Micronutrient iron regulates the plant circadian clock through a chloroplast retrograde signaling pathway.
Abstract
The homeostasis of iron (Fe) in plants is strictly regulated to maintain an optimal level for plant growth and development but not cause oxidative stress. About 30% of arable land is considered Fe deficient because of calcareous soil that renders Fe unavailable to plants. Under Fe-deficient conditions, Arabidopsis (Arabidopsis thaliana) shows retarded growth, disordered chloroplast development, and delayed flowering time. In this study, we explored the possible connection between Fe availability and the circadian clock in growth and development. Circadian period length in Arabidopsis was longer under Fe-deficient conditions, but the lengthened period was not regulated by the canonical Fe-deficiency signaling pathway involving nitric oxide. However, plants with impaired chloroplast function showed long circadian periods. Fe deficiency and impaired chloroplast function combined did not show additive effects on the circadian period, which suggests that plastid-to-nucleus retrograde signaling is involved in the lengthening of circadian period under Fe deficiency. Expression pattern analyses of the central oscillator genes in mutants defective in CIRCADIAN CLOCK ASSOCIATED1/LATE ELONGATED HYPOCOTYL or GIGANTEA demonstrated their requirement for Fe deficiency-induced long circadian period. In conclusion, Fe is involved in maintaining the period length of circadian rhythm, possibly by acting on specific central oscillators through a retrograde signaling pathway.
Metals such as iron (Fe), copper (Cu), zinc (Zn), manganese (Mn), molybdenum, and nickel are essential for the various biological processes that govern plant growth and development (Marschner, 1995). For example, Fe is required for DNA synthesis, photosynthesis, nitrogen fixation, hormone synthesis, and electron transport in the respiratory chain (Briat and Lobreaux, 1997). Similarly, Cu is an important component of electron-transfer reactions mediated by proteins such as superoxide dismutase, cytochrome oxidase, and plastocyanin (Clemens, 2001). Zn is a cofactor for many enzymes, and many proteins contain Zn-binding structural domains (Clarke and Berg, 1998). Although only minimal quantities of these micronutrients are required by plants, their limited availability in soils can significantly hinder crop production and affect nutritional quality (Grotz and Guerinot, 2002). In the case of Fe, about 30% of arable land worldwide is considered calcareous, rendering Fe in these soils unavailable to plants (Mori, 1999). Understanding of the fundamental processes involving metal uptake and sequestration has increased in recent years, but how the availability of particular metals interacts with internal signals to govern the growth and development of plants is largely unknown.
The daily biological rhythms of many organisms are regulated by a near 24-h circadian clock that is synchronized by environmental changes such as light and temperature (Harmer, 2009; Imaizumi, 2010). The circadian clock regulates diverse aspects of plant growth and development. The operation of the circadian clock in plants can basically be divided into three main parts, input, central oscillator, and output pathways, and each part has its own complex networks. In Arabidopsis (Arabidopsis thaliana), the central oscillator is composed of a network of multiple feedback loops that can be divided into the morning, central, and evening loops (Harmer, 2009). The central feedback loop is composed of the morning-expressed genes CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) and the evening-expressed gene TIMING OF CAB EXPRESSION1 (TOC1; Schaffer et al., 1998; Wang and Tobin, 1998; Strayer et al., 2000; Alabadí et al., 2001). Although TOC1 is genetically required for the activation of morning genes (Schaffer et al., 1998; Wang and Tobin, 1998; Strayer et al., 2000), it acts as a repressor and directly regulates the expression of CCA1 and LHY (Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012). In the morning loop, CCA1/LHY form another negative feedback loop with the morning genes PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9, with PRR9/PRR7 directly repressing the expression of CCA1 and LHY (Farré et al., 2005; Nakamichi et al., 2010). In the evening loop, TOC1 forms a negative feedback loop with GIGANTEA (GI) by repressing its expression, and GI in turn activates the expression of TOC1 through an unknown component, Y (Huq et al., 2000; Mizoguchi et al., 2005). After receiving input signals in the form of environmental cues, the central oscillator of the Arabidopsis circadian clock generates various rhythmic outputs that control various physiological events (Hotta et al., 2007; de Montaigu et al., 2010).
The central oscillator controls a range of important physiological output processes such as flowering, stress and hormone responses, and regulation of nutrient acquisition (Haydon et al., 2011). Although the uptake of nutrition in plants is known to be influenced by light and temperature (Lahti et al., 2005; Baligar et al., 2006), the interaction between nutritional status and the circadian clock is less well studied. The homeostasis of Cu is known to influence the regulation of oscillator genes (Andrés-Colás et al., 2010; Peñarrubia et al., 2010). Arabidopsis under excess Cu or overexpressing Cu transporters COPT1 and COPT3 showed increased Cu accumulation and reduced expression of CCA1, LHY, and circadian clock output genes. Defective developmental phenotypes were also observed in these plants. Spatial and temporal control of Cu homeostasis, therefore, may be important for plant environmental fitness (Andrés-Colás et al., 2010). It has also been reported that disordered circadian rhythm affects Fe homeostasis. Tight regulation of Fe homeostasis to maintain an optimal Fe level in plants has been found to be associated with circadian clock regulators such as TIME FOR COFFEE (TIC) that modulates the expression of the ferritin gene AtFer1 (Duc et al., 2009). The expression of AtFer1 was up-regulated with excess Fe. TIC could repress AtFer1 expression under low-Fe conditions in photoperiodic light and dark cycles (Duc et al., 2009). However, whether Fe status feeds back to regulate the circadian clock is uncertain.
Although Fe homeostasis in terms of uptake and translocation has been studied for decades, Fe availability is still an agricultural problem worldwide. Revealing the interplay between Fe homeostasis and internal cues such as modulation of the circadian clock can help increase understanding of their contributions to overall plant development. In this work, we investigated the effect of Fe deficiency on the circadian clock and found that it lengthened the circadian period. Our data suggest that the functional status of chloroplasts under Fe deficiency may play a key role in the lengthened circadian period.
