1/noninducible immunity1 interacting protein1; AT1G02450), (2) Pathogenesis-Related Protein5 (PR-5; AT1G75040), and (3) PR-1 (AT2G14610). The qPCR results show that NIMIN1, PR-5, and PR-1 expression levels are 4-, 12-, and 2,000-fold higher, respectively, in rop6DN relative to wild-type plants (Fig. 7, A–C), confirming the microarray results. Importantly, the expression levels were the same when RNA was extracted from 14-d-old seedlings (Supplemental Fig. S2), like the plants analyzed by microarrays or from mature flowering plants (Figs. 4 and 7).
To examine whether the changes in gene expression in the rop6DN plants are indeed associated with SA, we measured the levels of total and free SA in wild-type, ROP6, rop6CA, and two groups of rop6DN plants. One group is characterized by strong expression of the rop6DN protein, similar to the plants shown in Figures 3, 4, and 7, while the second group is characterized by very low expression levels of rop6DN and has a mild developmental phenotype (these plants are resistant to both kanamycin and basta, confirming that both transfer DNA (T-DNA) inserts of the transactivation expression system are present). In each of the lines, SA levels were compared with those in plants that were inoculated with spores of Golovinomyces orontii, a host-adapted virulent powdery mildew fungus of Arabidopsis (Spanu et al., 2010). As expected from the microarray and qPCR analyses, the levels of both total and free SA in noninoculated plants is markedly elevated in rop6DN plants (Fig. 6, I and J). In addition, noninoculated and G. orontii-infected rop6DN plants with high transgene expression accumulate similar SA levels, but these levels are considerably higher compared with pathogen-inoculated wild-type plants. In contrast, in wild-type, ROP6, and rop6CA plants, SA levels only increase following inoculation with G. orontii. SA levels are also increased in the rop6DN plants with low transgene expression levels but at lower levels compared with rop6DN plants with high expression levels of the transgene (Fig. 6, I and J). The higher SA levels confirmed that the changes in gene expression detected in rop6DN plants are indeed associated with the SA-induced defense response.
To examine whether the developmental phenotype of the rop6DN plants is linked to the constitutive SA-mediated response, the rop6DN plants were crossed to mutants in NPR1, which is required for SA-mediated signaling (Cao et al., 1997), and Salicylic Acid Induction Deficient2 (SID2)/Enhanced Disease Susceptibility16 (EDS16), which is required for pathogen-inducible SA biosynthesis (Nawrath and Métraux, 1999; Supplemental Figs. S4 and S5). It has been shown that both npr1 and sid2 plants fail to induce the expression of PR genes and display enhanced disease susceptibility to pathogens (Cao et al., 1997; Nawrath and Métraux, 1999; Kinkema et al., 2000; Stein et al., 2006). The expression of NIMIN1, PR-5, and PR-1 is suppressed in the rop6DN npr1 and rop6DN sid2 double mutant plants, resembling their expression in naive wild-type plants (Fig. 7, A–C). The mutant analysis confirmed that the expression of PR genes in the rop6DN mutant plants requires SA biosynthesis and NPR1 function, as is known to occur in authentic pathogen-induced SA-mediated defense responses (Durrant and Dong, 2004).
The macroscopic phenotype of both rop6DN npr1 and rop6DN sid2 double mutants resembles the rop6DN single mutants. The plants were small, the rosette and cauline leaves were uneven and twisted, and the plants lost their apical dominance and developed numerous adventitious inflorescence stems (Fig. 7, D–G). Qualitative (Fig. 7, H–K) and quantitative (Fig. 7, L–N) analyses of leaf epidermis pavement cell structure showed that cells of rop6DN npr1 and rop6DN sid2 double mutants resembled the rop6DN single mutant and are significantly (P ≤ 0.001, ANOVA, Tukey-Kramer test) different from wild-type plants. The phenotypic analysis of the rop6DN npr1 and rop6DN sid2 double mutant plants indicates that the developmental phenotype of the rop6DN plants can be uncoupled from the constitutive SA-dependent defense response. Hence, the overall small plant size, loss of apical dominance, twisted leaves, and deformed cells observed in the rop6DN plants are not associated with the constitutive SA-dependent defense response.
