Cotton fiber initiation and elongation may be affected by an arabinogalactan protein that alters the integrity of the primary cell wall matrix.
Abstract
Arabinogalactan proteins (AGPs) are involved in many aspects of plant development. In this study, biochemical and genetic approaches demonstrated that AGPs are abundant in developing fibers and may be involved in fiber initiation and elongation. To further investigate the role of AGPs during fiber development, a fasciclin-like arabinogalactan protein gene (GhFLA1) was identified in cotton (Gossypium hirsutum). Overexpression of GhFLA1 in cotton promoted fiber elongation, leading to an increase in fiber length. In contrast, suppression of GhFLA1 expression in cotton slowed down fiber initiation and elongation. As a result, the mature fibers of the transgenic plants were significantly shorter than those of the wild type. In addition, expression levels of GhFLAs and the genes related to primary cell wall biosynthesis were remarkably enhanced in the GhFLA1 overexpression transgenic fibers, whereas the transcripts of these genes were dramatically reduced in the fibers of GhFLA1 RNA interference plants. An immunostaining assay indicated that both AGP composition and primary cell wall composition were changed in the transgenic fibers. The levels of glucose, arabinose, and galactose were also altered in the primary cell wall of the transgenic fibers compared with those of the wild type. Together, our results suggested that GhFLA1 may function in fiber initiation and elongation by affecting AGP composition and the integrity of the primary cell wall matrix.
Arabinogalactan proteins (AGPs) are one type of glycoproteins that are universally distributed in the plant kingdom. AGP consists of a core-protein backbone O-glycosylated by type II arabinogalactan glycans. Based on the core protein sequences, AGPs are divided into six classes: classical AGPs with an N-terminal signal sequence, a central Pro/Hyp-rich domain (also named the AGP motif), and a hydrophobic C terminus; classical AGPs with Lys-rich domains; arabinogalactan peptides with short protein backbones; fasciclin-like AGPs (FLAs), a class of chimeric AGPs that contain both AGP motifs and one or two fasciclin domains; chimeric plastocyanin AGPs; and chimeric nonclassical AGPs (Gaspar et al., 2001; Showalter et al., 2010).
In Arabidopsis (Arabidopsis thaliana), the AGP family has 85 distinct members, including 22 classical AGPs, three Lys-rich AGPs, 16 arabinogalactan peptides, 21 FLAs, 17 plastocyanin AGPs, and six other nonclassical AGPs (Showalter et al., 2010). AGPs are usually located on the cell surface, such as the plasma membrane, the membrane-cell wall interspace, the cell wall, and the intercellular matrix or space. The N-terminal secretion signal sequence and hydrophobic C terminus of AGPs determine where the AGPs are localized in the cell (Showalter, 2001; Ellis et al., 2010). The hydrophobic C terminus of most AtAGPs can be posttranslationally modified by adding a glycosylphosphatidylinositol (GPI) membrane anchor that can be cleaved by phosphatidylinositol-specific phospholipase C and D (Borner et al., 2003; Elortza et al., 2006). GPI-anchored proteins are targeted to the plant cell surface and are likely to be involved in extracellular matrix remodeling and signaling (Borner et al., 2003; Lalanne et al., 2004; Gillmor et al., 2005). In Arabidopsis, 14 FLAs are identified as GPI-anchored proteins, eight of which are targets of phosphatidylinositol-specific phospholipase C and D (Borner et al., 2003; Elortza et al., 2003, 2006).
Previous studies demonstrated that AGPs are involved in various aspects of plant growth and development (Knox, 1999; Majewska-Sawka and Nothnagel, 2000; Gaspar et al., 2001; Showalter, 2001; Seifert and Roberts, 2007; Ellis et al., 2010; Tan et al., 2012). It has been reported that AGPs are involved in pollen tube growth (Jauh and Lord, 1996; Wu et al., 2000), cell differentiation and proliferation (Knox, 1997; Langan and Nothnagel, 1997), programmed cell death (Gao and Showalter, 1999), root epidermal cell expansion (Willats and Knox, 1996), xylem development (Zhang et al., 2003), somatic embryogenesis (Chapman et al., 2000), as well as interaction with cytoskeleton (Nguema-Ona et al., 2007) and pectin (Immerzeel et al., 2006). Knockout of the PpAGP1 gene resulted in a reduction in cell length of moss (Physcomitrella patens) protonema (Lee et al., 2005). Studies revealed that AtAGP18 was essential for the initiation of female gametogenesis (Acosta-García and Vielle-Calzada, 2004), while AtAGP19 functioned in plant development and reproduction (Yang et al., 2007). Overexpression of LeAGP1, a tomato (Solanum lycopersicum) Lys-rich AGP, promoted lateral branching but hampered reproductive development (Sun et al., 2004). AtAGP30 was required for root regeneration and seed germination (van Hengel and Roberts, 2003). Cucumber (Cucumis sativus) CsAGP1 was responsive to GA, and overexpression of CsAGP1 resulted in taller stature and earlier flowering compared with the wild type (Park et al., 2003). Proteins containing a fasciclin domain have been shown to function as adhesion molecules (Elkins et al., 1990). In Arabidopsis, FLA3 is involved in microspore development (Li et al., 2010a). Knockout of AtFLA4 resulted in abnormal cell expansion, thinner cell walls, increased sensitivity to salt, and a reduction in the rays of cellulose across the seed mucilage inner layer (Shi et al., 2003; Harpaz-Saad et al., 2011). AtFLA11 and AtFLA12 contribute to stem strength by regulating cellulose deposition and to stem elasticity by affecting the integrity of the cell wall matrix (MacMillan et al., 2010).
Cotton (Gossypium hirsutum) is the most important fiber crop for the textile industry in the world. Cotton fibers are the trichomes differentiated from the outer integuments of the ovule, and they undergo several distinctive but overlapping developmental stages: initiation (from 2 d before anthesis to 5 DPA), elongation (3–20 DPA), secondary cell wall deposition (16–40 DPA), and maturation (40–50 DPA; Basra and Malik, 1984). Each cotton fiber is a single cell that elongates without the complication of cell division and multicellular development. Thus, cotton fibers may serve as an excellent system for studying fundamental biological processes such as cell elongation and cell wall biogenesis (Kim and Triplett, 2001). In the past decade, some genes that are specifically/preferentially expressed in fibers have been identified in cotton. Until now, however, the limited numbers of cotton genes involved in fiber development have been characterized, and the molecular mechanism controlling fiber development still remains unclear. A previous study revealed that GhACT1, expressed predominantly in fiber cells, participates in fiber elongation (Li et al., 2005). GhDET2 plays a role in the initiation and elongation of fiber cells (Luo et al., 2007). Cotton MYB transcription factors function in fiber initiation and elongation. Knockdown of GhMYB109 in cotton dramatically reduced fiber elongation (Pu et al., 2008). Similarly, suppression of Suc synthase gene expression hindered fiber initiation and elongation (Ruan et al., 2003). Furthermore, GhMYB25 and GhMYB25-like regulate early fiber development (Machado et al., 2009; Walford et al., 2011). A study reported that GhAGP4 plays a role in fiber initiation and elongation of cotton, but its molecular mechanism in regulating fiber development still remains unclear (Li et al., 2010b). In our previous study, several fiber-preferential genes encoding FLAs were identified in cotton (Huang et al., 2008). In this study, we further demonstrate that cotton FLA1 is involved in fiber initiation and elongation.