RESULTS
Fe Deficiency Prolongs the Circadian Period of Oscillator Genes
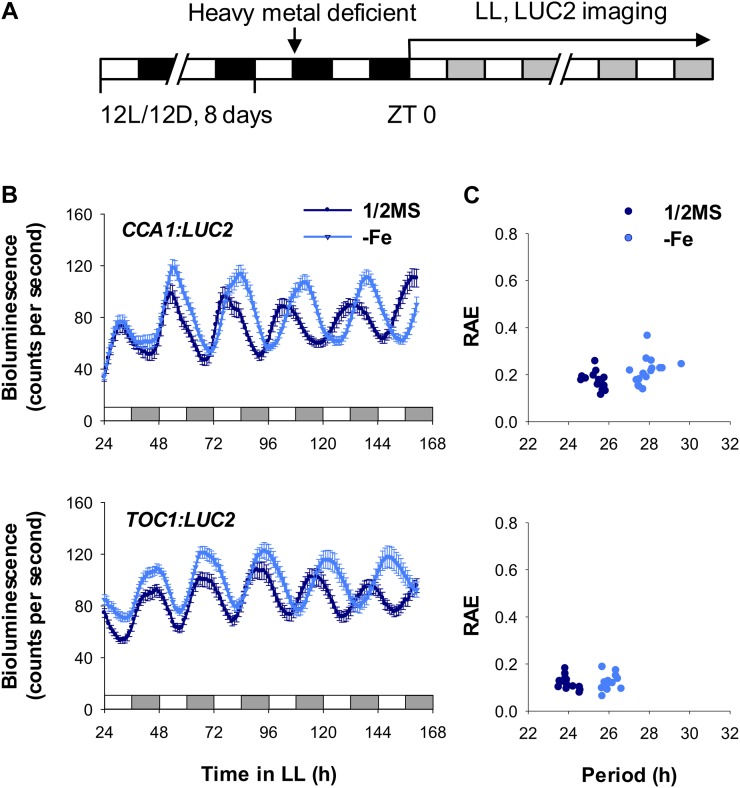
To monitor the circadian rhythm in plants under essential metal-deficient conditions, bioluminescence assays were performed on the oscillator reporter line CCA1:LUC2 (Wang et al., 2011) grown under Fe-, Zn-, Cu-, or Mn-deficient and constant-light (free-running) conditions. Circadian period lengthened in CCA1:LUC2 plants deficient in Fe (Fig. 1) but not in plants deficient in Zn, Cu, or Mn (Supplemental Fig. S1). The oscillator reporter line TOC1:LUC2 also showed a long circadian period under Fe-deficient conditions (Fig. 1) but not under Zn, Cu, or Mn deficiency (Supplemental Fig. S2). In addition, we detected a Fe deficiency-mediated long circadian period in the central oscillator reporter lines LHY:LUC2, PRR9:LUC2, PRR7:LUC2, PRR5:LUC2, GI:LUC2, and EARLY FLOWERING4 (ELF4):LUC2 (Supplemental Fig. S3). The degree of period lengthening in these lines was most obvious in CCA1:LUC2 and TOC1:LUC2 lines (Table I).
Figure 1.
Effect of Fe deficiency on the expression rhythm of central oscillator genes. A, Scheme of plant entrainment and treatments. Eight-day-old seedlings were placed in Fe-deficient medium; after 36 h, seedlings were transferred to continuous light (LL) at ZT 0. White and black bars denote the light and dark intervals, respectively. Gray bars indicate the subjective dark periods under continuous light. B, Promoter:LUC2 seedlings, indicated as CCA1 and TOC1, were placed in Fe-sufficient (1/2MS) or Fe-deficient (−Fe) medium, and bioluminescence was imaged every hour under continuous light. Data are expressed as means ± se (n ≥ 12). C, FFT-NLLS analysis of period length and relative amplitude error (RAE) from ZT 24 to ZT 162. Two or more independent experiments showed similar results.
Table I. Period length of oscillator genes under Fe-deficient (–Fe) conditions.
| Type | Genes | 1/2MS | –Fe | Period Increase in –Fe |
|---|---|---|---|---|
| h | ||||
| Promoter | pCCA1 | 25.42 ± 0.39 | 27.95 ± 0.61 | 2.53 |
| pTOC1 | 23.97 ± 0.38 | 26.03 ± 0.32 | 2.06 | |
| pLHY | 25.28 ± 0.33 | 25.96 ± 0.40 | 0.69 | |
| pPRR9 | 25.01 ± 0.37 | 25.79 ± 0.38 | 0.78 | |
| pPRR7 | 24.35 ± 0.27 | 25.29 ± 0.48 | 0.94 | |
| pPRR5 | 23.96 ± 0.29 | 24.51 ± 0.72 | 0.54 | |
| pGI | 24.76 ± 0.36 | 25.49 ± 0.37 | 0.73 | |
| pELF4 | 24.41 ± 0.32 | 25.09 ± 0.29 | 0.68 | |
| Transcripts | CCA1 | 22.77 ± 0.36 | 26.90 ± 0.5 | 4.13 |
| TOC1 | 22.86 ± 0.27 | 25.46 ± 0.41 | 2.60 | |
| LHY | 22.84 ± 0.25 | 25.46 ± 0.37 | 2.62 | |
| PRR9 | 22.79 ± 0.43 | 26.18 ± 0.39 | 3.39 | |
| PRR7 | 23.36 ± 0.38 | 25.41 ± 0.56 | 2.05 | |
| PRR5 | 22.86 ± 0.27 | 24.42 ± 0.51 | 1.56 | |
| GI | 22.99 ± 0.33 | 24.94 ± 0.63 | 1.95 | |
| ELF4 | 22.96 ± 0.43 | 25.75 ± 0.45 | 2.79 | |
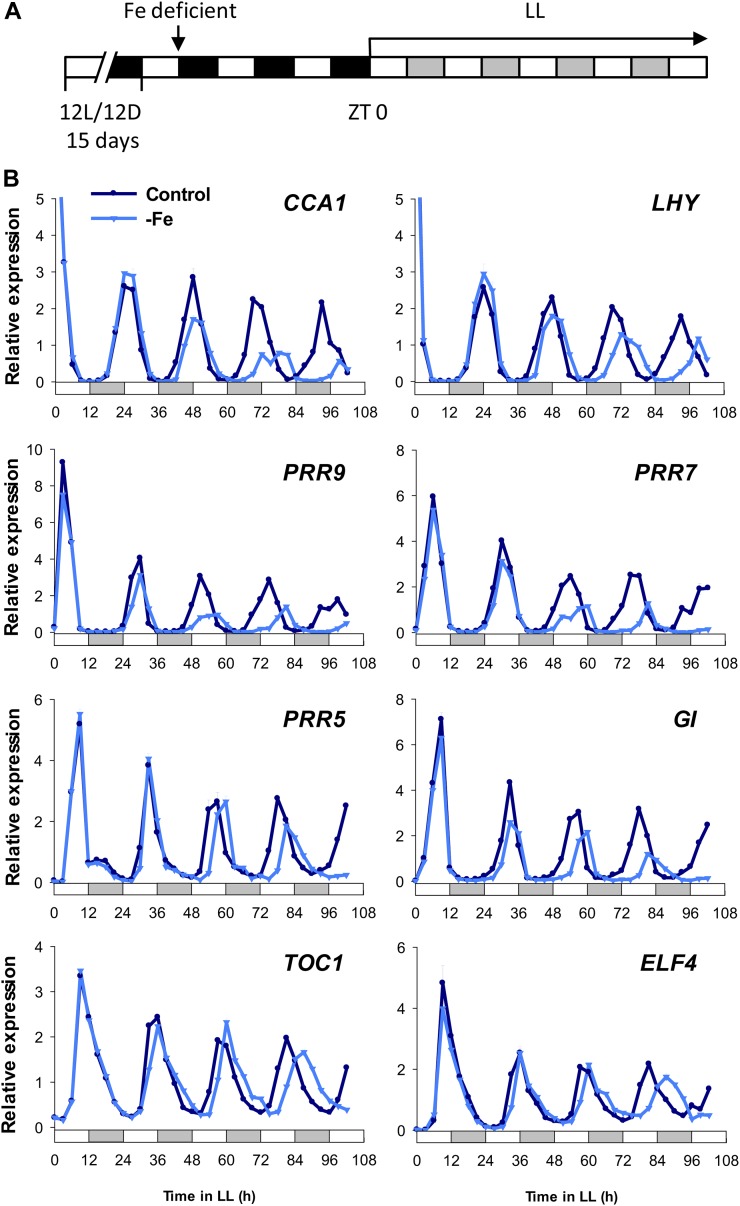
To confirm the effect of Fe deficiency on the expression of the central oscillator genes, quantitative reverse transcription (RT)-PCR was used to analyze the steady-state transcripts of CCA1, LHY, PRR9, PRR7, PRR5, GI, TOC1, and ELF4. The data showed long circadian periods under Fe deficiency (Fig. 2; Table I), and transcript levels of CCA1, LHY, PRR9, and PRR7 were reduced under Fe-deficient conditions in comparison with controls (Fig. 2). Therefore, Fe is essential for maintenance of the circadian oscillation period in Arabidopsis. It is worth noting that a Fe deficiency-mediated long period was more pronounced in the reporter line and in mRNA expression of CCA1 than in the other central oscillator genes examined. Our data suggested that Fe deficiency may have different impacts on the expression of different oscillator genes, possibly through the influence of promoter activity and also mRNA stability.