Pathogen Response of rop6DN Plants
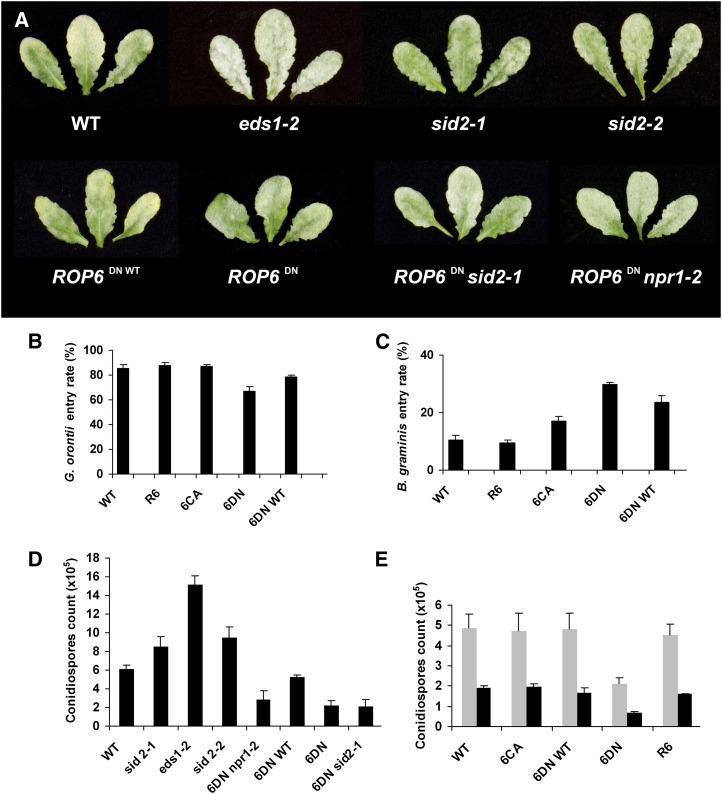
Mutants in SA biosynthesis or signaling enhance disease susceptibility to pathogens, while the induction of SA-dependent defense responses reduces pathogen growth, especially against biotrophic pathogens such as powdery mildews (Durrant and Dong, 2004). Hence, it was expected that the rop6DN plants would display increased resistance to G. orontii due to the high levels of SA and PR gene expression that greatly exceed the SA levels seen in wild-type plants after G. orontii infection (Fig. 6, I and J). To test this, wild-type, ROP6, rop6CA, rop6DN, rop6DN WT (plants that phenotypically resembled wild-type plants and in which no visible GFP-rop6DN expression could be detected, although they were resistant to both the kanamycin and basta selection markers), eds1-2, sid2-1, sid2-2, rop6DN sid2-1, and rop6DN npr1-2 plants were inoculated with G. orontii. Five days after conidiospore inoculation, patches of sporulating powdery mildew colonies were macroscopically detectable on the leaf surface of all lines (Fig. 8; Supplemental Fig. S6). Consistent with this, similar growth of epiphytic fungal hyphae was observed microscopically on leaves of wild-type, rop6CA, high-expressing, and low-expressing rop6DN plants at early stages of pathogenesis (48 h post inoculation; Supplemental Fig. S7). As expected, enhanced disease susceptibility was detected in the eds1-2 mutant that lacks a key regulatory node of plant immune responses known to limit the growth of host-adapted virulent pathogens (Fig. 8A; Wiermer et al., 2005).
Figure 8.
Susceptibility to the powdery mildew fungus. A, Wild-type (WT), GFP-rop6DN (ROP6DN), GFP-rop6DN with low GFP-rop6DN expression and weak phenotype (ROP6DN wild-type), SA biosynthesis mutants eds1-2, sid2-1, and sid2-2, and double mutants of GFP-rop6DN with sid2-1 (ROP6DN sid2-1) and with the SA signaling mutant npr1-2 (ROP6DN npr1-2) plants are susceptible to the powdery mildew fungal pathogen G. orontii. B, Fungal entry rates of G. orontii on GFP-rop6DN (6DN) and into GFP-rop6DN plants with low expression levels and weak phenotype (6DN WT) are lower compared with the entry rate on wild-type, GFP-ROP6 (R6), and GFP-rop6CA (6CA) plants. C, Fungal entry rates of Bgh on GFP-rop6DN (6DN) and on GFP-rop6DN plants with low expression levels and weak phenotype (6DN WT) are higher compared with the entry rate on wild-type, GFP-ROP6 (R6), and GFP-rop6CA (6CA) plants. The entry rates of Bgh on the GFP-rop6CA plants are higher than on wild-type and GFP-ROP6 plants. D, G. orontii conidiospore formation was significantly lower on GFP-rop6DN (6DN), GFP-rop6DN sid2-1 (6DN sid2-1), and GFP-rop6DN npr1-2 (6DN npr1-2) compared with wild-type and GFP-rop6DN WT (6DN WT) plants. Conidiospore formation was strongly enhanced compared with the wild type on eds1-2 and slightly enhanced on sid2-1 and sid2-2 SA biosynthesis mutants. E, Treatments with the SA analog BTH (black bars) reduced G. orontii conidiospore formation and further enhanced resistance of GFP-rop6DN plants. Gray bars show mock-treated (water) plants.
To obtain a more accurate account of infection phenotypes on the different lines, fungal entry into plant cells from each of the different lines was determined (as percentage entry at single plant-powdery mildew interaction sites). The results of this quantitative analysis show that 48 h after inoculation, the fungal entry rates into the rop6DN plants were around 65% compared with 90% in wild-type, ROP6, and rop6CA plants. The fungal entry rates on rop6DN plants with low expression of the transgene were around 80%, slightly lower compared with wild-type, ROP6, and rop6CA plants and higher compared with rop6DN plants with high transgene expression levels (Fig. 8B). These data indicate that the constitutive SA-dependent defense response in the rop6DN plants results in a moderate increase in preinvasive defense responses to the powdery mildew fungus, despite even higher constitutive SA levels than in wild-type plants found after pathogen challenge (Fig. 6, I and J).
We also determined preinvasion infection phenotypes after inoculation with Bgh, a nonadapted powdery mildew that fails to colonize Arabidopsis due to effective extracellular (preinvasive) resistance responses at early stages during fungal pathogenesis (Collins et al., 2003). At 48 h post inoculation, the entry rates of Bgh sporelings into wild-type Columbia (Col-0) leaf epidermal cells were around 10% (Fig. 8C). In comparison, the entry rates of virulent G. orontii at 48 h post inoculation were approximately 90% (Fig. 8B). The entry rates of Bgh into epidermal cells expressing a constitutively active rop6CA were higher, averaging between 16% and 20% (Fig. 8C). Interestingly, in a compatible interaction in barley expressing constitutively active ROP mutants, entry rates of Bgh were also higher compared with the wild type (Schultheiss et al., 2002, 2003; Opalski et al., 2005; Hoefle et al., 2011). Significantly, Bgh entry rates of rop6DN epidermal cells were even higher, reaching approximately 30% (Fig. 8C). Taken together, these data suggest that the disruption in cytoskeleton organization impaired extracellular disease resistance responses of the rop6DN plants toward Bgh, albeit the SA levels in these plants were constitutively elevated.