RESULTS
β-Yariv Inhibits Cotton Fiber Elongation
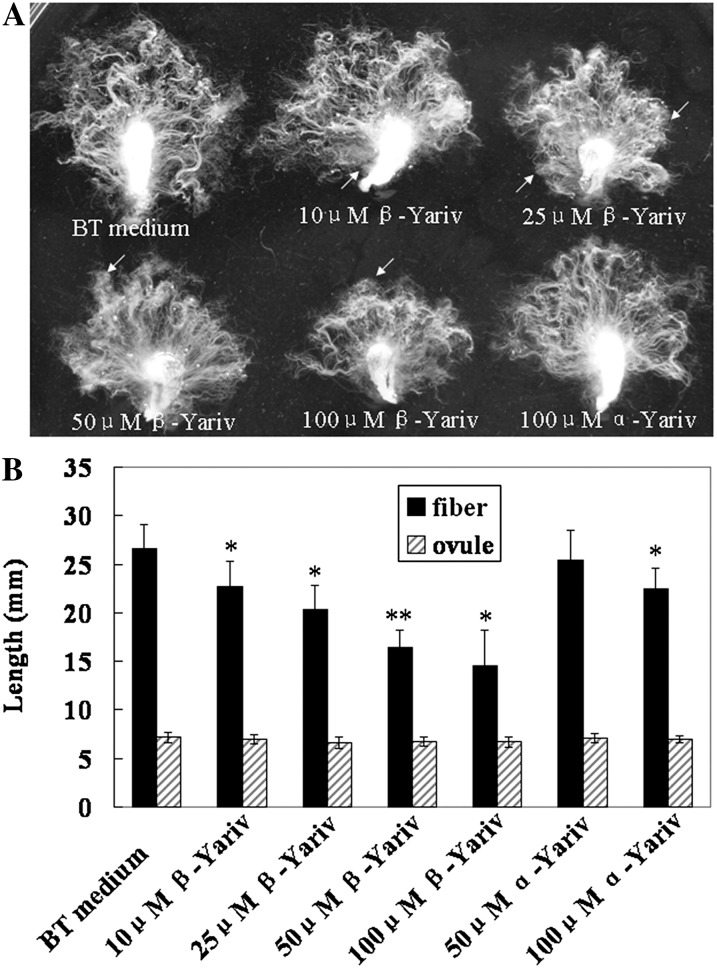
As β-glucosyl Yariv reagent (β-Yariv) binds AGPs, 1-DPA ovules of cotton were cultured in Beasley and Ting (BT) liquid medium supplemented with 0 (control), 10, 25, 50, and 100 μm β-Yariv, using α-glycosyl Yariv reagent (α-Yariv) as a negative control (α-Yariv is incapable of binding AGPs). As shown in Figure 1A, fiber elongation was remarkably inhibited when the ovules were cultured in the medium containing different concentrations of β-Yariv for 14 d. Fiber length was 22.9, 20.0, 17.0, and 14.9 mm as the cultures were treated with 10, 25, 50, and 100 μm β-Yariv, while fiber length was 27 mm in BT medium and 25.8 and 22.8 mm in BT medium with 50 and 100 μm α-Yariv, respectively. Statistical analysis revealed that there was a significant difference in fiber length between controls (BT medium and α-Yariv) and β-Yariv treatments (Fig. 1B). Although high concentration (100 μm) of α-Yariv slightly inhibited fiber elongation, no influence on fiber growth was found when the fibers were treated with 50 μm α-Yariv (Supplemental Fig. S1). In addition to changes in fiber length, the surface of partial ovules and fibers had taken up the β-Yariv reagent in a dose-specific manner. On the other hand, growth of cotton ovules was little interrupted by β-Yariv or α-Yariv.
Figure 1.
Inhibition of cotton fiber elongation in BT liquid medium by β-Yariv. A, Cotton ovules at 1 DPA were cultured in vitro in BT medium with different concentrations of β-Yariv reagent for 14 d, using α-Yariv as a negative control. B, Statistical analysis of ovule and fiber length in BT medium without or with β-Yariv and α-Yariv reagents. The data shown were based on three repeats of the experiment (n ≥ 50 cotton ovules in each repeat). Error bars represent sd. Independent Student’s t tests demonstrated that there was a significant difference (*P < 0.05) or a very significant difference (**P < 0.01) between β-Yariv-treated fibers and controls.
Immunolocalization of AGPs and Polysaccharides during Cotton Fiber Initiation and Elongation
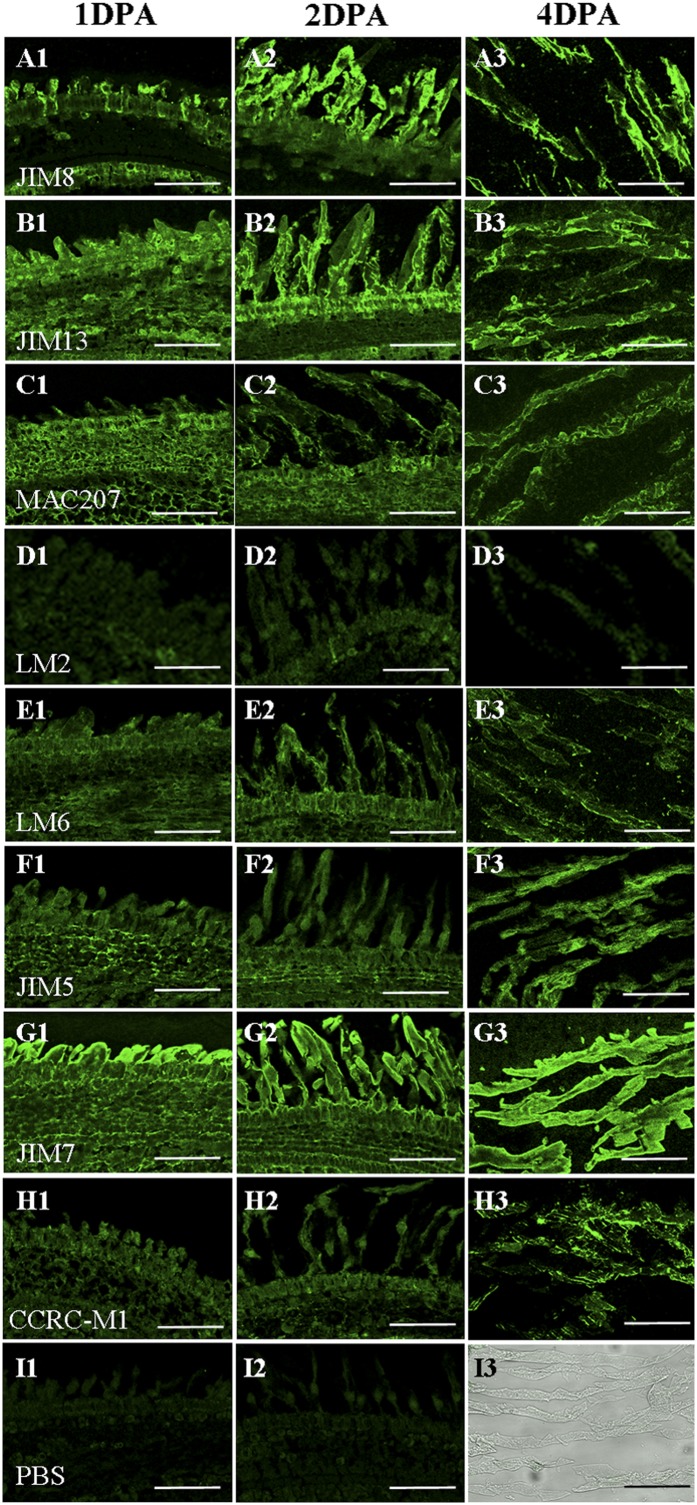
To investigate the presence of AGPs in initiating and elongating fibers, we immunolabeled 1-, 2-, and 4-DPA fibers with five anti-AGP monoclonal antibodies (MAbs): JIM8 (for arabinogalactan), JIM13 (for arabinogalactan and AGP), LM2 (for AGP), MAC207 (for AGP), and LM6 (for 1,5-Ara linkage AGPs or pectin). As shown in Figure 2, A1 to B3, very strong signals of JIM8 and JIM13 epitopes were found in fibers, especially at the 4-DPA stage. Strong signals of MAC207 epitopes were detected in 1-, 2-, and 4-DPA fibers (Fig. 2, C1–C3), although its signals were slightly weaker than those of JIM8 and JIM13. LM2 epitopes showed no or weak signals in 1-, 2-, and 4-DPA fibers (Fig. 2, D1–D3), while relatively strong signals of LM6 epitopes were observed in 1-, 2-, and 4-DPA fibers (Fig. 2, E1–E3). These data indicated that fiber cells contain significant amounts of AGPs during fiber initiation and elongation of cotton.
Figure 2.
Immunohistochemical analysis of cotton fiber cells. Paraffin sections of 1-, 2-, and 4-DPA ovules of cotton were used for immunohistochemical assay. Immunolocalization was carried out with AGP, pectin, and hemicellulose antibodies JIM8 (A1–A3), JIM13 (B1–B3), MAC207 (C1–C3), LM2 (D1–D3), LM6 (E1–E3), JIM5 (F1–F3), JIM7 (G1–G3), and CCRC-M1 (H1–H3) using PBS as a negative control (I1–I3). Bars = 75 µm.
Previous studies showed that AGPs bind to cell wall pectins and hemicelluloses (Kohorn, 2000). To investigate the correlation between AGPs and pectins or hemicelluloses, two methyl-esterified homogalacturonan (Me-HG)-recognized MAbs (JIM5/JIM7) and one α-l-fucosylated xyloglucan-recognized MAb (CCRC-M1) were used to check the pectin and hemicellulose localization during fiber initiation and elongation. As shown in Figure 2, F1 to F3, relatively strong signals of JIM5 epitopes were detected in 1-, 2-, and 4-DPA fibers, like LM6 and MAC207. JIM7 epitopes displayed very strong signals in all 1-, 2-, and 4-DPA fibers, especially in tips of fibers (Fig. 2, G1–G3), while CCRC-M1 shared a similar signal pattern with JIM5 (Fig. 2, H1–H3). In contrast, no signal was found in the control experiments with only the secondary antibody (Fig. 2, I1–I3).