Figure 2.
Effect of Fe deficiency on the expression of central oscillator genes. A, Scheme of plant entrainment and treatments. Fifteen-day-old seedlings were treated with Fe-deficient medium (−Fe); after 60 h, seedlings were transferred to continuous light (LL) at ZT 0. Samples were harvested every 3 h for 102 h from ZT 0. B, Quantitative RT-PCR of mRNA expression of CCA1, LHY, PRR9, PRR7, PRR5, GI, TOC1, and ELF4. The white and gray bars indicate subjective light and dark periods, respectively, under continuous light. UBC21 was used as an internal control. Relative expression was calculated by normalization to the mean of total data. Data are expressed as means ± sd from three technical repeats. Two independent experiments showed similar results.
A Long Circadian Period Is Not Controlled by Nitric Oxide-Mediated Fe-Deficient Signaling
Fe deficiency triggers an uptake response in plants, a process that depends on the rapid synthesis of nitric oxide (NO). NO was found to be a critical factor in the signaling pathway for Fe acquisition under Fe-deficient conditions (Graziano et al., 2002; Graziano and Lamattina, 2007). Depleting NO content with NO biosynthesis inhibitors was found to block the induction of the Fe-deficiency response, whereas NO donor treatment stimulated Fe-acquisition-related genes (Graziano et al., 2002; Graziano and Lamattina, 2007; Besson-Bard et al., 2009; Chen et al., 2010).
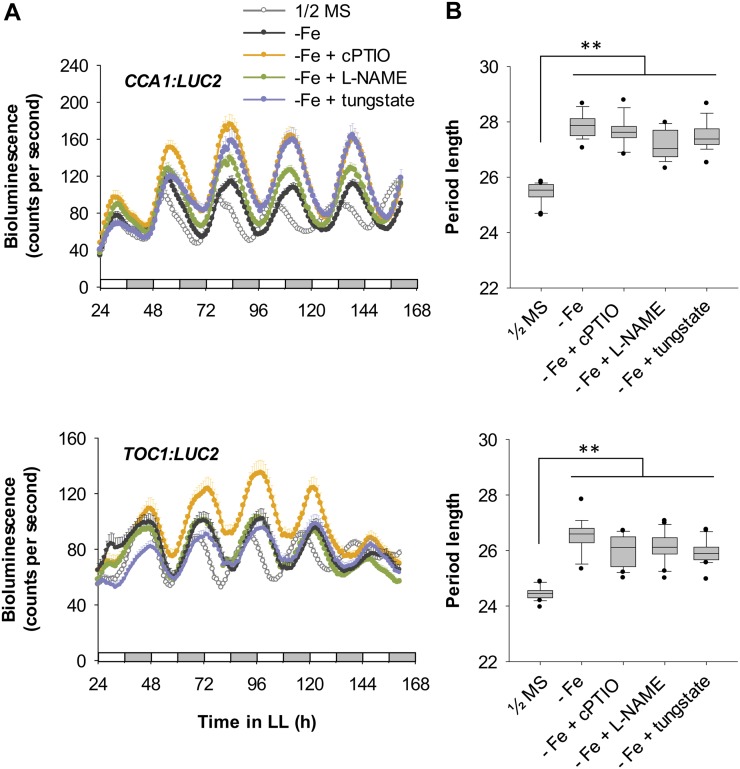
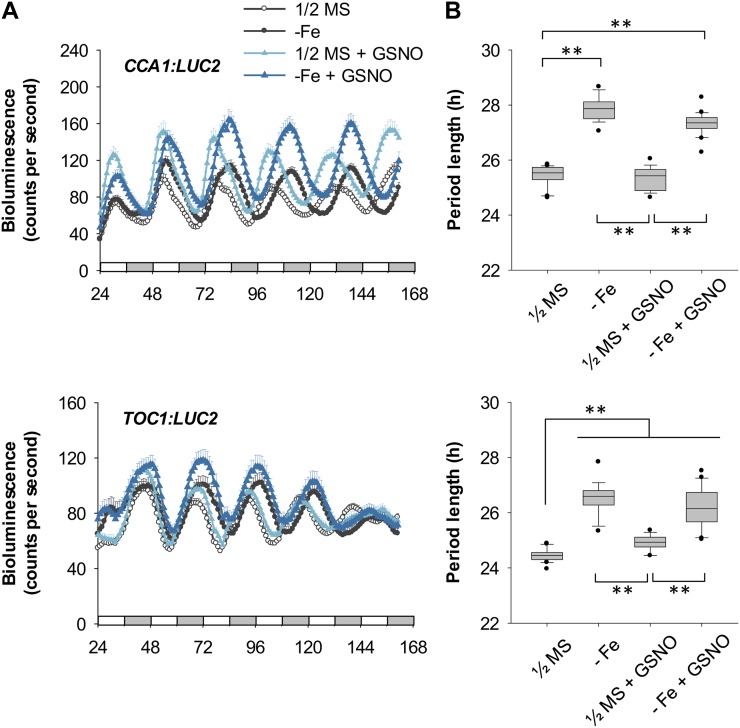
In light of these previous findings, we next examined whether the NO-mediated Fe-deficient signaling pathway was involved in modulating the Arabidopsis circadian clock. We used the inhibitors of NO production 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO), NG-nitro-l-arginine methyl ester (l-NAME), and tungstate to block the NO signaling pathway induced by Fe deficiency (Chen et al., 2010). Interestingly, none of these chemicals affected the Fe deficiency-mediated lengthened circadian period in CCA1:LUC2 and TOC1:LUC2 reporter lines (Fig. 3). Moreover, treatment with the NO donor S-nitrosoglutathione (GSNO) alone did not cause a long period of CCA1 expression, although it slightly increased TOC1 period length, nor did the donor have any additive effect on period length of either gene under Fe deficiency (Fig. 4). These results suggest that NO signaling is not involved in the long circadian period induced by Fe deficiency. Therefore, the signal for a slow-running circadian clock in Fe-deficient plants is not caused by typical Fe-deficiency signaling for Fe acquisition. We suggest that other signaling pathway(s) communicating the Fe deficiency to the central oscillator or other Fe-dependent component(s) may be responsible for the altered period length.
Figure 3.