Next, we quantified the reproductive fitness of the host-adapted G. orontii powdery mildew by counting fungal conidiospores at a late stage after pathogen challenge (7 d after inoculation) in wild-type, rop6DN, rop6DN WT (low levels of rop6DN expression), sid2-1, eds1-2, sid2-2, rop6DN sid2-1, and rop6DN npr1-2 plants. As expected, G. orontii reproductive fitness on eds1-2 plants was greatly increased compared with the wild type and was moderately increased on sid2-1 and sid2-2 plants (Fig. 8D; Supplemental Fig. S8). In contrast, G. orontii reproductive fitness on rop6DN plants was significantly lower compared with the wild type (Fig. 8D; Supplemental Fig. S8). Importantly, reproductive fitness of G. orontii remained significantly lower than wild-type levels in rop6DN sid2-1 and rop6DN npr1-2 double mutants and was comparable to or only slightly higher than in rop6DN single mutants (Fig. 8D; Supplemental Fig. S8). To further substantiate these findings, we tested G. orontii reproductive fitness following pretreatments with the SA analog benzothiadiazole (BTH). The BTH treatments caused decreased G. orontii reproductive fitness in wild-type, ROP6, rop6CA, rop6DN, and rop6DN WT plants, demonstrating that SA can confer enhanced disease resistance to G. orontii in Arabidopsis. In rop6DN plants, BTH treatments had an additive effect, further reducing the already lower reproductive fitness of the fungus (Fig. 8E; Supplemental Fig. S9). Taken together, the extended data set provides evidence that rop6DN mediates an enhanced disease resistance phenotype to host-adapted G. orontii independently of the SA-mediated defense pathway.
DISCUSSION
Plant Development and Hormonal Response and Transport
In the absence of other families of signaling small GTPases in plants, ROPs were suggested to function as regulators of diverse signaling pathways (Yang, 2002). The results in our work are consistent with earlier findings and implicate ROP6 in both developmental and pathogen response regulation. The expression pattern of ROP6 is highly specific and closely resembles that of auxin-regulated genes both in leaves and roots. Correspondingly, the small plant size, loss of apical dominance, and twisted leaf phenotypes of plants expressing a dominant negative rop6DN under the regulation of the ROP6 promoter resemble mutant plants that are affected in auxin signaling, transport, and biosynthesis. Because the phenotype of rop6 T-DNA knockout mutants is indistinguishable from wild-type plants, it is likely that the rop6DN mutant affected functions of other ROPs, which are expressed in the same tissue as ROP6. Hence, by expressing rop6DN under the regulation of it native promoter, we overcame functional redundancy between ROPs while maintaining specificity and analyzing ROP function in a developmental context.
It has recently been proposed that ROPs are activated by auxin through a mechanism that involves ABP1 (Xu et al., 2010). Activated ROPs undergo transient S-acylation and partition into lipid rafts, a process that is required for their function (Sorek et al., 2007, 2010) and that induces localized inhibition of endocytosis (Bloch et al., 2005). In lipid rafts, ROPs interact with their effector ICR1 (Sorek et al., 2010), which regulates the recruitment of PIN auxin efflux transporters to polar domains in the plasma membrane and is required for directional auxin transport (Hazak et al., 2010). ICR1 transcription is induced by auxin, and similar to ROP6, it is also expressed in LRI (Hazak et al., 2010). Our results (Fig. 3, Q–S) indicate that the dominant negative rop6DN accumulates in the cytoplasm and nuclei, where it presumably interacts with ROPGEFs, inhibiting their function in ROP activation (Feig, 1999; Berken et al., 2005; Berken and Wittinghofer, 2008). In addition, the altered cytoskeleton organization in the rop6DN plants (Fig. 5) may interfere with auxin transport. It has been demonstrated that actin is required for PIN recycling (Geldner et al., 2001) and MTs are required for polar PIN localization (Boutté et al., 2006; Kleine-Vehn et al., 2008; Heisler et al., 2010). Hence, the expression of rop6DN could have inhibited ROP function in regulating auxin transport.
Epidermal Cell Structure and Growth
The changes in rosette leaf pavement cell morphology, which were detectable in independent transgenic lines, suggest that although ROP6 is primarily expressed at the leaf tip and hydathodes (Fig. 1, A–C), it may also be expressed in pavement cells at levels that are below detection with GUS and GFP reporter genes. Alternatively, the dominant negative rop6DN may affect pavement cell morphology indirectly. For example, it has recently been shown that perturbations in auxin biosynthesis or distribution affect pavement cell morphology (Xu et al., 2010). Because ROP6 expression in rosette leaves corresponds to sites of high auxin response, it is possible that suppression of ROP function at these sites affects auxin response and/or distribution. Furthermore, the overall small size of the rop6DN plants and the development of multiple inflorescence stems indicate that the dominant negative rop6DN has non-cell-autonomous effects.
