Wheat oxophytodienoate reductase gene TaOPR1 enhances salinity tolerance by promoting an abscisic acid-dependent stress response pathway.
Abstract
The 12-oxo-phytodienoic acid reductases (OPRs) are classified into the two subgroups OPRI and OPRII. The latter proteins participate in jasmonic acid synthesis, while the function of the former ones is as yet unclear. We describe here the characterization of the OPRI gene TaOPR1, isolated from the salinity-tolerant bread wheat (Triticum aestivum) cultivar SR3. Salinity stress induced a higher level of TaOPR1 expression in the seedling roots of cv SR3 than in its parental cultivar, JN177. This induction was abolished when abscisic acid (ABA) synthesis was inhibited. The overexpression of TaOPR1 in wheat significantly enhanced the level of salinity tolerance, while its heterologous expression in Arabidopsis alleviated root growth restriction in the presence of salinity and oxidants and raised the sensitivity to ABA. In Arabidopsis, TaOPR1 promoted ABA synthesis and the ABA-dependent stress-responsive pathway, partially rescued the sensitivity of the Arabidopsis aba2 mutant defective in ABA synthesis to salinity, and improved the activities of reactive oxygen species scavengers and the transcription of their encoding genes while reducing malondialdehyde and reactive oxygen species levels. TaOPR1 did not interact with jasmonate synthesis or the jasmonate signaling pathway. Rather than serving purely as an antioxidant, we believe that TaOPR1 acts during episodes of abiotic stress response as a signaling compound associated with the regulation of the ABA-mediated signaling network.
Soil salinity represents an important constraint on plant growth and crop productivity, imposing a stress that encourages the production of reactive oxygen species (ROS). Plants have developed a complex ROS-scavenging system, the major components of which are a series of low-Mr nonenzymatic compounds such as ascorbate and glutathione and a set of enzymes including superoxide dismutase, catalase, and ascorbate peroxidase (Mittler, 2002). ROS-induced lipid peroxidation leads to the formation of a variety of cytotoxic molecules, some of which can be neutralized by members of the “Old Yellow Enzyme” (OYE) protein family, which is well represented in the yeast genome (Williams and Bruce, 2002). The overexpression of OYE2, for example, lowers ROS levels and thereby protects the cell against apoptosis (Odat et al., 2007).
Plant 12-oxo-phytodienoic acid reductases (OPRs) share some sequence similarity with OYE proteins and have been classified as FMN-dependent oxidoreductases. They are thought to catalyze the reduction of double bonds in α,β-unsaturated aldehydes and ketones. When presented with 12-oxo-phytodienoic acid (OPDA) as a substrate, their activity generates 3-oxo-2(2′-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0; Schaller et al., 1998). Four isomers of OPDA have been described, namely cis-(+), cis-(−), trans-(+), and trans-(−) (Schaller et al., 1998). On the basis of their substrate specificity, the OPRs have split into two subgroups (Schaller and Weiler, 1997; Schaller et al., 1998, 2000; Strassner et al., 1999). Subgroup II members (OPRIIs; e.g. AtOPR3) can reduce all four OPDA isomers, but in planta their activity is largely focused on the reduction of cis-(+)-OPDA (Schaller et al., 1998). Since OPC8:0 is a precursor of jasmonic acid (JA), the OPRIIs are thought to be implicated in JA synthesis (Schaller et al., 1998). Subgroup I members (OPRIs; e.g. AtOPR1 and AtOPR2), in contrast, are involved in the reduction of cis-(−)-OPDA (Strassner et al., 1999; Schaller et al., 2000), but their in vivo substrate(s) is unknown. It has been commonly suggested that the OPRI proteins are not enzymes in the octadecanoid pathway but rather serve some as yet unknown enzymatic function (Schaller et al., 1998). OPRI genes are typically up-regulated by pathogen invasion, wounding, and oxidative stress (Biesgen and Weiler, 1999; Strassner et al., 2002; Agrawal et al., 2003), events associated with ROS acceleration, so OPRIs are claimed to be concerned with antioxidant activity (Fitzpatrick et al., 2003; Trotter et al., 2006; Beynon et al., 2009).
Notably, in addition to cis-(+)-OPDA, enzyme preparations from some plants, such as flax (Linum usitatissimum), may yield a substantial amount of the cis-(−) isomer (Laudert et al., 1997), and the latter may be an in vivo substrate for the OPRIs. The allene oxide cyclase (AOC)-catalyzed enolization of 12,13-epoxyoctadecatrienoic acid to form OPDA generates a slow conversion of cis- to trans-isomer (Mueller and Brodschelm, 1994), and the generated trans-(+)-OPDA is also the substrate of OPRIs (Schaller et al., 1998). Thus, the in vivo fate of OPR-mediated OPDA metabolism is likely to be much more diverse than simply the formation of JA. However, whether OPRI activity is responsible for JA synthesis and signaling pathways is an open question. OPRI genes are transiently regulated by wounding, low temperature, salinity, pathogen attack, and other environmental cues (Biesgen and Weiler, 1999; Strassner et al., 2002; Agrawal et al., 2003). In Arabidopsis (Arabidopsis thaliana), the OPRII gene AtOPR3 is constitutively expressed throughout the plant, while the expression of the OPRIs AtOPR1 and AtOPR2 is largely confined to the root (Biesgen and Weiler, 1999). It has been shown that plants are able to regulate the levels of OPDA and JA independently of one another (Parchmann et al., 1997; Stelmach et al., 1998), and evidence is accumulating that OPDA is a signaling compound in its own right (Weiler et al., 1994; Stelmach et al., 1998). OPRI activity, therefore, may regulate the presence of its own substrates and thereby serve as an active regulatory component.
Bread wheat (Triticum aestivum) is one of the most important of our food crops, but it shows only limited tolerance to abiotic stress. Wheat ‘SR3’, with remarkable salinity tolerance, is bred from an asymmetric somatic hybrid between cv JN177 and tall wheatgrass (Thinopyrum ponticum; Xia et al., 2003; Xia, 2009). Transcriptomic and proteomic analyses have suggested that much of it can be ascribed to a superior level of ROS-scavenging capacity (Wang et al., 2008; Peng et al., 2009). Among the many genes differentially up-regulated by salinity stress in cv SR3 is one that encodes an OPRI protein (Liu et al., 2012). Here, we show that this gene, denoted TaOPR1, responds to salinity stress in an abscisic acid (ABA)-dependent manner and that its overexpression in wheat and heterologous expression in Arabidopsis improve tolerance to salinity. This effect appears to operate through the regulation of ROS and ABA signaling pathways and is quite independent of JA synthesis and signaling.