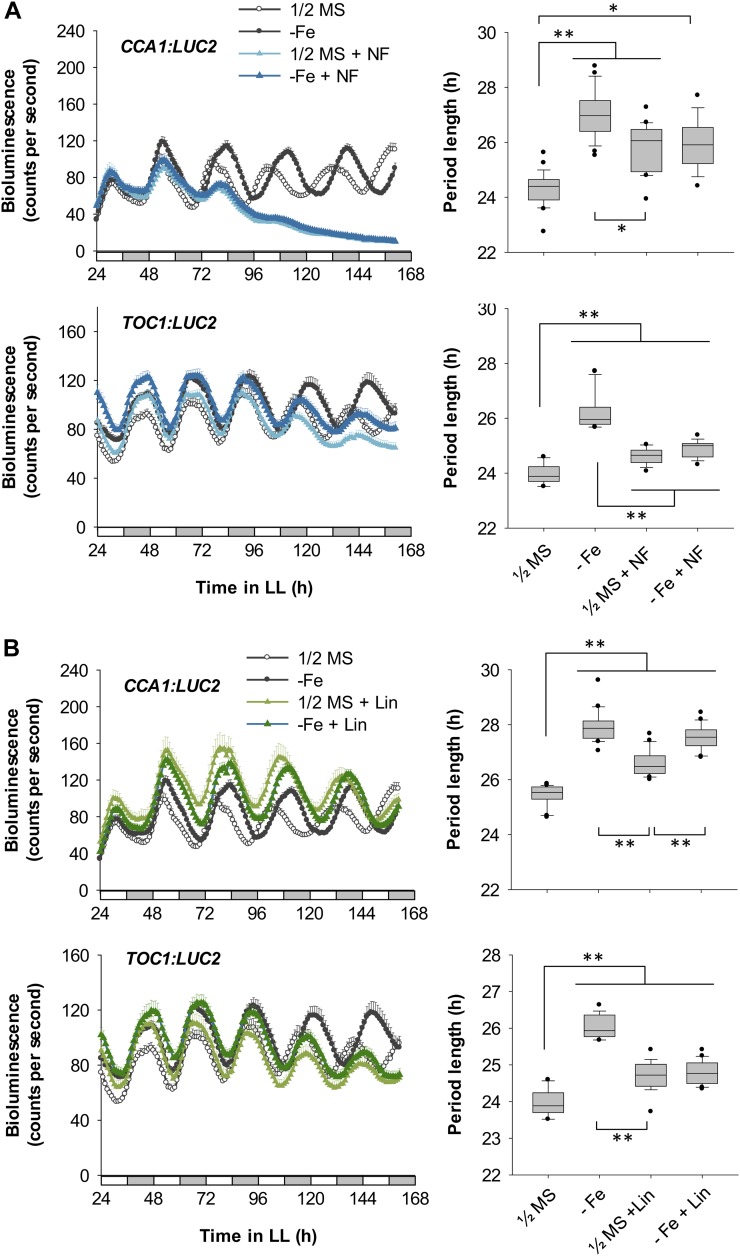
Circadian rhythm under Fe-deficient conditions with inhibition of Fe-deficiency signaling. A, CCA1:LUC2 and TOC1:LUC2 reporter plants were grown under Fe sufficiency (1/2MS), Fe deficiency (−Fe) alone, or Fe deficiency with 0.5 mm cPTIO, 1 mm l-NAME, or 0.25 mm tungstate. Data are expressed as means ± se (n ≥ 12). White and gray bars indicate subjective light and dark periods, respectively, under continuous light (LL). Seedlings were imaged for bioluminescence every hour from ZT 24 to ZT 162. B, FFT-NLLS analysis of period length (n ≥ 12). Box-plot analysis shows median and 25th and 75th percentiles. Error bars indicate the 90th and 10th percentiles. Black dots indicate the outlying points. **P < 0.0001. Two independent experiments showed similar results.
Figure 4.
Effect of GSNO. A, CCA1:LUC2 and TOC1:LUC2 reporter plants were grown under Fe-sufficient or Fe-deficient conditions (1/2MS or −Fe) with or without 50 μm GSNO. Data are expressed as means ± se of at least 12 seedlings. White and gray bars indicate subjective light and dark periods, respectively, under continuous light. Seedlings were imaged for bioluminescence every hour from ZT 24 to ZT 162. B, FFT-NLLS analysis of period length (n ≥ 12). Box-plot analysis shows median and 25th and 75th percentiles. Error bars indicate the 90th and 10th percentiles. Black dots indicate the outlying points. **P < 0.0001. Two independent experiments showed similar results.
Chloroplasts May Mediate the Fe-Deficiency Signal Resulting in a Long Circadian Period
Fe is an essential element for chloroplast development and function. In previous studies, plants grown under Fe deficiency showed disordered chloroplast structure and function (Stocking, 1975). Chloroplasts contain 80% to 90% of the Fe in leaf cells, and chlorosis and reduced photosynthetic activity are obvious physiological symptoms of Fe deficiency in plants (Landsberg, 1984; Terry and Abadia, 1986; Fig. 5, A and B). Circadian rhythm affects the expression of many chloroplast genes (Misquitta and Herrin, 2005). By contrast, the effect of chloroplast function on the circadian system has only been reported once in a mutant defective in a putative chloroplast RNA-binding protein (Hassidim et al., 2007).
Figure 5.
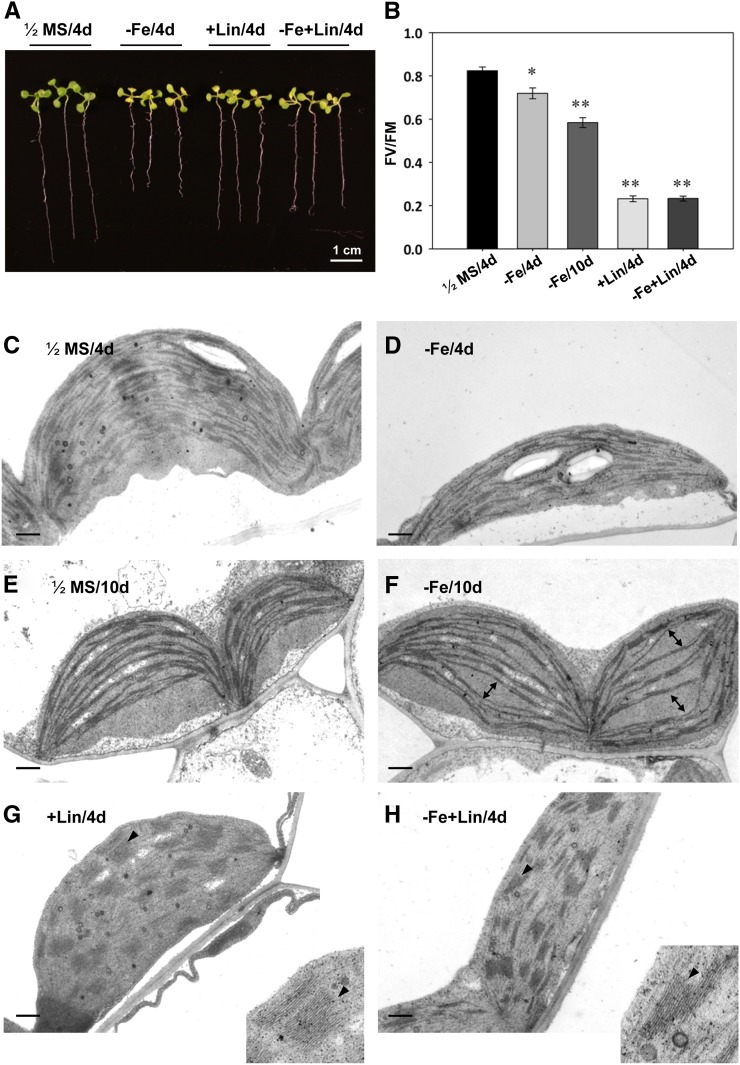
Effects of Fe deficiency and lincomycin treatment on Arabidopsis. A, Eight-day-old seedlings were placed on 1/2MS, 1/2MS containing 100 μm Ferrozine (−Fe), 0.5 mm lincomycin (+Lin), or lincomycin plus Ferrozine (−Fe+Lin) plates for 4 d under continuous light. B, Photosynthetic activity assay. Maximum photochemical efficiency of PSII in the dark-adapted state (FV/FM) was measured by use of a portable chlorophyll fluorometer (PAM-2100; Heinz Walz). *P < 0.05, **P < 0.01 compared with the control under the same conditions. C to H, Ultrastructure of chloroplasts in cotyledons or true leaves examined by transmission electron microscopy. C and D, Eight-day-old cotyledon under Fe sufficiency (1/2MS) or deficiency (−Fe) for 4 d. E and F, True leaf section of an 8-d-old plant under Fe-sufficient (1/2MS) or Fe-deficient (−Fe) conditions for 10 d. The thylakoid membranes became thinner in chloroplasts of Fe-deficient plants. Misaligned thylakoid membranes were also observed (marked by double-headed arrows). G and H, Cotyledon section of an 8-day-old plant treated with lincomycin under Fe-sufficient (+Lin) or Fe-deficient (−Fe+Lin) conditions for 4 d. The grana (black arrowheads) became thicker than the control (C). Bars = 0.5 μm.