The rosette and cauline leaves of the rop6DN plants have a wavy or twisted appearance (Fig. 4), likely owing to uneven cell growth rates in different parts of the leaf. Analysis of pavement cell growth dynamics in developing Arabidopsis cotyledons showed that cell growth is coordinated between groups of neighboring cells, and it was suggested that this coordination is required for the maintenance of flattened leaf surface (Zhang et al., 2011). Close comparison of the scanning electron microscopy images of wild-type and rop6DN rosette and cauline leaf epidermal cells shows that the pavement cells are flatter in the wild type compared with rop6DN (Fig. 4, compare G with H and I with J). Furthermore, the cells of the rop6DN plants range in size and shape: some cells are rectangular or cubical, while other cells develop more lobes (Fig. 7). The bulging of the rop6DN epidermal cells probably occurred due to isotropic and uncoordinated cell growth that may have altered the mutual pressures that cells exert on their neighbors. Pavement cells of plants expressing constitutively active ROP mutants also display isotropic growth, but they are usually more rectangular and even in their size. The leaf margins of the ropCA mutants usually bend downward, but the leaves are not wavy or twisted (Li et al., 2001; Fu et al., 2002; Bloch et al., 2005; Sorek et al., 2010). This suggests that the isomorphic cell growth cannot fully explain the uncoordinated cell growth that leads to leaf waviness and twisting. It could be that the expression of rop6DN disrupted a non-cell-autonomous mechanism that is responsible for coordinating cell growth in leaves.
Cytoskeleton Organization
Thick GFP-mTalin-labeled F-actin bundles have previously been observed in plants expressing a dominant negative ROP2 (rop2DN) under the regulation of the 35S promoter. It was further shown that the abundance of diffuse F-actin was reduced following transient expression of rop2DN. Interestingly, the same study showed that the expression of rop2CA had the opposite effect, causing an increase in the abundance of diffuse F-actin (Fu et al., 2002). Using a different experimental system, our data similarly show that the abundance of diffuse F-actin was low and that F-actin was primarily organized in thick bundles in the rop6DN plants (Fig. 5; Supplemental Fig. S3). The effect of constitutively active rop6CA on actin in pavement cells was not studied in this work. However, previous studies in root hairs showed that the expression of constitutively active rop6CA or rop11CA led to the formation of thick actin bundles and eliminated the zone of fine F-actin at the root hair tip (Molendijk et al., 2001; Bloch et al., 2005, 2011). This may indicate that activated ROPs possibly have different effects on actin organization in different cell types. An alternative and tempting explanation is that ROP cycling between active and inactive states is required for F-actin turnover and the formation of fine F-actin. The constitutively active mutants are locked in the GTP-bound state (Berken and Wittinghofer, 2008; Sorek et al., 2010). Hence, when signaling requires the binding and release of effectors, it should be impaired when the dissociation of the GTPase from target proteins is compromised. Thus, the bundling of actin filaments may reflect an inhibition of the ROP switch mechanism. How ROPs regulate actin nucleation and stability is yet an open question (Hussey et al., 2006; Deeks and Hussey, 2009).
The MT analysis in this work demonstrates that in many fully developed pavement cells, MTs were organized in transverse or oblique orientation relative to the long cell axis (Fig. 5; Supplemental Fig. S3). Earlier studies with plants expressing rop2DN showed that the cortical MTs have transverse orientation during early stages of pavement cell growth but are randomly organized in mature cells (Fu et al., 2002). The discrepancy between our data and the previous study could be explained by differences in the experimental system, such as the different ROPs used in each experiment, different expression systems, the level of transgene expression, and plant growth conditions. The observed changes in MT organization could have resulted from changes in tissue biomechanics or from the direct influence by ROPs or both. In cells of the shoot apical meristem, the orientation of cortical MTs was shown to correspond to biomechanical stress from the surrounding tissue (Hamant et al., 2008). It was further shown that both cortical MT orientation and the localization of PIN1 auxin efflux carrier likely respond to the same biomechanical regulator (Heisler et al., 2010). In developing metaxylem cells, the ICR1 homolog MIDD1/RIP3/ICR5 is recruited to specific plasma membrane domains by ROP11, where it interacts with and induces the severing of cortical MTs, leading to secondary cell wall pit formation (Oda et al., 2010; Oda and Fukuda, 2012). Interestingly, adaxial pavement cells of icr1 mutant plants have a cubical shape lacking lobes and indentations, resembling ROP mutants (Lavy et al., 2007). During pavement cell growth, ROP6 is activated by auxin and stabilizes MTs via its effector RIC1 (Fu et al., 2005; Xu et al., 2010). Hence, ROPs may modulate MTs via ICR and RIC family proteins. Analysis of pavement cell growth dynamics showed little correlation between MT organization and orientation and cell structure (Zhang et al., 2011). Clearly, more work is required to understand the complex relation between ROP function, cell growth, MT organization, and cell wall formation and flexibility.
The SA Response
The microarray studies were carried out on young seedlings before the developmental abnormalities induced by the expression of rop6DN became apparent (Supplemental Fig. S2). The logic behind this experimental design was that by looking at changes in gene expression prior to the stage where the phenotype of the plant becomes apparent, it should be possible to define whether the dominant negative ROP induces changes in gene expression and possibly identify the key genes whose alteration leads to the developmental aberrations. However, the microarray analysis revealed that the most significant changes in gene expression induced by rop6DN are associated with SA-dependent defense responses. Down-regulation of lignin biosynthesis by silencing the expression of hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT) in alfalfa (Medicago sativa) and Arabidopsis led to retarded plant growth, which has been associated with a constitutive SA response. Growth of HCT-silenced plants was regained in HCT-RNAi sid2 but not in HCT-RNAi npr1 double mutants (Gallego-Giraldo et al., 2011a, 2011b). In contrast to the HCT-silenced plants, the phenotype of the rop6DN npr1 and rop6DN sid2 double mutants, in which the SA-dependent defense response is blocked, resembled that of rop6DN single mutant plants, indicating that the developmental perturbations induced by rop6DN are unrelated to the constitutive SA-dependent response.