RESULTS
TaOPR1 Transcription Was Induced by Various Stresses
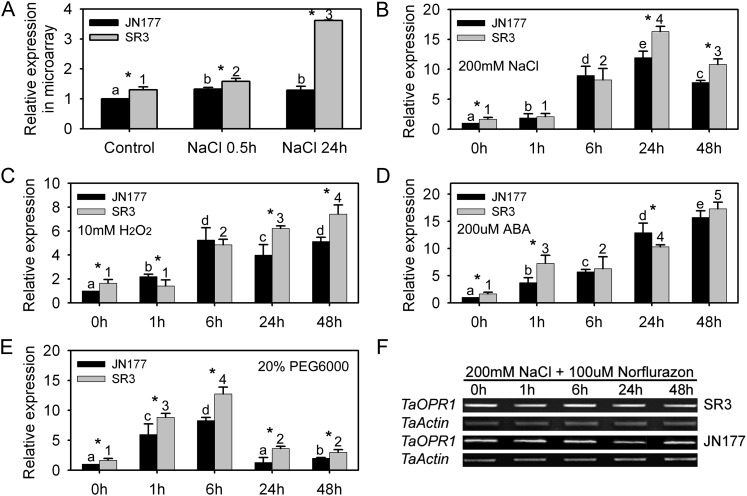
The microarray analysis indicated that the level of TaOPR1 transcription in cv SR3 plants was nearly 4-fold that in cv JN177 plants challenged with 200 mm NaCl for 24 h, but the levels were identical after just 0.5 h (Fig. 1A). Real-time PCR analysis confirmed that TaOPR1 transcript accumulated significantly over the course of 200 mm NaCl treatment and to a greater degree in cv SR3 than in cv JN177 after 24 and 48 h of treatment (Fig. 1B). High salinity often stimulates the production of ROS and ABA: the former causes severe oxidative damage, and the latter launches downstream stress-responsive pathways (Mittler et al., 2011). Here, the temporal transcription profiles of TaOPR1 after exposure to 10 mm hydrogen peroxide (H2O2) and 100 μm ABA almost mirrored those under salinity stress in the two cultivars, and it also had stronger responsive patterns in cv SR3 than in cv JN177 (Fig. 1, C and D). Apart from ionic toxicity, high salinity also consists of osmotic stress (Munns and Tester, 2008). In the presence of 20% polyethylene glycol 6000 (PEG6000) to simulate osmotic stress, TaOPR1 transcription rose markedly at an early stage, declining thereafter to a level that was still above the pretreatment one (Fig. 1E). These results demonstrate that TaOPR1 is a stress-responsive gene, and its higher expression level in cv SR3 implies its positive role in stress tolerance. The accumulation of transcript induced by salinity stress was counteracted by the provision of norflurazon, an inhibitor of ABA synthesis (Fig. 1F), indicating that the gene’s salinity-induced transcription was mediated by ABA.
Figure 1.
TaOPR1 is induced by various abiotic stresses in wheat roots. A, cDNA microarray assay of TaOPR1 transcription in cv SR3 and JN177 as affected by exposure to salinity stress. B to E, Real-time analysis of TaOPR1 expression in cv SR3 and JN177 roots subjected to 200 mm NaCl (B), 200 μm ABA (C), 10 mm H2O2 (D), and 20% (w/v) PEG6000 (E). F, Transcription of TaOPR1 in cv SR3 and JN177 roots exposed to 200 mm NaCl and 100 μm norflurazon (an inhibitor of ABA synthesis). Wheat seedlings at the three-leaf stage were used for analysis. Data are presented as means ± sd. Bars marked with different letters (cv JN177) or numbers (cv SR3) indicate significantly different means using the one-way ANOVA lsd analysis (P < 0.05). The column at each time point marked with an asterisk indicates a significant difference between cv JN177 and SR3 using Student’s t test analysis (P < 0.05).
TaOPR1 Is an OPRI Gene Mapping to Wheat Chromosome 2B
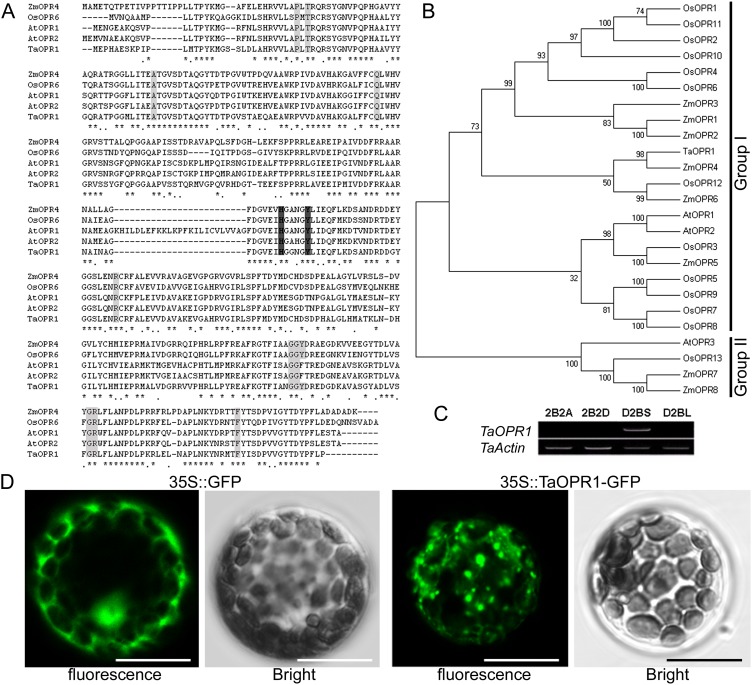
The full-length TaOPR1 complementary DNA (cDNA) comprises 1,347 bp, consisting of a 60-bp 5′ untranslated region, a 1,110-bp open reading frame, and a 69-bp 3′ untranslated region. The deduced peptide product comprises 369 residues and has an inferred molecular mass of 46 kD and a pI of 6.52. The phylogenetic analysis of the gene product indicated that TaOPR1 belongs to the OPRI subgroup and shares homology with rice (Oryza sativa) OPR6 (76%), maize (Zea mays) OPR4 (79%), and Arabidopsis OPR2 (67%; Fig. 2, A and B). The TaOPR1 sequence retains the conserved residues that account for the NADPH- and FMN-binding sites of the OYE family (Fig. 2A). In order to know the chromosomal location of TaOPR1, aneuploid stocks of cv Chinese Spring were used as template for genomic PCR with TaOPR1-specific primers. The results showed that TaOPR1 was not amplified from templates of stocks 2B2A, 2B2D, and D2BL, the former two of which lack the whole of chromosome 2B (2B is replaced by 2A and 2D, respectively) and the last of which lacks the short arm of 2B, while it was produced from the template of D2BS, which lacks the long arm of 2B (Fig. 2C). Thus, TaOPR1 must map to the short arm of the second chromosome of the B genome (2BS). The location of the fluorescent signal produced by the transient expression of a TaOPR1::GFP fusion in Arabidopsis mesophyll protoplasts indicated that the cytoplasm is the site of TaOPR1 deposition in planta (Fig. 2D). The cytoplasm location is due to the fact that TaOPR1, like other OPRI proteins, lacks the OPRII-specific peroximal signal peptide at its C terminus (Fig. 2A), which is thought to determine the protein’s localization to the peroxisome (Hayashi et al., 1996; Stintzi and Browse, 2000).
Figure 2.
TaOPR1 encodes an OPRI protein that locates in the cytoplasm. A, Peptide alignment of TaOPR1 and other OPR subgroup I members. Residues shown in black or gray are conserved. B, Phylogenetic analysis indicates that TaOPR1 is a subgroup I member. C, Aneuploid analysis shows that TaOPR1 maps to the short arm of wheat chromosome 2B. 2B2A and 2B2D are wheat nullitetrasomic lines whose chromosome 2 of the B genome (2B) is replaced by chromosome 2 of the A (2A) and D (2D) genomes, respectively; D2BS and D2BL are wheat ditelocentric lines that lack the long arm and short arm of chromosome 2B, respectively. D, TaOPR1 is deposited in the cytoplasm, as indicated by the fluorescent signal generated from a transiently expressed TaOPR1::GFP fusion protein in the Arabidopsis protoplast. The control experiment employed a simple GFP transgene. Bars = 10 μm. [See online article for color version of this figure.]