AGPs Are Abundant in Developing Fibers of Cotton
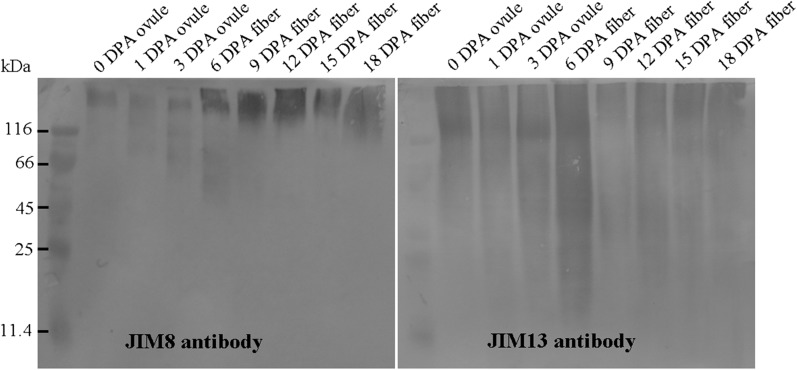
Total proteins were extracted from cotton fibers and separated by SDS-PAGE, followed by probing with JIM8 and JIM13, respectively. As shown in Figure 3, the smear bands with molecular masses of 25 to 300 kD were detected in the lanes with protein samples from fibers at different developmental stages, using JIM13 as a probe. On the contrary, fewer bands with greater size (114–250 kD) were found in the same protein samples when probed with JIM8. These results indicated that developing fibers contain a number of AGPs.
Figure 3.
Western-blot analysis of cotton fiber AGPs recognized by JIM8 and JIM13. An equal amount of total proteins was loaded and separated by 12% SDS-PAGE. The left panel shows expression of JIM8 epitopes in ovules and fibers, and the right panel shows expression of JIM13 epitopes in ovules and fibers. Molecular mass (kD) is indicated on the left.
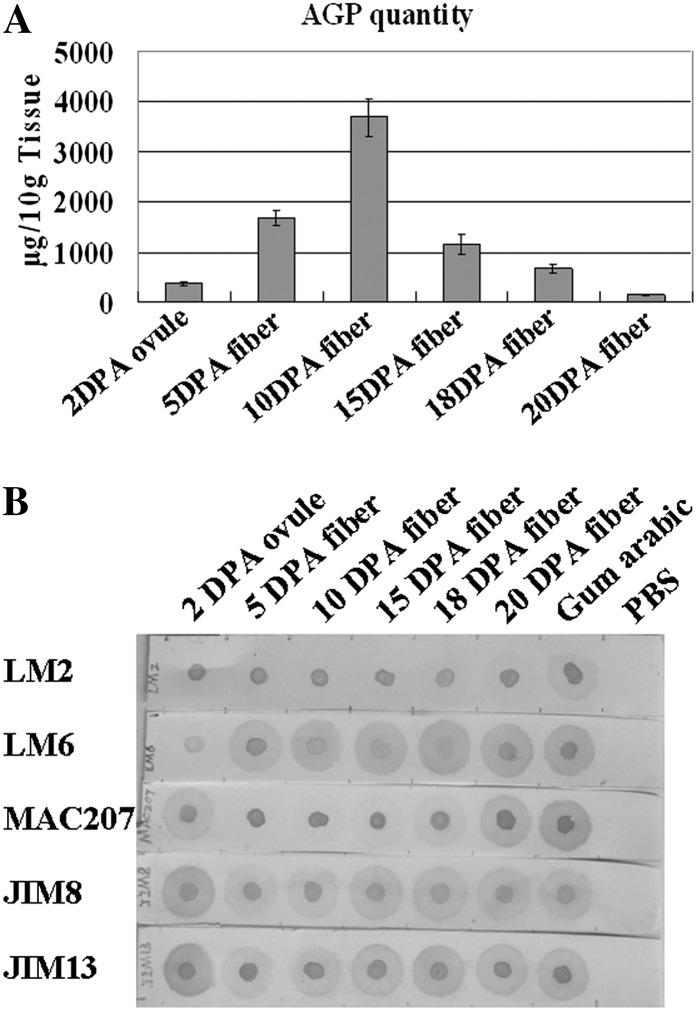
Furthermore, we purified AGPs from fibers at different developmental stages and dotted 2-µg AGPs in nylon membrane. As shown in Figure 4A, the amounts of total AGPs were highly changed during fiber development. At the very early stage, a small quantity of AGPs was found in 2-DPA ovules with fibers. As fibers further developed, AGPs were gradually increased and reached a peak value in 10-DPA fibers. Thereafter, AGPs remarkably declined in 15- to 20-DPA fibers. Dot-blot analysis showed that only weak signals of LM2 epitopes were detected in developing fibers. Strong LM6 signals were found in AGP samples of 5- to 20-DPA fibers, but weak signals were found in 2-DPA ovule/fiber AGPs. MAC207 labeling was strong in the AGP samples isolated in 2-DPA ovules/fibers and 20-DPA fibers but weak in the AGP samples isolated in fibers at other developmental stages. In addition, JIM8- and JIM13-bound AGPs were abundant and widespread in different fiber developmental stages (Fig. 4B).
Figure 4.
Quantitative and immunodot-blot analyses of AGPs in cotton ovules and fibers. A, Quantitative analysis of the purified AGPs in ovules and fibers. B, Immunodot-blot analysis of AGPs. AGPs extracted from fibers and ovules were spotted (2 μg of AGPs for each sample) on nylon membranes using 2 μg of gum arabic as a positive control and PBS as a negative control. Primary antibodies tested are indicated at left. LM2, MAC207, JIM8, LM6, and JIM13 are AGP antibodies.
Isolation and Characterization of Cotton GhFLA1
Through screening the fiber complementary DNA (cDNA) library and the public cotton EST database, seven FLA genes that are specifically or predominantly expressed in fibers were identified in cotton (Huang et al., 2008). To elucidate the role of the FLA proteins in fiber development, we selected GhFLA1 (GenBank accession no. EF672627), a fiber-specific gene, for further study. GhFLA1 protein shows 61.3% identity with AtFLA11, which belongs to group A (Johnson et al., 2003), and contains a signal peptide sequence, two AGP domains, one fasciclin-like domain, and a C-terminal hydrophobic transmembrane domain (Supplemental Fig. S2).
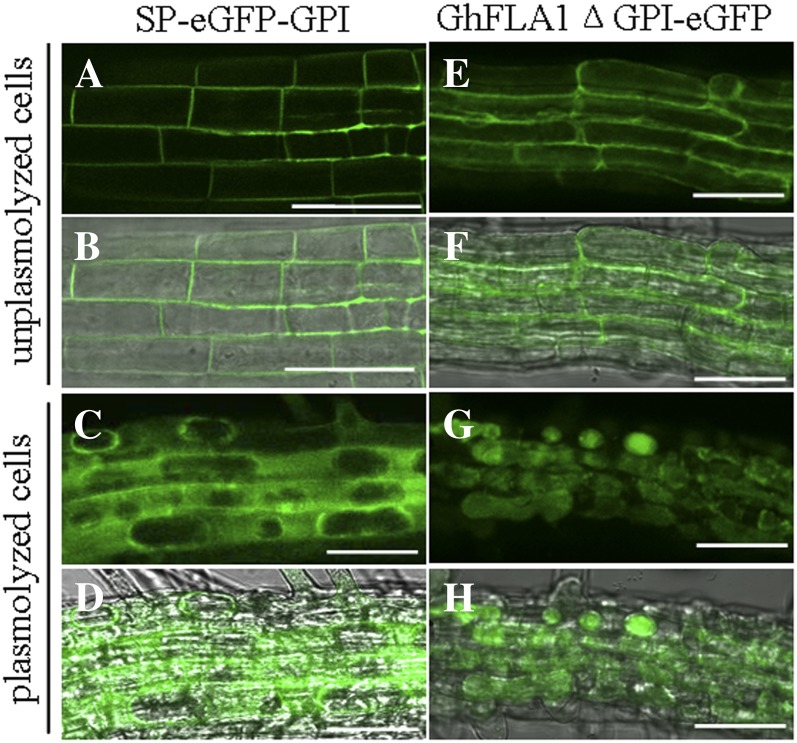
GhFLA1 Is Localized in the Cell Wall
AGPs are usually located in the cell wall and plasma membrane or secreted outside of the cell. To investigate the subcellular localization of GhFLA1, an enhanced GFP (eGFP) gene was inserted between the two sequences encoding GhFLA1 signal peptide and GPI-anchored signal or the truncated GhFLA1 GPI anchor to form GhFLA1(SP)-eGFP-GhFLA1(GPI) and GhFLA1ΔGPI-eGFP (lacking the putative GPI anchor addition signal sequence) constructs under the control of the constitutive cauliflower mosaic virus 35S promoter. The constructs were stably transferred in Arabidopsis. As shown in Figure 5, A and B, GFP fluorescence was localized on the peripheral (cell wall or plasma membrane) of 5-d-old transgenic GhFLA1(SP)-eGFP-GhFLA1(GPI) Arabidopsis root cells. And in plasmolyzed root cells of GhFLA1(SP)-eGFP-GhFLA1(GPI) seedlings, GFP fluorescence was localized between the cell wall and the plasma membrane (Fig. 5, C and D), indicating that GhFLA1 protein is a cell wall protein. On the contrary, the localization of GFP fluorescence in GhFLA1ΔGPI-eGFP transgenic seedlings was different from that in GhFLA1(SP)-eGFP-GhFLA1(GPI) seedlings. Although fluorescence seemed to be localized on the peripheral of the untreated cells (Fig. 5, E and F), GFP fluorescence was localized in cytoplasm of the cells when GhFLA1ΔGPI-eGFP transgenic seedlings were plasmolyzed with 4% NaCl for 20 min (Fig. 5, G and H). Collectively, these data suggested that GhFLA1 protein is a cell wall protein and that the GPI-anchoring signal is important for its localization.