ROPs and Pathogen Response
As described in the introduction, ROP function was linked to plant defense responses in several studies. Results in this work (Figs. 6–8) suggest that the possible suppression of SA-dependent defense response execution by activated ROPs or by loss of MT-associated ROPGAP1 function (Hoefle et al., 2011; Huesmann et al., 2011) may also contribute to the enhanced susceptibility of the respective mutants. Loss-of-function mlo mutant alleles in Arabidopsis and barley are resistant to host-adapted powdery mildews, and this preinvasive resistance depends on polarized exocytosis regulated by PEN1/ROR2 syntaxins (Collins et al., 2003; Consonni et al., 2006; Kwon et al., 2008). Infection by the Bgh powdery mildew was shown to induce a redistribution of MLO, ROR2, and other proteins involved in the defense response to plasma membrane microdomains, which could be lipid rafts, just below sites of attempted fungal ingress (Bhat et al., 2005). The actin and MT cytoskeleton are disorganized in dominant negative ROP mutants (Fu et al., 2002; this work), possibly affecting directed secretion and, in turn, plant defense responses to the powdery mildew fungus. Hence, impaired extracellular resistance responses of the rop6DN plants toward Bgh may have resulted from the disorganized cytoskeleton. Consistent with this interpretation, pharmacological inhibitors and genetic interference by ectopic expression of an actin-depolymerizing factor-encoding gene, HvADF, impaired extracellular resistance responses to Bgh entry in barley (Miklis et al., 2007), and silencing of the HvRAB- and HvRAC1-interacting protein kinase HvRBK1 disrupted MT organization and enhanced susceptibility to Bgh (Huesmann et al., 2012). Of note, however, preinvasive resistance responses to Bgh at early stages during pathogenesis are essentially independent of an intact SA defense pathway (Consonni et al., 2006; Stein et al., 2006). This could explain why extracellular resistance responses to Bgh in rop6DN plants are impaired despite constitutively elevated SA levels.
The BTH treatments (Fig. 8; Supplemental Fig. S9) demonstrated that SA-dependent defense responses can confer resistance to G. orontii in Arabidopsis. Hence, we cannot exclude a small contribution of the SA-dependent defense response to the reduced reproductive fitness of G. orontii seen on rop6DN plants. However, the double mutants of rop6DN with sid2 or npr1 show that the reduced reproductive fitness of G. orontii on rop6DN plants was largely a rop6DN-dependent and SA-independent defense response.
CONCLUSION
Our results imply that ROPs regulate highly complex defense responses to powdery mildews that can be detected at early and late stages of the infection process. ROPs either enhance or reduce early preinvasive disease resistance responses to the tested host-adapted and nonadapted powdery mildews, presumably via their effects on the cytoskeleton and intracellular trafficking. At later stages of fungal pathogenesis, interference with ROP function reduces fungal reproductive success by mechanisms that remain to be elucidated. The mechanism underlying the constitutive activation of SA accumulation/signaling in rop6DN plants remains unclear. One possibility is that perturbation of the cytoskeleton or default secretion by rop6DN mimics a pathogen attack. Alternatively, similar to HCT-RNAi plants (Gallego-Giraldo et al., 2011b), release of cell wall materials or changes in cell wall integrity may activate SA responses in rop6DN plants. Changes in cell wall integrity may have also contributed to the susceptibility of the rop6DN plants to Bgh. Irrespective of this, our results implicate ROPs as important regulators of plant development as well as in SA-dependent and SA-independent defense execution.
MATERIALS AND METHODS
Molecular Cloning
All plasmids and primers are listed in Supplemental Tables S6 and S7, respectively. Plasmid construction was carried out using standard molecular cloning techniques. AtROP6 was cut from pSY700 with SacI and then was subcloned to pGFP-MRC to create pSY811. PCR-based site-directed mutagenesis (QuikChange kit; Stratagene) was used to create a constitutively active mutant (Atrop6CA/DN) by substituting Gly-15 with Val and Thr-30 with Asn. pSY811 was used as template with the following primer sets: SYP189 + SYP190 to create pSY812 (GFP-rop6CA) and SYP602 + SYP603 to create pSY807 (GFP-rop6DN). To subclone GFP-ROP6, GFP-rop6CA, and GFP-rop6DN into the pOP effector plasmids, pSY811, pSY812, and pSY807 were digested with XhoI to isolate the GFP-ROP6, GFP-rop6CA, and GFP-rop6DN fragments. The three fragments were, in turn, subcloned into pOP-Bj36 to create pSY836, pSY819, and pSY835, respectively. For expression in plants, pSY836, pSY819, and pSY835 were digested with NotI, and the resulting pOP-GFP-ROP6/rop6CA/rop6DN fragments were subcloned into pMLBART plant binary vector to create pSY832, pSY818, and pSY831, respectively. For construction of the ROP6 promoter LhG4 driver plasmid, a 1,047-bp fragment upstream of AtROP6 (pROP6) was amplified by PCR from Arabidopsis (Arabidopsis thaliana Col-0) genomic DNA using SYP615 and SYP616. The amplified AtROP6 was subcloned into pGEM-T (Promega) and, in turn, into pBj36 (Eshed et al., 2001) with SalI and KpnI (Acc65I) to create the pSY810 promoter LhG4 driver plasmid. For expression in plants, pSY810 was digested with NotI, and the LhG4-pAtROP6 cassette was subcloned into a pART27 plant binary vector to create pSY813. All fragments were fully sequenced to verify that no PCR-generated errors were introduced.
