Smooth petal elongation requires the WS/DGAT family gene FOP1 in Arabidopsis.
Abstract
Flowering plants bear beautiful flowers to attract pollinators. Petals are the most variable organs in flowering plants, with their color, fragrance, and shape. In Arabidopsis (Arabidopsis thaliana), petal primordia arise at a similar time to stamen primordia and elongate at later stages through the narrow space between anthers and sepals. Although many of the genes involved in regulating petal identity and primordia growth are known, the molecular mechanism for the later elongation process remains unknown. We found a mutant, folded petals1 (fop1), in which normal petal development is inhibited during their growth through the narrow space between sepals and anthers, resulting in formation of folded petals at maturation. During elongation, the fop1 petals contact the sepal surface at several sites. The conical-shaped petal epidermal cells are flattened in the fop1 mutant, as if they had been pressed from the top. Surgical or genetic removal of sepals in young buds restores the regular growth of petals, suggesting that narrow space within a bud is the cause of petal folding in the fop1 mutant. FOP1 encodes a member of the bifunctional wax ester synthase/diacylglycerol acyltransferase family, WSD11, which is expressed in elongating petals and localized to the plasma membrane. These results suggest that the FOP1/WSD11 products synthesized in the petal epidermis may act as a lubricant, enabling uninhibited growth of the petals as they extend between the sepals and the anthers.
Floral organs usually develop sequentially from the outermost whorl toward the inner, in the order of sepals, petals, stamens, and carpels. Sepals arise first and form a tight external covering, which functions as a barrier to protect the developing internal floral organs from physical or biological attacks from the outside. Petals initiate when sepals cover the flower bud and grow rapidly at a later stage. Petal development can be divided into several stages, each of which has been well described at a molecular level in Arabidopsis (Arabidopsis thaliana; Smyth et al., 1990; Irish, 2008). Petal and stamen primordia arise simultaneously at developmental stage 5. Stamens grow faster than petals until stage 8, and the anthers fill the upper internal space created by the protective dome-like closed sepals. After stage 9, petal growth is accelerated, and petals elongate through a narrow space generated by the sepals and anthers.
Floral organ identity is established in a concentric pattern by MADS and APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) transcription factors, which is described by the floral ABCE or quartet model (Theissen and Saedler, 2001). Petal identity is fixed in the second whorl by the combined function of class A, B, and E genes (Bowman et al., 1989; Weigel and Meyerowitz, 1994; Krizek and Fletcher, 2005). Suppression of AGAMOUS activity in the perianth whorls is important for petal growth, and this process is controlled by AP2, AINTEGUMENTA, LEUNIG, SEUSS, RABBIT EARS, ROXY1, and STERILE APETALA (Liu and Meyerowitz, 1995; Byzova et al., 1999; Conner and Liu, 2000; Krizek et al., 2000, 2006; Franks et al., 2002; Sridhar et al., 2004; Xing et al., 2005; Grigorova et al., 2011). Petal primordia arise at four loci in the second whorl, and this positioning is established independently of the process that determines organ identity (Griffith et al., 1999; Brewer et al., 2004; Takeda et al., 2004; Xing et al., 2005; Lampugnani et al., 2013). After initiation, the growth of petals depends on the activity of cell division and expansion along the proximal-distal axis, which is partly regulated by JAGGED (Dinneny et al., 2004; Ohno et al., 2004). Final petal size is determined by the balance of cell proliferation and expansion (Mizukami and Fischer, 2000; Szécsi et al., 2006). The coordinated growth of petals and other floral organs leads to flower opening, which is regulated by auxin, jasmonic acids, and transcription factors such as MYB21, MYB24, AUXIN RESPONSE FACTOR6 (ARF6), ARF8, and BIGPETALp (Brioudes et al., 2009; Tabata et al., 2010; Varaud et al., 2011; Reeves et al., 2012). Most of the known genes involved in petal development encode transcription factors and act in the early developmental processes; however, regulators involved in later stages of the petal elongation process remain unidentified.
We isolated a mutant of Arabidopsis, folded petals 1 (fop1), in which petals are folded at the mature stage, but whose overall size and shape are normal. We found that the petals become stuck in the narrow space between the sepals and stamens in the bud, causing the petal to fold at flower opening, and that this defect is rescued by removal of sepals. Characterization and expression analysis of FOP1 suggests that FOP1 products, synthesized in the petal epidermis, play a lubricant role during petal elongation.
RESULTS
fop1 Petals Do Not Elongate Normally through the Narrow Space between Sepals and Anthers
The fop1-1 mutant was identified from a screen of floral organ-defective mutants in Arabidopsis. Petals of wild-type flowers were straight or slightly curved outwards (Fig. 1A), whereas the mutant petals were folded twice in the shape of a letter N (Fig. 1B), with an outward fold in the medial region and an inward fold in the distal portion. When fully expanded, the size, shape, color, and vascular pattern of a mature petal of the fop1-1 mutant were almost the same as those of the wild type, except for its folded pattern (Supplemental Fig. S1, A, B, O, and P), suggesting that the fop1 mutation is responsible for the physical process of growth rather than cell proliferation or expansion. This is supported by the expression pattern of a cell proliferation marker gene, HISTONE4, in petals, which was not significantly different from the wild type (Supplemental Fig. S1, C–J; Krizek, 1999; Gaudin et al., 2000; Dinneny et al., 2004). These data suggest that the overall petal shape and growth are not altered in fop1-1. We examined the phenotype of two transfer DNA (T-DNA) insertion mutants for the FOP1 gene after gene identification (Supplemental Fig. S3, see below) and found that they showed a similar phenotype to fop1-1 (Fig. 1, C and D). On the basis of this phenotypic and genetic similarity, we used fop1-1 for further analysis.