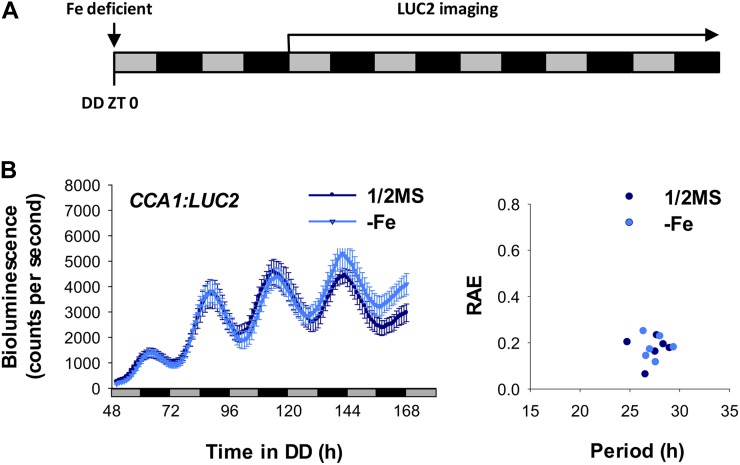
To examine the role of chloroplasts in a Fe deficiency-mediated long circadian period, we used the chloroplast biogenesis inhibitors norflurazon and lincomycin to create dysfunctional chloroplasts and monitored circadian behavior (Ruckle et al., 2007). Norflurazon or lincomycin alone caused a long circadian period. However, the degree of lengthening was less than with Fe deficiency alone (Fig. 6), despite their stronger impacts on the photosynthetic activity and structural integrity of membrane systems in chloroplasts (Fig. 5, C–H). Therefore, reduced photosynthetic activity is not the key determinant of the long circadian period in Fe-deficient plants. Consistent with this notion, growing the norflurazon-treated plants in Fe-deficient conditions did not further lengthen the circadian period (Fig. 6A). A slight increase in the period length of CCA1 expression was observed in lincomycin-treated plants grown under Fe-deficient conditions, but it was not an additive effect (Fig. 6B). The expression of TOC1 in the lincomycin-treated plants agrees with the observation of the requirement of functional chloroplasts for the Fe deficiency-mediated long circadian period (Fig. 6B). Interestingly, a long circadian period was also observed in etiolated seedlings with no mature chloroplasts (Pogson and Albrecht, 2011), and no further change was seen under Fe deficiency (Fig. 7). Therefore, functional chloroplasts may be involved in the mechanism by which the Fe-deficiency signal alters circadian period length. Alternatively, the delay in circadian rhythm may be caused by dysfunctional chloroplasts. In either case, the functional status of chloroplasts is important for transduction of the signal for altering circadian period length.
Figure 6.
Effect of norflurazon and lincomycin treatment on circadian period length. CCA1:LUC2 and TOC1:LUC2 reporter plants were grown under Fe-sufficient or Fe-deficient conditions with or without 5 μm norflurazon (NF; A) or 0.5 mm lincomycin (Lin; B). Data are means ± se (n ≥ 10). White and gray bars indicate subjective light and dark periods, respectively, under continuous light (LL). Seedlings were imaged for bioluminescence every hour from ZT 24 to ZT 162. Right panels show FFT-NLLS analysis of period length (n ≥ 10), except the first panel, which shows data from ZT 24 to ZT 120 (n ≥ 10). Box-plot analysis shows median and 25th and 75th percentiles. Error bars indicate the 90th and 10th percentiles. Black dots indicate the outlying points. *P < 0.0005, **P < 0.0001. Two independent experiments showed similar results.
Figure 7.
Effect of Fe deficiency on the circadian rhythm of etiolated seedlings. A, Seedlings were germinated on Fe-sufficient (1/2MS) or Fe-deficient (−Fe) medium in continuous dark (DD). Measurement of bioluminescence started at day 2. Gray and black bars indicate the subjective light and dark, respectively. B, Bioluminescence of pCCA1:LUC2 seedlings was measured every hour from ZT 50 to ZT 168 under continuous dark. Data are expressed as means ± se (n = 6). The right panel shows FFT-NLLS analysis with data from ZT 50 to ZT 168. Two independent experiments showed similar results. RAE, Relative amplitude error.
CCA1, LHY, and GI Are Involved in Fe-Deficiency Signaling for a Long Circadian Period
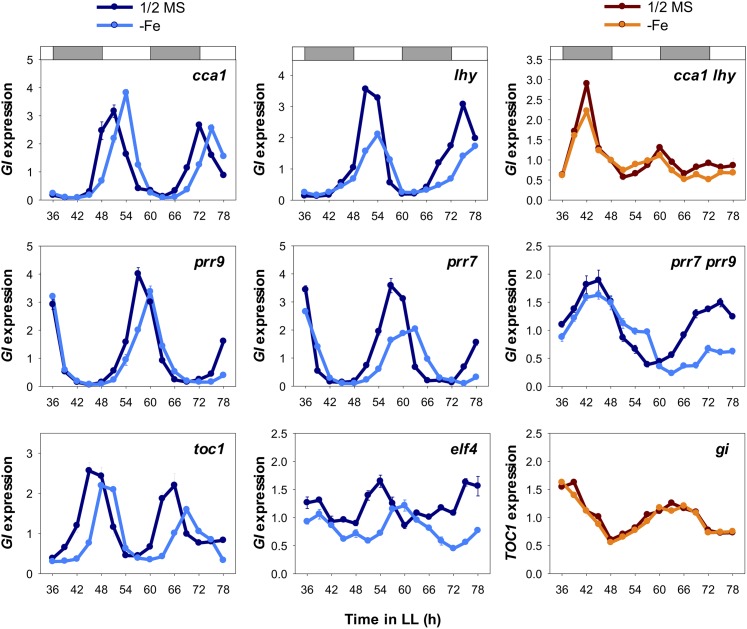
Because the results above suggested that Fe deficiency-mediated change in circadian period length may depend on functional chloroplasts, we next investigated how central oscillators receive the signal for a long circadian period induced by Fe deficiency. We examined the circadian rhythm by monitoring mRNA levels of GI as a reporter under Fe deficiency in oscillator mutants cca1-1 (Yakir et al., 2009), lhy-101 (Khanna et al., 2006), cca1-1 lhy-101 and prr9-10 (Ito et al., 2003), prr7-11 (Yamamoto et al., 2003), prr7-11 prr9-10 and toc1-101 (Kikis et al., 2005), and elf4-101 (Khanna et al., 2003) and with TOC1 as a reporter in the gi-2 mutant (Park et al., 1999). The long circadian period induced by Fe deficiency was ameliorated in the cca1 lhy double mutant and the gi mutant but not in the cca1, lhy, prr9, prr7, prr7 prr9, toc1, and elf4 mutants (Fig. 8). To validate these results, we further analyzed the expression of an additional gene, PRR7 (Supplemental Fig. S4). Together, these findings suggest that CCA1, LHY, and GI are required to relay the Fe-deficiency signal originating from chloroplasts for lengthened circadian period. PRR7 showed noticeable amplitude change under Fe deficiency (Fig. 2). It is difficult to clearly decipher the amplitude change in PRR7 in the cca1 lhy double mutant; however, the amplitude change in PRR7 was clearly ameliorated in the gi mutant (Supplemental Fig. S4). Therefore, we suggest that GI may also be responsible for the Fe deficiency-dependent amplitude change.