TaOPR1 Enhanced the Salinity Tolerance of Wheat and Arabidopsis
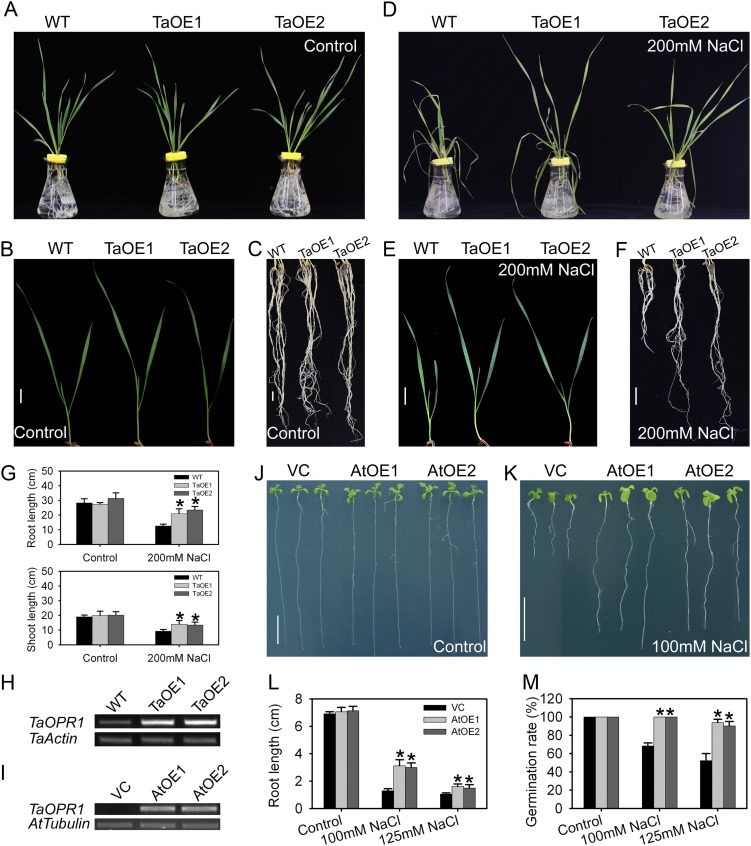
To ascertain whether the expression of TaOPR1 has any effect on the level of salinity tolerance, it was transformed into both a highly sensitive wheat (‘JN17’) and into Arabidopsis ecotype Columbia (Col-0). Of the 12 independent transgenic overexpression (TaOE) lines of TaOPR1 generated in wheat ‘JN17’, two (TaOE1 and TaOE2) were selected on the basis of the level of transgene expression (Fig. 3H). The two TaOE lines and the wild-type cv JN17 grew equally well in one-half-strength Hoagland liquid medium, and their seedling phenotype was indistinguishable (Fig. 3, A–C). However, after exposure to 200 mm NaCl, the wild-type seedlings became very wilted while the TaOE ones remained fully turgid (Fig. 3, D and E). The TaOE line shoots and roots were longer than those of the wild type (Fig. 3, E–G).
Figure 3.
TaOPR1 contributes to the salinity tolerance of wheat and Arabidopsis. A to F, Seedling phenotypes of wild-type (WT) wheat (the salinity-sensitive cultivar JN17) and TaOE1 and TaOE2 (transgenic wheat lines overexpressing TaOPR1) in response to 200 mm NaCl for 4 d. The treatment involved a pretreatment of the daily addition of 50 mm NaCl up to 200 mm. G, Root and shoot lengths of salinity-stressed wheat seedlings. H and I, RT-PCR analysis indicated that the TaOPR1 transgene was successfully transcribed in both wheat and Arabidopsis. Wheat Actin and Arabidopsis Tubulin genes were used as internal references. J and K, Arabidopsis seedling response to a 12-d exposure to salinity stress. AtOE1 and AtOE2, Transgenic lines heterologously expressing TaOPR1; VC, transgenic line carrying an empty pSTART vector. L, Root length of salinity-stressed Arabidopsis seedlings. M, Germination rate of Arabidopsis. Data are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line under the control or in each treatment using Student’s t test analysis (P < 0.05). Bars = 1 cm.
When the gene was heterologously expressed in Arabidopsis, there was no obvious phenotypic difference between the two highest expressors (AtOE1 and AtOE2) and the empty vector control (VC), either when the plants were plated on Murashige and Skoog (MS) agar (Fig. 3, J and L) or when the seed was sown in soil (data not shown). However, in the presence of 100 or 125 mm NaCl, the Arabidopsis overexpressor (AtOE) lines were more vigorous than the VC line, forming larger leaves and longer primary roots (57% longer at 100 mm NaCl and 33% longer at 125 mm NaCl; Fig. 3, K and L). The germination rate also responded positively: in the absence of salinity, the AtOE and VC lines behaved indistinguishably, but in the presence of salinity, the AtOE lines were superior (Fig. 3M). Inconsistent with the positive effect of TaOPR1, the double RNA interference (RNAi) knockdown mutant opr1opr2, from two homologous OPRI genes in Arabidopsis (Beynon et al., 2009), did not increase the sensitivity to NaCl treatment, and their phenotypes were comparable to the wild type (Supplemental Fig. S1, A and C).
TaOPR1 Reduced the Sensitivity to H2O2 in Arabidopsis
Tolerance to salinity stress has frequently been associated with tolerance to oxidative stress. Specifically, TaOPR1 overexpressor lines exhibited superior growth status in comparison with the VC line in the presence of 1.0 and 1.5 mm H2O2; after 10 d of exposure to H2O2, the primary roots of overexpressor lines were significantly longer than those of the VC line (Fig. 4, A and B), indicating that TaOPR1 overexpression reduced sensitivity to H2O2. Additionally, similar results were also obtained when the plants were supplied with higher levels (2, 3, and 5 mm) of H2O2 for 3 weeks (data not shown). Unlike TaOPR1-overexpressing lines, the sensitivity to H2O2 was not altered in Arabidopsis opr1opr2 mutants (Supplemental Fig. S1, A and B).
Figure 4.
TaOPR1 confers H2O2 tolerance by promoting ROS-scavenging capacity. A, Phenotypes of seedlings exposed to H2O2 for 10 d. AtOE1 and AtOE2, Transgenic lines heterologously expressing TaOPR1; VC, transgenic line carrying an empty pSTART vector. B, Root length of Arabidopsis. C, POD and CAT activities of Arabidopsis. D, MDA content of Arabidopsis. E, POD and CAT activities of wheat. WT, Wild type. F, Relative transcription levels of genes encoding ROS-scavenging enzymes in Arabidopsis (real-time PCR data). G, ROS levels of Arabidopsis by DAB staining. For C, D, F, and G, 2-week-old Arabidopsis seedlings cultured on medium plates were used; for E, three-leaf-stage wheat seedling leaves harvested from plants raised in one-half-strength Hoagland liquid medium were sampled. Data are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line under the control or in each treatment using Student’s t test analysis (P < 0.05). Bars = 1 cm. [See online article for color version of this figure.]
The tolerance to oxidative stress closely links to the homeostasis between ROS scavenging and production, so we further measured the transcription of some ROS scavengers and producers as well as the activities of their products. In the absence of stress, the activities of peroxidase (POD) and catalase (CAT) in the AtOE lines were 20% and 40%, respectively, above that in the VC line (Fig. 4C), and these two ROS-scavenging enzymes were similarly more active in the TaOE lines than in the wild type (Fig. 4E). The content of malondialdehyde (MDA), an indicator of intracellular ROS damage, in the AtOE seedlings was 20% less than in the VC ones (Fig. 4D). The real-time PCR analysis confirmed that the transcription of AtCAT1, AtCAT2, AtAPX1 (for ascorbate peroxidase1), and AtAPX2 was about 5-, 2-, 1.5-, and 5-fold higher, respectively, in the AtOE lines than in VC (Fig. 4F). The gene encoding AtZAT10, a protein that enhances the expression of AtAPX2 (Mittler et al., 2006), was also up-regulated (Fig. 4F). The transcription of AtSOD and AtGPX, encoding superoxidase dismutase and glutathione peroxidase, respectively, behaved similarly between AtOE and VC lines (Supplemental Fig. S2A). ROS is majorly produced by the catalysis of NADPH oxidase. AtRBOHC, AtRBOHD, and AtRBOHF, encoding three subunits of NADPH oxidase, were unaffected by the presence of TaOPR1 (Supplemental Fig. S2A). The ROS levels of the overexpressor and VC lines were visualized by 3,3′-diaminobenzidine (DAB) staining, and the staining signals in 3-week-old seedlings of two overexpressor lines were distinguishably weaker than those in the VC line (Fig. 4G). These results demonstrate that TaOPR1 can reduce intracellular levels of ROS by promoting the capacity of its removal.
