Trade-off between main and lateral root growth dominates the genetic plasticity of plant responses to potassium deficiency.
Abstract
Root architecture is a highly plastic and environmentally responsive trait that enables plants to counteract nutrient scarcities with different foraging strategies. In potassium (K) deficiency (low K), seedlings of the Arabidopsis (Arabidopsis thaliana) reference accession Columbia (Col-0) show a strong reduction of lateral root elongation. To date, it is not clear whether this is a direct consequence of the lack of K as an osmoticum or a triggered response to maintain the growth of other organs under limiting conditions. In this study, we made use of natural variation within Arabidopsis to look for novel root architectural responses to low K. A comprehensive set of 14 differentially responding root parameters were quantified in K-starved and K-replete plants. We identified a phenotypic gradient that links two extreme strategies of morphological adaptation to low K arising from a major tradeoff between main root (MR) and lateral root elongation. Accessions adopting strategy I (e.g. Col-0) maintained MR growth but compromised lateral root elongation, whereas strategy II genotypes (e.g. Catania-1) arrested MR elongation in favor of lateral branching. K resupply and histochemical staining resolved the temporal and spatial patterns of these responses. Quantitative trait locus analysis of K-dependent root architectures within a Col-0 × Catania-1 recombinant inbred line population identified several loci each of which determined a particular subset of root architectural parameters. Our results indicate the existence of genomic hubs in the coordinated control of root growth in stress conditions and provide resources to facilitate the identification of the underlying genes.
The ability of plants to actively respond to nutrient scarcity with changes in root architecture is a fascinating phenomenon. Advances in root research and breeding efforts that focus on the enhancement of root traits have been recognized as principal goals to ensure those high yields necessary to feed an ever-growing human population (Hammer et al., 2009; Den Herder et al., 2010). Indeed, understanding the adaptations of root systems to environmental factors has been pointed out as a key issue in modern agriculture (Den Herder et al., 2010).
Potassium (K) is the quantitatively most important cation for plant growth, as it serves as the major osmoticum for cell expansion (Leigh and Wyn Jones, 1984; Amtmann et al., 2006). Moreover, K is essential for many cellular and tissue processes, such as enzymatic activity, transport of minerals and metabolites, and regulation of stomatal aperture (Amtmann et al., 2006). Even in fertilized fields, rapid K uptake by plants can lead to K shortage in the root environment, especially early in the growth season. Root adaptations to K deficiency (low K) take place at the physiological (Armengaud et al., 2004; Shin and Schachtman, 2004; Alemán et al., 2011), metabolic (Armengaud et al., 2009a), and morphological levels. In a classic study, Drew (1975) showed an increase in overall lateral root (LR) growth of barley seedlings, even when K was supplied only to parts of the root system. Conversely, a typical response of Arabidopsis (Arabidopsis thaliana) Columbia (Col-0) seedlings to low K is the drastic reduction of LR elongation (Armengaud et al., 2004; Shin and Schachtman, 2004). Conflicting data have been published on the effect of low K on main root (MR) growth in the same species, ranging from no effect (Shin and Schachtman, 2004) to impaired MR elongation (Jung et al., 2009; Kim et al., 2010). Some components involved in K starvation responses have been identified, such as jasmonates (Armengaud et al., 2004, 2010), reactive oxygen species (Shin and Schachtman, 2004), and ethylene (Jung et al., 2009). However, the molecular identity of a root K sensor acting at the base of the signaling cascade is so far unknown.
Genetic variation within species is a useful resource to dissect the genetic components determining phenotypes (Koornneef et al., 2004; Trontin et al., 2011; Weigel, 2012). Natural variation within Arabidopsis has been the basis for many studies on plant morphology, physiology, and development as well as stress response (Alonso-Blanco et al., 2009; Weigel, 2012). Natural variation of root traits such as primary root length (Mouchel et al., 2004; Loudet et al., 2005; Sergeeva et al., 2006), LR length (Loudet et al., 2005), and total root size (Fitz Gerald et al., 2006) have pinpointed genomic regions underlying the phenotypic variation via mapping of quantitative trait loci (QTLs) as a first step toward the identification of novel regulatory genes (Mouchel et al., 2004). This strategy has also been applied to environmental responses, such as growth responses to phosphate starvation (Reymond et al., 2006; Svistoonoff et al., 2007). However, despite their importance for plant growth and their strong effect on overall root architecture, root responses to K deficiency have not been genetically dissected.
Here, we show that Arabidopsis accessions follow different strategies to adapt to K starvation. We present the quantification of a comprehensive set of root architectural parameters of Arabidopsis grown in K-sufficient and K-deficient media and the identification of genetic loci, each of which determines the response of a distinct subset of root architectural parameters to K starvation.
RESULTS
Genotype and K Supply Cause Phenotypic Variation of Root Architecture in Arabidopsis
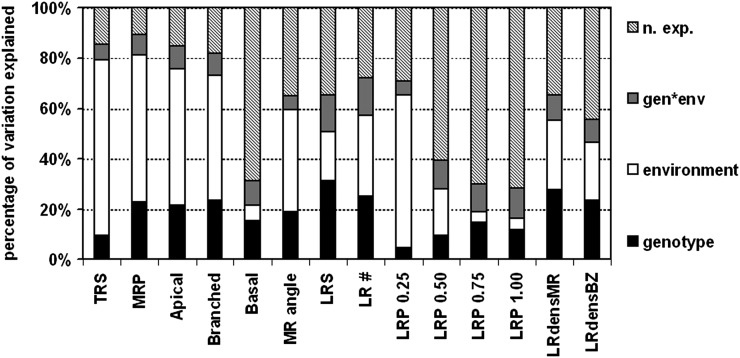
Seedlings of 26 natural accessions of Arabidopsis (Supplemental Table S1; McKhann et al., 2004) were grown in two contrasting environments: control ([K] = 2 mm) and low K ([K] = 0.01 mm). Quantitative analysis of 14 root architectural traits was performed 12 d after germination (DAG) with EZ Rhizo software (Armengaud et al., 2009b; Supplemental Data Set S1). For abbreviations and definitions of traits, see Table I. Across all accessions, low K supply resulted in a reduction of the trait value as compared with the control for 11 of the 14 quantified traits (Table I). Only LR path length in the third quartile from the root-shoot junction (LRP 0.75) and LR densities normalized to the length of the MR (LRdensMR) or branched zone (LRdensBZ) were, on average, increased in low K. We detected significant correlations between most traits (Supplemental Table S2). However, some correlations were inverse between control and low K, changing from positive to negative, most notably for LR path length, or from negative to positive, as for LR density. Moreover, accession-specific correlations of traits between control and low K were generally low (Table I; Supplemental Fig. S1), suggesting significant variation of K deficiency responses within the genotype pool. Exceptions were the angle of the MR from full verticality (MR angle) and LR densities with r2 values between 0.41 and 0.49. Hence, low-K responses of these traits are less variable between genotypes. Global ANOVA of the whole data set revealed the individual contributions of genotype, environment (low K versus control), and genotype-environment interactions to the total variation explained for each parameter (Fig. 1). The extent to which each of these three factors contributed to individual root parameters varied considerably, ranging from 4.5% to 31.2% for environment, from 3.8% to 69.8% for genotype, and from 5.2% to 14.8% for genotype-environment interaction. The highest percentage explained by genotype was found among LR parameters, such as LR number (LR #), LR system size, LRdensMR, and LRdensBZ. The environment (media composition) strongly influenced total root size (TRS) and MR parameters, such as MR path length (MRP), lengths of the apical zone (Apical) and branched zone, as well as MR angle. The environment also strongly affected LR path length in the first quartile (LRP 0.25). Although genotype-environment interactions were generally less important (5.2% for MR angle to 14.8% for LR #), genotype-environment interaction accounted for a higher proportion variation in LR path length in the second quartile (LRP 0.50), third quartile (LRP 0.75), and fourth quartile (LRP 1.00).