Figure 5.
Subcellular localization of GhFLA1 protein in root cells of 7-d-old Arabidopsis transgenic seedlings. A to D, Transgenic root cells expressing GhFLA1(SP)-eGFP-GhFLA1(GPI). E to H, Transgenic root cells expressing GhFLA1(noGPI)-eGFP. A, C, E, and G are GFP fluorescence images, and B, D, F, and H are the fluorescence images merged with their respective bright-field images. A, B, E, and F, Unplasmolyzed root cells. C, D, G, and H, Root cells plasmolyzed with 4% NaCl. Bars = 50 µm. [See online article for color version of this figure.]
GhFLA1 Functions in Fiber Cell Initiation and Elongation
To investigate the role of GhFLA1 in fiber development, we constructed both GhFLA1 overexpression and RNA interference (RNAi) vectors (Fig. 6A). The constructs were introduced into cotton via Agrobacterium tumefaciens-mediated transformation. A total of 27 GhFLA1 overexpression and 32 GhFLA1 RNAi transgenic cotton plants (T0 generation) were generated. Among them, nine GhFLA1 overexpression and 11 GhFLA1 RNAi transgenic plants have only a single insertion in the genome, while the others have two to six copies of the transgenes in the genome. Real-time reverse transcription (RT)-PCR analysis showed that the GhFLA1 expression level in fibers of transgenic plants (T1–T3 generations) overexpressing GhFLA1 was 2- to 7-fold higher than that of the wild type (Fig. 6B), whereas its expression was dramatically repressed in GhFLA1 RNAi transgenic lines (Fig. 6C) compared with the wild type. Further phenotypic observation indicated that cotton fiber initiation was postponed in the RNAi transgenic plants at 1 to 2 DPA, whereas fiber length in GhFLA1 overexpression transgenic plants was longer than that of the wild type during early fiber development (Fig. 6, D and E). Measurement and statistical analysis revealed that the average length (31.5–35.3 mm) of mature fibers of the GhFLA1 overexpression transgenic lines (T1–T3 generations) was significantly longer than that of the wild type (30.4–31.1 mm; Fig. 6, F and H). In contrast, fibers of the RNAi transgenic lines (T1 and T2 generations) were much shorter (24.8–25.2 mm) than those of the wild type (Fig. 6, F and G). In addition, GhFLA1 overexpression transgenic lines showed a similar phenotype to the wild type at the vegetative growth stage but had bigger ovules, higher stigmas, and lower rates of fruit setting than the wild type. In contrast, flowers and bolls of GhFLA1 RNAi lines were relatively smaller compared with the wild type (Supplemental Fig. S3). These data indicated that GhFLA1 may play an important role in reproductive growth, especially in fiber development of cotton.
Figure 6.
Analysis of GhFLA1 expression and function in overexpression and RNAi transgenic cotton plants. A, Maps of the GhFLA1 overexpression construct (top) and the RNAi construct (bottom). LB, Left border; ORF, open reading frame; RB, right border. B, Real-time RT-PCR analysis of the expression of GhFLA1 in overexpression plants (T2 generation). RNA was isolated from 10-DPA fibers. C, Real-time RT-PCR analysis of the expression of GhFLA1 in RNAi plants (T2 generation). RNA was isolated from 10-DPA fibers. D, Measurement of fiber length at 1 and 2 DPA (n ≥ 50 cotton ovules per line). E, Paraffin sections of 1- and 2-DPA cotton ovules. F, Measurement of fiber length at maturation (n ≥ 50 cotton ovules per line). G, Mature fibers of GhFLA1 RNAi plants. H, Mature fibers of GhFLA1 overexpression plants. Bars = 75 μm in E and 1 cm in G and H. Values shown are means ± sd. Independent Student’s t tests demonstrated that there was a significant difference (*P < 0.05) or a very significant difference (**P < 0.01) between transgenic fibers and the wild type (WT). OL, GhFLA1 overexpression transgenic lines; RL, GhFLA1 RNAi transgenic lines.
AGP Composition Is Changed in Initiating and Elongating Fibers of Transgenic Cotton
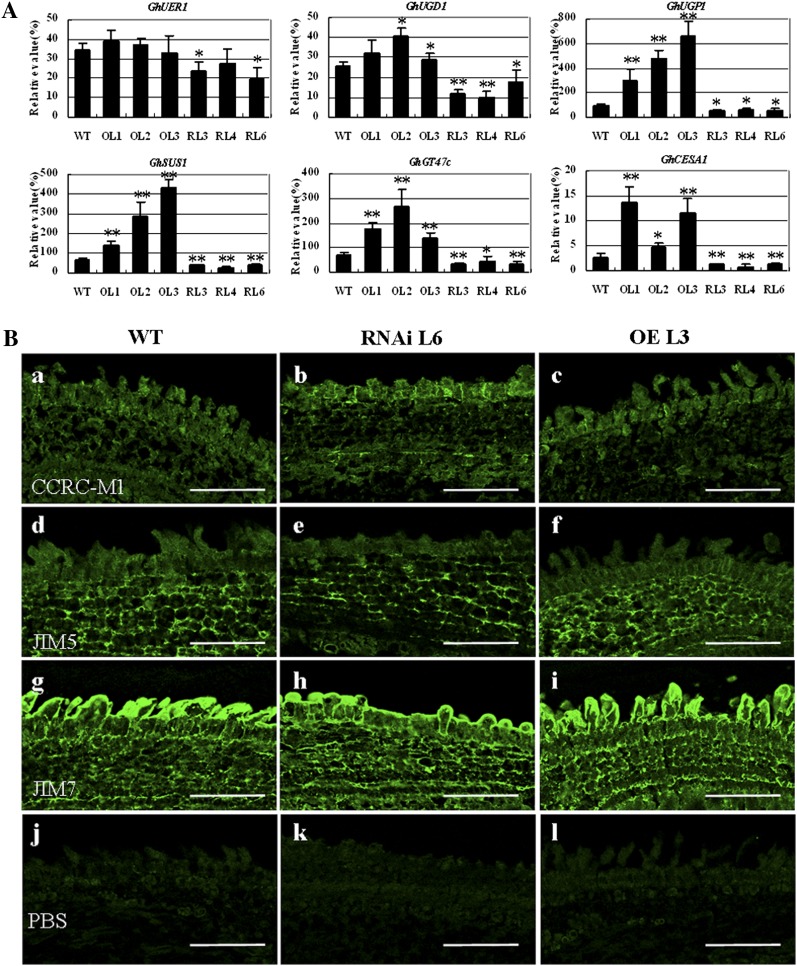
To understand whether the up-regulation and down-regulation of GhFLA1 expression impact the expression of the other GhFLAs, we further analyzed the expression levels of all the fiber-preferential GhFLAs in 10-DPA fibers of the overexpression and RNAi transgenic plants by quantitative RT-PCR analysis using gene-specific primers (Supplemental Table S1). As shown in Figure 7A, more transcripts of the fiber-preferential GhFLAs were accumulated in the fibers of GhFLA1 overexpression transgenic lines, but expression levels of these genes declined in the RNAi transgenic fibers. As a result, total AGP content and composition were altered in the transgenic fibers (Fig. 7B; Supplemental Fig. S4A). As shown in Figure 7B, a to c, MAC207 labeling signals were mainly found in ovule epidermis cells and fiber cells. Strong signals were observed in GhFLA1 overexpression transgenic ovules, whereas weak signals were detected in the RNAi transgenic ovules. Similarly, strong JIM8 labeling signals were detected in the overexpression transgenic fibers, but very weak signals were seen in the RNAi plants (Fig. 7B, d–f). On the contrary, strong JIM13 labeling signals were found in ovule epidermis cells and fibers of the RNAi transgenic plants, but weak signals were observed in fibers of the overexpression transgenic lines (Fig. 7B, g–i). In addition, signals of LM6 antibody were little altered in the GhFLA1 overexpression and RNAi transgenic ovules and fibers compared with wild-type plants (Fig. 7B, j–l). Collectively, immunofluorescence analysis indicated that various types of AGPs, including GhFLA1, are required for rapid fiber development.