Plant Material
All transgenic Arabidopsis lines used in this study are listed in Supplemental Table S8. Arabidopsis plants were grown as described previously (Lavy et al., 2002). For plant transformation, Arabidopsis plants were transformed with Agrobacterium tumefaciens strain GV3101/pMp90 harboring the appropriate plasmids using the floral dip method (Clough and Bent, 1998). A minimum of three independent transgenic lines for each plasmid were analyzed. Transcription-transactivation lines were obtained by crossing pROP6::LhG4 driver lines with pOp effector lines to obtain pROP6>>GUS, pROP6>>DsRed, pROP6>>GFP-ROP6, pROP6>>GFP-rop6CA, and pROP6>>GFP-rop6DN. An Arabidopsis T-DNA insertion line (SALK_91737) in the ROP6 gene was obtained from the Arabidopsis Biological Research Center at the University of Ohio. The existence of the insertion was verified with SYP179 and SYP163 ROP6-specific primers and a T-DNA left border primer (Lba1; SYP179).
Auxin Induction
For auxin induction, seeds were germinated in the presence of the auxin transport inhibitor N-1-naphthylphthalamic acid followed by transfer to growth medium containing 10 μm auxin (NAA), as described previously (Himanen et al., 2004). Alternatively, seeds were germinated on medium lacking N-1-naphthylphthalamic acid and were, in turn, treated with 10 μm NAA.
GUS Staining
GUS staining was carried out as described previously (Weigel and Glazebrook, 2002).
Whole-Mount in Situ Hybridization
Tissue preparation and whole-mount in situ hybridization were performed essentially as described previously (Brewer et al., 2006; Hejátko et al., 2006). A 244-bp fragment from the 3′-untranslated region of ROP6 was amplified by PCR from Arabidopsis genomic DNA and was used as template to create gene-specific digoxigenin-labeled antisense/sense RNA probes. The digoxigenin-labeled probes were prepared by in vitro transcription according to the manufacturer’s instructions (Roche).
Fixation and Immunostaining
Fixation and immunostaining were carried out essentially as described previously (Chaimovitsh et al., 2012). To stabilize actin and MT, specimens were fixed for 5 min with 500 mm m-maleimidobenzoyl-hydroxylsuccinimide ester in MT-stabilizing buffer (MTSB) containing 100 mm PIPES, pH 6.89, 5 mm EGTA, 5 mm MgSO4·7H2O, and 1% Triton X-100 (v/v) buffer and, in turn, for 1 h in freshly prepared 8% paraformaldehyde in MTSB. Fixation was carried out at room temperature in the dark. Samples were then rinsed three times in MTSB and treated with the following enzyme mixture: 2% cellulase Onzuka R-10 and 1% (w/v) pectinase, protease inhibitor cocktail (Sigma), and 20 μm phenylmethylsulfonyl fluoride for 10 min. After washing in MTSB for 30 min, the samples were squashed on coverslips coated with poly-l-Lys (1 mg mL−1; Sigma-Aldrich). Then, the cells were retreated for 10 min with enzyme mixture (2% cellulase Onzuka R-10 and 1.5% [w/v] pectinase) with protease inhibitors. This step was followed by a rinse in phosphate-buffered saline (PBS) with 1% Triton X-100 (v/v) for 30 min and then incubation in PBS containing 1% (w/v) bovine serum albumin for 20 min. To reduce aldehyde-induced autofluorescence, samples were treated for 5 min with 10 mg mL−1 sodium borohydride in PBS and then washed in PBS for 20 min. For immunofluorescence detection of MTs, specimens were incubated overnight at room temperature with primary sheep anti-αβ-tubulin polyclonal antibodies (Cytoskeleton) and mouse anti-actin monoclonal (clone C4) antibodies (ICN Biomedical) at a dilution of 1:100. Samples were then washed in PBS and incubated with anti-sheep secondary antibodies conjugated to Alexa Fluor 488 and anti-mouse secondary antibodies conjugated to Alexa 555 (Molecular Probes) diluted 1:100 for 1 h at room temperature. Mounting was done in 50% glycerol/water (v/v).
Plant DNA and RNA Isolation and qPCR
Plant genomic DNA was isolated with the GenElute Plant Genomic DNA miniprep kit according to the manufacturer's instructions (Sigma). Total RNA was isolated with the SV Total RNA isolation kit according to the manufacturer’s instructions (Promega). The RNA from the qPCR experiments was extracted from both 14-d-old seedlings and mature flowering plants. Complementary DNA first-strand synthesis was carried out using Moloney murine leukemia virus reverse transcriptase (Promega). Quantifications with the primer sets (Supplemental Table S7) were carried out by qPCR using an ABI Prism 7700 StepOnePlus Instrument (Applied Biosystems). Study samples were run in triplicate on eight-well optical PCR strips (Applied Biosystems) in a final volume of 10 μL. Primers were designed using the Roche Universal Probe Library (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). The PCR cycles were run as follows: 10-min initial denaturation at 95°C, followed by 40 subsequent cycles of 15 s of denaturation at 95°C, and 1 min of annealing and elongation at 60°C. The specificity of the unique amplification product was determined by melting-curve analysis according to the manufacturer’s instructions (Applied Biosystems). Relative quantities of RNA were calculated by the comparative threshold cycle method (Applied Biosystems User Bulletin 2: ABI PRISM 7700 Sequence Detection System; http://www.appliedbiosystems.com). DNA dilution series were prepared to calculate an amplification efficiency coefficient for each gene. The relative levels of RNA were calculated according to the amplification efficiency coefficient and normalized against ACTIN8 and UBQ21 gene standards (Czechowski et al., 2005), the level of which was taken as 1. The stability of the standards in each experiment was verified with the geNorm analysis tool (http://medgen.ugent.be/jvdesomp/genorm/) and was calculated as M ≤ 0.7. The analysis was repeated with three independent biological replicates.