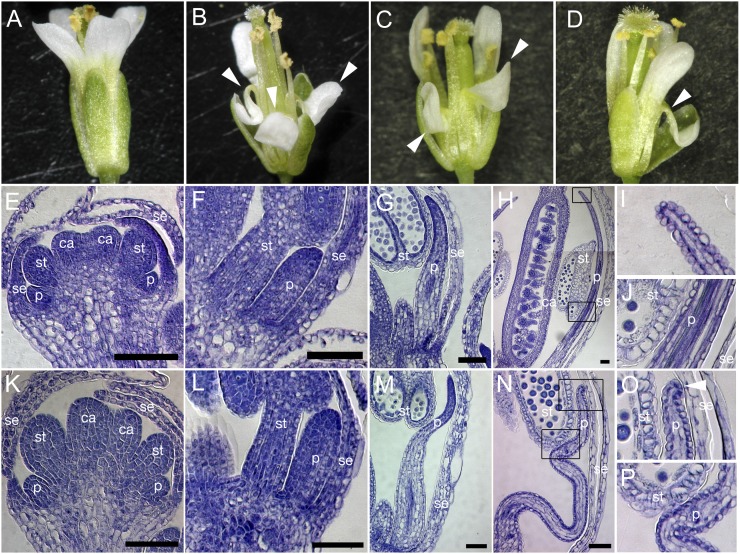
Figure 1.
fop1 petal phenotype. Flowers of the wild type (A), fop1-1 (B), fop1-2 (C), and fop1-3 (D). Arrowheads (B–D) show folded petals. E to P, Histological analysis of wild-type (E–J) and fop1-1 (K–P) flowers. E and K, Stage 7. F and L, Stage 9. G and M, Stage 10. H to J and N to P, Stage 12. I, J, O, and P, High-magnification images of the squares in H and N, respectively. Arrowhead in O shows the contacting petal tip to sepal. se, Sepal; p, petal; st, stamen; ca, carpel. Bars = 50 μm.
Morphological changes in developing petals were examined by sectioning of the bud at several developmental stages. Petal primordia of the fop1-1 mutant formed at stage 5 grew normally up to stage 9, when the petal tip reached the base of an anther (Fig. 1, E, F, K, and L). At stage 10, when wild-type petals elongate through the space between a sepal and an anther (Fig. 1G), the mutant petals did not show smooth elongation, becoming stuck between an anther and a sepal (Fig. 1M). Dissection of unopened buds revealed that when the tip of wild-type petals reached the top of the long stamens (Fig. 1H), the mutant petals failed to undergo straight elongation (Fig. 1N). The degree of folding increased at points where the mutant petal was stuck between the anther and sepal (Fig. 1, O and P), whereas wild-type petals remained straight (Fig. 1I), even though their middle part could contact the anther and sepal (Fig. 1J). These observations suggest that the FOP1 gene is involved in the smooth elongation of petals as they grow though the narrow space in a floral bud.
fop1 Petals Make Contact with the Sepal Surface
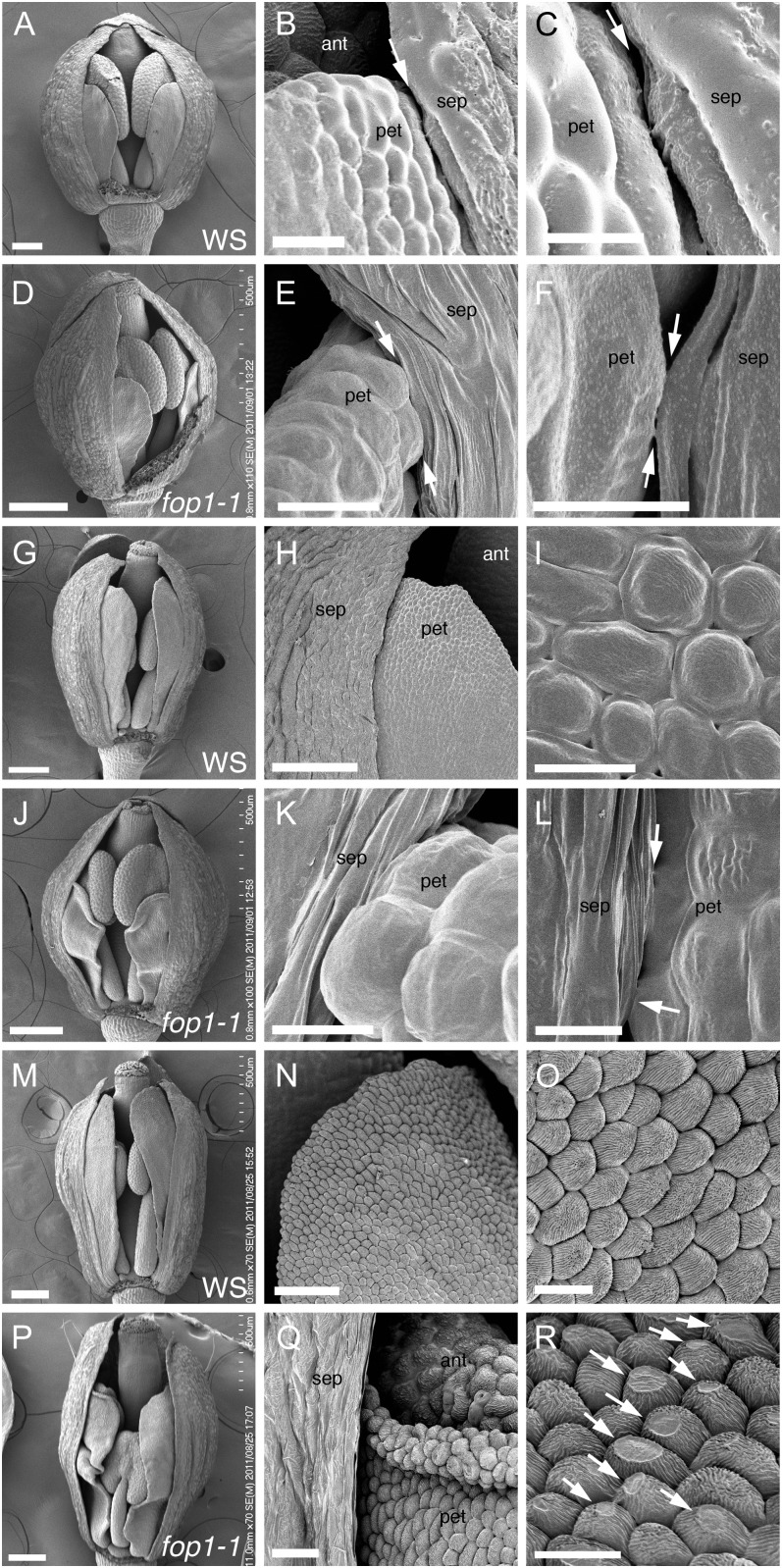
Next, we examined the contact region between petals and sepals using scanning electron microscopy to identify where the folding starts in the mutant. In the wild type, at the stage where the petals are equal in height to the anthers, no direct contact with the sepals was observed (Fig. 2, A–C). At a similar stage in fop1-1, the petal tip made contact with the sepal surface, and the middle part of the petal touched the sepal surface at several sites (Fig. 2, D–F). When wild-type petals grow over the top of anthers, epicuticular nanoridges started to deposit on the surface of the petal epidermis (Fig. 2, G–I). At this stage in fop1-1, petals were seen to have started folding (Fig. 2J), with the petal tip being in contact with the sepal surface (Fig. 2K) and contacted sepals at several sites (Fig. 2L). Before flower opening, epidermal cells of wild-type petals are covered with epicuticular nanoridges and become conical in shape (Fig. 2, M–O). At this stage, petal folding of fop1-1 was apparent (Fig. 2P), the petal apex turned outward (Fig. 2Q), and the surface of the conical epidermal cells were flattened, as if they had been pressed and rubbed from above (Fig. 2R). The flattened cells were frequent on the abaxial surface in the