Figure 7.
Expression of GhFLA genes and immunohistochemical localization of AGPs in fibers of the transgenic cotton plants. A, Quantitative RT-PCR analysis of the expression of six fiber-specific/preferential FLA genes in 10-DPA fibers of GhFLA1 overexpression and RNAi transgenic cotton plants (T2 generation). Three individual T2 plants were sampled for each transgenic line. Error bars show se (n = 3). Independent Student’s t tests demonstrated that there was a significant difference (*P < 0.05) or a very significant difference (**P < 0.01) between transgenic lines and the wild type (WT) . Accession numbers in GenBank are as follows: GhFLA1 (EF672627), GhFLA2 (EF672628), GhFLA4 (EF672630), GhFLA6 (EF672632), GhFLA14 (EF672640), GhFLA15 (EF672641). B, Paraffin sections of 1-DPA cotton ovules used for immunohistochemical assay. Immunolocalization was carried out with antibodies MAC207, JIM8, JIM13, and LM6, using PBS as a negative control. OL1, OL2, and OL3 (OE L3), GhFLA1 overexpression transgenic lines; RL3, RL4, and RL6 (RNAi L6), GhFLA1 RNAi transgenic lines. Bars = 75 µm.
GhFLA1 Participates in Modulating Primary Cell Wall Composition in Fibers
To evaluate if AGPs are correlated to the biosynthesis of cell wall polysaccharides, we analyzed the expression levels of six genes involved in cell wall biosynthesis in fibers by quantitative RT-PCR using gene-specific primers (Supplemental Table S1). The six genes were GhGT47c (for glycosyltransferase47), GhCESA1 (for cellulose synthase1), GhSUS1 (for Suc synthase1), GhUGP1 (for UDP-d-Glc pyrophosphorylase), GhUGD1 (for UDP-d-Glc dehydrogenase1), and GhUER1 (for UDP-4-keto-6-deoxy-d-Glc 3,5-epimerase 4-reductase1) and were all expressed at high levels in elongating fibers. Compared with those in the wild type, GhGT47c, GhCESA1, GhSUS1, and GhUGP1, involved in hemicellulose and cellulose biosynthesis, were up-regulated in GhFLA1 overexpression transgenic plants but down-regulated in the RNAi transgenic lines. Similarly, transcripts of GhUGD1 genes related to pectin biosynthesis were decreased in the RNAi transgenic plants but increased in GhFLA1 overexpression transgenic lines. However, GhUER1 expression was altered only in the RNAi plants (Fig. 8A).
Figure 8.
Expression of the genes related to cell wall biosynthesis and immunohistochemical localization of pectin and hemicellulose in fibers of the transgenic cotton plants. A, Quantitative RT-PCR analysis of genes related to cell wall biosynthesis in 10-DPA fibers of GhFLA1 overexpression and RNAi transgenic cotton plants (T2 generation). Three individual T2 plants were sampled for each transgenic line. Error bars show se (n = 3). Independent Student’s t tests demonstrated that there was a significant difference (*P < 0.05) or a very significant difference (**P < 0.01) between transgenic lines and the wild type (WT). Accession numbers in GenBank are as follows: GhUER1 (FJ415167), GhUGD1 (FJ415166), GhUGP1 (FJ415164), GhSUS1 (U73588), GhGT47c (ES807557), GhCESE1 (ES812699). B, Paraffin sections of 1-DPA cotton ovules were used for immunohistochemical assay. Immunolocalization was carried out with antibodies CCRC-M1, JIM5, and JIM7, using PBS as a negative control. OL1, OL2, and OL3 (OE L3), GhFLA1 overexpression transgenic lines; RL3, RL4, and RL6 (RNAi L6), GhFLA1 RNAi transgenic lines. Bars = 75 µm.
We further assayed pectin and hemicellulose in 1-DPA ovules/fibers of the transgenic plants and the wild type (Fig. 8B; Supplemental Fig. S4B). The experimental results revealed that signals of CCRC-M1 antibody, which recognizes α-l-fucosylated xyloglucan, were slightly stronger in the RNAi transgenic fibers than in wild-type and GhFLA1 overexpression transgenic fibers (Fig. 8B, a–c). JIM5 labeling of Me-HG showed weak signals in fibers but relatively stronger signals in ovules. Furthermore, higher level of JIM5 signals was detected in ovules of GhFLA1 overexpression transgenic lines compared with RNAi ovules (Fig. 8B, d–f). On the other hand, strong signals of JIM7 antibody that partially labeled Me-HG were found in 1-DPA fiber cells of both transgenic plants and the wild type (Fig. 8B, g–i).
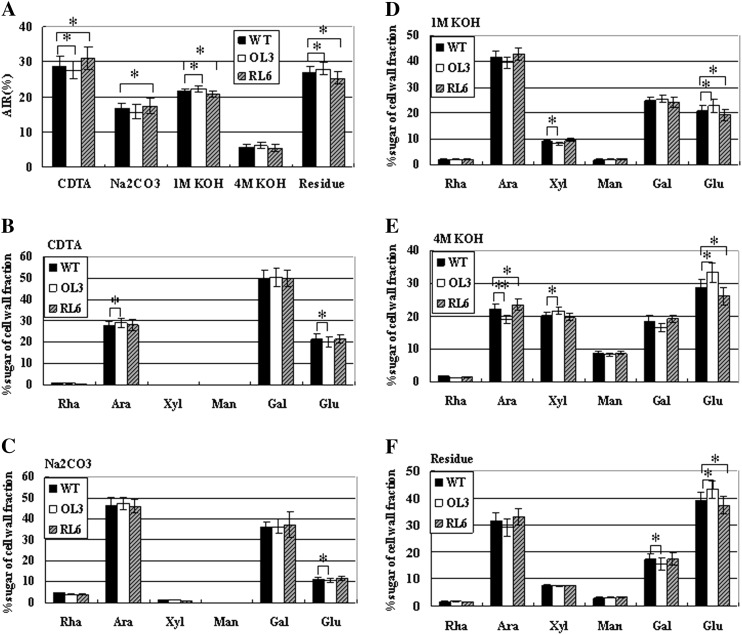
Additionally, fractions of pectin and hemicellulose were chemically separated in GhFLA1 transgenic fibers. Cell walls derived from 10-DPA cotton fiber cells were sequentially extracted in 1,2-cyclohexanediaminetetraacetic acid (CDTA), Na2CO3, and 1 and 4 m KOH. The CDTA and Na2CO3 fractions are generally considered to be enriched for ionically and covalently bound pectin, respectively. The 1 and 4 m KOH fractions represent hemicellulose and hemicellulose-rich polymers, respectively, that are tightly bound to the cell wall. Residues are mainly composed of cellulose and associated polysaccharides. The experimental results showed that a higher rate of pectin and lower rates of hemicellulose and cellulose were seen in the GhFLA1 RNAi lines, whereas a lower rate of pectin and higher rates of hemicellulose and cellulose were found in the GhFLA1 overexpression transgenic lines compared with the wild type (Fig. 9A). We further analyzed the sugar composition of the five fractions by measurement of alditol acetates using gas chromatography. The proportion of sugar composition in CDTA and Na2CO3 fractions of the transgenic lines was similar to that in the wild type (Fig. 9, B and C). In contrast, the proportion of Glc in the 1 and 4 m KOH fractions and residues was significantly reduced in GhFLA1 RNAi transgenic lines but increased in GhFLA1 overexpression transgenic lines compared with the wild type. The proportion of Ara and Gal showed a contrary pattern in the transgenic fibers, compared with that of Glc in the 1 and 4 m KOH fractions and residues (Fig. 9, D–F). These data suggested that GhFLA1 is involved in modulating the biosynthesis of cell wall polysaccharides during fiber development.
Figure 9.
Analysis of the sugar composition of cell wall fractions in 10-DPA fibers of transgenic cotton plants. The sugar composition of cell wall fractions was sequentially extracted with CDTA, Na2CO3, and 1 and 4 m KOH. Individual sugars Rha, Ara, Xyl, Man, Gal, and Glu are expressed as percentages of the cell wall fraction. Error bars show se (n = 3). OL3, GhFLA1 overexpression transgenic line 3; RL6, GhFLA1 RNAi transgenic line 6; WT, wild type. Independent Student’s t tests demonstrated that there was a significant difference (*P < 0.05) or a very significant difference (**P < 0.01) between transgenic lines and the wild type.