Table I. Means ± se of 14 root parameters across all accessions quantified in control and K deficiency.
The K response ratio (low K/control) was calculated as mean in low K divided by mean in control. Pearson correlation coefficients (r2) are shown for correlation of low-K with control values, computed from averages of each accession in each condition. se values are given in parentheses. n/a, Not available.
| Trait Identifier | Trait Description | Unit | Control | Low K | Low K/Control | r2 |
|---|---|---|---|---|---|---|
| % | ||||||
| TRS | Total root size | cm | 16.48 (0.33) | 4.58 (0.17) | 28 | 0.05 |
| MRP | MR path length | cm | 7.11 (0.12) | 2.72 (0.11) | 38 | 0.23 |
| Apical | Apical zone length | cm | 3.19 (0.07) | 1.08 (0.06) | 34 | 0.18 |
| Branched | Branched zone length | cm | 3.65 (0.08) | 1.44 (0.06) | 39 | 0.21 |
| Basal | Basal zone length | cm | 0.25 (0.01) | 0.19 (0.01) | 76 | 0.07 |
| MR angle | Angle of MR from full verticality | ° | 15.18 (0.5) | 2.42 (0.48) | 16 | 0.41 |
| LRS | LR system size (as proportion of total root size) | % | 55 (1) | 40 (1) | 73 | 0.18 |
| LR # | First-order LR number | 12.65 (0.26) | 7.17 (0.24) | 57 | 0.07 | |
| LRP 0.25 | Mean LR path length in the uppermost quartile of the MR | cm | 1.15 (0.03) | 0.29 (0.02) | 25 | 0.01 |
| LRP 0.50 | Mean LR path length in the second quartile from the top of the MR | cm | 0.44 (0.02) | 0.23 (0.02) | 52 | 0.01 |
| LRP 0.75 | Mean LR path length in the third quartile from the top of the MR | cm | 0.08 (0.01) | 0.15 (0.01) | 188 | 0.03 |
| LRP 1.00 | Mean LR path length in the fourth quartile from the top of the MR | cm | 0 (0) | 0.08 (0.01) | n/a | 0.03 |
| LRdensMR | LR density along the MR | cm−1 | 1.82 (0.03) | 3.16 (0.09) | 174 | 0.49 |
| LRdensBZ | LR density within the branched zone | cm−1 | 3.54 (0.06) | 5.86 (0.17) | 166 | 0.48 |
Figure 1.
Variation in root parameters explained by genotype and environmental conditions obtained through global ANOVA. Independently analyzed individual root parameters, as per Table I, are given on the x axis. Twenty-six Arabidopsis natural accessions (genotype), as per Supplemental Table S1, were grown on control and low-K media (environment), and root architecture parameters were quantified for 480 plants phenotyped 12 DAG (n = 7–12 per genotype per condition). ANOVA was computed using type III sums of squares at a significance level of P < 0.05. n. exp., Not explained; gen*env, genotype-environment interaction.
Accessions Adopt Different Strategies for Adjusting Their Root Architecture to Low K
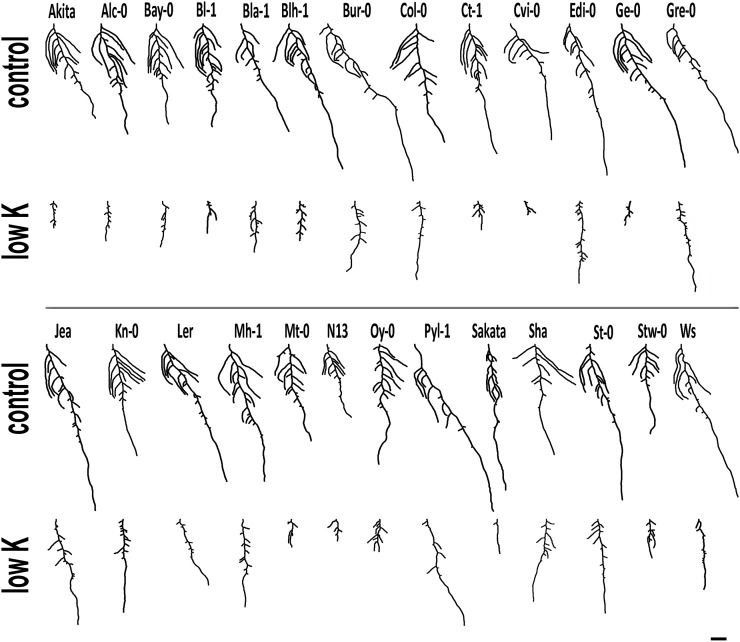
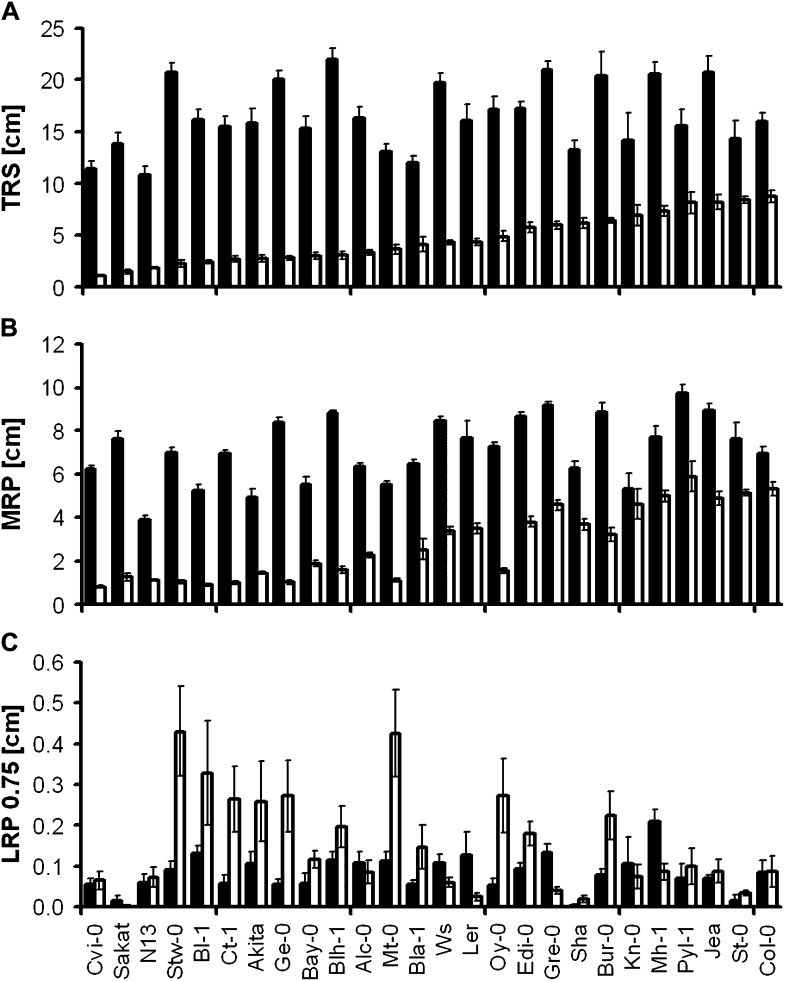
An overview of variation in root architecture is given in Figure 2. Differences between accessions were already visible in control conditions. Whereas some accessions grew long MRs while compromising LR elongation (e.g. Burren-0 [Bur-0], Landsberg erecta [Ler], Le Pyla-1 [Pyl-1]), others showed the opposite phenotype, i.e. short MRs but longer LRs (Bayreuth-0 [Bay-0], Stobowa-0 [Stw-0]). Low K generally reduced overall root growth (Fig. 3A), but dramatic differences occurred between genotypes in terms of MR and LR elongation (Fig. 3, B and C). Interestingly, the reference accession Col-0 had the largest total root system in the low-K condition, although it was average size in the control. Genotypes with higher TRS in low K were characterized by longer MRs. Whereas low K reduced LR length at the root base (LRP 0.25) in all accessions (Supplemental Fig. S2), some accessions also had a striking increase of LR length close to the root tip (LRP 0.75): Stw-0, Bologna-1 (Bl-1), Catania-1 (Ct-1), Akita, Geneva-0 (Ge-0), Martuba-0 (Mt-0), and Oystese-0 (Oy-0). Indeed, those LRs eventually outgrew the MR tip (Fig. 2). This suggests a tradeoff between MR and LR growth in K-deficient conditions, especially since these accessions were not the ones with the smallest TRS in low K.