TaOPR1 Heightened ABA Sensitivity in Arabidopsis
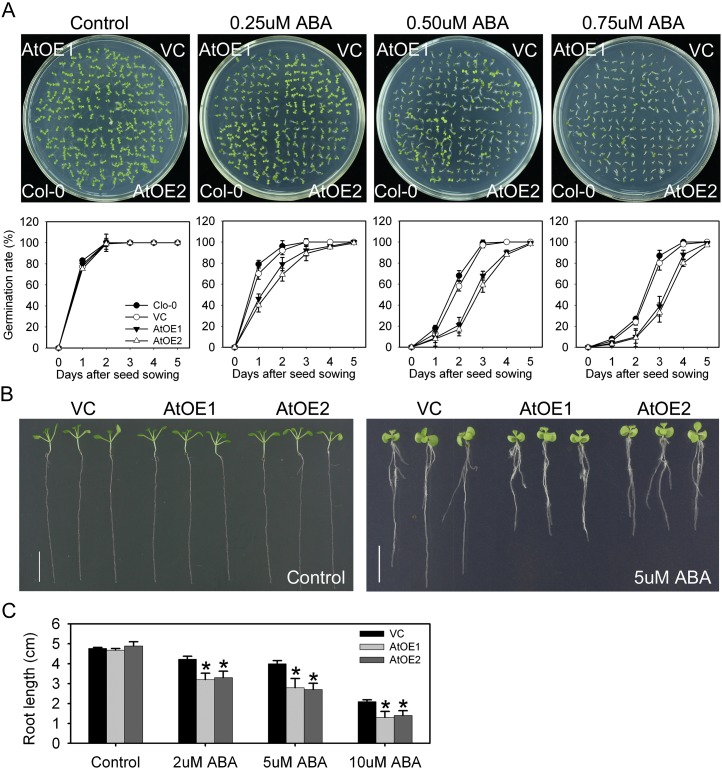
The AtOE lines were highly sensitive to the provision of exogenous ABA. In the absence of exogenous ABA, the germination rate of the AtOE and VC lines was indistinguishable; however, in the presence of 0.25 µm ABA, the germination rate of the AtOE lines was only 40% after 1 d and reached 90% after 3 d, whereas for both VC and wild-type Col-0, the equivalent proportions were nearly 80% and almost 100% (Fig. 5A). The effect of the higher levels of ABA (0.5 and 0.75 µm) was to suppress the germination over the first 4 d of the AtOE lines more severely than in either the VC or the wild-type line (Fig. 5A). By the second day, approximately 60% of Col-0 and VC seeds had germinated in the presence 0.5 µm ABA and approximately 25% had germinated in the presence of 0.75 µm ABA; at the same time point, respectively, just approximately 20% and approximately 10% of the AtOE seed had germinated. The germination rates of Col-0, VC, and AtOE all achieved approximately 100% germination by 5 d (Fig. 5A). The proportion of AtOE seedlings bearing green cotyledons was markedly lower than in VC and Col-0 seedlings (Fig. 5A). Under nonstressed conditions, both the leaf and root growth of the AtOE lines were similar to those of the VC line. In the presence of 2 µm ABA, in comparison with the VC line, root growth in the AtOE lines was reduced by approximately 24%, and higher concentrations (5 and 10 µm) reduced it by more than 30% (Fig. 5, B and C).
Figure 5.
TaOPR1 increases the ABA sensitivity of Arabidopsis. A, Delayed germination in the presence of ABA treatment. AtOE1 and AtOE2, Transgenic lines heterologously expressing TaOPR1; VC, transgenic line carrying an empty pSTART vector. B, TaOPR1 suppresses the growth of seedlings exposed to ABA for 10 d. C, Root lengths of seedlings shown in B. Data are presented as means ± sd, and columns marked with asterisks indicate significant differences from the VC line under the control or in each treatment using Student’s t test analysis (P < 0.05). [See online article for color version of this figure.]
TaOPR1 Stimulated the ABA-Dependent Stress-Responsive Pathway
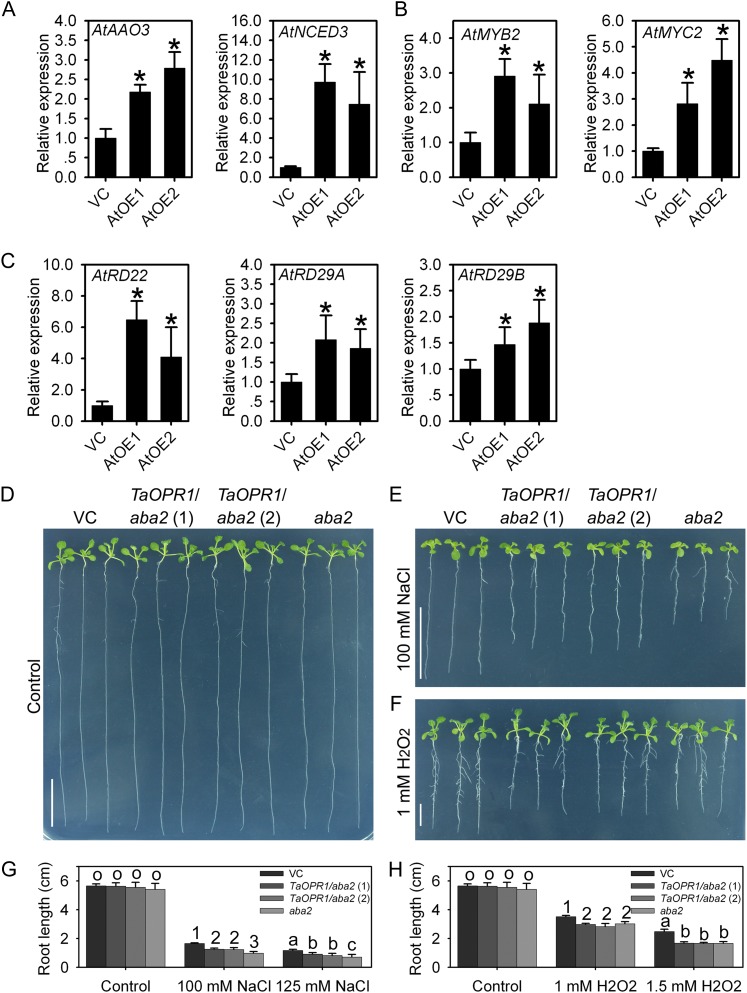
When the expression levels of a set of well-known stress response genes were monitored, AtRD22, AtRD29A, and AtRD29B proved to have been substantially up-regulated in the AtOE lines, as were the two upstream transcription factors AtMYB2 and AtMYC2 (Fig. 6, B and C). Since these genes are all key components of the ABA-dependent stress-responsive signaling pathway in Arabidopsis, the transcript abundance of AtNCED3 and AtAAO3 was monitored and shown to be up-regulated as well (Fig. 6A). In contrast, among stress-responsive genes acting independently of ABA, all known components had no obviously differential expression in the AtOE lines (Supplemental Fig. S2, B and C).
Figure 6.
TaOPR1 accounts for salinity tolerance by promoting the ABA signaling pathway and decreases H2O2 sensitivity in an ABA-dependent manner. A to C, Relative transcription levels in 2-week-old seedlings of genes involved in ABA synthesis and the ABA-dependent stress response pathway (real-time PCR data). D to F, Phenotypes after exposure to exogenous NaCl for 12 d and to H2O2 for 10 d. TaOPR1/aba2 (1) and TaOPR1/aba2 (2), aba2 mutant lines heterologously expressing TaOPR1; VC, transgenic line carrying an empty pSTART vector. G and H, Root lengths of seedlings shown in D to F. Data are presented as means ± sd. In A to C, columns marked with asterisks indicate significant differences from the VC line using Student’s t test analysis (P < 0.05); in G and H, bars in each column marked with different letters or numbers indicate significantly different means using the one-way ANOVA lsd analysis (P < 0.05). [See online article for color version of this figure.]