DISCUSSION
The data presented in this study indicate the presence of abundant AGP carbohydrate epitopes and AGP backbone peptides during fiber development of cotton. Several classes of AGPs might contribute to the same process of fiber initiation, elongation, and secondary cell wall biosynthesis, while each of these AGPs may play a unique role and act together in regulating fiber development. In Arabidopsis, 46 of all 85 AGPs coexpress with each other and with glycosyltrasferase, glycosylhydrolase, and peroxidase that function in cell wall development (Showalter et al., 2010). Our previous study demonstrated the several GhFLAs were expressed at high levels in 5- to 20-DPA fibers (Huang et al., 2008). In addition, bioinformatic analysis indicated the presence of at least five classical AGP sequences in cotton fibers. These AGPs were strongly expressed in elongating fiber cells and showed a coexpression pattern with GhFLAs and β-Yariv-binding AGPs (Fig. 4A; Supplemental Fig. S5). AGPs play important roles in cell elongation, and β-Yariv inhibited the extension of tip-growing lily (Lilium longiflorum) pollen tubes and extension of apical cells of P. patens (Roy et al., 1998; Lee et al., 2005). Similarly, our results indicated a concentration-dependent inhibitory effect of β-Yariv on fiber elongation, suggesting that AGPs function in fiber development. A previous study showed that some AGPs were located predominantly in the shoot apex and the basal part of the embryos, which also coincided with Yariv-binding sites using JIM8 and JIM13 antibodies (Tang et al., 2006). In this study, immunofluorescence assays indicated that MAC207, JIM8, and JIM13 epitopes were presented abundantly in initiating and elongating fibers, implying that AGPs may be required for normal fiber initiation and elongation of cotton.
Cell growth occurs in many dimensions, such as tip growth and diffuse growth. There are three intracellular components for tip growth of pollen tubes and root hairs: a tip high-calcium gradient, a polarized actin cytoskeleton, and tip-directed vesicle trafficking (Cole and Fowler, 2006). The components needed to add new plasma membrane and cell wall are delivered to the apex and added to the tip by secretory vesicles and the process of exocytosis. Microtubules, noncalcium ion gradients and fluxes, pectin methylesterases, and actin-binding proteins that regulate actin dynamics also participate the tip growth process (Cole and Fowler, 2006). Through labeling pollen tubes with monoclonal antibody MAC207, it has been demonstrated that AGP epitopes were localized at the tip of the emerging pollen tube in Arabidopsis (Pereira et al., 2006). Using LM6 antibody, the same results were also found in moss apical cells, suggesting that the localized movement of AGPs from plasma membrane to cell wall is a component of the mechanism of tip growth (Lee et al., 2005). In our study, however, the results indicated that the immunofluorescence signals of MAC207, JIM13, JIM8, LM6, JIM5, and JIM7 were diffusely distributed in whole fiber cells. Similarly, a previous study also reported that JIM13 and LM6 antibody labeling signals were diffusely distributed in fiber cells (Bowling et al., 2011). The above results revealed that the distributed patterns of AGPs and pectin in cotton fibers are different from those in pollen tubes and moss apical cells. It has been suggested that cotton fiber elongation occurs through the linear cell growth mode (Qin and Zhu, 2011). In this linear cell growth mode, high levels of Ca2+, reactive oxygen species, and even secretory vesicles are observed in the tip of the apical zone, which is normally associated with tip growth. However, in the apical zone of cotton fibers and other related cell types, a common linear cell growth mode has not been experimentally verified yet. Thus, our experimental data may further our understanding of the cotton fiber growth pattern.
Our results indicated that GhFLA1 was localized in cell walls and/or extracellularly. Deletion of the C-terminal domain (GPI signal sequence) leads to GFP fluorescence accumulation in the cytoplasm, demonstrating that the C-terminal hydrophobic region of GhFLA1 is essential for its cell surface localization. Likewise, previous studies also revealed that the subcellular localization of several AGPs depends on the GPI anchor at the C terminus (Zhao et al., 2002; Li et al., 2010a). In Arabidopsis, 40% of GPI-anchored proteins contain arabinogalactan glycomodules, suggesting that AGPs may fulfill roles in intercellular signaling (Borner et al., 2003). Moreover, distinct classes of AGPs are involved in the complex interactions among different cell types in suspension cultures and act in pollen tube tip elongation, cell expansion, and moss cell tip elongation (Roy et al., 1998; Shi et al., 2003; Lee et al., 2005). In our study, transgenic cotton plants that carry a constitutively expressed 35S-GhFLA1 transgene produced longer fibers than wild-type plants. Inheritance of the constitutively expressed GhFLA1 transgene cassette led to a 1- to 4-mm enhancement in fiber length under irrigated field conditions compared with the wild type. On the contrary, suppression of GhFLA1 expression retarded fiber initiation and elongation, ultimately leading to a “short-fiber” phenotype. These results suggested that GhFLA1 plays an important role in regulating fiber development of cotton.
Vissenberg et al. (2001) provided direct evidence that β-Yariv inhibits cellulose deposition on cultured tobacco protoplasts and prevents the elongation of these cells. The orientation of cellulose microfibrils controls the orientation of cell expansion. A previous study demonstrated that GPI-anchored AGPs may be secreted to the cell surface in parallel with cellulose synthase. When AGPs are bound to cellulose, they may be released from their GPI anchor and incorporate into the cell wall (Seifert and Roberts, 2007). Deficiency of BC10 (for brittle culm10, a DUF266-containing and Golgi-located type II membrane protein) causes a reduction in the levels of cellulose and AGPs, leading to inferior mechanical properties (Zhou et al., 2009). AtFLA3 is involved in microspore development and may affect pollen intine formation, possibly by participating in cellulose deposition (Li et al., 2010a). It has been proposed that a subset of group A FLAs contribute to the strength and stiffness of load-bearing plant materials (such as stems) in a partially redundant manner via their impact on cellulose synthesis and deposition and also as structural components of the extracellular matrix. The same study also revealed that Arabidopsis FLA11 and FLA12 affected cellulose deposition and the integrity of the cell wall matrix (MacMillan et al., 2010). Furthermore, a previous study revealed that some carrot (Daucus carota) AGPs from the medium and cell wall may be covalently linked to pectin containing a homogalacturonan structural element (Immerzeel et al., 2006). Pectin is an important constituent in the cell wall. It may account for 30% of the total sugar content in fiber cells but less than 18% in 10-DPA ovules (Pang et al., 2010). In our study, the expression of several GhFLAs and other genes related to hemicellulose and cellulose biosynthesis was dramatically up-regulated in fibers of GhFLA1 overexpression transgenic cotton plants but down-regulated in GhFLA1 RNAi transgenic lines. Similarly, the expression of GhUER1 and GhUGD1 was down-regulated in fibers of the RNAi transgenic cotton plants. Meanwhile, immunofluorescence analysis showed that the compositions of AGPs and hemicellulose were altered in transgenic fibers. The proportion of Glc, Ara, and Gal in 1 and 4 m KOH fractions and residues was also changed in the transgenic lines. These results suggested that GhFLA1 may be involved in modulating the biosynthesis of cell wall polysaccharides during fiber development of cotton.
MATERIALS AND METHODS
Plant Growth Conditions and Cotton Genetic Transformation
Cotton (Gossypium hirsutum ‘Coker312’) seeds were surface sterilized with 70% (v/v) ethanol for 1 min and 10% hydrogen peroxide for 2 h, followed by washing with sterile water. The sterilized seeds were germinated on one-half-strength Murashige and Skoog medium (12-h-light/12-h-dark cycle, 28°C). Hypocotyls were cut from sterile seedlings as explants for transformation as described previously (Li et al., 2002). Tissues for DNA, RNA, and protein extraction were collected from transgenic cotton plants (T1–T3 generations) grown in the field.
Treatment of In Vitro-Cultured Ovules with β-Yariv
Bolls at 1 DPA were harvested and surface sterilized by using 75% ethanol for 1 min, followed by washing with sterile water. Ovules were dissected from the ovaries under sterile conditions and immediately floated on BT liquid medium containing 5 μm indole-3-acetic acid and 0.5 μm GA3 (Beasley and Ting, 1974) supplemented with 0 (control), 10, 25, 50, or 100 μm β-Yariv. Additional control cultures were grown in the same BT medium containing 100 μm α-Yariv. The ovules were cultured at 32°C in the dark without agitation. Fiber length was measured after the ovules were cultured for 14 d. Experiments were repeated at least three times and run in three replicates each time. The total number of ovules counted in each group was 50, and P values were calculated according to the untreated series as controls.
DNA and Protein Sequence Analysis
Unless otherwise stated, nucleotide and amino acid sequences were analyzed using DNAstar software (DNA Star), and protein sequence homology analysis was performed with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The identification of protein domains and significant sites was performed with Motifscan (http://myhits.isb-sib.ch/cgi-bin/motif_scan).
Quantitative RT-PCR Analysis
Total RNA was extracted from roots, hypocotyls, cotyledons, leaves, petals, anthers, ovules, and developing fibers (2−20 DPA). RNAs were purified using the Qiagen RNeasy kit according to the manufacturer’s instructions. First-strand synthesis of cDNA was performed using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions.