Figure 2.
Typical root phenotypes of Arabidopsis accessions grown on control and low-K media. Representative root images, obtained in the analysis with EZ Rhizo, are shown for each accession in each condition (12 DAG). Bar = 1 cm.
Figure 3.
K deficiency response of selected root parameters for individual genotypes. Means of TRS (A), MRP (B), and LRP 0.75 (C) were calculated from plants grown in control (black bars) and low-K (white bars) conditions. Accessions are sorted according to mean TRS in low K. Error bars indicate se (n = 7–12 plants per genotype per condition).
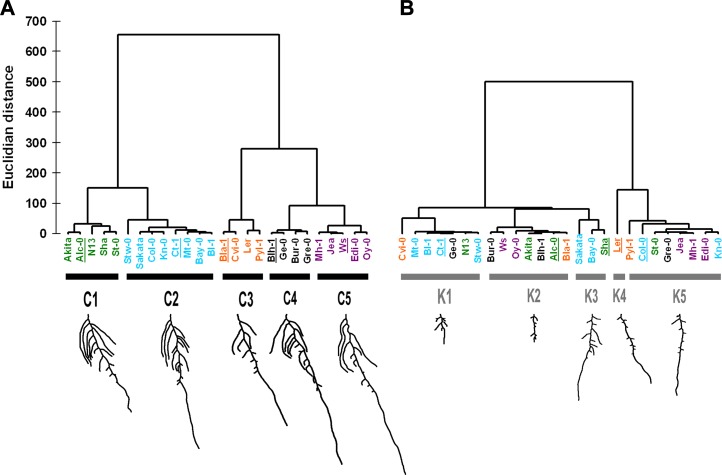
To identify common morphological responses among accessions, we performed an agglomerative hierarchical cluster (AHC) analysis on the accession means of all root parameters in each condition. In both conditions, clustering was limited to five classes. In control conditions (classes C1–C5), genotypes with a large TRS and generally longer MRP were found in C4 and C5 (Fig. 4). LR # was also high in C4 and C5, whereas lower LR # characterized C1 and C3. A distinguishing feature was MR angle, with lowest values in C2 and highest values in C3 and C4. Branched zones were short in C2 plants and long in C4 and C5. In addition, C3 members were distinguished by small LRP 0.25 and low LRdensMR. In low K (classes K1–K5), MRP and Apical were the main determinants of classification: low values were characteristic for all genotypes in K1, and high values were characteristic for all genotypes in K5. Since the opposite was observed for the length of LRs, especially those close to the tip, we defined two phenotypes as response strategies to low K: strategy I, long MRs with short LRs (classes K4 and K5); strategy II, short MRs with long LRs (K1). K2 accessions grouped in the middle of the spectrum. High LR density and high LRP 0.75 underlie the proximity of K1 and K2. The reference accession Ler was the only member of K4, characterized by its high positive MR angle, in contrast to K3 members, which had the lowest, and in fact negative, MR angles. AHC analysis provided a refined picture of correlations between root architecture traits in control and low K (Table I), showing that classes in the low-K condition consisted of combinations of genotypes that differed from those in the control condition. Indeed, some accessions were highly similar in the control but were positioned at opposite ends of the two main branches in low K, such as Col-0 and Ct-1. Due to their similarity in control and their opposite response to low K, this pair represents an interesting genetic resource to study adaptations to K deficiency.
Figure 4.
AHC of natural accessions according to their overall root architecture highlights different response strategies to low K. All 14 quantified root traits of plants grown on control (A) and low-K (B) media were taken into account. Unweighted clustering was performed using Ward’s method for agglomeration and Euclidian distance for dissimilarity. Genotype names are colored according to cluster composition in the control condition. For each cluster, the phenotype of a representative accession (underlined) is shown below.
Strategies I and II Are Characterized by Cell Death around the Apical Meristems of LRs and MRs, Respectively
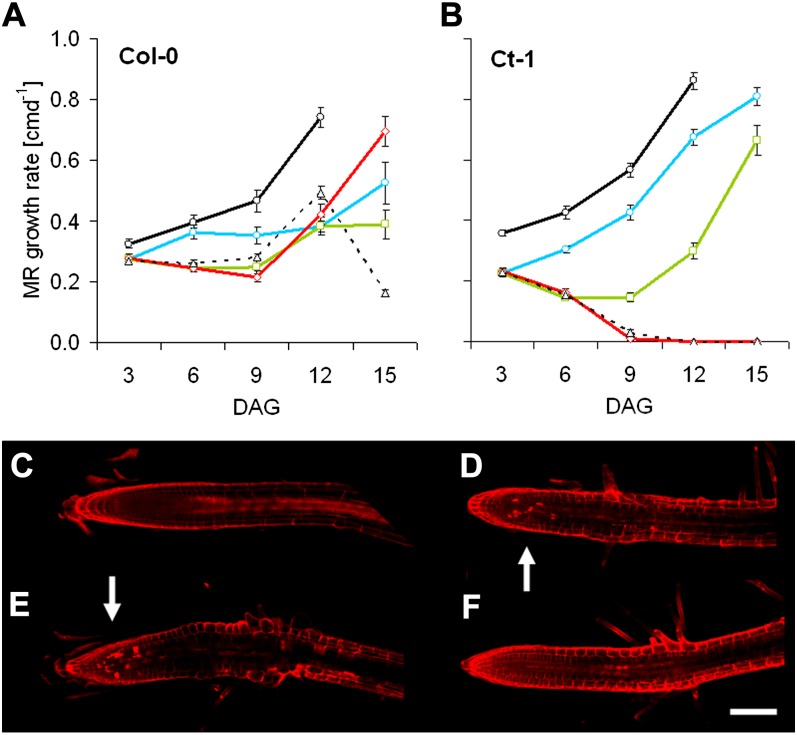
First, we followed Col-0 and Ct-1 MR growth over time. In the control, MR growth continuously accelerated in both accessions. Growth rates of Col-0 MRs also slightly increased in low K until 12 DAG, dropping significantly thereafter (Fig. 5A). In contrast, MR growth rates constantly decreased in Ct-1, reaching zero at about 9 DAG (Fig. 5B). To investigate whether this phenotype could be rescued by resupply of K, we treated plants that had been exposed to K starvation for an extended period of time with 2 mm KCl solution for 1 h. Col-0 MRs, resupplied on any DAG at low K, eventually showed increased growth rates after K resupply (Fig. 5A). MRs of Ct-1, however, only recovered when resupplied 6 DAG or earlier (Fig. 5B). Hence, after this point, MR growth in Ct-1 not only slowed down but came to an irreversible halt. In Col-0, LR elongation was not rescued if growth had already stopped due to K starvation (Fig. 3C; Supplemental Fig. S2). By contrast, in Ct-1, LRs close to the tip continued to grow in any condition. Since cell cycle marker lines are not readily available in the Ct-1 background, we performed propidium iodide staining on both genotypes to test for cytological changes under K starvation. Col-0 showed no obvious aberrations in the MR apex (Fig. 5C), but cell lesions were visible around the LR meristem (Fig. 5E), indicated by cells completely filled with propidium iodide. The opposite was observed for Ct-1: cell death was visible around the MR meristem (Fig. 5D), while LRs were undamaged (Fig. 5F). We conclude that K starvation causes cell death in the apical meristem of Ct-1 MRs, eventually abolishing MR growth completely. Similarly, cell death within the LR meristem inhibits LR elongation in Col-0.