distal part of a folded petal facing a sepal (Supplemental Fig. S1L). Similar flat-tip cells were also found in wild-type petals, but the number of such cells was comparatively few (Supplemental Fig. S1K). Cuticle formation is not altered in fop1 because petal cells had epicuticular nanoridges, although they were affected after its formation. The surface of epidermal cells of anthers or sepals was not altered in the mutant (data not shown). Taken together, the flattened surface of the epidermal cells in fop1-1 mutant petals could be the result of strong pressure and friction between the petals and sepals.
Figure 2.
Scanning electron microscopy images of floral organ surface. A to C, G to I, and M to O, Wild type. D to F, J to L, and P to R, fop1-1. A to F, Stage 10. G to L, Stage 11. M to R, Stage 12. B and C, Higher magnification of A, showing a space between petal and sepal (arrows). E and F, Higher magnification of D, showing direct contact between petal and sepal (arrows). H and I, Higher magnification of G, showing straight petal elongation (G) and abaxial epidermis of petals (I) starting the deposition of the epicuticular nanoridges on their surface. K and L, Higher magnification of J, showing direct contact of petal and sepal (arrows). N and O, Petal surface of the same flower as M. Abaxial epidermal cells of petals deposit nanoridges on cell surface. Q, Higher magnification of P, showing the petal edge turning outward. R, Higher magnification of petals in Q, showing the trace of friction (arrows). The front side sepal is removed in A, D, G, J, M, and P to show the inside of flowers. sep, Sepal; pet, petal; ant, anther. Bars = 200 μm (D, G, J, M, and P), 100 μm (A and H), 50 μm (N), 20 μm (Q), 10 μm (B, E, I, O, and R), 5 μm (C, K, and L), and 3 μm (F).
Sepal Removal Restores the Straight Growth of fop1 Petals
These data indicate that the petals on the fop1 mutant do not easily extend through the space between the sepals and the anthers. To confirm this further, we examined whether regular petal elongation is restored in open buds where sepals do not form a tight covering so that the physical contact between petals and sepals or anthers would not be strong. First, we removed one or two sepals from a fop1-1 mutant bud before petal folding started and let the buds grow. After 3 d, the petals grew flat and did not show the folding phenotype (n = 19; Fig. 3, A and B; Supplemental Fig. S2). Sepal removal also restored petal growth in individuals whose petals had already started to fold (n = 5; Fig. 3, C and D; Supplemental Fig. S2). The epidermal cells were flattened in the fop1-1 mutant, especially in the marginal regions of the folding site (Fig. 3E), although the petals adjacent to removed sepals had much less flattened cells (Fig. 3F), suggesting that sepal removal reduces the physical contact of petals and restores the straight growth.
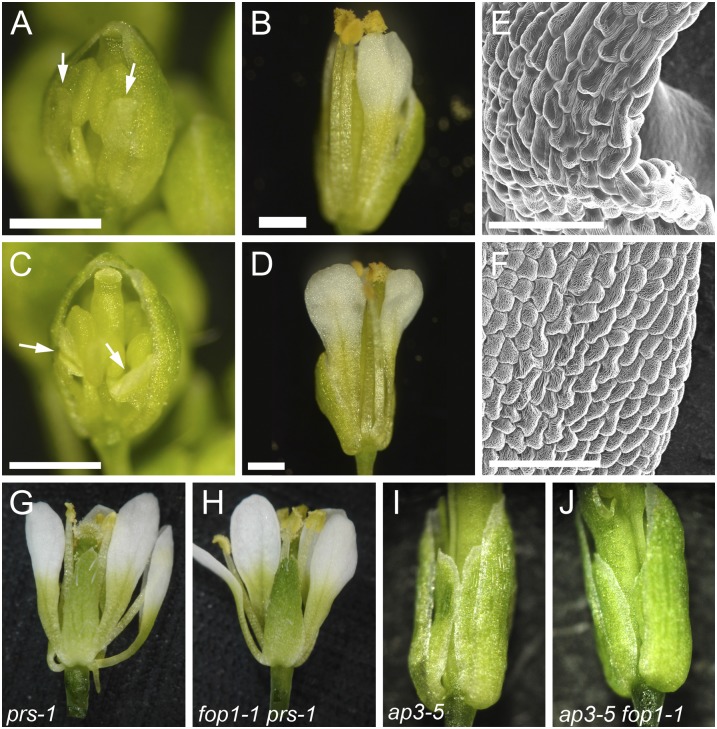
Figure 3.