Figure 8.
Effect of Fe deficiency on the circadian clock in central oscillator mutants. Quantitative RT-PCR of mRNA expression of the central oscillator genes GI and TOC1 is shown in central oscillator mutants. The white and gray bars indicate subjective light and dark periods, respectively, under continuous light (LL). Samples were harvested every 3 h from ZT 36 to ZT 78. UBC21 level was used as an internal control. Relative expression was calculated by normalization to the mean of total data. Data are expressed as means ± sd from three technical repeats. Two independent experiments showed similar results.
DISCUSSION
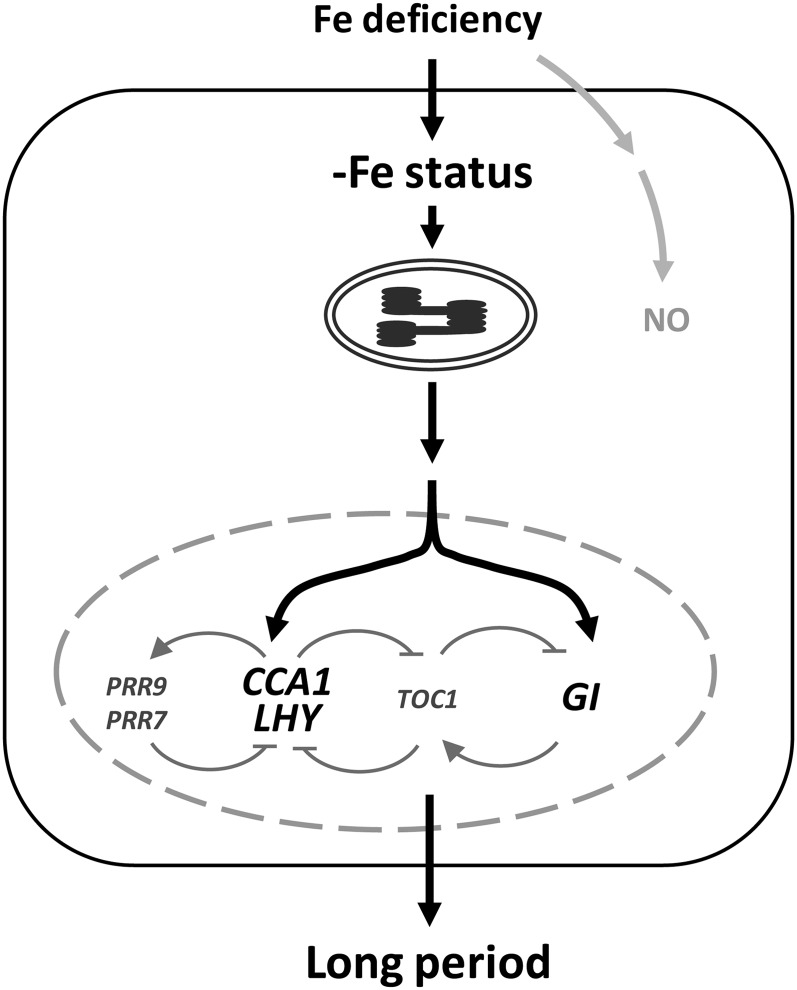
The expression of many micronutrient transporters is under the control of the circadian clock (Haydon et al., 2011). This suggests a close relationship between the circadian clock and the regulation of homeostasis of essential micronutrients in plants for nutrient acquisition, translocation to the shoot, and unloading to sink organs. Therefore, understanding how the availability of micronutrients in natural soil conditions affects the plant circadian clock is critical to increase our knowledge of both nutrient homeostasis and plant growth and development. In this study, using promoter:LUC2 reporter lines of oscillator genes and mRNA expression pattern analyses of the central oscillator genes in Arabidopsis, we revealed that Fe deficiency lengthens the period of the circadian oscillator. Interestingly, the key central clock components CCA1, LHY, and GI were required for the lengthened period under Fe deficiency. This finding indicates the existence of a feedback loop between Fe deficiency and the operation of the circadian clock. Our data ruled out NO as the signal communicating Fe deficiency to the circadian clock. However, chloroplasts, which depend heavily on Fe availability, were found to play an essential role in Fe deficiency-induced lengthening of the circadian period (Fig. 9).
Figure 9.
Working model showing the relationship between Fe deficiency and the circadian clock in Arabidopsis. When Fe is deficient, functional chloroplasts may be responsible for the transduction of signals to regulate the expression of oscillator genes in the nucleus. CCA1, LHY, and GI are required to relay signals to lengthen period length; however, NO may not be involved in the regulation of the circadian rhythm under Fe deficiency.
Chloroplasts accumulate high levels of Fe (Wollman et al., 1999). In chloroplasts, Fe is present in PSI and PSII, the cytochrome b6f complex, and ferredoxin for electron transfer in the light reactions of photosynthesis (Raven et al., 1999). Also, stroma-localized Fe and Cu/Zn superoxide dismutase can prevent the overaccumulation of reactive oxygen species (Asada, 1999). In Fe deficiency, the functional state of chloroplasts may change. In our study, treatment with norflurazon or lincomycin to produce dysfunctional chloroplasts lengthened the circadian period in Arabidopsis (Fig. 6). Fe-deficient conditions did not further lengthen the circadian period in norflurazon- or lincomycin-treated plants or in plants grown in the dark (Figs. 6 and 7). Therefore, functional chloroplasts are essential for plants to maintain an approximately 24-h circadian clock. In a previous study, dysfunctional chloroplasts were found to affect the circadian system in Arabidopsis defective in CHLOROPLAST RNA BINDING (CRB; Hassidim et al., 2007). The crb mutant showed altered chloroplast morphologic features and reduced photosynthesis as well as a phase delay in circadian rhythm (Hassidim et al., 2007). Our results lend further credence to the hypothesis that the functional state of chloroplasts is important for an approximately 24-h circadian rhythm.