Figure 5.
Irreversible MR growth arrest of Ct-1 in low K is caused by cell death in the apical meristem. A and B, Col-0 (A) and Ct-1 (B) seedlings (n = 8–15 per genotype per condition) were grown on control (black lines, circles) and low-K (dashed lines, triangles) media or on low-K medium resupplied with 2 mm KCl for 1 h at 3 DAG (blue lines, circles), 6 DAG (green lines, squares), or 9 DAG (red lines, diamonds). MR growth in Ct-1 could not be recovered when resupply occurred later than 6 DAG, whereas in Col-0, MRs continued to elongate in all conditions. C to F, Seedlings starved for K were stained with propidium iodide solution 6 DAG and observed with a confocal microscope. Representative images are shown for MR apices of Col-0 (C) and Ct-1 (D) as well as LR tips (Col-0, E; Ct-1, F). Cell lesions are highlighted with white arrows. Bar = 0.1 mm.
QTL Analysis of Col-0 × Ct-1 Recombinant Inbred Lines Identifies Genetic Loci Underlying K-Specific Root Architecture
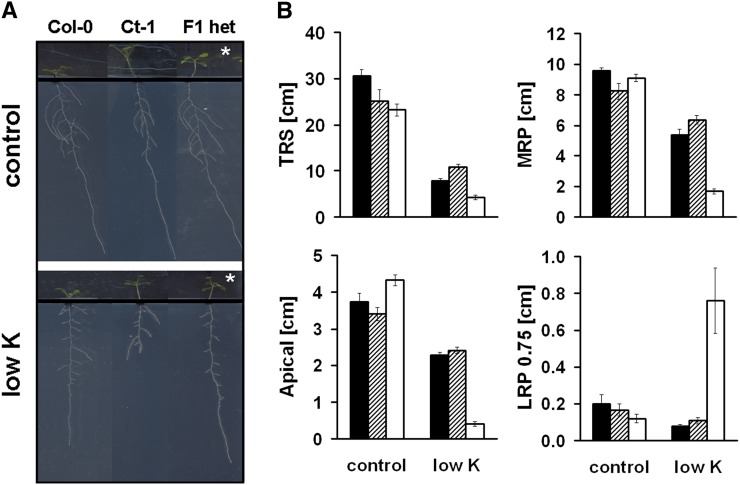
To get a better handle on the genetic structure of root architectural responses to low K, we crossed Col-0 and Ct-1, both with Col-0 and Ct-1 as either male or female, and phenotyped confirmed heterozygotes from the F1 generation. All heterozygous offspring showed the Col-0 root phenotype in low K (Fig. 6). As the direction of crossing (male versus female) did not make any difference, maternal effects can be ruled out. Interestingly, in both conditions, all heterozygotes also had an elongated hypocotyl, which is typical for Ct-1. Thus, Ct-1 is dominant for hypocotyl length, whereas Col-0 is dominant for root growth in low K.
Figure 6.
The K starvation response of Col-0 is dominant over Ct-1. A, Representative images of Col-0, Ct-1, and heterozygous offspring of a Col-0 × Ct-1 cross (F1 het) grown on control or low-K medium (12 DAG). Note that F1 heterozygotes have elongated hypocotyls (asterisks), which is typical for Ct-1. B, Quantitative root parameters of F1 heterozygotes (dashed bars) confirm the dominance of the Col-0 phenotype (black bars) over Ct-1 (white bars). Crosses were performed in both directions with each accession as either the male or female partner, and data of both offspring were pooled for F1 heterozygotes (n = 13–22). [See online article for color version of this figure.]
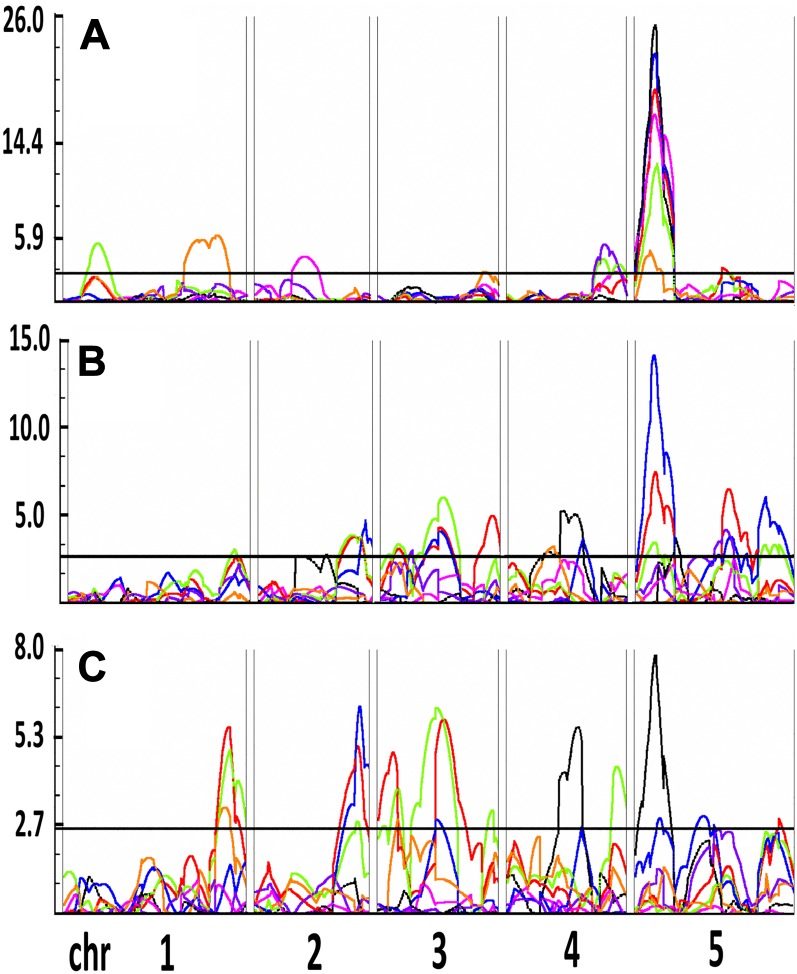
A recombinant inbred line (RIL) population of Col-0 and Ct-1 was obtained from the Versailles stock center (7 RV; Simon et al., 2008) as the basis for a quantitative genetics approach. A subset of 154 lines from the Core-Pop 164 were chosen and phenotyped together with the two parental accessions. All raw data are supplied as Supplemental Data Set S2. Heritabilities for individual traits ranged from 0 to 0.85 in the control and from 0.02 to 0.87 in low K (Supplemental Fig. S3). Log of the odds (LOD) scores were computed for each root parameter with Windows QTL Cartographer 2.5 (Wang et al., 2011) using the composite interval mapping function. A total of 1,000 permutations were performed to determine the LOD threshold. For each QTL, the position of the LOD peak and the QTL interval (determined as LOD − 1 and LOD − 2 drop) were recorded, and the phenotypic variation explained by each locus was calculated (for full lists of QTLs detected in all conditions, see Supplemental Tables S3–S5). A QTL located on top of chromosome 5 was detected for all traits measured in control conditions (Fig. 7A) and largely dominated the percentage of phenotypic variation explained (Fig. 8A). For all genomic intervals with multiple QTLs, we assigned a code composed of chromosome number and an incremental identifier, in this case, CHR5.1 (Fig. 8). Additional control loci included four QTLs for Apical (CHR1.1, CHR4.3, CHR5.1) that partly colocalized with QTLs for MR angle, LRdensMR, and LRdensBZ. Another MR angle QTL was located on chromosome 2 (CHR2.1). LR length QTLs were located at CHR1.2 (LRP 0.50 and LRP 0.75) and CHR3.3 (LRP 0.75). CHR5.1 was also the region with the highest percentage of phenotypic variation explained in low K (Fig. 7B; Fig. 8B). Low-K-specific MR QTLs were located at CHR1.3, CHR2.2, CHR3.1, and CHR3.2. Two MR QTLs in low K, CHR3.3 and CHR5.3, colocalized with QTLs of other traits in the control. Moreover, a low-K-specific LR path length QTL was located on chromosome 4 (CHR4.2). We also performed composite interval mapping using the ratio of average trait values from low K divided by control values (low K/control ratio; Fig. 7C; Fig. 8C). This resulted in the emphasis of low K- or control-specific loci while diminishing the effect of loci found in both conditions. Using low K/control ratio, for most traits no significant loci were found at CHR5.1, suggesting a putative role of this genomic region in general root development rather than in stress response.
Figure 7.