Surgical and genetic removal of sepals restores straight growth of petals. A, Flower bud of fop1-1 at stage 11. Sepal on the abaxial side is removed. Petals are shown with arrows. B, The same flower with A after 3 d, showing the side of the sepal removal. C, A sepal-removed flower at the stage when petals have already folded (arrows). D, The same flower with C after 3 d, with the straight growth of petals. E, Scanning electron microscopy image of petal epidermis in fop1-1. Surface of epidermal cells, especially those at the marginal region, are flattened. F, Scanning electron microscopy image of petal epidermis in fop1-1, where a sepal has been removed and petals grow straight. G, prs-1 flower. H, fop1-1 prs-1 double mutant flower. Petals of the double mutant are not folded. I, ap3-5 flower. J, ap3-5 fop1-1 double mutant flower. Bars = 500 μm (A–D) and 50 μm (E and F).
Next, we tested the petal phenotype of a double mutant carrying both fop1 and pressed flower (prs). The prs mutant is defective in sepal formation and lacks two sepals in the lateral positions, thus forming an open floral bud (Fig. 3G; Matsumoto and Okada, 2001). As we expected, petals of the fop1-1 prs-1 double mutant did not fold; instead, they elongated normally (Fig. 3H). Next, we examined the structure of the second whorl organ in the ap3-5 fop1-1 double mutant. In ap3-5, which is one of the mutants of homeotic transition of floral organs, petals and stamens are converted to sepals and carpels, respectively (Fig. 3I; Bowman et al., 1989). The sepaloid second whorl organs in the ap3-5 fop1-1 double mutant did not show the folded phenotype (Fig. 3J). Rescue of the mutant phenotype may be due to the change of the floral organ identity or to the increased space provided by the loss of anthers. Together, elimination of the tightness in a floral bud restored the regular petal elongation, suggesting that physical contact of floral organs is the cause of the folded petals in the mutant.
FOP1 Encodes WSD11, a Member of the WAX SYNTHASE/DIACYLGLYCEROL ACYLTRANSFERASE Family
We mapped the FOP1 gene on chromosome 5 and identified a mutation in At5g53390 (Supplemental Fig. S3A). We found that G at nucleotide 820 of the coding sequence was substituted with A in fop1-1, replacing Gly with Arg in the mutant protein (Supplemental Fig. S3B). We examined the border sequences of three T-DNA insertion lines found in the SIGnAL database (http://signal.salk.edu/cgi-bin/tdnaexpress) within or near the gene (Supplemental Fig. S3B; Alonso et al., 2003) and confirmed that the three lines had a small deletion at the T-DNA insertion sites (Supplemental Fig. S3B). Two of them, SALK_093133 (named fop1-2) and SALK_137481 (fop1-3), showed a similar phenotype to fop1-1 (Fig. 1, B–D). The SALK_149804 line, in which T-DNA was inserted at 31 bp upstream from the ATG initiation codon, was indistinguishable from the wild type and did not show the petal-folding phenotype.
To confirm whether At5g53390 is FOP1, a genomic fragment comprising the 2.2-kb promoter, 2.2-kb open reading frame, and 1.6-kb 3′ region was transformed into the fop1-1 mutant. We obtained 80 independent primary transgenic plants carrying the FOP1 genomic fragment, and all lines complemented the petal defects, indicating that At5g53390 is FOP1. We cloned a full-length FOP1 complementary DNA (cDNA) using a RACE strategy and identified the region covering the 48-bp 5′ untranslated region, 217-bp 3′ untranslated region, and seven exons (Supplemental Fig. S3B).
FOP1 corresponds to WSD11, a member of the bifunctional WAX SYNTHASE/DIACYLGLYCEROL ACYLTRANSFERASE (WS/DGAT) family (Kalscheuer and Steinbüchel, 2003; Li et al., 2008). Ten genes encode similar proteins in Arabidopsis, and a gene located next to FOP1 on chromosome 5, At5g53380, is the closest homolog of the same family (Supplemental Fig. S3A; Li et al., 2008). The amino acid sequence of FOP1 showed 17.3% identity with that of Acinetobacter spp. WS/DGAT (GenBank accession no. AAO17391; Kalscheuer and Steinbüchel, 2003). Alignment of the FOP1, At5g53380, and Acinetobacter spp. WS/DGAT proteins revealed three domains of high homology, and the first domain (amino acids 106–155 of FOP1) includes a putative active-site HHxLxDGxS box, which is conserved among Mycobacterium spp. and plant WS/DGAT proteins (Supplemental Fig. S3C; Kalscheuer and Steinbüchel, 2003; Li et al., 2008), suggesting that FOP1 is involved in the synthesis of wax esters and triacylglycerols.