Many chloroplast proteins are encoded in the nucleus, synthesized in the cytoplasm, and posttranslationally imported into the chloroplasts to carry out their biological functions. A well-known example of a nucleus-encoded chloroplast protein is the small subunit of Rubisco, a membrane-bound protein complex that performs the dark reactions of photosynthesis (Rochaix, 1987). In contrast, the large subunit of Rubisco is encoded by the chloroplast genome. That different genomes inside a cell encode separate subunits of a protein complex suggests that functionally related protein synthesis in these organelles must be well coordinated (Misquitta and Herrin, 2005). Coordination of gene expression in the two organelles may allow plants to respond effectively to environmental stimuli such as the light/dark cycle and the availability of nutrition. Our results strongly support the relationship between the circadian clock, chloroplasts, and Fe status in Arabidopsis. Under Fe deficiency, chloroplasts may have an integral function to coordinate the signals and delay the circadian rhythm via retrograde signaling pathways. Chloroplasts may act as sensors of environmental changes and help integrate cellular signaling networks in the plant cell for effective responses (Fernández and Strand, 2008).
Fe is one of the most important elements for vegetative and reproductive plant growth (Le Jean et al., 2005; Briat et al., 2007; Chu et al., 2010; Roschzttardtz et al., 2011). Because chloroplasts are the major Fe storage sink in plants (Briat et al., 2007), they should be the most sensitive to Fe deficiency. Our study suggests that functional chloroplasts signal Fe deficiency to the central oscillator. CCA1, LHY, and GI are likely entry points or sites of sensing the chloroplast signal under Fe deficiency (Fig. 8). The long circadian period in cca1 and lhy, but not the cca1 lhy double mutant, is consistent with the partially redundant functions of CCA1 and LHY in the circadian clock of Arabidopsis (Alabadí et al., 2002; Mizoguchi et al., 2002). With lengthened circadian oscillation, plants under Fe deficiency may be able to continue to accumulate Fe by remaining in the vegetative stage, thereby reducing the chances of forming immature fruits and seeds. Therefore, this system may be an adaptive strategy that allows increases in progeny fitness under Fe deficiency.
Whether the circadian clock plays a gating role in Fe acquisition is of interest. Fe acquisition-related genes IRON REGULATED TRANSPORTER1 (IRT1), FERRIC CHELATE REDUCTASE, and FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR may be regulated by the circadian clock (Vert et al., 2003; Meiser and Bauer, 2012). By analyzing public transcriptome data, we found that the expression of IRT1 was regulated by the circadian clock http://diurnal.mocklerlab.org; data not shown). Chloroplasts may be one of the sensors of Fe deficiency that coordinate the control of Fe acquisition over an approximately 24-h period through the circadian clock. These lines of research are worthy of further exploration.
In conclusion, in Arabidopsis, Fe deficiency may induce disordered chloroplast development or functional state, which is a signal to the central oscillator interpreted by CCA1, LHY, or GI to modulate circadian period length for plant growth and development.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) Columbia ecotype was used for all experiments. The luciferase reporter lines CCA1:LUC2, LHY:LUC2, TOC1:LUC2, PRR9:LUC2, PRR7:LUC2, PRR5:LUC2, ELF4:LUC2, and GI:LUC2 for bioluminescence measurement were generated by Wang et al. (2011). The circadian clock mutants used in this study were cca1-1 (Yakir et al., 2009; Wang et al., 2011), lhy-101 (Khanna et al., 2006), toc1-101 (Kikis et al., 2005), elf4-101 (Khanna et al., 2003), gi-2 (Park et al., 1999), prr7-11 (Yamamoto et al., 2003), prr9-10 (Ito et al., 2003), and cca1-1 lhy-101 and prr7-11 prr9-10 double mutants from this work.
Heavy Metal-Deficient Conditions
For bioluminescence assay, CCA1:LUC2 and TOC1:LUC2 lines were grown on one-half-strength Murashige and Skoog (1/2MS) agar medium (Murashige and Skoog, 1962) under a 12-h-light/12-h-dark period with white light illumination (70 μmol m−2 s−1). After 8 d, seedlings were transferred to 1/2MS as a control or medium with −Fe (Fe dropout and with 100 μm Ferrozine [P9762; Sigma-Aldrich]), −Zn (Zn dropout and with 50 μm N,N,N′,N′-tetrakis[2-pyridylmethyl] ethylenediamine [P4413; Sigma-Aldrich]), −Cu (Cu dropout and with 10 μm bathocuproine sulfonate [B1125; Sigma-Aldrich]), or −Mn (Mn dropout) for 36 h and moved to continuous light (50 μmol m−2 s−1) at Zeitgeber time (ZT) 0.
Bioluminescence Measurement and Data Analyses
Promoter:LUC2 lines were grown on 1/2MS agar in 12-h-light/12-h-dark conditions with white light illumination (70 μmol m−2 s−1). After 8 d, seedlings were treated with different reagents for 36 h and then moved to continuous light (50 μmol m−2 s−1) at ZT 0. For −Fe experiments, seedlings were transferred to −Fe medium. To inhibit −Fe signaling, seedlings were treated with −Fe plus 0.5 mm cPTIO (C221; Sigma-Aldrich), plus 1 mm l-NAME (N5751; Sigma-Aldrich), or plus 0.25 mm tungstate (72069; Sigma-Aldrich). To induce −Fe signaling, seedlings were treated with 1/2MS plus 50 μm GSNO (N4148; Sigma-Aldrich) or −Fe plus 50 μm GSNO. To block chloroplast biogenesis or induce photobleaching of chloroplasts, seedlings were treated with 1/2MS plus 0.5 mm lincomycin (L6004; Sigma-Aldrich) or plus 5 μm norflurazon (34364; Fluka) or −Fe plus 0.5 mm lincomycin or plus 5 μm norflurazon.
Bioluminescence assays used black 96-well microplates as described previously (Wang et al., 2011) with minor modifications. Luciferin was added to seedlings (final concentration, 0.5 mm [RE1605; Promega]) before transfer to continuous light. Bioluminescence activity was measured every hour from 24 to 162 h. To obtain etiolated seedlings, CCA1:LUC2 seeds were sown on 96-well microplates. Each well contained approximately 50 to 100 seeds and 100 μL of solid medium with or without 100 μm Ferronine. Plates were placed at 4°C for 4 d to synchronize the germination, and seeds were treated with light for 3 h at 23°C on day 2. After cold stratification, seedlings were grown in darkness. Luciferin (final concentration, 2.5 mm) was added to 2-d-old etiolated seedlings, and bioluminescence was measured from ZT 50 to ZT 168 in continuous darkness.
Data were imported into the Biological Rhythms Analysis Software System (available from http://www.amillar.org; Southern et al., 2006), and period length and relative amplitude error were calculated for each promoter by fast Fourier transform-nonlinear least squares (FFT-NLLS) analysis (Plautz et al., 1997).
Real-Time Quantitative RT-PCR
Wild-type plants and clock mutant plants were grown on 1/2MS medium under a 12-h-light/12-h-dark cycle for 12 d and then transferred to hydroponic culture. After adaptation for 3 d, plants were washed and transferred to −Fe medium for 60 h and then to continuous light at ZT 0. The shoots of plants were harvested every 3 h for 102 h.
RNA extraction and RT were performed as described (Chen et al., 2011). Quantitative RT-PCR analyses involved use of the KAPA SYBR FAST qPCR Kit (Kapa Biosystems) with the StepOnePlus Real-Time PCR System (Applied Biosystems) using the programs recommended by the manufacturer (20 s at 90°C, 40 cycles at 95°C for 3 s and 60°C for 30 s). UBC21 was used as an internal control (James et al., 2008). For relative expression of genes of interest, the value at each time point was normalized to the mean value for all time points. Sequences of primers are listed in Supplemental Table S1.