Expression of cotton genes in 10-DPA fibers of cotton was analyzed by real-time quantitative RT-PCR using the fluorescent intercalating dye SYBR Green in the detection system (MJ Research; Option 2). A cotton polyubiquitin gene (GhUBI1; GenBank accession no. EU604080) was used as a standard control in the RT-PCR. A two-step RT-PCR procedure was performed in all experiments using a method described earlier (Li et al., 2005). In brief, total RNA was reverse transcribed into cDNA and used as templates in real-time PCR with gene-specific primers (Supplemental Table S1). PCR was performed using SYBR Green Real-Time PCR Master Mix (Toyobo) according to the manufacturer’s instructions. The relative target gene expression was determined using the comparative cycle threshold method. To achieve optimal amplification, PCR conditions for every primer combination were optimized for annealing temperature and Mg2+ concentration. PCR products were confirmed on an agarose gel.
Subcellular Localization
A pBI121-eGFP vector was constructed, and then the GhFLA1 N-signal peptide sequence was cloned into pBI121-eGFP to form pBI121-GhFLA1SP-eGFP. Part of the GPI signal of GhFLA1 was also cloned into pBI121GhFLA1SP-eGFP to form pBI121-GhFLA1SP-eGFP-GhFLA1GPI. Also, a pBI121-GhFLA1ΔGPI-eGFP vector (GhFLA1 lacking the putative GPI anchor addition signal sequence) was constructed. The constructs were introduced into Arabidopsis (Arabidopsis thaliana) by the floral dip method. Homozygous transgenic plants were used for GFP observation. eGFP fusion protein expression was visualized in root cells of 7-d-old seedlings using a Leica SP5 confocal laser scanning microscope. To visualize the eGFP distribution, root cells were plasmolyzed in 4% NaCl for 20 min. All primers used are listed in Supplemental Table S2.
Construction of GhFLA1 Overexpression and RNAi Recombinant Plasmids
For the overexpression construct, the open reading frame of GhFLA1 was cloned in pBI121 vector. For the RNAi construct, a 265-bp specific sequence of the GhFLA1 gene was cloned into a pBluescript SK+ vector to create an inverted repeat transgene and then cloned into pBI121 vector. All primers used are listed in Supplemental Table S2.
Immunofluorescence Labeling with Anti-AGP, Anti-Pectin, and Anti-Hemicellulose MAbs
The distribution of AGPs, pectins, and hemicelluloses was investigated with the rat MAbs CCRC-M1, JIM5, JIM7, JIM8, JIM13, LM2, MAC207, and LM6. Samples were fixed overnight in 4% paraformaldehyde and 0.1 m phosphate-buffered saline (PBS), pH 7.5. After washing twice for 5 min each time in PBS buffer, the samples were blocked in 5% bovine serum albumin (BSA) in culture medium for 1 h at room temperature. Subsequently, they were incubated with monoclonal antibodies (diluted 1:20 with 0.1 m PBS containing 0.1% BSA) at room temperature for 2 h. After rinsing in PBS three times (5 min each), the samples were then incubated with fluorescein isothiocyanate-labeled goat anti-rat IgG antiserum (Sigma) diluted 1:20 in the same buffer at room temperature for 2 h. After a final rinse series in PBS, the samples were mounted on slides with 0.1 m PBS for observation. The controls were prepared following the same procedure but omitting the primary antibody incubation.
A collection of MAbs (JIM8, JIM13, MAC207, and LM2) directed against glycosyl moieties specific to AGPs was obtained as a gift from Dr. P. Knox (University of Leeds). The other MAbs (CCRC-M1, JIM5, and JIM7) were bought from the Complex Carbohydrate Research Center. Fluorescein isothiocyanate-conjugated anti-rat IgG (F-1763; Sigma) was used as a secondary antibody.
Extraction of Total Proteins and Purification of AGPs from Cotton Ovules and Cotton Fibers
Extraction of total proteins and purification of AGPs from cotton ovules and fiber cells were performed by the method described by Schultz et al. (2000). Briefly, 10 g (fresh weight) of freeze-dried ovule or fiber tissue was ground to fine powders in liquid nitrogen, and then 10 mL of extraction buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 0.1% β-mercaptoethanol, and 1% [w/v] Triton X-100) was added. After incubation at 4°C for 3 h, the samples were centrifuged for 10 min at 14,000g. The supernatant was collected and precipitated with 5 volumes of ethanol at 4°C overnight. The pellet was resuspended by vortex mixing in 5 mL of 50 mm Tris-HCl, pH 8.0. The insoluble material was removed by centrifugation, and the supernatant was retained. The pellet was resuspended in an additional 5 mL of 50 mm Tris-HCl, pH 8.0, and the supernatant was pooled with the first supernatant. The buffer-soluble material was freeze dried overnight to concentrate the sample (total proteins). The dried total proteins were resuspended in 300 μL of 1% (w/v) NaCl and transferred to 1.5-mL microcentrifuge tubes. AGPs were precipitated with 300 μL of β-Yariv by mixing the resuspended samples and leaving them overnight at 4°C. The insoluble Yariv-AGP complex was collected by centrifugation at 14,000g for 1 h. The β-Yariv was removed by washing the pellet three times in 1% (w/v) NaCl and then twice in methanol. The purified pellet was dissolved in a minimum volume of dimethyl sulfoxide and mixed with solid sodium dithionite. Water was added with vortex mixing until the mixture became a clear yellow color. The resulting yellow solution was then desalted on a PD-10 column (Pharmacia) that had been equilibrated with water, and the eluate was freeze dried.
SDS-PAGE and Western-Blot Analysis for AGPs
Proteins were quantified using a Bradford protein assay kit (Bio-Rad). Twenty micrograms of total protein extract was loaded per lane, separated by 10% SDS-PAGE, and electroblotted using semidry transfer to polyvinylidene difluoride membranes. Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) overnight at 4°C and then incubated with the specific anti-AGP JIM8 and JIM13 antibodies (1:10 dilution) in Tris-buffered saline containing 0.1% Tween 20 (TBST) with 5% nonfat dry milk for 2 h at room temperature. Membranes were washed three times with TBST and then incubated with goat anti-rat IgG secondary antibodies conjugated to horseradish peroxidase (Bio-Rad). Signals were visualized with a diaminobenzidine kit.
Dot-Blot Analysis for AGPs
AGPs were also characterized in terms of the binding capacity of several antibodies. Since AGPs frequently coprecipitate with some pectin and may share common epitopes with pectin, we used MAC207, LM2/6, JIM8, and JIM13 antibodies. Antibody-binding tests were performed by the dot-blot method. Briefly, 2 µg of AGP sample from different cotton fibers (2-DPA ovules/fibers and 5-, 10-, 15-, 18-, and 20-DPA fibers) were dotted onto nylon membranes (Hybond Trans-Blot Transfer Medium; Bio-Rad) and air dried. The nylon membrane were then blocked in TBST buffer supplemented with 1% BSA and 1% dry, nonfat milk for 2 h at room temperature. The incubation with primary antibodies (all diluted 1:200 in TBST with 1% BSA and 1% milk) was carried out for 12 h at room temperature, followed by washing in TBST. The blots were subsequently incubated for 2.5 h with secondary anti-rat antibody coupled to alkaline phosphatase (Sigma-Aldrich) that was diluted 1:1,500 (v/v) in TBST with 1% milk. After washing in TBST and alkaline phosphatase buffer (100 mm Tris, 100 mm NaCl, and 5 mm MgCl2, pH 8.5), the blots were incubated with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium in alkaline phosphatase buffer for 18 min. The reaction was stopped by dipping the blots in distilled water. Controls were performed by omitting the incubation with primary antibodies. Gum arabic was used as a positive control.
The MAbs used in this project included (1) MAC207, which recognizes α-GlcA-(1,3)-α-GalA-(1,2)-α-Rha; (2) LM2, which recognizes AGP; (3) JIM8, which recognizes the sugar part of AGPs; (4) JIM13, which recognizes AGPs [recognizes the β-GlcA(1-3)-α-GalA(1-2)-Rha-region]; (5) LM6, which recognizes 1,5-α-l-arabinan epitopes of type I rhamnogalacturonans; (6) JIM5 and JIM7, which recognize Me-HG; and (7) CCRC-M1, which recognizes α-l-fucosylated xyloglucan.