To further confirm the role of ABA in TaOPR1-enhanced salinity stress, TaOPR1 was transformed into the Arabidopsis aba2 mutant, which is defective in ABA synthesis, to construct TaOPR1/aba2 lines. TaOPR1/aba2, aba2, and VC seedlings appeared comparable with each other under nonstress conditions (Fig. 6D). Salinity stress seriously restricted the growth of aba2 seedlings, and in comparison with the VC line, the leaves were much smaller and root lengths were shorter by more than 50% under two levels of NaCl treatment (Fig. 6, E and G). TaOPR1 overexpression obviously rescued the phenotype of aba2 when challenged with NaCl treatments: the root lengths of TaOPR1/aba2 lines were about 70% and 60% of those of the VC line, and the leaf sizes between TaOPR1/aba2 and VC lines had no significant difference (Fig. 6, E and G). However, TaOPR1 did not alleviate the sensitivity of aba2 to H2O2: the root lengths of TaOPR1/aba2 and aba2 seedlings were similar, which was significantly shorter than the VC line (Fig. 6, F and H).
TaOPR1 Did Not Disturb JA Synthesis and Signaling
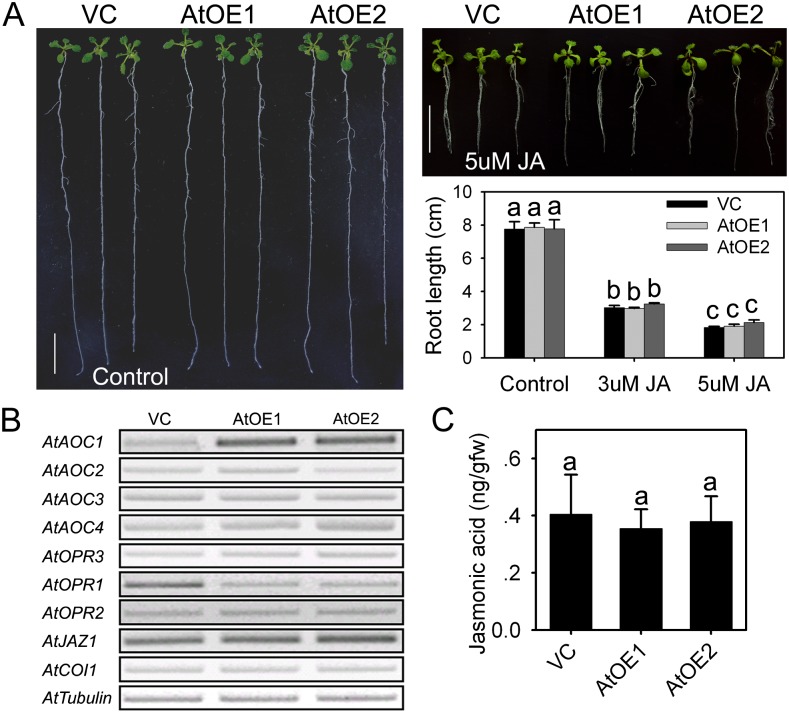
The effect of heterologous TaOPR1 expression on the response to exogenously supplied JA and on JA synthesis and signaling genes was then investigated. The growth of both the AtOE and VC root and leaf was restricted by the presence of either 3 or 5 µm JA (Fig. 7A). Among genes encoding enzymes involved in JA synthesis, AtAOC1 was clearly up-regulated in the AtOE lines, but AtAOC2, AtAOC3, and AtAOC4 were not differentially transcribed; AtOPR3 expression was not influenced by the heterologous expression of TaOPR1 (Fig. 7B). Following the transcription of JA synthesis genes, JA levels of AtOE lines were not altered in comparison with the VC line (Fig. 7C). Moreover, no transcriptional difference was found in either AtCOI1 or AtJAZ1 (both of which are components of the JA signaling pathway; Fig. 7B). Within two OPRI genes, AtOPR1 but not AtOPR2 was reduced in the AtOE lines (Fig. 7B).
Figure 7.
TaOPR1 in Arabidopsis influences neither JA synthesis nor the JA signaling pathway. A, Seedling phenotypes and root length comparison following JA treatment for 16 d. AtOE1 and AtOE2, Transgenic lines heterologously expressing TaOPR1; VC, transgenic line carrying an empty pSTART vector. B, Transcription of genes involved in JA synthesis and signaling pathways in 2-week-old seedlings (RT-PCR data). C, JA levels (ng g−1 fresh weight) of 2-week-old seedlings detecting by LC-MS/MS. Data are presented as means ± sd, and bars marked with different letters indicate significantly different means using the one-way ANOVA lsd analysis (P < 0.05). [See online article for color version of this figure.]
DISCUSSION
TaOPR1 Confers Salinity Tolerance by Enhancing Antioxidation Capacity
Salinity stress is associated with the production of ROS, so a component of tolerance is represented by the plant’s ability to cope with higher levels of these harmful molecules. Excess ROS in the cell causes extensive lipid peroxidation, which generates a range of toxic breakdown products, such as the α,β-unsaturated aldehydes (Esterbauer, 1993; Trotter et al., 2006). The antioxidation property of the OPRIs is thought to operate via the reduction of double bonds in these products (Esterbauer, 1993; Trotter et al., 2006). For example, AtOPR1 can efficiently reduce the double bond of trinitrotoluene to form reduced trinitrotoluene derivatives, both in vitro and in vivo (Beynon et al., 2009). The reactive aldehyde MDA has been identified to reflect the extent of ROS-induced lipid peroxidation (Nankivell et al., 1994), so the decrease in MDA contents in AtOE lines (Fig. 4D) indicates that TaOPR1 certainly alleviates ROS damage. However, TaOPR1 overexpression in the aba2 genetic background did not reduce the sensitivity to H2O2 (Fig. 6, F and H), suggesting that the role of TaOPR1 in ROS damage alleviation is not achieved through its direct reduction ability on the reactive products of lipid peroxidation.
In addition to counteracting the toxicity of ROS-induced lipid peroxidation, the direct neutralization of ROS has been proposed as a component of stress tolerance (Mittler, 2002). The OYZ family is believed to protect the cell against the damaging effects of lipid peroxidation products, and the function of OYE in yeast also appears to be to reduce the level of ROS present (Fitzpatrick et al., 2003). However, yeast strains deficient for OYE2 are hypersensitive to acrolein, a ubiquitous reactive aldehyde formed from lipid peroxidation, but show little sensitivity to exogenous H2O2 (Trotter et al., 2006). This demonstrates a diverse role for OYE family proteins in the response to direct oxidative stress. Here, TaOPR1 overexpression elevated the activities of several ROS-scavenging enzymes and the transcription of their encoding genes (Fig. 4, C–F). AtZAT10, which has been shown to promote the transcription of AtAPX2 (Mittler et al., 2006), was also up-regulated in the AtOE lines (Fig. 4F). Thus, there is some evidence that TaOPR1 promotes the efficiency of ROS scavenging to affect ROS removal rather than protects cells from ROS-induced lipid peroxidation. ABA enhances the transcription and activity of ROS network genes, and defects in this network can also disrupt the expression of ABA and stress-responsive genes (Miao et al., 2006). Along with the fact that TaOPR1/aba2 did not rescue the sensitivity of aba2 to H2O2 (Fig. 6, F and H), it could be concluded that the enhancement of ROS scavenging promoted by TaOPR1 expression may be, at least in part, mediated by an acceleration of ABA synthesis and an up-regulation of relevant signaling pathways.