QTL mapping of the K deficiency response in a Col-0 × Ct-1 RIL population. LOD profiles of selected parameters, MRP (red), MR angle (pink), Apical (green), TRS (blue), LRdensBZ (purple), LRP 0.25 (black), and LRP 0.75 (orange), are shown using root parameters quantified in control (A) and low-K (B) media and low K/control ratio (C) as input. The QTL threshold was determined with 1,000 random permutations of the phenotypic data set and is shown as a horizontal black line. Chromosomes 1 to 5 (chr 1– 5) are shown from left to right, separated by double lines. LOD score values are shown on the y axis.
Figure 8.
Positions and effects of root architecture QTLs for control and low-K conditions. Results of multiple interval mapping analysis of 12 root parameters are shown for control (top panel) and low-K (middle panel) media and for the low K/control ratio (bottom panel). The five Arabidopsis chromosomes are shown as gray bars with physical distances (Mb) on the left and genetic distances (centimorgan) on the right. Colored bars give the percentage of phenotypic variation explained by the QTLs within a certain chromosome region (indicated by circles and sticks). Each color corresponds to a root parameter according to the legend provided at the bottom of the graph. Stacked bars of the same color show individual contributions from multiple QTLs within the region. Identifiers of low-K-specific regions (e.g. CHR1.3) are in red. White arrows inside the chromosomes highlight areas for which a QTL was confirmed by HIF analysis. The numbers identify the HIF for which significant (**P < 0.01, ***P < 0.001) phenotypic segregation was achieved, as shown in the box at the bottom right of the graph (Col-0 allele in black, Ct-1 allele in white).
To validate the obtained loci, we developed heterogeneous inbred families (HIFs; Tuinstra et al., 1997) from RILs with residual heterozygosity within QTL intervals (for all HIFs, see Supplemental Table S6; for primers used, see Supplemental Table S7). Three low-K-specific MR loci were validated with HIFs 49, 178, and 479 and one low-K-specific LR locus was validated with HIF 434 (Fig. 8). These HIFs, therefore, can be used for future fine-mapping. The low-K-specific locus CHR2.2 could not be confirmed by any HIF used. We also observed phenotypic segregation at loci CHR3.3 and CHR5.3. However, although multiple interval mapping analysis identified both loci as low K specific for most traits, phenotypic segregation of HIFs 116, 297, and 309 also persisted in the control (Supplemental Table S6).
DISCUSSION
Here, we investigated the response of Arabidopsis root architecture to changes in external K supply in a set of 26 natural accessions. We quantified 14 root traits of seedlings grown on control and low-K media and found significant contributions of genotype, environment, and genotype-environment interactions to the total variation within each root parameter (Fig. 1). Analysis of individual accessions (Fig. 2), correlation analysis (Table I), and cluster analysis based on phenotypic data (Fig. 4) revealed a gradient of sensitivity toward low K. This gradient links two opposite low-K response strategies at either end of the spectrum. Response strategy I consists of the maintenance of MR growth accompanied by a dramatic reduction of LR elongation (Figs. 2, 3, 4, and 5E). This response has been reported previously for Col-0, the most widely used laboratory wild-type accession (Armengaud et al., 2004; Shin and Schachtman, 2004). In contrast, strategy II accessions respond to low K with a drastic reduction of MR growth. In fact, MR growth is completely eliminated under prolonged K deficiency (Fig. 5B) as a consequence of cell death around the apical meristem (Fig. 5D). At the same time, LR elongation is maintained, so that the MR tip is eventually outgrown by LRs originating close to the root tip (Fig. 6, Ct-1). Since this response does not occur in the commonly used reference accessions, it has so far been unknown. Both strategies allow plants to maintain elongation of at least certain root parts, and as a result, no differences in shoot growth were obvious in low K. Therefore, we conclude that they constitute viable strategies to overcome K deficiency. In K-starved plants, reactive oxygen species are formed in an area close to the MR tip (Shin and Schachtman, 2004; Kim et al., 2010). However, the peak of reactive oxygen species production was detected in the elongation zone rather than the meristem (Shin and Schachtman, 2004), making reactive oxygen species toxicity not a prime suspect for MR cessation observed in strategy II accessions. Nevertheless, phosphate starvation has been shown to elicit a root phenotype similar to the strategy II low-K response (Williamson et al., 2001; López-Bucio et al., 2002; Pérez-Torres et al., 2008), and reactive oxygen species are also up-regulated in the root apex in low phosphorus (Tyburski et al., 2009). This suggests that root architecture responses elicited by low K and low phosphorus share a common regulatory pathway.