Noncellulosic Neutral Monosaccharide Analysis
Cell wall fractionation was based upon the methods described by Brown et al. (2007) with minor modification. In brief, 10-DPA cotton fiber materials were harvested, lyophilized, and ground into fine powders. Then, the samples were washed three times with 70% ethanol, three times with 1:1 methanol:chloroform, and two times with acetone to obtain alcohol-insoluble residue. The alcohol-insoluble residue was subsequently destarched with amylase (A6380; Sigma-Aldrich) and extracted using (1) 50 mm CDTA containing 1% NaBH4, (2) 50 mm Na2CO3 containing 1% NaBH4 (Na2CO3-soluble fraction), (3) 1 m KOH containing 1% NaBH4 (1 m KOH-soluble fraction), and (4) 4 m KOH containing 1% NaBH4 (4 m KOH-soluble fraction) for 24 h at room temperature. All fractions were filtered through a GF/C glass filter (Whatman). The alkali fractions were neutralized with acetic acid. All cell wall fractions were then dialyzed extensively against deionized water for 5 d and lyophilized. The fractionation was repeated three times on three sets of plants grown independently, and the mean of these three independent replicates was calculated. All cell wall fractions were subjected to 2 m trifluoroacetic acid at 120°C for 2 h to produce neutral monosaccharides and subsequent derivatization of the solubilized monosaccharides into their corresponding alditol acetates. Finally, the different fractions were run on a gas chromatograph (6890N; Agilent Technologies) with nitrogen as the carrier gas to determine their sugar composition by the method described previously (Pang et al., 2010).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GhFLA1 (EF672627).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. No inhibition of 50 μm α-Yariv on fiber elongation.
Supplemental Figure S2. Analyses of the cotton FLA1 protein sequence and phylogenetic relationship.
Supplemental Figure S3. Morphological alterations as a result of GhFLA1 RNAi and overexpression transgenic plants.
Supplemental Figure S4. Bright-field micrographs of the images in Figures 7B and 8B.
Supplemental Figure S5. Analysis of the expression of five classical AGP genes in cotton tissues.
Supplemental Table S1. Gene-specific primers used in quantitative RT-PCR analysis.
Supplemental Table S2. Gene-specific primers used in vector construction.
Acknowledgments
We thank Dr. P. Knox for the gift of LM2, JIM8, JIM13, LM6, and MAC207 antibodies.
Glossary
- AGP
arabinogalactan proteins
- GPI
glycosylphosphatidylinositol
- α-Yariv
α-glycosyl Yariv reagent
- MAb
monoclonal antibody
- Me-HG
methyl-esterified homogalacturonan
- RNAi
RNA interference
- RT
reverse transcription
- CDTA
1,2-cyclohexanediaminetetraacetic acid
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- TBST
Tris-buffered saline containing 0.1% Tween 20
- cDNA
complementary DNA
- β-Yariv
β-glucosyl Yariv reagent
- BT
Beasley and Ting
References
- Acosta-García G, Vielle-Calzada JP. (2004) A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra AS, Malik CP. (1984) Development of the cotton fiber. Int Rev Cytol 89: 65–113 [Google Scholar]
- Beasley CA, Ting IP. (1974) Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Amer J Bot 61: 188–194 [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P. (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling AJ, Vaughn KC, Turley RB. (2011) Polysaccharide and glycoprotein distribution in the epidermis of cotton ovules during early fiber initiation and growth. Protoplasma 248: 579–590 [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. (2007) Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J 52: 1154–1168 [DOI] [PubMed] [Google Scholar]
- Chapman A, Blervacq AS, Vasseur J, Hilbert JL. (2000) Arabinogalactan-proteins in Cichorium somatic embryogenesis: effect of β-glucosyl Yariv reagent and epitope localisation during embryo development. Planta 211: 305–314 [DOI] [PubMed] [Google Scholar]
- Cole RA, Fowler JE. (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9: 579–588 [DOI] [PubMed] [Google Scholar]
- Elkins T, Zinn K, McAllister L, Hoffmann FM, Goodman CS. (1990) Genetic analysis of a Drosophila neural cell adhesion molecule: interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell 60: 565–575 [DOI] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elortza F, Mohammed S, Bunkenborg J, Foster LJ, Nühse TS, Brodbeck U, Peck SC, Jensen ON. (2006) Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J Proteome Res 5: 935–943 [DOI] [PubMed] [Google Scholar]
- Elortza F, Nühse TS, Foster LJ, Stensballe A, Peck SC, Jensen ON. (2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol Cell Proteomics 2: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Gao M, Showalter AM. (1999) Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J 19: 321–331 [DOI] [PubMed] [Google Scholar]
- Gaspar YM, Johnson KL, McKenna JA, Bacic A, Schultz CJ. (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol 47: 161–176 [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C. (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ. (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Huang GQ, Xu WL, Gong SY, Li B, Wang XL, Xu D, Li XB. (2008) Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol Plant 134: 348–359 [DOI] [PubMed] [Google Scholar]
- Immerzeel P, Eppink MM, de Vries SC, Schols HA, Voragen AGJ. (2006) Carrot arabinogalactan proteins are interlinked with pectins. Physiol Plant 128: 18–28 [Google Scholar]
- Jauh GY, Lord EM. (1996) Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta 199: 251–261 [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiol 133: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD. (2000) Plasma membrane-cell wall contacts. Plant Physiol 124: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP. (1997) The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int Rev Cytol 171: 79–120 [DOI] [PubMed] [Google Scholar]
- Knox JP. (1999) Intriguing, complex and everywhere: getting to grips with arabinogalactan-proteins. Trends Plant Sci 4: 123–125 [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GHH, Lilley KS, Dupree P, Grossniklaus U, Twell D. (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan KJ, Nothnagel EA. (1997) Cell surface arabinogalactan proteins and their relation to cell proliferation and viability. Protoplasma 196: 87–98 [Google Scholar]
- Lee KJ, Sakata Y, Mau SL, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. (2005) Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17: 3051–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu M, Geng LL, Zhao J. (2010a) The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J 64: 482–497 [DOI] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW. (2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol 130: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC. (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Liu DQ, Tu LL, Zhang XL, Wang L, Zhu LF, Tan JF, Deng FL. (2010b) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep 29: 193–202 [DOI] [PubMed] [Google Scholar]
- Luo M, Xiao YH, Li XB, Lu XF, Deng W, Li DM, Hou L, Hu MY, Li Y, Pei Y. (2007) GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J 51: 419–430 [DOI] [PubMed] [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52–62 [DOI] [PubMed] [Google Scholar]
- MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG. (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62: 689–703 [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA. (2000) The multiple roles of arabinogalactan proteins in plant development. Plant Physiol 122: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Bannigan A, Chevalier L, Baskin TI, Driouich A. (2007) Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. Plant J 52: 240–251 [DOI] [PubMed] [Google Scholar]
- Pang CY, Wang H, Pang Y, Xu C, Jiao Y, Qin YM, Western TL, Yu SX, Zhu YX. (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol Cell Proteomics 9: 2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Suzuki Y, Chono M, Knox JP, Yamaguchi I. (2003) CsAGP1, a gibberellin-responsive gene from cucumber hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiol 131: 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LG, Coimbra S, Oliveira H, Monteiro L, Sottomayor M. (2006) Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta 223: 374–380 [DOI] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan XP, Yang WC, Xue YB. (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Zhu YX. (2011) How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol 14: 106–111 [DOI] [PubMed] [Google Scholar]
- Roy S, Jauh GY, Hepler PK, Lord EM. (1998) Effects of Yariv phenylglycoside on cell wall assembly in the lily pollen tube. Planta 204: 450–458 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Shi HZ, Kim YS, Guo Y, Stevenson B, Zhu JK. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153: 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WX, Kieliszewski MJ, Showalter AM. (2004) Overexpression of tomato LeAGP-1 arabinogalactan-protein promotes lateral branching and hampers reproductive development. Plant J 40: 870–881 [DOI] [PubMed] [Google Scholar]
- Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS, Bacic A. (2012) Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front Plant Sci 3: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XC, He YQ, Wang Y, Sun MX. (2006) The role of arabinogalactan proteins binding to Yariv reagents in the initiation, cell developmental fate, and maintenance of microspore embryogenesis in Brassica napus L. cv. Topas. J Exp Bot 57: 2639–2650 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. (2003) AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J 36: 256–270 [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Feijó JA, Weisenseel MH, Verbelen JP. (2001) Ion fluxes, auxin and the induction of elongation growth in Nicotiana tabacum cells. J Exp Bot 52: 2161–2167 [DOI] [PubMed] [Google Scholar]
- Walford SA, Wu Y, Llewellyn DJ, Dennis ES. (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65: 785–797 [DOI] [PubMed] [Google Scholar]
- Willats WG, Knox JP. (1996) A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J 9: 919–925 [DOI] [PubMed] [Google Scholar]
- Wu HM, Wong E, Ogdahl J, Cheung AY. (2000) A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J 22: 165–176 [DOI] [PubMed] [Google Scholar]
- Yang J, Sardar HS, McGovern KR, Zhang YZ, Showalter AM. (2007) A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. Plant J 49: 629–640 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brown G, Whetten R, Loopstra CA, Neale D, Kieliszewski MJ, Sederoff RR. (2003) An arabinogalactan protein associated with secondary cell wall formation in differentiating xylem of loblolly pine. Plant Mol Biol 52: 91–102 [DOI] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DT, Kieliszewski MJ. (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]
- Zhou YH, Li SB, Qian Q, Zeng DL, Zhang M, Guo LB, Liu XL, Zhang BC, Deng LW, Liu XF, et al. (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57: 446–462 [DOI] [PubMed] [Google Scholar]