The Interplay between TaOPR1 and the ABA-Mediated Salinity-Responsive Pathway
ABA is a vital component of the abiotic stress response, and a boost in ABA synthesis and/or a limitation to its degradation is a frequently observed plant response to abiotic stress (Jakab et al., 2005). ABA is often recruited as the primary signal activating the transcription of stress-responsive genes (Lee and Luan, 2012). TaOPR1 is induced by both salinity and ABA, while ABA synthesis inhibition largely counteracted the salinity-induced accumulation of TaOPR1 transcript (Fig. 1, B and C). Therefore, there is a reasonably strong case that the induction of TaOPR1 expression operates via an increase in the rate of ABA synthesis. ABA triggers a signaling cascade that up-regulates a suite of abiotic stress-responsive genes (Nakashima et al., 2009). The Arabidopsis genes AtRD22, AtRD29A, and AtRD29B are all up-regulated via an ABA-dependent pathway (Fujita et al., 2011). AtRD22 is known to be regulated by the transcription factors AtMYB2 and AtMYC2 (Abe et al., 2003). The enhancement of ABA sensitivity (Fig. 5), mirroring the effect of AtMYB2 and AtMYC2 overexpression (Abe et al., 2003), and the transcription elevation of these genes in AtOE lines (Fig. 6, B and C) provide firm evidence for a positive role of TaOPR1 in stress-responsive ABA signaling.
TaOPR1 expression also markedly increased the transcript abundance of AtNCED3 and AtAAO3 (Fig. 6A), which encode proteins responsible for the catalysis of two key steps in the ABA synthesis pathway. The suggestion is that an acceleration in the ABA-dependent pathway caused by TaOPR1 expression is initiated from a burst in ABA synthesis. Given the partially rescued phenotype of aba2 under salinity stress (Fig. 6, E and G), it is reasonable to claim that TaOPR1 regulates ABA synthesis and ABA signaling pathways separately. Thus, we speculate that, in addition to the well-documented cross talk between OPRII-generated JA and ABA (Kazan and Manners, 2011), OPRIs can also interact with ABA-mediated pathways.
TaOPR1 Does Not Disturb JA Synthesis and Signaling Machinery
Although the in planta OPRI substrates have yet to be identified, they are known to be able to reduce, to a moderate extent at least, trans-(+)-OPDA, a stereoisomer converted from cis-(+)-OPDA, the intermediate precursor of JA (Mueller and Brodschelm, 1994). The AOCs catalyze the formation of cis-(+)-OPDA from 12,13-epoxyoctadecatrienoic acid, a reaction regarded as important in both octadecanoid and JA synthesis (Mueller and Brodschelm, 1994). Of the four AOC genes present in Arabidopsis, AtAOC2 is the most effective in the formation of cis-(+)-OPDA, and this copy also shows the strongest response to wounding (Stenzel et al., 2003). Neither the transcript of JA synthesis genes nor the level of endogenous JA was dependent on TaOPR1 expression (Fig. 7, B and C). These observations indicate that JA synthesis is out of TaOPR1, a conclusion supported by the lack of any differential transcription of the JA receptor gene AtCOI1 or of JAZ1 (Fig. 7B). AtMYC2 is activated by JA, which allows it to be a pivotal mediator in both the ABA and JA signaling pathways (Katsir et al., 2008; Yan et al., 2009). The behavior of the AtOE lines allows the conclusion to be drawn that the induction of AtMYC2 was due to a promotion of the ABA but not of the JA signaling pathway and, hence, that TaOPR1 exerts no regulatory activity over the JA synthesis and signaling machinery.
AtAOC1 was induced in the AtOE lines (Fig. 7B), which appears inconsistent with the lack of interaction with JA synthesis and the JA signaling pathway. The distinct functions of the various AOCs are not as yet well defined, because no clear correlation has been established between their transcription and the synthesis of JA (Kramell et al., 2000; Miersch and Wasternack, 2000; Ziegler et al., 2001; Stenzel et al., 2003; Delker et al., 2006). A possible scenario is that the enolization that allows the slow conversion of cis- to trans-OPDA isomers is catalyzed by AtAOC1. If the heterologous expression of TaOPR1 utilizes more trans-(+)-OPDA, this could provide a positive feedback loop to promote the expression of AtAOC1. AOCs are known to differ in their substrate specificity, either arising from α-linolenic acid and leading to the synthesis of OPDA or from hexadecatrienoic acid and leading to the synthesis of dinor-OPDA (Weber et al., 1997; Reymond et al., 2000; Stenzel et al., 2003; Delker et al., 2006). Whether the OPRIs, along with AOC1, participate in the synthesis and/or metabolism of other lipid fatty acid derivatives is a question that deserves some attention.
The AtOE lines accumulated less AtOPR1 transcript than the VC line, while the transcript abundance of AtOPR2 was independent of TaOPR1 expression (Fig. 7B). AtOPR1 is up-regulated during leaf senescence by some 2-fold, but AtOPR2 appears to be constitutively expressed during the course of leaf development (He et al., 2002). Thus, although these two OPRI gene sequences are highly similar to one another and are both predominantly expressed in the root (Biesgen and Weiler, 1999), their regulation, if not their function as well, appears to have diverged. The repression of AtOPR1 by the TaOPR1 transgene suggests a negative feedback regulatory effect, perhaps based on functional redundancy between TaOPR1 and AtOPR1.
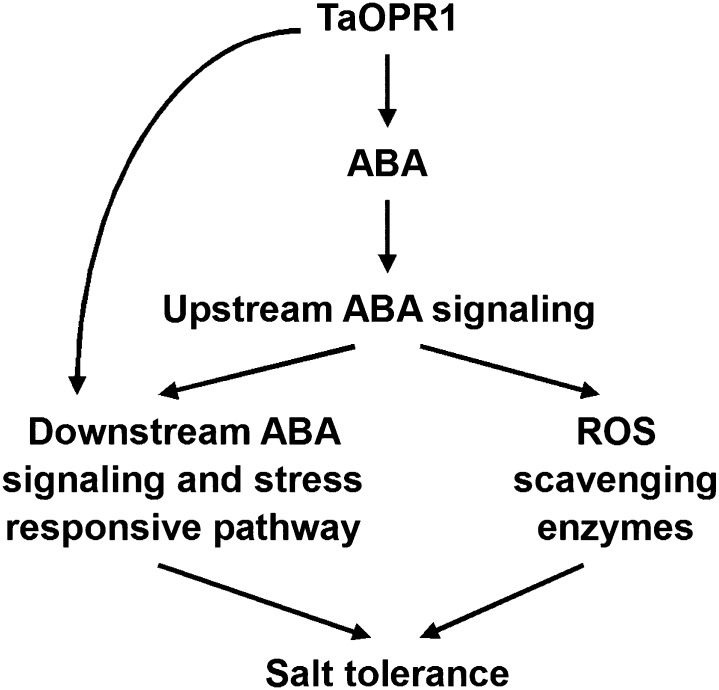
Our overall proposition is that TaOPR1 expression is induced by a boost in ABA synthesis prompted by abiotic stress and that this, in turn, promotes ABA synthesis. TaOPR1 promotes ABA synthesis and signaling pathways, which elevates the level of activity within the ABA-dependent abiotic stress-responsive signaling pathway and increases the level of ROS-scavenging activity, thereby explaining the positive effect of TaOPR1 on the salinity (and other abiotic stress) tolerance of Arabidopsis; besides, TaOPR1 can also accelerate the ABA-dependent abiotic stress-responsive signaling pathway in ABA synthesis-independent behavior (Fig. 8). The function of TaOPR1 may lie in the metabolism of trans-(+)-OPDA, with consequent effects on the workings of the (ABA-dependent-responsive and/or ROS) signaling pathway that this molecule mediates. At the same time, since the slow conversion to trans-(+)-OPDA would likely not sufficiently alter the size of the cis-(+)-OPDA pool, the presence of TaOPR1 does not induce any change in JA synthesis or in the JA signaling pathway. We have assembled a body of evidence to show that the products of various OPDA isomers catalyzed by the two OPR subgroups can cross talk with ABA, while there is little sign of any interplay within the two subgroups. How TaOPR1 performs these roles, especially whether TaOPR1 operates via the catalysis of trans-(+)-OPDA, has yet to be elucidated.