The set of natural accessions used in this study was largely based on a nested core collection widely used in the field (McKhann et al., 2004). The genotypes Ct-1, Stw-0, and Mt-0 have also been shown to cluster in response to nitrogen availability (Chardon et al., 2010; Ikram et al., 2012). In fact, Mt-0 and Ct-1 have been described as “ideotypes” for seed production in suboptimal nitrogen conditions, whereas Stw-0 and Bl-1 were among the accessions with highest dry matter production in nitrogen deficiency (Chardon et al., 2012). In addition, Ge-0 clustered with the aforementioned genotypes due to low nitrate uptake efficiency and nitrogen content in contrasting environments (Chardon et al., 2010). Yet, strategy II accessions do not display high overall genetic similarity (Ostrowski et al., 2006; Simon et al., 2012), suggesting a central role for only a few polymorphisms in this low-K response. Moreover, Mt-0, Bl-1, Ct-1, Ge-0, and N13 were also included in a study on the natural variation of drought responses (Bouchabke et al., 2008), but no clear clustering of these accessions was observed. Thus, their phenotype in low K is probably not the result of similar overall stress responses. A direct connection to K transport might be suggested when looking at shoot K measured by Buescher et al. (2010). Mt-0, the only strategy II member in the study, had the highest shoot K concentration among 12 accessions. Interestingly, although no cluster analysis was provided, all low-K strategy II ecotypes also appear among the less zinc-tolerant ecotypes in a study by Richard et al. (2011). Supplement of surplus zinc also elicited changes in root architecture, namely a decrease in MRP at higher zinc levels and a slight increase of LR path length in the lower range of concentrations. Changes in the homeostasis of other metals might also be important in the root architectural response to low K. It would be interesting to investigate the soil conditions occurring at the origin of strategy I and strategy II accessions, as Poormohammad Kiani et al. (2012) have recently demonstrated a correlation between the activity of the molybdenum transporter MOT1 and the molybdenum availability in the native range of natural accessions used. Unfortunately, data on the exact locations of sampling sites are sparsely available, making it very difficult to draw such conclusions from the accession set used here. As an example, the sampling site coordinates of Ct-1 are only estimated (Simon et al., 2012). The origin described is Catania, Sicily, which encompasses quite different ecosystems and soils, such as the Mediterranean seashore, alluvial floodplains (Capaccioni et al., 2005), and the volcanic slopes of Mount Etna. It would be interesting to resample those sites and compare the root response.
Our study included the accessions Bay-0 and Sha, which have been extensively used in natural variation and QTL studies, among others for flowering time (Loudet et al., 2002), root architecture (Loudet et al., 2005), partial resistance to Pseudomonas syringae (Perchepied et al., 2006), phosphate starvation (Reymond et al., 2006; Svistoonoff et al., 2007), sulfate content (Loudet et al., 2007), shoot mineral content (Buescher et al., 2010), and growth on acidic soil (Poormohammad Kiani et al., 2012). As they cluster very closely in low K (Fig. 4), it appears that adaptive responses to K availability are very similar between these accessions.
As representatives for each low-K response strategy, we chose Col-0 and Ct-1 for a quantitative genetics approach. Although the overall Ct-1 phenotype in low K is recessive (Fig. 6), several QTLs contribute to it (Figs. 7 and 8). To date, there is only one published record of QTL studies performed on the Col-0 × Ct-1 RIL set, with flowering time, rosette diameter, and total seed weight as traits of interest (Simon et al., 2008). All seven loci mapped in this study overlap with ours. Six of them were loci we identified in the control, plus the CHR2.2 locus (low-K specific in our study but not confirmed via HIF). Since the authors of that study mapped a considerably different set of traits to similar loci, most notably CHR5.1 and CHR5.3, these intervals are likely to contain genes that control development rather than stress response. However, none of our other four low-K-specific loci was previously detected in Col-0 × Ct-1. Two loci, one in the upper part of chromosome 3 and one in the lower part of chromosome 4, affected total root size in Col-0 × Ler (Fitz Gerald et al., 2006). In the same regions, Reymond et al. (2006) mapped QTLs of root growth responses to phosphate deficiency in a Bay-0 × Sha RIL population. To date, only the gene corresponding to the third locus identified in this study has been mapped as LPR1 (Svistoonoff et al., 2007). For the same RIL population, LR QTLs were mapped on the bottom of chromosome 1, the top of chromosome 3, and the middle of chromosome 4 (Loudet et al., 2005), potentially equal to CHR1.2, CHR3.1, and CHR4.2. A recent study, using 18 accessions to produce 17 F2 populations, confirmed once more the importance of flowering loci for plant growth and development (Salomé et al., 2011). Indeed, three of the control QTLs might correspond to major flowering loci such as FT (CHR1.2), FLC (CHR5.1), and MAF2 (CHR5.3). Another flowering locus, FLM, is in the low-K-specific CHR1.3 interval. However, although many QTL efforts have resulted in mapping flowering loci (Salomé et al., 2011; Strange et al., 2011), there is no guarantee that the corresponding flowering genes are indeed crucial here.
In Arabidopsis, QTLs have been mapped for K accumulation in seeds (Vreugdenhil et al., 2004) and shoots (Harada and Leigh, 2006), using a Cvi × Ler population on both occasions. One seed QTL overlaps with CHR2.2, two with CHR3.1, and one with CHR5.1 (Vreugdenhil et al., 2004). The most important QTL for shoot K in dry matter also coincides with CHR3.1 (Harada and Leigh, 2006), and the remaining two correspond to CHR4.1 and CHR5.3. Moreover, loci for shoot K per fresh weight colocalize with CHR2.1, CHR4.1, CHR5.2, and CHR5.3. Harada and Leigh (2006) provided a list of candidate cation transporter genes within their intervals, including important K transporters such as AKT1 (CHR2.1), SKOR (CHR3.1), HAK5 (CHR4.1), and several members of the TPK family (CHR5.2 and CHR5.3). Recently, ionomics have become a standard tool in the study of natural variation and stress responses, providing researchers with data for multiple elements at a time (Salt et al., 2008; Buescher et al., 2010; Prinzenberg et al., 2010). Buescher et al. (2010) not only compared natural accessions but also mapped shoot mineral contents for several RIL populations in various environments. In a large Bay-0 × Sha population, loci for K content were mapped to regions corresponding to CHR1.2, CHR1.3, CHR2.2, and CHR5.3. A K locus equivalent to CHR3.1 was also found in a smaller population of Bay-0 × Sha and in Cvi × Ler. For all these loci, significant QTLs for several ions at a time were detected, suggesting homeostatic “hubs.” These findings are backed up by Prinzenberg et al. (2010), who measured shoot and root growth traits combined with element profiles in a Ler × Kashmir-2 RIL population in three conditions: control, low K, and low phosphorus. They identified among others multielemental loci at CHR1.3 (K, iron, magnesium), CHR3.1 (K, phosphorus, magnesium, cobalt, manganese, zinc), CHR3.2 (K, phosphorus, zinc), and CHR5.3 (multiple elements). We compared publicly available ionomic data of Col-0 and Ct-1 (Purdue Ionomics Information Management System database; Baxter et al., 2007). The accessions show differences in manganese and cobalt content, but all other ions, including K, are present at similar levels. However, these data are derived from shoots of plants grown in (K-sufficient) soil and therefore are mirrored by similar root architectures of Col-0 and Ct-1 in control conditions in our study. To date, no ionomic data are available for Col-0 and Ct-1 in K-deficient conditions. Future experiments should test whether the Ct-1 phenotype in low K is caused by direct sensing effects at the root tip, comparable to the local response to phosphate starvation (Svistoonoff et al., 2007), or by larger scale changes in nutrient/metal homeostasis, potentially controlled by ionomic hubs.