FOP1 Is Expressed in the Epidermis of Growing Petals
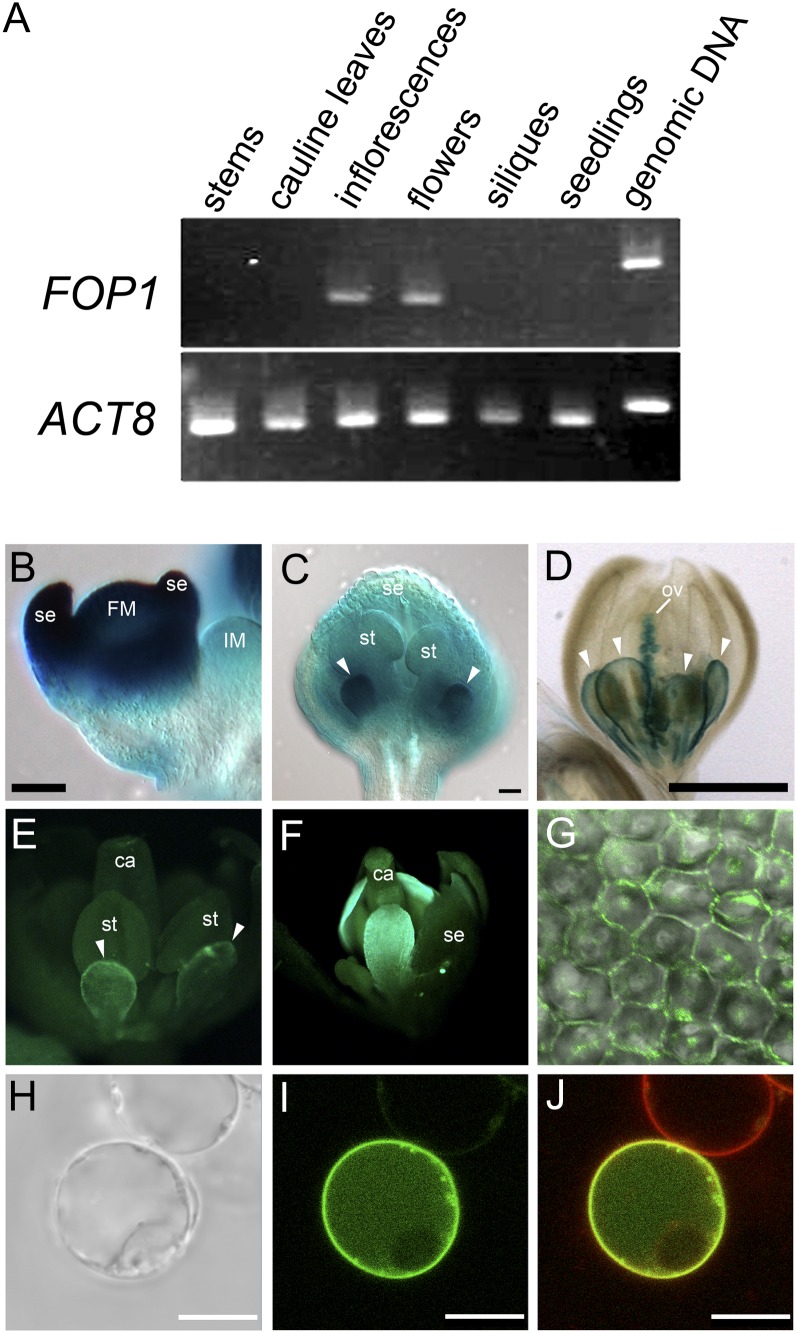
We investigated the spatiotemporal expression pattern of FOP1 during flower development. Reverse transcription (RT)-PCR analysis revealed that FOP1 was preferentially expressed in inflorescences, including young floral buds, and in open flowers (Fig. 4A). We performed in situ hybridization with FOP1 probes, but no signals were obtained, probably due to low expression levels. We then generated transgenic plants carrying FOP1p:GUS, in which the GUS gene was expressed under the control of the 2.2-kb FOP1 promoter. Out of 29 independent transgenic lines obtained by screening, 26 lines showed GUS signals. In floral buds at stage 2, when sepal primordia initiated, GUS signal was detected in the floral meristem and sepal primordia (Fig. 4B). The GUS expression was high in petal primordia at stage 9, with weak expression in the other floral organs (Fig. 4C). At stage 11, FOP1 is expressed in the marginal region of petals and in ovules (Fig. 4D). We also generated a translational fusion line by transforming the FOP1p:FOP1-GFP transgene into the fop1-1 mutant. The petal phenotype of the mutant was rescued, indicating that the fusion protein was functional in plants (Fig. 4F). GFP signal was detected at the margin of elongating petals, when they grew through the space between anthers and sepals (Fig. 4E), and the expression expanded widely in the distal part of petals (Fig. 4F). FOP1 had lower expression in sepals and anthers than in petals, indicating that function of FOP1 in petals is required for straight elongation. The FOP1-GFP fusion protein was localized at the periphery of petal epidermal cells (Fig. 4G). We further examined the cellular localization of FOP1 protein by expressing the GFP-FOP1 fusion gene transiently in suspension-cultured cells of Arabidopsis (Fig. 4H). The GFP signal was detected in the periphery of the cell, which overlapped with the FM4-64 signal (Fig. 4, I and J), indicating that FOP1 is localized to the plasma membrane. Together, these data suggest that FOP1 is involved in wax ester synthesis at the plasma membrane in the petal epidermis.
Figure 4.
FOP1 is expressed in elongating petals. A, RT-PCR analysis of FOP1 transcripts. FOP1 is expressed in inflorescences, including young floral buds, and in open flowers. ACT8 was used as a control. B to D, GUS expression in inflorescences and flowers carrying the FOP1p:GUS transgene. B, GUS is expressed strongly in floral meristem and sepal primordia. C, Strong GUS expression in petal primordia of a stage 9 flower (arrowheads). D, GUS expression is observed at petal margin (arrowheads) and ovules in a stage 11 flower. E to G, GFP fluorescence in fop1-1 flowers of plants carrying the FOP1p:FOP1-GFP transgene. The fusion protein is expressed preferentially at the margin of petals of a stage 10 flower (E) and in the apical part of petals of a stage 12 flower (F). G, A merged image of bright-field and GFP images of petal epidermal cells. The FOP1-GFP fusion protein is localized at the cell periphery. H to J, Suspension cell expressing GFP-FOP1 transiently. H, Bright-field image. I, GFP image. J, Merged image of the GFP and FM4-64 images, showing that GFP-FOP1 is localized to plasma membrane. Note that the upper cell, which is supposed not to carry the construct, shows only FM4-64 signal. IM, Inflorescence meristem; FM, floral meristem; se, sepal; st, stamen; ov, ovule; ca, carpel. Bars = 50 μm (B and C), 500 μm (D), and 10 μm (H–J).