Figure 8.
Hypothetical model of TaOPR1 action in Arabidopsis. TaOPR1 promotes ABA synthesis and signaling pathways, which elevates the activity occurring within the ABA-dependent abiotic stress-responsive pathway and increases the level of ROS-scavenging activity, thereby accounting for salinity tolerance. TaOPR1 can also directly improve downstream ABA signaling and the ABA-dependent stress-responsive pathway in an ABA synthesis-independent manner.
MATERIALS AND METHODS
Isolation of the TaOPR1 Full-Length cDNA and Characterization of Its Sequence
The fragment of the OPRI homolog identified via microarray analysis was used as a BLASTN query against the wheat (Triticum aestivum) EST database held at the National Center for Biotechnology Information. All matching ESTs were assembled using CAP3 software, and a pair of gene-specific primers designed from this assembly (Supplemental Table S1) was used to amplify a full-length cDNA from a cv SR3 cDNA library. The PCR consisted of a 3-min denaturation at 94°C, followed by 35 cycles of 94°C/30 s, 59°C/30 s, and 72°C/90 s, with a final extension of 72°C/5 min. The amplicon was inserted into the pMD18-T vector (Takara), following the supplier’s instructions, and submitted for sequencing. The sequence, designated TaOPR1, was used to construct a phylogeny using a neighbor-joining method implemented within ClustalX and MEGA5 software (Kumar et al., 2004; Larkin et al., 2007), and its predicted peptide product TaOPR1 was aligned with other OPRIs using ClustalX software (Larkin et al., 2007).
Plant Materials and Treatments
Seedlings of cv SR3 and of its parental wheat cultivar JN177 were raised to the three-leaf stage in one-half-strength Hoagland liquid medium under a 16-h photoperiod at 22°C, after which they were transferred for up to 48 h to the same medium containing 200 mm NaCl, 20% (w/v) PEG6000, 10 mm H2O2, 100 mm ABA, or 100 mm ABA/100 mm norflurazon/200 mm NaCl. A pCam23A::TaOPR1 construct driven by the ubiquitin promoter was introduced into the salinity-sensitive wheat ‘JN17’ using the shoot apical meristem method described by Zhao et al. (2006). Ten-day-old seedlings of both transgenic and wild-type plants were raised in one-half-strength Hoagland liquid medium with or without NaCl under a 16-h photoperiod at 22°C. The NaCl treatment involved the daily addition of 50 mm NaCl until the concentration had reached 200 mm. The seedlings were held in this solution for a further 4 d. All measurements were carried out with three repeats.
Transgenic forms of Arabidopsis (Arabidopsis thaliana) Col-0 were made by introducing a pSTART::TaOPR1 construct driven by the cauliflower mosaic virus 35S promoter (or just an empty vector) using the floral dip method (Clough and Bent, 1998). Surface-sterilized seeds were plated on one-half-strength MS agar medium, kept in the dark at 4°C for 2 d to break dormancy, and subsequently transferred to a 16-h photoperiod at 22°C for 2 d. They were then replated on one-half-strength MS agar medium supplemented with 0, 100, or 125 mm NaCl for 12 d, with 0, 1.0, or 1.5 mm H2O2 for 10 d, with 0, 2, 5, or 10 μm ABA for 10 d, or with 0, 3, or 5 μm JA for 16 d and held under a 16-h photoperiod at 22°C. A germination assay was conducted by plating surface-sterilized seeds on one-half-strength MS agar medium containing 0, 125, or 175 mm NaCl as well as 0, 0.25, 0.5, or 0.75 μm ABA. After holding for 48 h at 4°C in the dark, the plates were transferred to a 16-h photoperiod at 22°C for 5 d. The emergence of the radicle was taken as representing a successfully germinated seed. Germination rates were expressed as the proportion of seeds that had successfully germinated. The experiment was replicated three times.
Reverse Transcription-PCR and Real-Time PCR Analyses
Total RNA was extracted from both wheat roots and Arabidopsis seedlings using the Trizol Reagent (Invitrogen) and treated with DNase I. The first cDNA strand synthesized using a Moloney murine leukemia virus reverse transcription (RT) system kit (Invitrogen) according to the manufacturer’s instructions was used as the template for 20-μL RT-PCRs containing 1× Easy Taq buffer (Transgene), 1 unit of EasyTaq (Transgene), 100 μm deoxyribonucleotide triphosphates, 0.5 μm forward and 0.5 μm reverse primers (sequences given in Supplemental Table S1), and 1 μL of a 1:10 dilution of the cDNA. The PCR regime consisted of a 5-min denaturation at 94°C, followed by 28 cycles of 94°C/30 s, 55°C/30 s, and 72°C/30 s, completed by an extension step of 10 min at 72°C. The 20-μL real-time PCRs contained 10 μL of 2× SYBR Premix Ex Taq mix (Takara), 0.2 μm forward and 0.2 μm reverse primers (sequences given in Supplemental Table S1), and 1 μL of a 1:10 dilution of the cDNA first strand, and the cycling regime was composed of a denaturation step of 95°C/2 min, followed by 45 cycles of 95°C/10 s, 60°C/20 s, and 72°C/20 s. A melting-curve analysis was performed over the range 80°C to 95°C at 0.5°C intervals. Relative gene expression levels were detected using the comparative threshold cycle method2-ΔΔCT (Livak and Schmittgen, 2001). A positive control was provided by a parallel analysis based on a fragment of the wheat Actin gene (AB181991) or the Arabidopsis Tubulin gene (AT1G04820), with three independent replicates per experiment. The specificity of the real-time PCR was confirmed by agarose gel electrophoresis of the amplicon. The analysis was confirmed three times.
Chromosomal Location of TaOPR1
The chromosomal location of TaOPR1 was obtained via a PCR strategy based on the amplification of genomic DNA extracted from a set of wheat cultivar Chinese Spring nullitetrasomic lines (Sears, 1954) and a partial set of ditelocentric lines (Sears and Sears, 1978). Each 20-μL PCR contained 1× EasyTaq buffer, 0.2 μm deoxyribonucleotide triphosphates, 0.5 μm TaOPR1-specific primers (Supplemental Table S1), 0.5 unit of Easy Taq, and 100 ng of DNA, and the amplification regime consisted of an initial denaturation of 94°C/5 min, followed by 35 cycles of 94°C/40 s, 60°C/40 s, and 72°C/50 s, ending with a final extension of 72°C/10 min. The resulting amplicons were separated by agarose electrophoresis.
Subcellular Localization of TaOPR1
The TaOPR1 sequence, lacking its stop codon, was ligated into the XbaI and BamHI cloning sites of the 326-GFP vector. Either this recombined plasmid or the empty 326-GFP was transformed into Arabidopsis mesophyll protoplasts following the method described by Yoo et al. (2007). After a 16-h incubation at 22°C in the dark, GFP signal was detected by bright-field and fluorescence microscopy.
Physiological Characterization
Three-leaf-stage wheat seedling leaves harvested from plants raised in one-half-strength Hoagland liquid medium and whole 2-week-old Arabidopsis seedlings were analyzed for the activity of the ROS-scavenging enzymes superoxide dismutase, CAT, and POD, following Sequeira and Mineo (1966), and for MDA content as described by Heath and Packer (1968). The experiment was performed three times.