CONCLUSION
Our study provides quantitative evidence for phenotypic plasticity in the root architectural response of Arabidopsis to K deficiency. A phenotypic gradient within 26 Arabidopsis accessions of root morphological responses linked two extreme adaptive low-K response strategies that consisted of opposite patterns of growth arrest and growth prioritization between MRs and LRs. Subsequent quantitative genetics revealed several loci determining specific subsets of root architectural traits. Some of these overlapped with loci previously identified in different nutritional and phenotypic contexts, suggesting a role for so far unknown genomic hubs in coordinating root growth with environmental stresses. The results and genomic tools presented here are a new resource for the identification of genes underlying nutrient sensing and root architecture.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions Akita, Alc-0, Bay-0, Bl-1, Bla-1, Bur-0, Blh-0, Col-0, Ct-1, Cvi-0, Edi-0, Ge-0, Gre-0, Jea, Kn-0, Mh-1, Mt-0, N13, Oy-0, Pyl-1, Sakata, Sha, St-0, and Stw-0 were obtained from the Versailles Resource Centre (Institut National de la Recherche Agronomique Versailles; http://dbsgap.versailles.inra.fr/vnat/). Ler and Wassilewskija from seed stocks maintained at the University of Glasgow were also included in the analysis. For QTL analysis, a Col-0 × Ct-1 core population (7 RV; Simon et al., 2008) of 164 RILs was obtained from the Versailles Resource Centre (Supplemental Table S1). Ten lines were excluded from the analysis due to missing data at some markers (Simon et al., 2008).
Seeds were surface sterilized with absolute ethanol for 1 min and in 2% sodium hypochlorite containing 0.1% Tween 20 for 5 min, then rinsed with sterile double distilled water. Seeds were stratified for 3 d in the dark at 4°C.
For phenotypic analysis of Arabidopsis accessions, five or six seeds were sown in equal distance on 12- × 12-cm2 square petri dishes containing 35 mL of control or low-K growth medium, respectively (for media, see below). Each accession was represented by two plates. Seeds that had not germinated at 2 DAG were discarded from further analysis, resulting in seven to 12 seedlings analyzed per genotype per condition. For QTL analysis, seeds of 154 RILs and of the two parental ecotypes Col-0 and Ct-1 were treated and sown as described above. Each plate contained four seeds of two genotypes (two seeds per genotype). Three batches of all lines were grown independently. Pairing of the seeds on the plates and positions of the plates in the growth chamber followed a random design.
The control growth medium consisted of 0.5 mm CaCl2, 0.25 mm MgSO4, 2 mm KNO3, 1 mm NaCl, 0.5 mm NaH2PO4, 42.5 μm Fe(III)Na-EDTA, 1.8 μm MnSO4, 45 μm H3BO3, 0.38 μm ZnSO4, 0.015 μm (NH4)6Mo7O24, 0.16 μm CuSO4, and 0.01 μm CoCl2 as micronutrients. In the low-K medium, 0.5 mm CaCl2 was replaced with 0.5 mm Ca(NO3)2, 2 mm KNO3 with 0.01 mm KCl, and 1 mm NaCl with 1 mm NaNO3. All media were buffered with 2 mm MES, adjusted to pH 5.6 with TRIS-HCl, and supplied with 0.5% Suc and 1% agar (A-1296; Sigma-Aldrich; http://www.sigmaaldrich.com).
After autoclaving, 35 mL of medium was poured into 120- × 120-mm2 petri dishes. Before application of the seeds, 2 cm of agar was removed from the top of the plate. Seeds were placed just below the rim of the agar so that developing leaves did not touch the agar. Plates with seeds were placed vertically into racks made of cardboard boxes so that only the upper (agar-free) parts protruded from the boxes. Throughout the experiments, the plate position within the box and box position in the growth chamber were rerandomized every day. Plants were grown in short-day conditions (9/15 h light/dark) at 22°C/18°C with a light intensity of 160 μmol m−2 s−1.
Quantification of Root Architecture
Plates were scanned at 6, 8, 10 and 12 DAG using a conventional flatbed scanner and a resolution of 200 dpi. All scanned images were stored at Glasgow and can be made available to interested researchers for further analysis. Root phenotypes of different accessions were compared 12 DAG. For QTL analysis, control plants were scored 10 DAG because some lines reached the bottom of the plate before day 12. Image analysis was performed with EZ Rhizo software (Armengaud et al., 2009b). In addition to the parameters measured by EZ Rhizo, the mean LR length within MR quartiles was calculated for each root.
Propidium Iodide Staining
Plants grown on control or low-K medium were immersed for 10 min in 10 μg mL−1 propidium iodide (Fluka 81845; Sigma-Aldrich) solution and observed with a confocal microscope.
Crossing
Pollen from Col-0 plants was transferred to Ct-1 flowers and vice versa. Offspring of the crosses were grown on control and low-K media and phenotyped 12 DAG. Tissue samples were taken from individual plants, and heterozygous F1 individuals were identified with PCR using custom TaqMan single-nucleotide polymorphism assays (Applied Biosystems, Life Technologies) at markers c2_17606 and c3_05141 (Simon et al., 2008) according to the supplier’s protocols. Only confirmed heterozygotes were used for the quantification of root architecture.
QTL Analysis
Twelve root architecture traits (Fig. 8) were included in the analysis. Outliers were removed from the data set, and genotype averages were calculated for all quantified root parameters. Heritability of traits was estimated as h2 = σ2g/(σ2g + [σ2e/r]), with σ2g being the genetic variance, σ2e the residual variance, and r the number of replicates. LOD scores were computed for each root parameter with Windows QTL Cartographer 2.5 (Wang et al., 2011) using the composite interval mapping function and 1,000 permutations to determine the LOD threshold. QTL effects were subsequently calculated using the multiple interval mapping function.
Generation of HIFs
RILs with residual heterozygosity in QTL intervals were selected to produce HIFs (Tuinstra et al., 1997). All RILs were grown on soil, and seeds from individuals harboring the homozygous Col-0 or Ct-1 allele at the corresponding locus were harvested and phenotyped on control and low-K plates. Simple sequence length polymorphism markers (Supplemental Table S4) were designed on the basis of insertions/deletions detected between whole genomic sequences of Col-0 and Ct-1 (Weigel and Mott, 2009) and respective genomic fragments amplified using standard laboratory protocols. Multiple simple sequence length polymorphism markers were used for HIFs corresponding to low-K-specific loci (i.e. HIFs 49, 178, 434, and 479) to determine the area of heterozygosity with higher precision.
Statistical Analysis
All data were cleared from outliers, and statistical analysis was performed with the XLSTAT (Addinsoft) add-in for Excel. ANOVA was calculated using type III sums of squares, and Tukey’s t test was used for pairwise comparisons. In both cases, the significance threshold was set at P < 0.05. Ward’s agglomeration method and Euclidian distance for dissimilarity were used for AHC analysis based on the phenotypic traits of natural accessions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Accession means in low K plotted against control.
Supplemental Figure S2. K deficiency response of LRP 0.25.
Supplemental Figure S3. Heritability of traits quantified in the QTL analysis.
Supplemental Table S1. List of Arabidopsis accessions used.
Supplemental Table S2. Correlation analysis of root traits.
Supplemental Table S3. Positions of all QTLs identified in control condition.
Supplemental Table S4. Positions of all QTLs identified in the low-K condition.