DISCUSSION
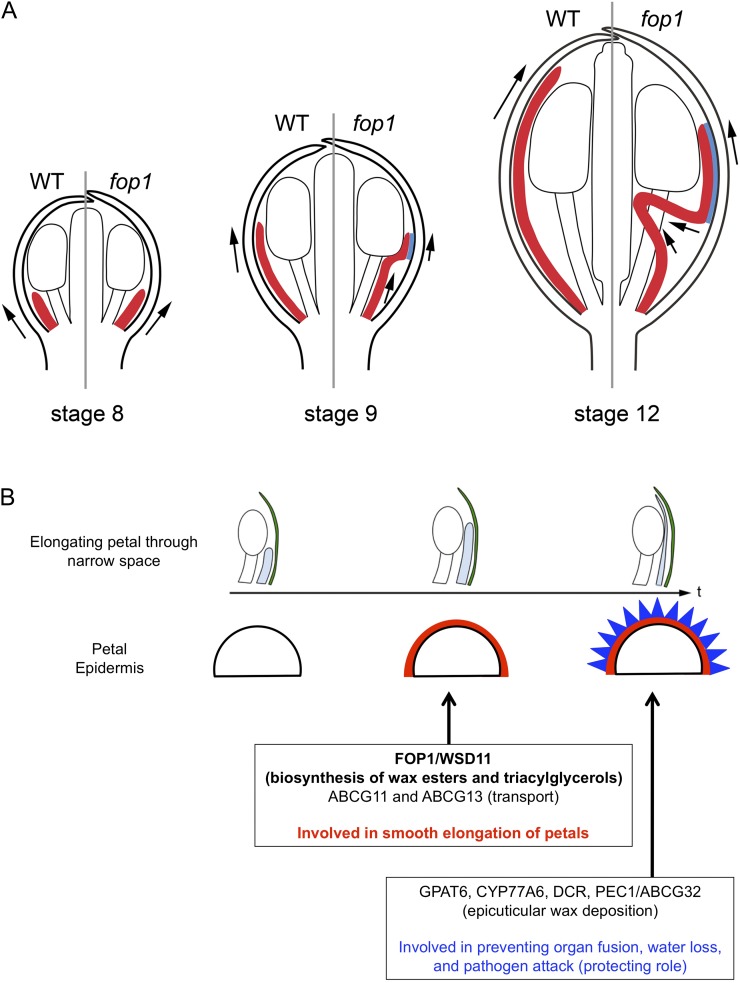
Study of the fop1 mutant directed attention to an aspect of petal growth in a floral bud, namely to how petals elongate through the narrow space between sepals and anthers. Figure 5A describes the schematic model highlighting the differences in the petal elongation process between wild type and fop1. Growth of petal primordia is normal up to stage 9; however, after stage 9, the petal tip becomes stuck between sepals and anthers in the mutant, while in the wild type, petals elongate straight. Our data suggest that the products of FOP1 act as a lubricant on the petal surface, enabling petals to elongate smoothly through the narrow space in a floral bud. It is also possible that the FOP1 products are involved in making cuticle rigid, so that in the fop1 mutant, the petal surface intensity is lower and easy to get rubbed off, enhancing the petal folding.
Figure 5.
Schematic model of the FOP1 function. A, Model explaining the process of petal folding in fop1 floral buds at stage 8 (left), stage 9 (center), and stage 12 (right). Red represents petals. Left and right halves show the petal structure in the wild type and fop1, respectively. The blue area represents the region of contact between petals and sepals. Arrows indicate the direction of petal elongation. B, Model explaining the process of petal surface modification. During petal elongation between sepal and anther, FOP1 products may be secreted to the surface, enabling smooth elongation through the narrow space. Two ATP-binding cassette transporters, ABCG11 and ABCG13, may be involved in a similar process (center). Before maturation, epicuticular nanoridges deposit the outermost surface of petal epidermis, mediated by GPAT6, CYP77A6, DCR, and PEC1/ABCG32, preventing organ fusion and protecting petals from water loss and pathogen attack. WT, Wild type.
FOP1 encodes a homolog of the WS/DGAT enzymes, suggesting that biosynthesis of wax esters and/or triacylglycerols mediated by FOP1 is required for the smooth petal elongation. Several mutants defective in wax biosynthesis or secretion show a similar petal-folding phenotype, and their petals are sensitive to dye immersion due to the lower repellency of the petal surface (Panikashvili et al., 2009; Li-Beisson et al., 2009). We immersed the fop1-1 petals in the dye toluidine blue (Tanaka et al., 2004), but they were not stained (Supplemental Fig. S1, M and N). We also examined the difference of the hexane-soluble surface contents of floral buds between wild type and fop1-1, but found no significant change (Supplemental Fig. S4). Arabidopsis WSD1, a homolog of FOP1, has high wax synthase activity and lower but significant DGAT activity (Li et al., 2008), suggesting that FOP1 has the same activities as WSD1. Identification of substrates and products of FOP1 will elucidate the role of petal surface components in petal elongation.
The petal surface, as with the entire aerial plant body, is covered by cuticle (Pollard et al., 2008; Samuels et al., 2008; Domínguez et al., 2011). Cuticle mainly consists of cutin and cuticular wax. Cutin is an insoluble lipid polymer, consisting of aliphatics (C16 and C18 fatty acids), aromatics, and glycerols, and covers the external surface of the cell wall. The outer layer of the cutin is covered with cuticular wax, a complex of C20 to C60 aliphatics, aldehydes, ketones, and wax esters, coating the outermost surface of the plant body (Pollard et al., 2008). A series of Arabidopsis mutants defective in cuticle synthesis and secretion show the biological roles of cuticles as a barrier to biotic or abiotic stresses, osmotic stress, water loss, and damage from UV radiation, and in preventing the fusion of leaves and floral organs (Yephremov et al., 1999; Pruitt et al., 2000; Krolikowski et al., 2003; Aharoni et al., 2004; Kurdyukov et al., 2006; Bessire et al., 2007; Shi et al., 2011; Wang et al., 2011). Some mutants are known to be involved in petal morphogenesis: Lack of nanoridges of petals, due to a mutation in DEFECTIVE IN CUTICULAR RIDGES (DCR, encoding a BAHD acyltransferase), CYP77A6 (cytochrome P450 family), GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE6 (GPAT6), or PERMEABLE CUTICLE1 (PEC1), result in increased permeability of petals to a dye and cause organ fusion (Li-Beisson et al., 2009; Panikashvili et al., 2009; Bessire et al., 2011). Compared with these mutants, fop1 petals form nanoridges on the petal epidermis, and the degree of organ fusion is, if any, only subtle (Fig. 2). Because the petal folding starts before the nanoridges deposit on the surface of petal epidermis, we propose that FOP1 is involved in the synthesis of wax-related products in the petal epidermis before nanoridge deposition (Fig. 5B). Similar petal-folding defects are found in mutants of the DESPERADO/AtWBC11/ABCG11 and ABCG13 genes, both of which encode members of ATP-binding cassette transporters (Panikashvili et al., 2007, 2011), suggesting that they are involved in a similar process to that of FOP1 (Fig. 5B).