Photosynthetic Activity Assay and Transmission Electron Microscopy
Photosynthetic activity assay was as described previously (Shin et al., 2012). The maximum quantum yield was measured by use of a portable chlorophyll fluorometer (PAM-2100; Heinz Walz). Transmission electron microscopy was performed in the Plant Cell Biology Core Laboratory at the Institute of Plant and Microbial Biology, Academia Sinica, according to Chang et al. (2008).
Sequence data from this study can be found in the Arabidopsis Genome Initiative data library with the following locus identifiers: CCA1 (At2g46830), LHY (At1g01060), TOC1 (At5g61380), ELF4 (At2g40080), PRR5 (At5g24470), PRR7 (At5g02810), PRR9 (At2g46790), GI (At1g22770), and UBC21 (At5g25760).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Circadian rhythm of CCA1 under metal-deficient conditions.
Supplemental Figure S2. Circadian rhythm of TOC1 under metal-deficient conditions.
Supplemental Figure S3. Circadian rhythm of LHY, PRR9, PRR7, PRR5, GI, and ELF4 under Fe-deficient conditions.
Supplemental Figure S4. Effect of Fe deficiency on the circadian clock in central oscillator mutants.
Supplemental Table S1. Primers for quantitative RT-PCR.
Note Added in Proof
An extended period length in Arabidopsis under iron deficiency was also observed in a recently published report (Hong S, Kim SA, Guerinot ML, McClung CR [2013] Reciprocal interaction of the circadian clock with the iron homeostasis network in Arabidopsis. Plant Physiol 161: 893–903).
Acknowledgments
We thank Dr. Ming-Hsiun Hsieh for valuable discussion and Miranda Loney and Laura Smales for manuscript editing. We also thank Wann-Neng Jane for technical assistance with transmission electron microscopy.
Glossary
- Fe
iron
- Cu
copper
- Zn
zinc
- Mn
manganese
- RT
reverse transcription
- NO
nitric oxide
- GSNO
S-nitrosoglutathione
- 1/2MS
one-half-strength Murashige and Skoog
- ZT
Zeitgeber time
- FFT-NLLS
fast Fourier transform-nonlinear least squares
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- l-NAME
NG-nitro-L-arginine methyl ester
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Andrés-Colás N, Perea-García A, Puig S, Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol 153: 170–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Baligar VC, Fageria NK, Paiva AQ, Silveira A, Pomella AWV, Machado RCR. (2006) Light intensity effects on growth and micronutrient uptake by tropical legume cover crops. J Plant Nutr 29: 1959–1974 [Google Scholar]
- Besson-Bard A, Astier J, Rasul S, Wawer I, Dubreuil-Maurizi C, Jeandroz S, Wendehenne D. (2009) Current view of nitric oxide-responsive genes in plants. Plant Sci 177: 302–309 [Google Scholar]
- Briat JF, Curie C, Gaymard F. (2007) Iron utilization and metabolism in plants. Curr Opin Plant Biol 10: 276–282 [DOI] [PubMed] [Google Scholar]
- Briat JF, Lobreaux S. (1997) Iron transport and storage in plants. Trends Plant Sci 2: 187–193 [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chen CC, Chen YY, Tang IC, Liang HM, Lai CC, Chiou JM, Yeh KC. (2011) Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol 156: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Yang JL, Qin C, Jin CW, Mo JH, Ye T, Zheng SJ. (2010) Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol 154: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL. (2010) Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ND, Berg JM. (1998) Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science 282: 2018–2022 [DOI] [PubMed] [Google Scholar]
- Clemens S. (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 475–486 [DOI] [PubMed] [Google Scholar]
- de Montaigu A, Tóth R, Coupland G. (2010) Plant development goes like clockwork. Trends Genet 26: 296–306 [DOI] [PubMed] [Google Scholar]
- Duc C, Cellier F, Lobréaux S, Briat JF, Gaymard F. (2009) Regulation of iron homeostasis in Arabidopsis thaliana by the clock regulator time for coffee. J Biol Chem 284: 36271–36281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fernández AP, Strand A. (2008) Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr Opin Plant Biol 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Beligni MV, Lamattina L. (2002) Nitric oxide improves internal iron availability in plants. Plant Physiol 130: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Lamattina L. (2007) Nitric oxide accumulation is required for molecular and physiological responses to iron deficiency in tomato roots. Plant J 52: 949–960 [DOI] [PubMed] [Google Scholar]
- Grotz N, Guerinot ML. (2002) Limiting nutrients: an old problem with new solutions? Curr Opin Plant Biol 5: 158–163 [DOI] [PubMed] [Google Scholar]
- Harmer SL. (2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hassidim M, Yakir E, Fradkin D, Hilman D, Kron I, Keren N, Harir Y, Yerushalmi S, Green RM. (2007) Mutations in CHLOROPLAST RNA BINDING provide evidence for the involvement of the chloroplast in the regulation of the circadian clock in Arabidopsis. Plant J 51: 551–562 [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Bell LJ, Webb AA. (2011) Interactions between plant circadian clocks and solute transport. J Exp Bot 62: 2333–2348 [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA. (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30: 333–349 [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. (2010) Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol 13: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T. (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, Nimmo HG. (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Khanna R, Kikis EA, Quail PH. (2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133: 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH. (2006) Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Lahti M, Aphalo PJ, Finér L, Ryyppö A, Lehto T, Mannerkoski H. (2005) Effects of soil temperature on shoot and root growth and nutrient uptake of 5-year-old Norway spruce seedlings. Tree Physiol 25: 115–122 [DOI] [PubMed] [Google Scholar]
- Landsberg EC. (1984) Regulation of iron-stress-response by whole-plant activity. J Plant Nutr 7: 609–621 [Google Scholar]
- Le Jean M, Schikora A, Mari S, Briat JF, Curie C. (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44: 769–782 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London [Google Scholar]
- Meiser J, Bauer P. (2012) Looking for the hub in Fe signaling. Plant Signal Behav 7: 688–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta R, Herrin DL. (2005) Circadian regulation of chloroplast transcription: a review. Plant Tissue Culture 15: 83–101 [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G. (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S. (1999) Iron acquisition by plants. Curr Opin Plant Biol 2: 250–253 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Peñarrubia L, Andrés-Colás N, Moreno J, Puig S. (2010) Regulation of copper transport in Arabidopsis thaliana: a biochemical oscillator? J Biol Inorg Chem 15: 29–36 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V. (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Evans MCW, Korb RE. (1999) The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth Res 60: 111–149 [Google Scholar]
- Rochaix JD. (1987) Molecular genetics of chloroplasts and mitochondria in the unicellular green alga Chlamydomonas. FEMS Microbiol Rev 46: 13–34 [Google Scholar]
- Roschzttardtz H, Séguéla-Arnaud M, Briat JF, Vert G, Curie C. (2011) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Shin LJ, Lo JC, Yeh KC. (2012) Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiol 159: 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern MM, Brown PE, Hall A. (2006) Luciferases as reporter genes. Methods Mol Biol 323: 293–305 [DOI] [PubMed] [Google Scholar]
- Stocking CR. (1975) Iron deficiency and the structure and physiology of maize chloroplasts. Plant Physiol 55: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Terry N, Abadia J. (1986) Function of iron in chloroplasts. J Plant Nutr 9: 609–646 [Google Scholar]
- Vert GA, Briat JF, Curie C. (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH. (2011) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wollman FA, Minai L, Nechushtai R. (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim Biophys Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM. (2009) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T. (2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130 [DOI] [PubMed] [Google Scholar]