Supplemental Table S5. Positions of all QTLs identified using low K/control ratio.
Supplemental Table S6. HIFs used for validation of QTLs.
Supplemental Table S7. Simple sequence length polymorphism primers used to genotype HIF.
Supplemental Data Set S1. Root parameters of natural accessions grown in control and low-K conditions.
Supplemental Data Set S2. Phenomic and genomic data used in the QTL analysis.
Acknowledgments
We thank A. Ruiz-Prado and L. O’Donnell for technical assistance and P. Armengaud for organizational help.
Glossary
- K
potassium
- LR
lateral root
- MR
main root
- Col-0
Columbia
- QTL
quantitative trait locus
- DAG
days after germination
- TRS
total root size
- AHC
agglomerative hierarchical cluster
- Ler
Landsberg erecta
- RIL
recombinant inbred line
- LOD
log of the odds
- HIF
heterogeneous inbred family
- LRdensBZ
LR density within the branched zone
- LRdensMR
LR density along the MR
- LRP 0.75
mean LR path length in the third quartile from the top of the MR
- LRP 0.50
mean LR path length in the second quartile from the top of the MR
- LRP 0.25
mean LR path length in the uppermost quartile of the MR
- LR #
first-order LR number
- MR angle
angle of MR from full verticality
- Apical
apical zone length
- MRP
MR path length
- Bur-0
Burren-0
- Pyl-1
Le Pyla-1
- Bay-0
Bayreuth-0
- Stw-0
Stobowa-0
- Bl-1
Bologna-1
- Ct-1
Catania-1
- Ge-0
Geneva-0
- Mt-0
Martuba-0
- Oy-0
Oystese-0
References
- Alemán F, Nieves-Cordones M, Martínez V, Rubio F. (2011) Root K(+) acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol 52: 1603–1612 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Hammond JP, Armengaud P, White PJ. (2006) Nutrient sensing and signalling in plants: potassium and phosphorus. Adv Bot Res 43: 209–257 [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. (2010) Coronatine-insensitive 1 (COI1) mediates transcriptional responses of Arabidopsis thaliana to external potassium supply. Mol Plant 3: 390–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y. (2009a) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 150: 772–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009b) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. (2007) Purdue ionomics information management system: an integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardif M. (2008) Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS ONE 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher E, Achberger T, Amusan I, Giannini A, Ochsenfeld C, Rus A, Lahner B, Hoekenga O, Yakubova E, Harper JF, et al. (2010) Natural genetic variation in selected populations of Arabidopsis thaliana is associated with ionomic differences. PLoS ONE 5: e11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaccioni B, Didero M, Paletta C, Didero L. (2005) Saline intrusion and refreshening in a multilayer coastal aquifer in the Catania Plain (Sicily, southern Italy): dynamics of degradation processes according to the hydrochemical characteristics of groundwaters. J Hydrol (Amst) 307: 1–16 [Google Scholar]
- Chardon F, Barthélémy J, Daniel-Vedele F, Masclaux-Daubresse C. (2010) Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J Exp Bot 61: 2293–2302 [DOI] [PubMed] [Google Scholar]
- Chardon F, Noël V, Masclaux-Daubresse C. (2012) Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot 63: 3401–3412 [DOI] [PubMed] [Google Scholar]
- Den Herder G, Van Isterdael G, Beeckman T, De Smet I. (2010) The roots of a new green revolution. Trends Plant Sci 15: 600–607 [DOI] [PubMed] [Google Scholar]
- Drew MC. (1975) Comparison of effects of a localized supply of phosphate, nitrate, ammonium and potassium on growth of seminal root system, and shoot, in barley. New Phytol 75: 479–490 [Google Scholar]
- Fitz Gerald JN, Lehti-Shiu MD, Ingram PA, Deak KI, Biesiada T, Malamy JE. (2006) Identification of quantitative trait loci that regulate Arabidopsis root system size and plasticity. Genetics 172: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GL, Dong ZS, McLean G, Doherty A, Messina C, Schusler J, Zinselmeier C, Paszkiewicz S, Cooper M. (2009) Can changes in canopy and/or root system architecture explain historical maize yield trends in the US Corn Belt? Crop Sci 49: 299–312 [Google Scholar]
- Harada H, Leigh RA. (2006) Genetic mapping of natural variation in potassium concentrations in shoots of Arabidopsis thaliana. J Exp Bot 57: 953–960 [DOI] [PubMed] [Google Scholar]
- Ikram S, Bedu M, Daniel-Vedele F, Chaillou S, Chardon F. (2012) Natural variation of Arabidopsis response to nitrogen availability. J Exp Bot 63: 91–105 [DOI] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP. (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Ciani S, Schachtman DP. (2010) A peroxidase contributes to ROS production during Arabidopsis root response to potassium deficiency. Mol Plant 3: 420–427 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG. (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. (2005) Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet 110: 742–753 [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- McKhann HI, Camilleri C, Bérard A, Bataillon T, David JL, Reboud X, Le Corre V, Caloustian C, Gut IG, Brunel D. (2004) Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J 38: 193–202 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS. (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M-F, David J, Santoni S, McKhann H, Reboud X, Le Corre V, Camilleri C, Brunel D, Bouchez D, Faure B, et al. (2006) Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol 15: 1507–1517 [DOI] [PubMed] [Google Scholar]
- Perchepied L, Kroj T, Tronchet M, Loudet O, Roby D. (2006) Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS ONE 1: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poormohammad Kiani S, Trontin C, Andreatta M, Simon M, Robert T, Salt DE, Loudet O. (2012) Allelic heterogeneity and trade-off shape natural variation for response to soil micronutrient. PLoS Genet 8: e1002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzenberg AE, Barbier H, Salt DE, Stich B, Reymond M. (2010) Relationships between growth, growth response to nutrient supply, and ion content using a recombinant inbred line population in Arabidopsis. Plant Physiol 154: 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Richard O, Pineau C, Loubet S, Chalies C, Vile D, Marquès L, Berthomieu P. (2011) Diversity analysis of the response to Zn within the Arabidopsis thaliana species revealed a low contribution of Zn translocation to Zn tolerance and a new role for Zn in lateral root development. Plant Cell Environ 34: 1065–1078 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RAE, Yant L, Mott R, Weigel D. (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE, Baxter I, Lahner B. (2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59: 709–733 [DOI] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, van der Plas LH, Koornneef M, Vreugdenhil D. (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA 103: 2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Loudet O, Durand S, Bérard A, Brunel D, Sennesal FX, Durand-Tardif M, Pelletier G, Camilleri C. (2008) Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Simon A, Martins F, Botran L, Tisné S, Granier F, Loudet O, Camilleri C. (2012) DNA fingerprinting and new tools for fine-scale discrimination of Arabidopsis thaliana accessions. Plant J 69: 1094–1101 [DOI] [PubMed] [Google Scholar]
- Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C. (2011) Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6: e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Trontin C, Tisné S, Bach L, Loudet O. (2011) What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol 14: 225–231 [DOI] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95: 1005–1011 [Google Scholar]
- Tyburski J, Dunajska K, Tretyn A. (2009) Reactive oxygen species localization in roots of Arabidopsis thaliana seedlings grown under phosphate deficiency. Plant Growth Regul 59: 27–36 [Google Scholar]
- Vreugdenhil D, Aarts MGM, Koorneef M, Nelissen H, Ernst WHO. (2004) Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ 27: 828–839 [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. (2011) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Mott R. (2009) The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HMO. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]