Is the petal elongation mechanism conserved among flowering plants? A similar petal-folding phenotype is also known in a breed of Japanese morning glory (Ipomoea nil). Corollas of the crepe (cp) mutant fold twice like those of fop1, forming an additional tube-like structure surrounding the stamens and carpels (Supplemental Fig. S5; Miyake and Imai, 1927). Similar to our results, the removal of calyx restored the formation of funnel-shaped corollas (Nishino and Gotoh, 2002). These characters suggest that the cp phenotype might be caused by a similar mechanism to that of fop1 petals. Molecular identification of the CP gene will provide an answer.
CONCLUSION
Here, we propose a mechanism for the smooth elongation of petals. After primordia initiation, petals elongate through the narrow space generated by anthers and sepals. FOP1/WSD11 is expressed in elongating petals and may catalyze the wax ester biosynthesis at the epidermal plasma membrane. The FOP1 products may act as a lubricant that enables petals to grow straight in a narrow space in a bud. Identifying the substrates and products of FOP1/WSD11 is the next challenge in the effort to understanding the relationship between the cell surface components and petal morphogenesis.
MATERIALS AND METHODS
Plant Growth Conditions
The Wassilewskija and Columbia ecotypes of Arabidopsis (Arabidopsis thaliana) were used as the wild type. The fop1-1 mutant was isolated from an M2 population of long hypocotyl5 (Wassilewskija ecotype background; Oyama et al., 1997) mutagenized by ethyl methanesulfonate. T-DNA insertion mutants were obtained from the SIGnAL Web site (http://signal.salk.edu) and the Arabidopsis Biological Resource Center (http://abrc.osu.edu). Seeds were sown on the surface of vermiculite in small pots and incubated for 1 week at 4°C. Plants were grown under continuous white light at 22°C to 24°C.
Histology and Microscopes
For scanning electron microscopy, samples were prepared as previously described (Matsumoto and Okada, 2001) and observed with S-3200N (Hitachi) and JSM-5800 (JEOL) microscopes. For histological analysis, inflorescences were fixed in formaldehyde-acetic acid (50% [v/v] ethanol, 3.7% [v/v] formaldehyde, 5% [v/v] acetic acid), dehydrated in an ethanol series, and embedded in Technovit 7100 resin (Heraeus Kulzer). Sections of 5-μm thickness were stained with 0.1% (w/v) toluidine blue. For petal clarification, flowers were fixed in acetic acid and ethanol mixture (1:9) and cleared in clearing solution (40 g chloral hydrate, 10 mL glycerol, 5 mL distilled water). The length and width of cleared petals were measured using the Image Pro-Plus 5.0 software. For whole-mount toluidine blue staining, petals were treated with 0.05% (w/v) toluidine blue for 2 min and washed in distilled water twice (Tanaka et al., 2004). Samples were visualized under an Axiophot 2 microscope (Carl Zeiss).
Mapping and Cloning of FOP1
F2 plants generated by crossing fop1-1 with the Columbia ecotype were used for mapping. Information about the RPS4NT, nga129, JV61/62, and EG7F2 markers was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org). Other markers were generated as sequence markers based on the polymorphisms revealed by genome sequencing. The sequences of the oligonucleotides used in mapping and gene cloning are listed in Supplemental Table S1. cDNA cloning was performed by both 5′ RACE and 3′ RACE using the SMART RACE cDNA Amplification Kit (Clontech). Sequencing was performed using the ABI BigDye Terminator Cycle Sequencing Ready Reaction Kit and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Complementation Test
The genomic fragment including the 2.2-kb promoter, 2.2-kb open reading frame, and 1.6-kb 3′ regions of FOP1 was cloned as follows. Fragments amplified by PCR using the 53390R2 and FOP3R primers were digested with SpeI and HindIII, and cloned into pBluescriptII SK+ (Stratagene) to generate pFg3SK. The promoter region was amplified using the FOP5SalIF and FOPpXbaIR primers, digested by SalI and XbaI, and cloned into pBluescriptII SK+ to generate pFpSK. The HindIII fragment from pFpSK was subcloned into pFg3SK to generate pFgfSK, and the KpnI-SacI fragment of pFgfSK was subcloned to pPZP211 (Hajdukiewicz et al., 1994) and pPZP211NP (a gift from T. Nishimura, Nagoya University) to generate pFgf21135 and pFgf211NP, respectively. These constructs were introduced into the fop1-1 mutant by a vacuum infiltration procedure with the Agrobacterium tumefaciens strain C58C1. Transgenic plants were screened on an agar medium containing 30 μg mL-1 kanamycin and 100 μg mL-1 carbenicillin. Sequences of oligonucleotide primers used for cloning are listed in Supplemental Table S1.
RNA Isolation and RT-PCR
Total RNA was isolated with the Isogen reagent (Nippon Gene) or RNeasy Plant Mini Kit (Qiagen). One microgram of total RNA was reverse transcribed with the SuperScript II reverse transcription kit (Invitrogen). The oligonucleotide primers used for RT-PCR are as follows: FOP1, 53390F1a and 53390R1, and ACTIN8, ACT8F and ACT8R. The sequences of the oligonucleotide primers are listed in Supplemental Table S1.
mRNA in Situ Hybridization
mRNA in situ hybridization was performed as previously described (Matsumoto and Okada, 2001). The HISTONE4 probe was prepared as previously described (Dinneny et al., 2004).