Array Hybridization and Evaluation
Seedlings of pAtROP6>>GFP-ROP6, pAtROP6>>GFP-ROP6CA, and Col-0 were grown on 0.5× Murashige and Skoog medium for 14 d in growth chambers at 21°C under long-day photoperiods (16 h of light, 8 h of darkness). A total of 100 mg of plant tissue was harvested from seedlings, and RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. The entire experiment was performed three times, providing independent biological replicates. Affymetrix Arabidopsis ATH1 GeneChips were used in the experiment. Labeling of samples, hybridizations, and measurements were performed as described (Hennig et al., 2004). Signal values were derived from the Affymetrix *.cel files using the GCRMA algorithm (Psarros et al., 2005). Data were processed with the statistical package R (version 1.9.1). Differentially expressed genes were determined using the rank-product method (Breitling et al., 2004) implemented in R. Genes were considered as being differentially expressed at P ≤ 0.05.
Light, Fluorescence, and Confocal Microscopy
Low-resolution imaging was performed with an SV-11 stereomicroscope (Zeiss). Wide-field fluorescence imaging was performed with an Axioplan-2 Imaging fluorescence microscope (Zeiss) equipped with an AxioCam cooled CCD camera, using either 20× dry or 63× water-immersion objectives with numerical aperture values of 0.5 and 1.2, respectively. Confocal imaging was performed with a Leica TCS-SL confocal laser scanning microscope equipped with 20× multi-immersion and 63× water-immersion objectives with numerical aperture values of 0.7 and 1.2, respectively. GFP was visualized by excitation with an argon laser at 488 nm, a 500-nm beam splitter, and a spectral detector set between 505 and 530 nm. DsRed was visualized by excitation with an argon laser set to 514 nm, a 456/514-nm double dichroic beam splitter, and a spectral detector set between 530 and 560 nm. Scanning electron microscopy was carried out with JEOL JSM-840A scanning electron microscope. Image analysis was performed with Zeiss AxioVision, ImageJ, Leica LCS, and Adobe Photoshop.
SA Measurements, Fungal Entry Rates, Conidiospore Counts, and BTH Treatments
Measurements of free and conjugated SA levels were carried out by analytical HPLC essentially as described previously (Bartsch et al., 2006). Golovinomyces orontii and Blumeria graminis f. sp. hordei conidiospore inoculations were carried out as described previously (Consonni et al., 2006). Conidiospore counts were carried out as described previously (Wessling and Panstruga, 2012). BTH treatments were carried out essentially as described previously (Lawton et al., 1996). Briefly, 4-week-old plants were sprayed with 42.6 mg L−1 water-solubilized BTH 24 h prior to inoculation by G. orontii conidiospores. Control plants were sprayed with water (BTH mock control). For all infection assays, plants were grown in a phytochamber with the following settings: 65% humidity; temperature of 22°C from 9 am to 7 pm and 20°C from 7 pm to 9 am; and 10/14-h light/dark cycles. Light intensity was 100 µE m−2 s−1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence of the AtROP6 promoter.
Supplemental Figure S2. Fourteen-day-old Col-0 (wild type), rop6CA (6CA), and rop6DN (6DN) seedlings grown on vertical-oriented plates.
Supplemental Figure S3. Immunostaining of actin and microtubule in wild-type and GFP-rop6DN leaf epidermis cells.
Supplemental Figure S4. Immunostaining of actin and microtubule in wild-type and GFP-rop6DN leaf epidermis cells.
Supplemental Figure S5. Molecular identification of rop6DN npr1 and rop6DN sid2 double mutants.
Supplemental Figure S6. Susceptibility to the powdery mildew fungus.
Supplemental Figure S7. G. orontii fungal hyphae.
Supplemental Figure S8. G. orontii conidiospores formation.
Supplemental Figure S9. Susceptibility to G. orontii following SA analog BTH treatments.
Supplemental Table S1. Differentially expressed genes in GFP-rop6DN versus the wild type (Col-0).
Supplemental Table S2. Differentially expressed genes in GFP-rop6DN versus GFP-ROP6.
Supplemental Table S3. Differentially expressed genes in GFP-rop6DN versus GFP-rop6CA.
Supplemental Table S4. Gene Ontology of differentially expressed genes by GoToolBox.
Supplemental Table S5. Strong differential expression of SAR associated genes in the GFP-rop6DN lines.
Supplemental Table S6. Plasmids used in this study.
Supplemental Table S7. Oligonucleotide primers used in this study.
Supplemental Table S8. Transgenic Arabidopsis lines used in this study.
Acknowledgments
We thank R. Fluhr for materials.