Measurement of ROS and JA Levels
ROS level was visualized by DAB staining according to Asselbergh et al. (2007). Briefly, 3-week-old Arabidopsis seedlings were placed under light conditions and floated in a solution of 1 mg mL−1 DAB-HCl (pH 4) at 28°C for 8 h, and then chlorophyll was removed with 95% ethanol in a boiling bath. JA content was measured by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as follows. A total of 1,000 mg of fresh 3-week-old Arabidopsis seedlings was frozen in liquid nitrogen and well ground with a small glass pestle in a 2-mL vial. Following the addition of 1,000 μL of methanol, homogenates were well mixed in an ultrasonic bath and then kept at 4°C overnight. After being centrifuged at 15,000g for 10 min, the supernatant was collected and then vacuumed to dryness in a Jouan RCT60 concentrator. Dried extract was dissolved in 200 μL of sodium phosphate solution (0.1 mol L−1, pH 7.8) and later passed through a Sep-Pak C18 cartridge (Waters). JA was eluted with 1,500 μL of 50% methanol and vacuumed to dryness again, redissolved in 50 μL of 20% acetonitrile, and injected (5 μL) into a LC20AD-MS8030 LC-MS/MS system (Shimadzu); a BEH C18 (100 × 2.1 mm, 1.7 μm) column (Waters) was used, column temperature was 30°C, the mobile phase comprised 55% acetonitrile and 45% water (v/v), and the flow rate was 0.30 mL min−1. Mass spectrography was operated in the negative mode under the following conditions: ionization, electrospray; capillary voltage, 3.5 kV; collision energy, 15 eV; desolvation temperature, 250°C; mass-to-charge ratio, 209.1/59.
The accession number of TaOPR1 in GenBank is JQ409278.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. opr1opr2 double RNAi knockdown mutation does not change the response of Arabidopsis to NaCl and H2O2.
Supplemental Figure S2. The transcription of a set of well-established abiotic stress-responsive genes, based on RT-PCR.
Supplemental Table S1. PCR primer sequences used for this research.
Acknowledgments
We thank Dr. Neil C. Bruce at the University of York for kindly providing Arabidopsis opr1opr2 RNAi knockdown seeds.
Glossary
- ROS
reactive oxygen species
- OYE
Old Yellow Enzyme
- OPDA
12-oxo-phytodienoic acid
- JA
jasmonic acid
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- PEG6000
polyethylene glycol 6000
- Col-0
ecotype Columbia
- TaCE
transgenic overexpression
- MS
Murashige and Skoog
- VC
empty vector control
- RNAi
RNA interference
- AtOE
Arabidopsis overexpressor
- MDA
malondialdehyde
- DAB
3,3′-diaminobenzidine
- RT
reverse transcription
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- cDNA
complementary DNA
- TaOE
wheat overexpressor
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Jwa NS, Shibato J, Han O, Iwahashi H, Rakwal R. (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochem Biophys Res Commun 310: 1073–1082 [DOI] [PubMed] [Google Scholar]
- Asselbergh B, Curvers K, Franca SC, Audenaert K, Vuylsteke M, Van Breusegem F, Höfte M. (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144: 1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon ER, Symons ZC, Jackson RG, Lorenz A, Rylott EL, Bruce NC. (2009) The role of oxophytodienoate reductases in the detoxification of the explosive 2,4,6-trinitrotoluene by Arabidopsis. Plant Physiol 151: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW. (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208: 155–165 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C. (2006) Jasmonate biosynthesis in Arabidopsis thaliana: enzymes, products, regulation. Plant Biol (Stuttg) 8: 297–306 [DOI] [PubMed] [Google Scholar]
- Esterbauer H. (1993) Cytotoxicity and genotoxicity of lipid-oxidation products. Am J Clin Nutr (Suppl) 57: 779S–785S, discussion 785S–786S [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Macheroux P. (2003) Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J Biol Chem 278: 19891–19897 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Aoki M, Kato A, Kondo M, Nishimura M. (1996) Transport of chimeric proteins that contain a carboxy-terminal targeting signal into plant microbodies. Plant J 10: 225–234 [DOI] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L. (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Métraux J-P, Mauch-Mani B. (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol 139: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2011) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kramell R, Miersch O, Atzorn R, Parthier B, Wasternack C. (2000) Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves: implications for different signaling pathways. Plant Physiol 123: 177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Laudert D, Hennig P, Stelmach BA, Müller A, Andert L, Weiler EW. (1997) Analysis of 12-oxo-phytodienoic acid enantiomers in biological samples by capillary gas chromatography-mass spectrometry using cyclodextrin stationary phases. Anal Biochem 246: 211–217 [DOI] [PubMed] [Google Scholar]
- Lee SC, Luan S. (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35: 53–60 [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Wang M, Xia GM. (2012) A transcriptomic analysis reveals the nature of salinity tolerance of a wheat introgression line. Plant Mol Biol 78: 159–169 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O, Wasternack C. (2000) Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: endogenous jasmonates do not induce jasmonate biosynthesis. Biol Chem 381: 715–722 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mueller MJ, Brodschelm W. (1994) Quantification of jasmonic acid by capillary gas chromatography-negative chemical ionization-mass spectrometry. Anal Biochem 218: 425–435 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nankivell BJ, Chen J, Boadle RA, Harris DC. (1994) The role of tubular iron accumulation in the remnant kidney. J Am Soc Nephrol 4: 1598–1607 [DOI] [PubMed] [Google Scholar]
- Odat O, Matta S, Khalil H, Kampranis SC, Pfau R, Tsichlis PN, Makris AM. (2007) Old yellow enzymes, highly homologous FMN oxidoreductases with modulating roles in oxidative stress and programmed cell death in yeast. J Biol Chem 282: 36010–36023 [DOI] [PubMed] [Google Scholar]
- Parchmann S, Gundlach H, Mueller MJ. (1997) Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol 115: 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Wang M, Li F, Lv H, Li C, Xia G. (2009) A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol Cell Proteomics 8: 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Biesgen C, Müssig C, Altmann T, Weiler EW. (2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210: 979–984 [DOI] [PubMed] [Google Scholar]
- Schaller F, Hennig P, Weiler EW. (1998) 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol 118: 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Weiler EW. (1997) Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana: structural and functional relationship to yeast old yellow enzyme. J Biol Chem 272: 28066–28072 [DOI] [PubMed] [Google Scholar]
- Sears ER. (1954) The aneuploids of common wheat. Mo Agric Exp Sta Res Bull 572: 1–58 [Google Scholar]
- Sears ER, Sears LMS (1978) The Telocentric Chromosomes of Common Wheat. Indian Agricultural Research Institute, New Delhi, India [Google Scholar]
- Sequeira L, Mineo L. (1966) Partial purification and kinetics of indoleacetic acid oxidase from tobacco roots. Plant Physiol 41: 1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW. (1998) Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry 47: 539–546 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C. (2003) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato: amplification in wound signalling. Plant J 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner J, Fürholz A, Macheroux P, Amrhein N, Schaller A. (1999) A homolog of old yellow enzyme in tomato: spectral properties and substrate specificity of the recombinant protein. J Biol Chem 274: 35067–35073 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, Schaller A. (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Trotter EW, Collinson EJ, Dawes IW, Grant CM. (2006) Old yellow enzymes protect against acrolein toxicity in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 72: 4885–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Peng ZY, Li CL, Li F, Liu C, Xia GM. (2008) Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8: 1470–1489 [DOI] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE. (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94: 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F. (1994) The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett 345: 9–13 [DOI] [PubMed] [Google Scholar]
- Williams RE, Bruce NC. (2002) “New uses for an old enzyme”: the Old Yellow Enzyme family of flavoenzymes. Microbiology 148: 1607–1614 [DOI] [PubMed] [Google Scholar]
- Xia GM. (2009) Progress of chromosome engineering mediated by asymmetric somatic hybridization. J Genet Genomics 36: 547–556 [DOI] [PubMed] [Google Scholar]
- Xia GM, Xiang FN, Zhou AF, Wang H, Chen HM. (2003) Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor Appl Genet 107: 299–305 [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao TJ, Zhao SY, Chen HM, Zhao QZ, Hu ZM, Hou BK, Xia GM. (2006) Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep 25: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Ziegler J, Keinänen M, Baldwin IT. (2001) Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58: 729–738 [DOI] [PubMed] [Google Scholar]