Histochemical Analysis of FOP1
The HindIII fragment from pFpSK was subcloned into pBI101 (Clontech) to generate pFpGUSBI. pFpGUSBI was transformed into Columbia plants and screened as described above. Samples were incubated in a staining buffer (100 mm NaPO4, pH 7.0, 10 mm EDTA, pH 8.0, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 0.1% Triton X-100 (w/v), and 0.5 mg mL–1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid) at 37°C for 6 h. The stained samples were treated with 70% ethanol for 10 min at room temperature, 100% ethanol for 30 min at 37°C, and 70% ethanol for 10 min at room temperature, and then cleared in clearing solution. Samples were visualized under a Leica M420 (Leica Microsystems) or an Axiophot 2 microscope with a differential interference contrast filter. For transient expression in suspension cells, the FOP1 cDNA was amplified by PCR with FOP1FSacI2nt and FOP1RKpnI primers (Supplemental Table S1), digested with SacI and KpnI, and cloned into 35S-GFP-NOS/pUC18 plasmid. The generated 35S:GFP:FOP1c construct was transformed into Arabidopsis suspension-cultured cells by polyethylene glycol-based method as has been previously described (Uemura et al., 2004). The fluorescent images were taken using a LSM710 confocal microscope (Carl Zeiss).
Generation of FOP1p:FOP1-GFP
The G3GFP and FOP1 cDNAs were cloned into pPZP211 and pPZP211NP to generate pFcG21135 and pFcG211NP, respectively. The HindIII fragment from pFpSK was subcloned into pFcG21135 and pFcG211NP to generate pFpFcG21135 and pFpFcG211NP, respectively. Transformation and screening were performed as described above.
Gas Chromatography
Surface fatty acids were extracted from flowers of 6-week-old plants with hexane. C19COOH (Fluka) and N,O-bis(trimethylsilyl)trifluoroactamide with 1% trimethylchlorosilane (Sigma) were added as internal controls. The solution was incubated at 80°C for 30 min and concentrated and analyzed with a GC-14A gas chromatograph (Shimadzu) with the use of a DB-1 column (J&W Scientific). The measurement conditions were 170°C to 265°C with a 2°C increase per minute, then to 310°C with a 4°C increase per minute, and hold for 10 min (alcohol-insoluble residue: 0.5 kg cm–2; hydrogen: 0.5 kg cm–2; carrier [P1]: 1 kg cm–2; carrier [P2]: 2 kg cm–2 [carrier: He]).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FOP1/At5g53390 and NM-124718.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of the fop1-1 petal.
Supplemental Figure S2. Sepal removal experiment in fop1-1.
Supplemental Figure S3. Molecular characterization of FOP1.
Supplemental Figure S4. GC analysis of floral buds.
Supplemental Figure S5. Petal phenotype of crepe mutant in Japanese morning glory.
Supplemental Table S1. Oligonucleotide primers used in this work.
Acknowledgments
We thank Eisho Nishino (Chiba University, Japan), Eiji Nitasaka (Kyushu University, Japan), Noriyoshi Yagi (Kyoto University, Japan), Mitsuhiro Aida (Nara Institute of Science and Technology, Japan), Koichi Toyokura (National Institute for Basic Biology), Maki Kondo (National Institute for Basic Biology), Mikio Nishimura (National Institute for Basic Biology), the Model Plant Research Facility (National Institute for Basic Biology), and the Functional Genomics Facility (National Institute for Basic Biology) for their help with this work; Rebecca Horn (John Innes Centre) for critical reading of the paper; and the Salk Institute and the Arabidopsis Biological Resource Center for providing T-DNA insertion mutants.
Glossary
- T-DNA
transfer DNA
- cDNA
complementary DNA
- RT
reverse transcription
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bessire M, Borel S, Fabre G, Carraça L, Efremova N, Yephremov A, Cao Y, Jetter R, Jacquat AC, Métraux JP, et al. (2011) A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petétot JM, Métraux JP, Nawrath C. (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Howles PA, Dorian K, Griffith ME, Ishida T, Kaplan-Levy RN, Kilinc A, Smyth DR. (2004) PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131: 4035–4045 [DOI] [PubMed] [Google Scholar]
- Brioudes F, Joly C, Szécsi J, Varaud E, Leroux J, Bellvert F, Bertrand C, Bendahmane M. (2009) Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J 60: 1070–1080 [DOI] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MG, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC. (1999) Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev 13: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Liu Z. (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 97: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. (2004) The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Domínguez E, Heredia-Guerrero JA, Heredia A. (2011) The biophysical design of plant cuticles: an overview. New Phytol 189: 938–949 [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z. (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263 [DOI] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, Coen E, Doonan JH. (2000) The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol 122: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith ME, da Silva Conceição A, Smyth DR. (1999) PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 126: 5635–5644 [DOI] [PubMed] [Google Scholar]
- Grigorova B, Mara C, Hollender C, Sijacic P, Chen X, Liu Z. (2011) LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Irish VF. (2008) The Arabidopsis petal: a model for plant organogenesis. Trends Plant Sci 13: 430–436 [DOI] [PubMed] [Google Scholar]
- Kalscheuer R, Steinbüchel A. (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278: 8075–8082 [DOI] [PubMed] [Google Scholar]
- Krizek BA. (1999) Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev Genet 25: 224–236 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Lewis MW, Fletcher JC. (2006) RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J 45: 369–383 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE. (2003) Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J 35: 501–511 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux JP, et al. (2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Kilinc A, Smyth DR. (2013) Auxin controls petal initiation in Arabidopsis. Development 140: 185–194 [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. (2008) Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM. (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Okada K. (2001) A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 15: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Imai Y. (1927) On the double flowers of the Japanese morning glory. J Genet 19: 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino E, Gotoh S. (2002) Reversed corolla tube formation in crepe mutant of Japanese morning glory. J Plant Res (Suppl) 115: 134 [Google Scholar]
- Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. (2004) The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Höfer R, Schreiber L, Chory J, Aharoni A. (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol 145: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. (2009) The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol 151: 1773–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. (2011) The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol 190: 113–124 [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ. (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97: 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101: 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25: 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K. (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. (2004) A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. (2001) Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. (2011) AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Xiong L, Li W, Zhu JK, Zhu J. (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 23: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. (1994) The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S. (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]