Glossary
- ABA
abscisic acid
- Bgh
Blumeria graminis f. sp. hordei
- SA
salicylic acid
- LRI
lateral root initials
- NAA
naphthaleneacetic acid
- MT
microtubule
- qPCR
real-time quantitative reverse transcription PCR
- Col-0
Columbia
- BTH
benzothiadiazole
- MTSB
microtubule-stabilizing buffer
- PBS
phosphate-buffered saline
- T-DNA
transfer DNA
References
- Aloni R, Schwalm K, Langhans M, Ullrich CI. (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216: 841–853 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Berken A, Thomas C, Wittinghofer A. (2005) A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436: 1176–1180 [DOI] [PubMed] [Google Scholar]
- Berken A, Wittinghofer A. (2008) Structure and function of Rho-type molecular switches in plants. Plant Physiol Biochem 46: 380–393 [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Hazak O, Lavy M, Yalovsky S. (2008) A novel ROP/RAC GTPase effector integrates plant cell form and pattern formation. Plant Signal Behav 3: 41–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Lavy M, Efrat Y, Efroni I, Bracha-Drori K, Abu-Abied M, Sadot E, Yalovsky S. (2005) Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell 16: 1913–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S. (2011) Nitrogen source interacts with ROP signalling in root hair tip-growth. Plant Cell Environ 34: 76–88 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B. (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Heisler MG, Hejátko J, Friml J, Benková E. (2006) In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat Protoc 1: 1462–1467 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chaimovitsh D, Rogovoy Stelmakh O, Altshuler O, Belausov E, Abu-Abied M, Rubin B, Sadot E, Dudai N. (2012) The relative effect of citral on mitotic microtubules in wheat roots and BY2 cells. Plant Biol (Stuttg) 14: 354–364 [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. (2010) Analysis of the Rac/Rop small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant Cell Physiol 51: 585–595 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2006) Systemic acquired resistance. Plant Signal Behav 1: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, et al (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ (2009) Plant actin biology. In Encyclopedia of Life Sciences. John Wiley & Sons, Chichester, UK [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Feig LA. (1999) Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol 1: E25–E27 [DOI] [PubMed] [Google Scholar]
- Fischer U, Ikeda Y, Ljung K, Serralbo O, Singh M, Heidstra R, Palme K, Scheres B, Grebe M. (2006) Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr Biol 16: 2143–2149 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z. (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z. (2009) A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol 19: 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA. (2011a) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. (2011b) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S. (2010) A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Yalovsky S. (2010) An auxin regulated positive feedback loop integrates Rho modulated cell polarity with pattern formation. Plant Signal Behav 5: 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM. (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J, Blilou I, Brewer PB, Friml J, Scheres B, Benková E. (2006) In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat Protoc 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C. (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135: 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T. (2004) Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R. (2011) A barley ROP GTPase activating protein associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 23: 2422–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendrop F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesmann C, Hoefle C, Hückelhoven R. (2011) ROPGAPs of Arabidopsis limit susceptibility to powdery mildew. Plant Signal Behav 6: 1691–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesmann C, Reiner T, Hoefle C, Preuss J, Jurca ME, Domoki M, Fehér A, Hückelhoven R. (2012) Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiol 159: 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JA, Vejlupkova Z, Luo A, Meeley RB, Sylvester AW, Fowler JE, Smith LG. (2011) ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 23: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Ketelaar T, Deeks MJ. (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J. (2008) Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S. (2007) A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol 17: 947–952 [DOI] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S. (2002) A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z. (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorek J, Panstruga R, Huckelhoven R. (2010) The role of seven-transmembrane domain MLO proteins, heterotrimeric G-proteins, and monomeric RAC/ROPs in plant defense. In S Yalovsky, F Baluska, A Jones, eds, Integrated G Protein Signaling in Plants. Springer-Verlag, Berlin, pp 197–220 [Google Scholar]
- Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B. (2004) GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol 5: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. (2007) Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K, Klessig DF. (2005) Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol Plant Microbe Interact 18: 116–124 [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. (2001) Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J 20: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K. (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP. (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. (2006) RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 11: 309–315 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H. (2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Oda Y, Iida Y, Kondo Y, Fukuda H. (2010) Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr Biol 20: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Hückelhoven R. (2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Pathuri IP, Zellerhoff N, Schaffrath U, Hensel G, Kumlehn J, Kogel KH, Eichmann R, Hückelhoven R. (2008) Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep 27: 1877–1887 [DOI] [PubMed] [Google Scholar]
- Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. (2005) RACE: remote analysis computation for gene expression data. Nucleic Acids Res 33: W638–W643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. (2002) A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol 128: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R. (2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36: 589–601 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Hensel G, Imani J, Broeders S, Sonnewald U, Kogel KH, Kumlehn J, Hückelhoven R. (2005) Ectopic expression of constitutively activated RACB in barley enhances susceptibility to powdery mildew and abiotic stress. Plant Physiol 139: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Gutman O, Bar E, Abu-Abied M, Feng X, Running MP, Lewinsohn E, Ori N, Sadot E, Henis YI, et al (2011) Differential effects of prenylation and S-acylation on type I and II ROPS membrane interaction and function. Plant Physiol 155: 706–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Bar E, Lewinsohn E, Yalovsky S. (2007) Activation status-coupled transient S acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sorek N, Segev O, Gutman O, Bar E, Richter S, Poraty L, Hirsch JA, Henis YI, Lewinsohn E, Jürgens G, et al (2010) An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Curr Biol 20: 914–920 [DOI] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ, et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Nibau C, Wu HM. (2005) RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell 17: 2369–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Wu HM. (2002) Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell 14: 2745–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Chen L, Nakashima A, Hara S, Umemura K, Takahashi A, Shirasu K, Kawasaki T, Shimamoto K. (2007) RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 19: 4035–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wessling R, Panstruga R. (2012) Rapid quantification of plant-powdery mildew interactions by qPCR and conidiospore counts. Plant Methods 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM. (1997) Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol 35: 483–495 [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, et al (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Hazak O, Cheung AY, Yalovsky S. (2011) RAC/ROP GTPases and auxin signaling. Plant Cell 23: 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL. (2005) Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol 139: 1350–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. (2010) Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Bloch D, Sorek N, Kost B. (2008) Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol 147: 1527–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2008) Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol 24: 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Halsey LE, Szymanski DB. (2011) The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol 11: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z. (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]